Abstract
Earlier studies have reported on both proinflammatory and anti-inflammatory activities of cholera toxin (CT). As CT is a powerful adjuvant, we were interested in identifying genes with a possible involvement in these functions. A global gene expression analysis in mouse B cells showed that CT regulated <100 annotated genes, which encoded transcription factors, G proteins, cell-cycle regulators, and immunoregulating molecules. Interestingly, CT regulated the expression of the signal transducer and activator of transcription (STAT)3 gene and influenced the level and activation of both isoforms STAT3α and STAT3β, in vitro in a B-cell line and in Peyer's patch (PP) B cells and in vivo in freshly isolated splenic B cells from CT-treated mice. This effect was cAMP dependent and was not seen with CTB. B cells pre-exposed to CT were significantly more susceptible to the activation of STAT3 by interleukin (IL)-6 and IL-10. This exerted a stronger inhibitory effect of IL-10 on lipopolysaccharide (LPS)-stimulated B-cell proliferation and cytokine production (IL-6). Moreover, IgG1 and IgA production induced by LPS and IL-10 were enhanced by the addition of CT to cultures of PP or splenic B cells. This is the first study to provide a molecular mechanism that can reconcile previous findings of proinflammatory and anti-inflammatory effects by CT adjuvant.
Similar content being viewed by others
Introduction
Most adjuvants have a microbial origin, and it is considered that much of their efficacy relates to the degree of inflammation induced at the site of administration.1 The struggle to identify novel vaccine adjuvants has resulted in a large plethora of candidates with very diverse functions. However, the mechanism by which these candidates act on the immune system, and in particular their stimulating effect on innate immunity, has been studied inadequately.2 In descriptive terms, their effects on costimulatory molecules, and cytokine and chemokine production have been documented, but their global effects on gene transcription and the complex regulatory machinery in the cell that leads to enhanced immune responses, are poorly understood.3 Some adjuvant formulations containing lipopolysaccharide (LPS) or bacterial CpG oligodeoxynucleotide, are known to act through Toll-like receptors, a family of pattern-recognition receptors present on dendritic cells (DCs), macrophages, and B cells, and in this respect, their global effects have been established or could be potentially predicted.4
Other adjuvants, such as the powerful enteroxins, cholera toxin (CT) and the closely related Escherichia coli heat labile toxin, which bind to ganglioside receptors on the cell membrane of all nucleated cells, have strong immunoenhancing effects on both systemic and mucosal immune responses.1 This makes them interesting candidates for mucosal vaccine development.5 These toxins are AB5 complexes that carry an adenosine diphosphate-ribosylating function in the A1 subunit and receptor-binding properties in B subunits.6 Thus, the optimal immunomodulating effect is dependent on both binding and enzymatic activities, although enzymatically killed mutants have been found to host partial adjuvant activity.7 The A1 subunit adenosine diphosphate ribosylates Gsα in the cell membrane, which results in adenylate cyclase activation and subsequent increase in intracellular cAMP. This intracellular second messenger is well known to regulate gene expression at the transcriptional level and could be important for the adjuvant function.8
We, along with others, have previously studied the immunomodulating function of CT on B lymphocytes and found evidence for direct effects on isotype-switch differentiation and expression of costimulatory molecules.9 Interestingly, CT strongly promoted sterile RNA transcripts and switching to IgG1 and IgE in interleukin (IL)-4 plus LPS-exposed murine IgM+ B cells, suggesting that CT affected gene transcription.10 Moreover, the expression of costimulatory molecules CD40, CD80, and CD86 was found to be strongly enhanced in B cells, macrophages, and DCs after stimulation with CT.11, 12 In addition, the holotoxin has been found to upregulate mRNA expression of several cytokine and chemokine and chemokine receptor genes, such as CXCR4, CCR7, IL-10, IL-6, and IL-1, in classical antigen presenting cell (APC).12, 13, 14 In contrast, IL-12 expression was reduced by CT in DCs or macrophages.15 Thus, there is evidence that both anti-inflammatory and proinflammatory response patterns are supported by CT. At present, we do not know how to reconcile these effects with the strong immunoenhancing effects on antibody production, CD4+ T-cell priming, and cytotoxic T lymphocytes (CTL) stimulation, seen with CT adjuvant in vivo.
The signal transducer and activator of transcription (STAT) family of proteins are key players in the regulation of many immune processes.16 These are the main mediators of cytokine signaling and are activated by the phosphorylation of tyrosine residues by Jak kinases associated with cytokine receptors. In particular, STAT3 has been found to not only host a critical function as a mediator of anti-inflammation affecting the function of DCs, macrophages, and neutrophils but also to be critically involved in the differentiation of both activated T and B cells.17 The fact that STAT3 could mediate very diverse functions in different tissues from driving inflammation by augmenting Th17 differentiation in one tissue to promoting tolerance and anti-inflammation in another tissue makes this regulatory pathway especially interesting from an adjuvant point of view.18, 19 We, therefore, speculated that the activation of members of such a gene family could account for the seemingly conflicting evidence of CT-mediated proinflammatory and anti-inflammatory effects found in the literature. Moreover, recent studies have shown that STAT3 is involved in not only the early development of B cells but also in the activation, survival, and terminal differentiation into plasma cells.20, 21
One strategy to dissect and study the complex interactions between adjuvants and innate immunity is to perform global gene expression profiling analysis.22, 23 Although Yang and co-workers have reported effects of CT on gene expression in mixed populations of human lymphocytes and monocytes, we lack a detailed analysis of gene expression profiles in well-defined cell populations after CT exposure.24 In fact, very few adjuvants, except for LPS and CpG, have been subjected to analysis of global gene expression in targeted cells. Such information would be a powerful tool in the search for adjuvant mechanisms and aid in the design of novel adjuvants. It would also allow us to categorize the different adjuvant substances better into functionally defined groups. On the basis of such knowledge, the rational development of novel adjuvants could be undertaken, and in this way meet the demands for safe and efficacious future vaccines, in particular for needle-free mucosal vaccines.
