Abstract
Fine-needle aspiration (FNA) of pancreatic solid masses can be significantly impacted by sampling variation. Molecular analysis of tumor DNA can be an aid for more definitive diagnosis. The aim of this study was to evaluate how molecular analysis of the cell-free cytocentrifugation supernatant DNA can help reduce sampling variability and increase diagnostic yield. Twenty-three FNA smears from pancreatic solid masses were performed. Remaining aspirates were rinsed for preparation of cytocentrifuged slides or cell blocks. DNA was extracted from supernatant fluid and assessed for DNA quantity spectrophotometrically and for amplifiability by quantitative PCR (qPCR). Supernatants with adequate DNA were analyzed for mutations using PCR/capillary electrophoresis for a broad panel of markers (KRAS point mutation by sequencing, microsatellite fragment analysis for loss of heterozygosity (LOH) of 16 markers at 1p, 3p, 5q, 9p, 10q, 17p, 17q, 21q, and 22q). In selected cases, microdissection of stained cytology smears and/or cytocentrifugation cellular slides were analyzed and compared. In all, 5/23 samples cytologically confirmed as adenocarcinoma showed detectable mutations both in the microdissected slide-based cytology cells and in the cytocentrifugation supernatant. While most mutations detected were present in both microdissected slides and supernatant fluid specimens, the latter showed additional mutations supporting greater sensitivity for detecting relevant DNA damage. Clonality for individual marker mutations was higher in the supernatant fluid than in microdissected cells. Cytocentrifugation supernatant fluid contains levels of amplifiable DNA suitable for mutation detection and characterization. The finding of additional detectable mutations at higher clonality indicates that supernatant fluid may be enriched with tumor DNA. Molecular analysis of the supernatant fluid could serve as an adjunct method to reduce sampling variability and increase diagnostic yield, especially in cases with a high clinical suspicion for malignancy and limited number of atypical cells in the smears.
Similar content being viewed by others
Main
Pancreatic cancer rates have been recently reported to be on the rise.1 Moreover, the lack of early symptomatic manifestation of disease usually results in lack of early detection. The overall dismal prognosis combined with the rising incidence make pancreatic cancer a growing public health concern.2 With the American Cancer Society estimating that there will be 45 220 newly diagnosed cases of pancreatic cancer in 2013 and an estimated 38 460 pancreatic cancer-related deaths in the same year,3 pancreatic cancer is becoming the fourth leading cause of cancer-related death among men and the third leading cause of cancer-related death among women in the United States.3 Moreover, the 5-year survival remains around 5%, essentially unchanged in the last three decades.4
Recent advancements in imaging techniques have improved the detection of pancreatic solid and cystic pancreatic lesions and have led to an increase in the absolute number of pancreatic cytological specimens obtained for evaluation.5, 6, 7 While fine-needle aspiration (FNA) cytology represents a minimally invasive, well-tolerated procedure that provides initial, and sometimes the only, pathological assessment, it is often limited by paucicellular specimens offering few or, at times, no cells suitable for cytomorphological evaluation.8 According to a large meta-analysis study evaluating 33 relative previous studies with a total population of 4984 patients, the negative predictive value for pancreatic solid mass fine-needle aspirates was only 65%.9
In FNA of solid pancreatic masses, the risk of underdiagnosis is present and is related to failure to sample the neoplastic cell population. Furthermore, the biological variability within the neoplastic cell population, makes for a spectrum of morphological and molecular changes, present in different topographical areas of a solid mass.10 This may represent a diagnostic challenge when there is morphological evidence of cellular atypia, but insufficient evidence for a definite diagnosis of cancer.
Unfortunately, cytological underdiagnosis of these lesions is not the only challenge, as false positive diagnosis can occur with atypical but reactive processes.11, 12 Although the false positive rates for pancreatic cytology are generally estimated to be in the range of 0–1%, more recent studies, comparing positive and suspicious cytology diagnosis with their histological counterpart, found the false positive rates to be ∼5%.13
Correlation of the cytomorphological findings with molecular biomarker tests may provide a more accurate evaluation and a means to surmount the limitations of cytopathology examination. For this purpose, panels of pancreatic cancer biomarkers have been developed.14, 15 Furthermore, the molecular analysis might prove to be of great help in addressing problems related to low sample volumes, few cells for evaluation and/or sampling inadequacy.
PCR-based methods are employed on pancreatic cytology specimens such as FNAs to evaluate the mutational profile of the cells following cell microdissection from the cytology slides, in an attempt to resolve indeterminate microscopic diagnosis and to provide clinically actionable information not otherwise obtained by microscopic cellular examination.16, 17, 18, 19, 20, 21, 22, 23 The use of these techniques nonetheless requires the presence of not only an adequate amount of cellular material, but also cellular material that shows cytomorphological atypia, in order to be selected for microdissection. In fact, paucicellular or acellular specimens not only do not provide optimal material for cytological evaluation but also, at times, fail to provide material for cell-based DNA mutational analysis. However, we believe that the neoplastic cell population, when compared with the normal surrounding tissue, can exhibit an increased cellular turnover and the progression of carcinogenesis reflects the progressive mutation acquisition that these cells undergo. The result of this increased activity is the shedding of DNA in the interstitial microenvironment of the tumor and ultimately, in the lymphatic and blood circulation. The concept of cell-free tumor DNA is well established and is at the base of the development of genetic humoral cancer markers in a multitude of cancer types.24, 25, 26, 27, 28, 29, 30, 31, 32
Moreover, it has been suggested that in the presence of a malignant tumor there are high amounts of cell-free DNA, a byproduct of malignant cell necrosis,33 whose release has different mechanisms when compared with non-tumor cell-free DNA that is a product of normal cellular apoptosis.34 An increased amount of cell-free DNA can be an indicator of malignancy.35, 36 The presence of circulating cell-free tumor DNA corroborates the shedding of this DNA into the local extracellular space that is sampled along with the cells in an FNA procedure. The concept of studying this local extracellular DNA is a novel one for the study of pancreatic cancer and can complement cellular DNA analysis especially in the cases where scarce or no cellular material is present in the smears. Therefore, we believe that cell-free DNA released from the tumor cells, even when the malignant cells are not sampled, can yield DNA suitable for evaluation.