Results
Global gene expression in CT-exposed B cells
We used the Affymetrix microarrays (Affymetrix, Santa Clara, CA) to investigate the pleiotropic effects of CT on gene expression in targeted B cells. To ensure a homogenous target cell population, we chose to work with a well-defined B cell that was selected from a panel of different murine B-cell lines. Cells were stimulated in vitro with CT for 2 and 8 h. Three separate experiments were carried out, and total RNA was isolated from the respective samples and converted into biotinylated cRNA and analyzed on Affymetrix GeneChip expression arrays. To identify differentially regulated genes after CT exposure, we used the dChip method. Genes were considered to be significantly regulated if they demonstrated fold changes ≥1.9 (P≤0.05) and a difference of the mean ≥100 as compared with that of untreated B cells in at least two separate experiments. These are considered stringent selection criteria. The initial screening of genes affected by CT showed a total of 99 annotated genes that were regulated, of which a majority (61 genes) were upregulated and a minority (38 genes) were negatively regulated. CT increased the expression of 27 genes in X16C8.5 B cells at 2 h of stimulation (Table 1), whereas at 8 h of CT stimulation, 50 upregulated annotated genes were recorded as compared with the gene expression levels of untreated B-cell controls (Table 2). Of the latter genes, 16 of these were also upregulated at 2 h. Thus, within the two time points of this study, CT appeared to induce kinetically distinct genes as well as genes with a more prolonged upregulated expression. It is noteworthy that CT-exposed B cells exhibited enhanced expression of genes encoding transcription factors, G proteins, cell-cycle regulators, and membrane-expressed molecules, such as the costimulatory molecule CD86. Hence, functionally very different gene families were represented among the upregulated genes, both at 2 and 8 h after CT stimulation.
Identification of functional clusters of CT-regulated genes in B cells
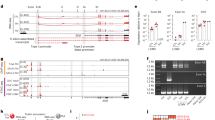
Next, we undertook hierarchical clustering of the CT-regulated genes. We used the UPGMA (Unweighted Pair-Group Method with Arithmetic mean) similarity method with correlation distance on the gene expression data from 2 and 8 h after CT exposure as compared with data from untreated B cells cultured only in medium for the same time periods. The analysis demonstrated that a total of nine clusters could be identified. Four of them were dominating, I, II, V, and IX, hosting genes of potential association with an adjuvant function (Figure 1). Genes in cluster I showed an upregulated expression level at both 2 and 8 h. In this cluster, genes encoding transcription factors/cofactors, G proteins/regulators, cell surface proteins, proteins affecting cell proliferation, and a structural protein were identified. It is noteworthy that some of these genes belong to the family of immediate-early genes, such as RGS2, CREM, and G-α13, which are rapidly induced after stimulation. In contrast, genes in cluster II appear to be only transiently induced by CT and were found exclusively upregulated at 2 h. Genes in this cluster included JunB, TIS21/BTG2, NR4A1/Nur77/N10/TIS1, and Nfil3/E4BP4. Cluster V genes were upregulated only at 8 h, and displayed a delayed onset (Figure 1). In addition, in this cluster, we found not only genes encoding transcription factors/cofactors, cell surface proteins, proteins affecting cell proliferation but also proteins involved in metabolism, cation transport, structural proteins, and a proteinase inhibitor. In contrast, genes in cluster IX varied in their expression at 2 h, but all were downregulated at 8 h. In this cluster, we found annotated genes encoding, e.g., immunoglobulin heavy chain 6 NFATc1 and Cyclin G2 (Figure 1). To validate the microarray data, we selected 23 genes that were distinctly upregulated and undertook real-time (RT)-PCR analysis of mRNA from CT-exposed or untreated control B cells (A Sjöblom-Hallén, unpublished observation). The expression pattern of these genes mimicked closely that observed in the microarray analysis and, thus, we conclude that the transcriptome data perfectly matched the RT-PCR data, confirming the high quality of the analysis.
Cluster analysis of gene expression in B cells exposed to CT. Hierarchical clustering of genes regulated by CT in X16C8.5 B cells at indicated time points. The criteria were a mean fold change >2.0 and P<0.05. Colors represent mean fold changes (red=upregulated) in gene expression at 2 and 8 h stimulation with 0.1 μg/ml CT compared with unstimulated B cells. Each row represents a single gene at the indicated time point. Nine different clusters of genes were identified (I–IX) and the annotated genes are given in the margin. These are the compiled data from three independent experiments. CT, cholera toxin.
CT promotes transcription of the STAT3 gene
As we searched for gene regulations that could reconcile the earlier findings of anti-inflammatory and proinflammatory effects by CT, we focused on the STAT3 gene expression. This finding was also confirmed by RT-PCR analysis using STAT3-specific primers. We consistently observed 7–8-fold higher STAT3 gene expression at 8 h compared with unstimulated X16C8.5 B cells at this time (Figure 2). At 2 h, we detected a two-fold (P<0.05) increase compared with unstimulated control cultures. As CT has been found to stimulate both IL-6 and IL-10 production in mononuclear cells and both these cytokines can induce STAT3 in B cells, we concomitantly analyzed whether IL-10 and IL-6 were produced in these cultures. However, we found no production of these cytokines by X16C8.5 B cells after CT exposure (Figure 2). In contrast, in LPS-stimulated X16C8.5 B cells, CT evoked significant IL-6 production, whereas IL-10 levels were dramatically reduced (Figure 2). These data agree well with the failure to annotate IL-6 or IL-10 in the transcriptome and with previous reports showing that CT alone fails to induce IL-6 or IL-10 production by B cells.13
CT stimulates STAT3 gene expression independently of IL-10 and IL-6. (a) RT-PCR on mRNA extracted from X16C8.5 B cells stimulated for 2 or 8 h with CT at 0.1 μg/ml. The fold change in STAT3 gene expression was determined relative to the expression level of the 36B4 housekeeping gene. In each experiment, three samples were analyzed for each time point and the average fold increase in STAT3 gene expression is given as mean±s.e.m. of three experiments. (b) Mean IL-6 and IL-10 cytokine production by X16C8.5 B cells of triplicate cultures after 24 h of stimulation w/wo CT at 0.1 μg/ml in the presence or absence of LPS at 10 μg/ml. The cytokine levels were assessed by ELISA and expressed in mean pg/ml±s.e.m. This is one representative experiment of three producing similar results. *Indicates statistical significance at P<0.05 above untreated control cells. CT, cholera toxin; ELISA, enzyme-linked immunosorbent assay; IL, interleukin; RT-PCR, real-time PCR; STAT, signal transducer and activator of transcription.
Augmented STAT3 responsiveness to IL-6 and IL-10 in CT-treated B cells
To also verify that, at the protein level, CT promoted STAT3 gene expression, we stimulated X16C8.5 B cells and assessed intracellular STAT3 levels using antibodies specific for the α- and β-isoforms in western blotting. A dose-response analysis with CT was performed and the results were compared with those of the enzymatically inactive CTB to address whether the effects observed were enzyme-dependent or independent. As shown in Figure 3a, we detected a strong dose-dependent augmented production of STAT3 protein, both α- and β-isoforms, in response to CT at 8 h, whereas no significant effect was seen at 2 h or with CTB. This result argued in favor of an adenosine diphosphate ribosyltransferase-dependent effect of CT (Figure 3).
CT induces STAT3α and STAT3β protein production in B cells in vitro. X16C8.5 B cells were untreated (Co) or treated with increasing amounts of CT (0.001–1 μg/ml) or CTB (1 μg/ml) for 2 or 8 h. (a) Expression levels of STAT3α and STAT3β were analyzed by western blot using specific Mabs, and filters were also stripped and reprobed with anti-PKB Mabs to control for equal loading. (b) STAT3α and STAT3β levels in X16C8.5 B cells treated with 0.1 μg/ml CT or 1 μg/ml CTB, respectively, were quantified and normalized to protein kinase B (PKB) levels and expressed as fold induction. (c) X16C8.5 B cells w/wo CT at 0.1 μg/ml for 8 h were extensively washed and subjected to IL-6 (40–5,000 U/ml) or IL-10 (1–200 ng/ml) for 15 min and the level of STAT3 activation (P-STAT3) and expression (pan-STAT3) were analyzed and equal loading is shown by anti-pan-PKB antibodies. Three independent experiments producing similar results were performed. *Indicates statistical significance at P<0.05 above unstimulated control cells. CT, cholera toxin; IL, interleukin; STAT, signal transducer and activator of transcription.
Next, we analyzed whether CT-exposed B cells were more responsive to limiting concentrations of IL-6 and IL-10. To this end, X16C8.5 B cells were pretreated with CT for 8 h, and after extensive washing, cells were exposed to a dose range of IL-6 or IL-10 for 15 min and subsequently analyzed for STAT3 phosphorylation as a sign of activation. We found increased sensitivity to both these cytokines and even low levels of IL-6 or IL-10 effectively activated STAT3 in CT-treated X16C8.5 B cells (Figure 3). This enhanced susceptibility to IL-10 and IL-6 was not due to enhanced receptor expression on B cells, as we failed to observe any change in either IL-10R or IL-6Rα expression in CT-treated B cells (not shown). Furthermore, CT also affected STAT3 levels in normal B cells after intravenous injections of 1 μg of CT in T-cell-deficient (nu/nu) mice. We found that sorted B cells from nu/nu mice showed significantly increased STAT3 phosphorylation as well as enhanced STAT3 levels, as compared with that found in B cells from untreated mice (Figure 4). This was most evident in B cells at 24 h, and was also found in B cells at 8 h after CT administration (not shown). Thus, we found evidence that CT affected levels and activation of STAT3 in both cultured B cells and in normal mouse B cells isolated from in vivo CT-treated mice. Importantly, CT promoted an enhanced sensitivity in B cells for STAT3 activation by limiting concentrations of IL-6 and IL-10.
CT activates STAT3α and STAT3β in B cells in vivo. Nu/nu mice were injected i.v. with PBS (Co) or 1 μg of CT (+CT). (a) After 24 h, mice were killed and splenic CD19+ B cells were enriched (>96% purity) and analyzed by western blot for STAT3α and STAT3β protein expression (STAT3α or STAT3β) or activation by anti-phosphorylated STAT3 Mab (P-STAT3α or P-STAT3β), and equal loading anti-PKB Mabs. (b) STAT3α and STAT3β mean expression levels or (c) activation levels (phosphorylation). Each lane represents an individual mouse (panel a) or pooled values from five mice (panels b, c). One representative experiment of three producing similar results. Statistical significance at P<0.05 (*). CT, cholera toxin; i.v., intravenous; PBS, phosphate-buffered saline; STAT, signal transducer and activator of transcription.
Functional impact of CT-induced STAT3 on B-cell differentiation
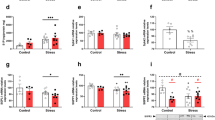
Whereas not all effects of IL-6 on B-cell differentiation have been linked to STAT3, in contrast, the effects of IL-10 are considered STAT3 dependent.25, 26 Therefore, we focused on IL-10 and assessed to what extent CT treatment of normal highly enriched splenic B cells affected the outcome of IL-10 treatment in LPS or anti-CD40 mAb-stimulated CD19+ B cells (>97% purity). We found that B cells treated with CT exhibited a higher level of phosphorylated STAT3 in response to IL-10 than did B cells cultured in the absence of CT (Figure 5a). Furthermore, we observed a more pronounced reduction in proliferation in response to IL-10 in LPS-stimulated cultures in the presence of CT, whereas CT and IL-10 treatment had no effect on anti-CD40 Mab-driven B-cell proliferation or IgG1 differentiation (Figure 5b and c). In contrast, CT and IL-10 used alone had only a minor effect in reducing LPS-driven B-cell proliferation (Figure 5b). Similar to what was shown in Figure 2, CT promoted significantly enhanced IL-6 production by LPS-treated B cells. In addition, other proinflammatory factors, such as MIP1α (macrophage inflammatory protein 1α) and RANTES were found to be upregulated in these cultures as well (not shown). However, the addition of IL-10 to the cultures reduced the IL-6 (as well as MIP1α or RANTES) levels by nearly 85% (Figure 5c). Thus, inhibition of cytokine production by IL-10 was more pronounced in the presence of CT compared with untreated LPS-stimulated B-cell cultures (Figure 5c). This inhibition of IL-6 production was IL-10 dose dependent in the presence of CT (not shown). The immunomodulating effect of CT on B-cell differentiation could also be assessed at the level of isotype differentiation. Our previous studies had documented an impact of CT on IgG1 differentiation, in particular, in the context of IL-4.10 In this study, we analyzed IgG1 production by LPS-stimulated B cells after addition of CT or IL-10 or the combination of the two. Whereas IL-10 promoted increased IgG1 production, the combination with CT was much more effective in enhancing IgG1 production in these cultures (Figure 5d). Thus, the regulatory role of IL-10 on B-cell isotype differentiation was augmented in CT-treated and LPS-stimulated B cells.
IL-10 modulation of B-cell functions is augmented in the presence of CT and correlates with increased STAT3 activation. (a) Freshly isolated and highly enriched (>97% purity) CD19+ splenic B cells were cultured in the absence (dotted line) or presence of IL-10 (10 ng/ml) plus CT (0.1 μg/ml) (broken line) or with IL-10 without CT (broken and dotted line), and the level of STAT3 activation (phosphorylation) was assessed by FACS. (b) B cells were cultured for 72 h with LPS (10 μg/ml) or anti-CD40 Mabs (10 μg/ml), and we determined the inhibition of B-cell proliferation (mean c.p.m.±s.d.) after adding IL-10 in the presence or absence of CT. (c) The inhibitory effect of IL-10 on IL-6 production (mean pg/ml±s.d.) by LPS-stimulated enriched B cells in the presence or absence of CT was assessed. (d) Finally, the augmenting effect of IL-10 on IgG1 production (mean ng/ml±s.d.) in LPS- (closed bars) or anti-CD40-stimulated (open bar) enriched B cell cultures in the presence or absence of CT was determined. These are representative experiments of three producing similar results and statistical significance at P<0.05 (*). CT, cholera toxin; FACS, fluorescence-activated cell sorting; IL, interleukin; LPS, lipopolysaccharide; STAT, signal transducer and activator of transcription.
CT also promotes STAT3 activation and gene transcription in mucosal B cells
Given that CT augmented IL-10 functions in splenic B cells concomitant with an increase in STAT3 gene transcription, we investigated whether this phenomenon also related to mucosal B cells. To this end, we isolated and highly enriched B cells from Peyer's patches (PPs) and cultured these cells in the presence of IL-10 and CT. In addition, with PP B cells, we observed significant STAT3 activation, and especially the combination of IL-10 with CT produced a strong upregulation of STAT3 phosphorylation in PP B cells as assessed by fluorescence-activated cell sorting (FACS) (Figure 6a). Furthermore, PP B cells cultured with IL-10 and CT had significantly higher expression of STAT3 mRNA than did cells cultured with either stimulant alone (Figure 6b). The membrane expression of IL-10R in PP B cells, as assessed by FACS, did not change after CT exposure (not shown). Finally, when PP B cells were cultured in the presence of IL-10 and CT, they produced more IgA than LPS-driven cultures with either factor alone (Figure 6c). Thus, CT affected mucosal B cells similar to that found with splenic B cells, arguing in favor of that STAT3 is a key element in CT's immunomodulating effects on B cells and could also be the critical denominator for the seemingly conflicting earlier reports on concomitant proinflammatory and anti-inflammatory effects by CT.
Peyer's patch PP B cells demonstrate enhanced STAT3 activation and gene transcription after CT treatment. (a) Freshly isolated and highly enriched (>97% purity) CD19+ PP B cells were cultured in the absence (dotted line) or presence of IL-10 (10 ng/ml) plus CT (0.1 μg/ml) (broken line) or with IL-10 without CT (broken and dotted line), and the level of STAT3 activation (phosphorylation) was assessed by FACS. PP B cells were cultured for 8 h with LPS (10 μg/ml), IL-10, and/or CT, and the fold change in STAT3 gene expression was determined relative to the expression level of the 36B4 housekeeping gene. (b) In each experiment, three samples were analyzed for each time point and the average fold increase in STAT3 gene expression is given as mean±s.e.m. of three experiments. (c) The augmenting effect of IL-10 on LPS-driven IgA production (log2-titer±s.d.) in enriched PP B cell cultures in the presence or absence of CT was determined. These are representative experiments of three producing similar results and statistical significance at P<0.05 (*). CT, cholera toxin; FACS, fluorescence-activated cell sorting; IL, interleukin; LPS, lipopolysaccharide; STAT, signal transducer and activator of transcription.
Discussion
This study is the first to investigate global gene regulation in B cells after exposure to CT adjuvant.27 Previous studies have documented effects by CT or E. coli labile toxin (LT) or derivatives of these holotoxins on mixed populations of human blood mononuclear cells or whole mouse lung tissues.28, 29 Our results both confirm and conflict with these reports, as will be discussed. However, a key and unique observation in this study is our finding that CT regulates STAT3 gene expression. A consequence of this is likely an increased susceptibility to regulatory cytokines such as IL-10 and IL-6, providing a molecular mechanism to partly explain the seemingly contradictory proinflammatory and anti-inflammatory effects of CT reported in earlier studies.15, 30 Whereas STAT3 is essential for all anti-inflammatory effects of IL-10, it has also gained increasing attention as a critical regulator of IL-6-driven functions in B-cell development and plasma cell differentiation, in particular.25, 31, 32, 33, 34 Moreover, apart from the well-known involvement of STAT3 in the generation of proinflammatory Th17 and anti-inflammatory Treg cells and as a mediator of IL-27-dependent proliferation of CD4+ T cells, it has also recently been shown to have a central role in IL-6-driven differentiation of CD4+ T cells into helper cells (Th cells) for B cells.19, 35, 36, 37, 38 Thus, by augmenting STAT3 gene expression, CT may critically modulate immune responses in both a proinflammatory and anti-inflammatory direction, depending on the circumstances and the types of cells involved.
Using Affymetrix global gene expression profiling, we found evidence that CT significantly upregulated STAT3 gene expression in murine B cells. This finding was confirmed by RT-PCR both in vitro and in vivo, showing that CT upregulated STAT3 gene expression in a B-cell line (X16C8.5) as well as in normal splenic or mucosal PP B cells after exposure to CT (P<0.05). This effect appeared to be cAMP dependent as no change in STAT3 protein levels was observed after treatment with CTB, the receptor-binding element of CT holotoxin, lacking the cAMP-inducing A1 subunit.39 This agrees well with a recent report showing that activators of Gsα-coupled receptors can directly stimulate STAT3 phosphorylation in human embryonic kidney cells (HEK293), a mechanism that was dependent on protein kinase A, JNK, and phophatidylinositol 3-kinase.40 As the A1 subunit of CT-holotoxin activates Gsα, we propose that CT increased intracellular STAT3 protein and phosphorylation levels in B cells through a Gsα-mediated pathway and that this process was cAMP dependent in analogy with the effects observed by Liu et al.,40 when using cAMP analogs or forskolin in HEK293 cells. In this study, we extend the information by showing that CT exposure also affects STAT3 gene transcription, arguing that CT probably mediates this effect by increasing intracellular cAMP. Whether there is a cAMP-responsive element in the promoter region of the STAT3 gene is currently debated.16 However, we favor this interpretation rather than secondary transcriptional effects on the STAT3 gene, such as those induced by, e.g., IL-6 or IL-10 receptor signaling, because, first, we found significantly enhanced STAT3 gene transcription despite no detectable IL-6 or IL-10 produced by X16C8.5 cells, and second, CT exposure affected gene transcription already after 2 h, when LPS-induced IL-6 and IL-10 production in B cells was undetectable or very low (ASH, unpublished observation).
The biological consequence of CT treatment of B cells could be enhanced susceptibility to IL-10-mediated regulation, as shown in this study by dramatically augmenting IL-10-induced inhibition of LPS-driven proliferation and cytokine production. In contrast, the IgG1- or IgA-promoting effect of IL-10 was further enhanced by exposure to CT, resulting in almost 100% increased IgG1 or even more for IgA production in culture. It is probable that this effect of CT relates to our previous documented effect of CT on IL-4 plus LPS-driven B-cell isotype differentiation from IgM to IgG1.10 Thus, it appeared that CT, by augmenting STAT3, significantly modulated both splenic and mucosal PP B-cell responses to IL-10.32 We also found that B cells pretreated with CT showed enhanced activation (phosphorylation) of STAT3 when exposed to even low levels of IL-10 in vitro. It is noteworthy that, in anti-CD40 Mab-driven B-cell proliferation, we observed no inhibitory effect of IL-10 and subsequently CT exerted no modulating effect in this system, suggesting that STAT3 was not involved. Moreover, we found no effect of CT and IL-10 on IgG1 production in the anti-CD40 Mab-driven cultures. This observation also supports our finding that CT did not simply upregulate IL-10R or IL-6Rα in B cells, but rather affected the intracellular signal transduction pathway by upregulating STAT3. Importantly, CT affected STAT3 gene expression and both alternatively spliced STAT3α and STAT3β isoforms were more abundant in murine B cells.41 What the functional implication of this is can presently only be speculated as the isoforms have been ascribed unique and specific functions.42 A characteristic of STAT3 is its capacity to activate different sets of genes in different cell types, and with the two isoforms, the complexity of this system is well appreciated.43 Several cytokines are known to signal through STAT3, among them are IL-6 and IL-10.17 In particular, the anti-inflammatory effects of the latter have been found to be critically dependent on STAT3, but whether this effect is mediated by the α- or β-isoform or by both isoforms is still debated.42, 43 On the other hand, other functions, such as responsiveness to IL-6 in macrophages, appear more restricted to STAT3α .41 The role of STAT3 in B cells is only beginning to emerge.25 We observed enhanced IL-6 production by LPS-stimulated B cells after exposure to CT, which is in line with previous reports using macrophages or DCs.12 Whether this enhanced production of LPS-stimulated IL-6 involved STAT3 cannot be concluded from this study. However, in analogy with what has been reported for macrophages, a differential effect of CT on STAT3β more than STAT3α (partly supported by our data in Figures 4 and 5) could possibly account for the augmenting effect on IL-6 production concurrent with a reduced IL-10 production (Figure 3, X16C8.5 cells) and the strengthened inhibitory effect of IL-10 that we observed in the presence of CT in LPS-stimulated B cells (Figure 6).41
Global gene profiling showed that CT upregulated several other genes that encode proteins with diverse and critical functions, impacting on the immune system, which could also be part of the immunoenhancing functions of CT in vivo. In this list of genes, we find early-response genes, and genes involved with transcription, cell-cycle control, and membrane signaling, which is in agreement with the studies by Royaee et al.29 However, at striking variance with their study, we found less than half (<100 genes) as many genes to be regulated by CT exposure. This may be because we analyzed B cells exclusively, whereas Royaee et al. investigated a mix of lymphocytes and monocytes. Moreover, we failed to detect a direct effect of CT on proinflammatory cytokine genes as reported by Royaee et al., but we clearly observed an upregulating effect on the costimulatory genes encoding CD83 and CD86, in agreement with previous reports.11, 29 This can perhaps also be explained on the basis of the selection of target cells, B cells, or the mix of cells, for assessing the effects of CT, as gene transcriptional effects of CT on proinflammatory cytokines genes, IL-1 and IL-6, have previously been reported with DCs as well as epithelial cells by others.13, 44 It is noteworthy although that the presence of contaminating small amounts of endotoxin could also have accounted for the differences observed.13 Cong et al.12 found that IL-6 production by macrophages was partly impaired by the concomitant production of IL-10, as anti-IL-10 antibody further enhanced IL-6 production. Given the role of CT on the regulation of STAT3 gene expression in B cells demonstrated in this study, we may speculate that the inhibitory effect of IL-10 in macrophages could also have been due to an increased sensitivity of the STAT3 pathway as a consequence of CT-enhanced STAT3 gene expression. Ongoing studies in macrophages will further evaluate this idea. Notwithstanding this, in this study, we showed that one important pathway for CT immunomodulation is mediated by STAT3 signaling in B cells. In addition, we clearly showed that this immunomodulating effect of CT results in enhanced susceptibility for STAT3-regulated actions of IL-10 both in splenic and mucosal B cells leading to augmented IgG1 and IgA antibody production. We expect that this finding together with further analysis of molecular events underlying the adjuvant action of CT will pave the way for a better understanding of key molecular components of immunomodulation and for the future development of much improved mucosal vaccines.
Methods
Mice. Six-week-old female nu/nu mice (C57BL/6) from the Jackson Laboratory (Maine, MA) were used.
Cell lines and criteria. Screening of B-cell lines was undertaken to select a candidate for further studies of the effects of CT (List Biological Labs, Campbell, CA) on global gene expression; A20 (ATCC, Manassas, VA (American Type Culture Collection) TIB-208),45 M12 4.1 (ATCC CRL-2826),45 CH12LX (a generous gift from Dr Geoffrey Haughton), X16C8.5 (ATCC TIB-209).46 The selection criteria were CT induction of intracellular cAMP increases and upregulated expression of membrane CD80 and CD86.47 Target cells were stimulated with CT at 0.1 μg/ml for 24 h and analyzed by FACS for expression of membrane molecules (BD Biosciences, San Jose, CA). CT-induced increases in intracellular cAMP were determined by ELISA (enzyme-linked immunosorbent assay) (Amersham Biosciences, Little Chalfont, England) according to the manufacturers’ instructions.
Cell cultures. Freshly isolated splenic or PP B cells or X16C8.5 cells were cultured in 96-well cultures (Nunc, Roskilde, Denmark) at 106 cells per ml in Iscove's modified Dulbecco's medium (Gibco-BRL, Invitrogen, Carlsbad, CA), containing 10% heat-inactivated fetal bovine serum (Gibco-BRL, Invitrogen), 100 μg/ml of gentamycin (Sigma-Aldrich, St Louis, MO) 2 mM L-glutamine (Biokrom KG, Berlin, Germany), and 50 μM β-mercaptoethanol. The cells were stimulated for the indicated time points (2 or 8 h for Affymetrix and RT-PCR, 16 h for STAT3 western blot analysis, 24 h for FACS analysis, 72 h for cytokine detection, and 5 days for IgG1 or IgA antibody production by ELISA assays) at 37°C with various amounts of CT (ranging from 1 to 0.0001 μg/ml CT) or 1 μg/ml CTB (both from List Biological Labs), in the presence or absence of recombinant IL-10 (R&D Systems, Minneapolis, MN) at indicated concentrations w/wo LPS (Sigma-Aldrich) or anti-CD40 Mab (BD Biosciences) at 10 μg/ml. CT was routinely tested for endotoxin and contained <70 EU/mg protein. Cell sorting was performed with AutoMacs using anti-CD19 microbeads according to the manufacturer's description (Miltenyi Biotech, Bergisch Gladbach, Germany). A purity >97% was routinely achieved as assessed by FACS. Cytokine IL-6 and IL-10 or chemokine MIP1α or RANTES were determined in indicated cultures using specific antibody kits (R&D Duoset, R&D Systems or BD Biosciences) according to the manufacturer's instructions.
FACS analysis, cell proliferation, IgG1, IgA, and cytokine ELISA. Cells were stimulated with CT at 0.1 μg/ml, IL-10 at 100 ng/ml, or the combination for 24 h and analyzed by FACS (BD Biosciences) for the level of phosphorylated STAT3 using the specific antibody (phospho-STAT3/Y705, BD Biosciences) and isotype-control antibodies (BD Biosciences). Membrane expressions of IL-10R and IL-6Rα were detected by phycoerythrin-labeled CD126- and CD210-specific antibodies (BD Biosciences), respectively. Highly enriched CD19+ splenic or PP B cells from nu/nu mice were cultured in 96-well microtiter plates (Nunc) for 3 days in medium or with LPS at 10 μg/ml w/wo CT at 0.1 μg/ml and w/wo IL-10 at 100 ng/ml (R&D Systems). B-cell proliferation was determined using a β-scintillation counter (Beckman Coulter, Bromma, Sweden) after addition of [3H]Thymidine (Amersham Biosciences) for the last 6 h of culturing. Supernatants were collected after 72 h of culturing and stored at −70°C until assayed. Cytokine and chemokine production was determined using ELISA specific for IL-6 or IL-10 using the Duoset (R&D Systems) or antibodies against MIP1α or RANTES (BD Biosciences) as described earlier.48 The concentrations of the cytokines or chemokines were calculated from standard curves generated with recombinant cytokines. The sensitivity for detection of IL-6 or IL-10 was 5 pg/ml. Cultures were allowed 5 days before assessing the IgG1 or IgA production.49
Expression profiling. Three separate experiments were performed with X16C8.5 B cells and CT at 0.1 μg/ml for 2 or 8 h. Anti-sense cRNA preparation was carried out as recommended by Affymetrix. In brief, total RNA was obtained by Trizol reagent (Life Technology, Invitrogen), RNA was then purified by RNeasy columns (Qiagen, Hilden, Germany), and thereafter converted into cDNA by using a T7 promoter-polyT primer and the reverse transcriptase Superscript II (Gibco-BRL/Invitrogen), followed by a second-stranded cDNA synthesis into ds cDNA (Gibco-BRL/Invitrogen). This ds cDNA was cleaned up in a phase-lock gel-phenol/chloroform extraction (Eppendorf, Bergman & Beving, Uppsala, Sweden), the ds cDNA was precipitated, in vitro transcribed into biotinylated cRNA (Enzo Diagnostics, NY, NY), and the resulting cRNA was passed over RNeasy columns (Qiagen). This cRNA was then fragmented into 50–200 bp length by incubating it for 35 min at 94°C in 40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, 30 mM magnesium acetate. The target for hybridization was prepared by combining 0.05 μg/μl fragmented cRNA, 50 pM control oligonucleotide B2, control cRNA cocktail containing BioB, BioC, BioD, and cAMP-responsive element at concentrations of 1.5, 5, 25, and 100 pM, respectively, 0.1 mg/ml of sonicated herring sperm DNA, 0.5 mg/ml of acetylated bovine serum albumin, 100 mM 2-(N-morpholino) ethanesulfonic acid buffer (MES), 1.0 M NaCl, 20 mM EDTA, and 0.01% Tween 20. The target was heated for 5 min at 99°C, then transferred to 45°C for 5 min, and spun down, and the liquid upper phase was allowed to hybridize for 16 h at 45°C and 60 r.p.m. to a set of either MG-U74A chips A, MG-U74B, and MG-U74C chips, or MG-U74Av2, MG-U74Bv2, and MG-U74Cv2 chips, or only MG-U74Av2. After hybridization, the chips were washed and stained with streptavidin–phycoerythrin (Molecular Probes, Eugene, OR) according to the EukGE-WS2v3 program (Affymetrix). The fluorescence intensity was read using a confocal scanner and analyzed with the MicroArray Suite 4.0 (MAS 4.0) Gene Expression analysis program (Affymetrix).
Data analysis. The resulting CEL files from the MAS 4.0 evaluation were further analyzed with the dChip method, which can be obtained at http://biosun1.harvard.edu/Idchip.exe. The dChip method is a software package implementing model-based expression analysis of oligonucleotide arrays, including comparative analysis and hierarchical clustering. The following filters were used to select the probe sets potentially influenced by CT: P≤0.05, fold change ≥1.9, difference of mean ≥100, and the probe set had to be present in at least two of the three experiments. The values listed in Tables 1 and 2 are means of the resulting fold-change values at 2 or 8 h of CT stimulation as compared with unstimulated controls.
In the hierarchical clustering, UPGMA was used with correlation distance, which was supplied with the Spotfire program (TIB, Sommerville, MA). Before clustering of the genes, the variables were standardized within the respective experiments in dChip and then selected for variables likely to be regulated by CT with the following selection criteria: P≤0.05, fold change ≥2.0, and difference of mean ≥200. The selected variable intensities were then transformed to the respective logarithmic values. The mean of the logarithmic intensities for unstimulated B cells at 2 and 8 h was calculated, and the mean fold change calculated for each gene for all the three experiments was then used for clustering.
RT-PCR. Total RNA that was prepared for DNA array investigations was also used in RT-PCR experiments. After Trizol extractions of the RNA DNase I treatment removed any contaminating DNA (DNA-free Ambion, Intermedica, Stockholm, Sweden), cDNA was obtained by the Superscript first-strand synthesis system for RT-PCR (Gibco-BRL/BRL). RT-PCR was then performed using the cDNA and SYBR Green PCR Master Mix on an ABI PRISM 7700 Sequence Detection System according to the manufacturer's recommendations (both from Applied Biosystems, Foster City, CA). All PCR products were then size determined by gel electrophoresis, and the relative amounts of PCR product in the different RNA preparations were evaluated by comparison with the expression of the housekeeping 36B4 gene in the same samples. The primer sequences for the STAT3 gene were as followswith forward primer followed by reverse primer; GGGCATTTTTATGGCTTTCAAT and GTTAACCCAGGCACACAGACTTC, respectively.
Detection of STAT3 by western blot. Cells samples were prepared and subjected to western blot analysis as described previously.50 For analysis of IL-6- and IL-10-stimulated (R&D Systems) activation of STAT3, cells were pretreated w/wo 0.1 μg/ml of CT for 8 h and after washing stimulated for 15 min by a dose range of IL-6 or IL-10 as indicated. Cell extractions were purified and subjected to western blot analysis. Anti-STAT3 antibody (#9132, Cell Signalling Technology, Stockholm, Sweden) and anti-phospho-STAT3 (Tyr705) antibody (#9131, Cell Signalling Technology) were used to detect STAT3α (86 kDa) and STAT3β (79 kDa) activation and expression levels, respectively. Data were quantified using a CCD camera and Chemidoc XRS system (Bio-Rad Laboratories, Hercules, CA) and analyzed by Quantity One software (Bio-Rad Laboratories). For quantification, specific signals were visualized by Immun-Star Chemiluminescence (Bio-Rad Laboratories).
Statistical analysis. Comparisons between treatment groups were performed with the Mann–Whitney test using the SPSS 13.0 software (SPSS, Chicago, IL). All reported P-values are two-sided and values <0.05 were considered to indicate statistical significance. P<0.05 (*).
References
Lycke, N. Mechanisms of adjuvant action. In Vaccine Adjuvants and Delivery Systems (Singh, M., ed) 53–79 ( John Wiley & Sons, Hoboken, NJ, 2007 ).
Levine, M.M. & Sztein, M.B. Vaccine development strategies for improving immunization: the role of modern immunology. Nat. Immunol. 5, 460–464 (2004).
Shaffer, A.L. et al. Signatures of the immune response. Immunity 15, 375–385 (2001).
Ricciardi-Castagnoli, P. & Granucci, F. Opinion: interpretation of the complexity of innate immune responses by functional genomics. Nat. Rev. Immunol. 2, 881–889 (2002).
Holmgren, J. & Czerkinsky, C. Mucosal immunity and vaccines. Nat. Med. 11, S45–S53 (2005).
Rappuoli, R., Pizza, M., Douce, G. & Dougan, G. Structure and mucosal adjuvanticity of cholera and Escherichia coli heat-labile enterotoxins. Immunol. Today 20, 493–500 (1999).
Fujihashi, K., Koga, T., van Ginkel, F.W., Hagiwara, Y. & McGhee, J.R. A dilemma for mucosal vaccination: efficacy versus toxicity using enterotoxin-based adjuvants. Vaccine 20, 2431–2438 (2002).
Torgersen, K.M., Aandahl, E.M. & Tasken, K. Molecular architecture of signal complexes regulating immune cell function. Handb. Exp. Pharmacol. 4, 327–363 (2008).
Elson, C.O. & Dertzbaugh, M.T. Mucosal adjuvants. In Mucosal Immunology, Vol 1 (Mestecky, J., et al. eds) 967–986 ( Elsevier Academic Press, San Diego, CA, 2005 ).
Lycke, N., Severinson, E. & Strober, W. Cholera toxin acts synergistically with IL-4 to promote IgG1 switch differentiation. J. Immunol. 145, 3316–3324 (1990).
Grdic, D. et al. Splenic marginal zone dendritic cells mediate the cholera toxin adjuvant effect: dependence on the ADP-ribosyltransferase activity of the holotoxin. J. Immunol. 175, 5192–5202 (2005).
Cong, Y., Oliver, A.O. & Elson, C.O. Effects of cholera toxin on macrophage production of co-stimulatory cytokines. Eur. J. Immunol. 31, 64–71 (2001).
Lavelle, E.C. et al. Effects of cholera toxin on innate and adaptive immunity and its application as an immunomodulatory agent. J. Leukoc. Biol. 75, 756–763 (2004).
Gagliardi, M.C. & De Magistris, M.T. Maturation of human dendritic cells induced by the adjuvant cholera toxin: role of cAMP on chemokine receptor expression. Vaccine 21, 856–861 (2003).
Braun, M.C., He, J., Wu, C.Y. & Kelsall, B.L. Cholera toxin suppresses interleukin (IL)-12 production and IL-12 receptor beta1 and beta2 chain expression. J. Exp. Med. 189, 541–552 (1999).
Brierley, M.M. & Fish, E.N. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 25, 733–744 (2005).
Fu, X.Y. STAT3 in immune responses and inflammatory bowel diseases. Cell Res. 16, 214–219 (2006).
Sakamori, R. et al. Signal transducer and activator of transcription 3 signaling within hepatocytes attenuates systemic inflammatory response and lethality in septic mice. Hepatology 46, 1564–1573 (2007).
Liu, X., Lee, Y.S., Yu, C.R. & Egwuagu, C.E. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J. Immunol. 180, 6070–6076 (2008).
Ding, B.B. et al. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 111, 1515–1523 (2008).
Fornek, J.L. et al. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood 107, 1085–1091 (2006).
Granucci, F. et al. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat. Immunol. 2, 882–888 (2001).
Tureci, O. et al. Cascades of transcriptional induction during dendritic cell maturation revealed by genome-wide expression analysis. Faseb. J. 17, 836–847 (2003).
Royaee, A.R. et al. Cholera toxin induced novel genes in human lymphocytes and monocytes. Mol. Immunol. 43, 1267–1274 (2006).
Hirano, T., Ishihara, K. & Hibi, M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 19, 2548–2556 (2000).
Nishinakamura, H. et al. An RNA-binding protein alphaCP-1 is involved in the STAT3-mediated suppression of NF-kappaB transcriptional activity. Int. Immunol. 19, 609–619 (2007).
Royaee, A.R. et al. Induction of immunomodulator transcriptional responses by cholera toxin. Mol. Immunol. 43, 1020–1028 (2006).
Tritto, E. et al. The acquired immune response to the mucosal adjuvant LTK63 imprints the mouse lung with a protective signature. J. Immunol. 179, 5346–5357 (2007).
Royaee, A.R., Mendis, C., Das, R., Jett, M. & Yang, D.C. Cholera toxin induced gene expression alterations. Mol. Immunol. 43, 702–709 (2006).
Burkart, V. et al. Cholera toxin B pretreatment of macrophages and monocytes diminishes their proinflammatory responsiveness to lipopolysaccharide. J. Immunol. 168, 1730–1737 (2002).
Calame, K. Activation-dependent induction of Blimp-1. Curr. Opin. Immunol. 20, 259–264 (2008).
Murray, P.J. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 6, 379–386 (2006).
Takeda, K. et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49 (1999).
Williams, L., Bradley, L., Smith, A. & Foxwell, B. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 172, 567–576 (2004).
Owaki, T. et al. STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J. Immunol. 180, 2903–2911 (2008).
Matsumura, Y. et al. Selective expansion of foxp3-positive regulatory T cells and immunosuppression by suppressors of cytokine signaling 3-deficient dendritic cells. J. Immunol. 179, 2170–2179 (2007).
Mathur, A.N. et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178, 4901–4907 (2007).
Eddahri, F. et al. Interleukin-6/STAT3 signalling regulates the ability of naive T cells to acquire B cell help capacities. Blood 113, 2426–2433 (2009).
Holmgren, J. Actions of cholera toxin and the prevention and treatment of cholera. Nature 292, 413–417 (1981).
Liu, A.M. et al. Activation of STAT3 by G alpha(s) distinctively requires protein kinase A, JNK, and phosphatidylinositol 3-kinase. J. Biol. Chem. 281, 35812–35825 (2006).
Maritano, D. et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 5, 401–409 (2004).
Dewilde, S., Vercelli, A., Chiarle, R. & Poli, V. Of alphas and betas: distinct and overlapping functions of STAT3 isoforms. Front Biosci. 13, 6501–6514 (2008).
Levy, D.E. & Lee, C.K. What does Stat3 do? J. Clin. Invest. 109, 1143–1148 (2002).
Blumberg, R.S., Pitman, R.S., Taylor, C.T. & Colgan, S.P. Cholera toxin potentiates influences of IFN-gamma through activation of NF-kappaB and release of tumor necrosis factor-alpha. J. Interferon Cytokine Res. 25, 209–219 (2005).
Kim, K.J., Kanellopoulos-Langevin, C., Merwin, R.M., Sachs, D.H. & Asofsky, R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 122, 549–554 (1979).
Kim, K.J., Chaouat, G., Leiserson, W.M., King, J. & De Maeyer, E. Characterization of T-cell-soluble factors modulating the expression of Ia and H-2 antigens on BALB/c B lymphoma cell lines. Cell Immunol. 76, 253–267 (1983).
Lycke, N. A molecular approach to the construction of an effective mucosal vaccine adjuvant: studies based on cholera toxin ADP-ribosylation and cell targeting. In Essentials of Mucosal Immunology (Kagnoff, M. & Kiyono, H., eds) 563–580 ( Academic Press Inc, San Diego, 1996 ).
Akhiani, A.A., Stensson, A., Schon, K. & Lycke, N. The nontoxic CTA1-DD adjuvant enhances protective immunity against Helicobacter pylori infection following mucosal immunization. Scand. J. Immunol. 63, 97–105 (2006).
Lycke, N.Y. Cholera toxin promotes B cell isotype switching by two different mechanisms. cAMP induction augments germ-line Ig H-chain RNA transcripts whereas membrane ganglioside GM1-receptor binding enhances later events in differentiation. J. Immunol. 150, 4810–4821 (1993).
Marklund, U., Lightfoot, K. & Cantrell, D. Intracellular location and cell context-dependent function of protein kinase D. Immunity 19, 491–501 (2003).
Acknowledgements
This study was supported by the Swedish Research Council, the Swedish Cancer Foundation, The Swedish Foundation for Strategic Research MIVAC grant, the Norwegian Research Council Globvac, The Wellcome Trust, the Sahlgrenska University Hospital Foundation, and EU Grant numbers QLK2-CT-2001-01702, LSHP-CT-2003-503240, COOP-CT-2005-017749.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declared no conflict of interest.
Rights and permissions
About this article
Cite this article
Sjöblom-Hallén, A., Marklund, U., Nerstedt, A. et al. Gene expression profiling identifies STAT3 as a novel pathway for immunomodulation by cholera toxin adjuvant. Mucosal Immunol 3, 374–386 (2010). https://doi.org/10.1038/mi.2010.16
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/mi.2010.16