Often, FNA samples of cystic and occasionally solid pancreatic masses undergo cytocentrifugation with the cellular component used for cytology and molecular analyses and the supernatant is usually discarded. However, our hypothesis is that the supernatant fluid that is left after cytocentrifugation does in fact contain cell-free tumor DNA, and this DNA may represent the extracellular space that surrounds the cancer cells and contains high-quality tumor cell-free DNA.23 Furthermore, the utilization of the cytocentrifugation supernatant, normally discarded, can prove to be of utmost value in those cases where the cellular material is scarce or even absent, as tumor cell-free DNA can still be analyzed, without compromising any of the material routinely used for other diagnostic studies.
These concepts prompted the examination of the cell-free supernatant fluid from cytocentrifugation of solid pancreatic mass fine-needle aspirates for the presence and characterization of cell-free DNA for pancreatic cancer-associated molecular changes. Cellular DNA from microdissected tumor cells of the same specimens underwent the same type of analysis for comparison.
Materials and methods
Study Population
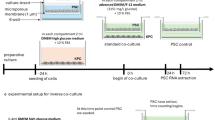
Following IRB approval, stained cytology slides and the corresponding supernatant fluid specimens were collected from 22 patients. Cytological adequacy assessment and diagnosis were carried out using established morphologic criteria.37 Standard cytology procedures for FNA biopsies were employed, involving fixation of the fine-needle aspirated cells with Shandon Cytospin® Collection Fluid (Thermo Scientific Anatomical Pathology USA, Pittsburgh, PA), followed by standard cytology slide preparation. The needle was then rinsed with Hank’s Balanced Salt Solution and the rinsings were centrifuged. For each case, the pelleted cells were used for the preparation of the cytology cell block, while the supernatant was saved, refrigerated at 4°C. The part of this supernatant underwent DNA extraction and mutational analysis within 48 h of the procedure (Figure 1).
The 23 specimens from 22 patients underwent cytological evaluation and diagnosed as follows: 5 pancreatic adenocarcinomas, 5 mucinous neoplasms, 5 cases of pancreatitis, 1 neuroendocrine neoplasm, 4 negative, and 3 specimens characterized as suboptimal (paucicellular or acellular) on cytomorphological evaluation. For all cases, including the ones diagnosed as negative, the indication for FNA was the diagnostic finding of a solid pancreatic mass (19/22 cases) or of a cystic mass with a solid component (3/22 cases) upon radiological examination. For the five cases diagnosed as pancreatic adenocarcinomas, surgical excision was performed in the follow-up and confirmatory histological diagnosis was used as the diagnostic gold standard. For the five cases diagnosed as mucinous neoplasms, surgical excision was performed and a histological diagnosis of either intraductal papillary mucinous neoplasms (4 out of 5 cases) or mucinous cystic neoplasm (1 out of 5 cases) was rendered and used as a diagnostic gold standard as well. The one case diagnosed as neuroendocrine neoplasm had a follow-up excision that confirmed the cytological diagnosis. One of the cases deemed as suboptimal (number 23), showed a mucinous neoplasm on repeat cytology, result that was confirmed by the excision specimen, histologically diagnosed as intraductal papillary mucinous neoplasm. The rest of the cases had no malignancy identified on subsequent clinical follow-up. The average negative follow-up duration for these 11 patients was 7.73 months (follow-up data range: 4–12 months).
Under microscopy guidance, cell targets were microdissected and underwent mutational analysis. The supernatant obtained from every specimen underwent molecular analysis, and the results were paired with the ones obtained from the corresponding microdissected slides.
Molecular Analysis
DNA was extracted from the microdissected stained cytology slides, molecular analysis was performed and the findings were compared with the results obtained from the DNA extracted from the corresponding cytocentrifugation supernatant fluid, for each of the samples. Supernatant fluid (2 ml) underwent DNA extraction (Qiagen, Valencia, CA). The resulting DNA was resuspended in hypotonic buffer and quantified by optical density (NanoDrop, Thermo Scientific, Wilmington, DE). Cytology-stained slide microdissected cells underwent the same DNA extraction and resuspension processing. DNA amplifiability was determined by quantitative PCR (qPCR) targeting a 150-bp length of the first coding exon of the KRAS oncogene. KRAS point mutation determination targeted codons 12 and 13 using dideoxy chain termination. Possible allelic imbalance was evaluated by loss of heterozygosity (LOH) investigation. A panel of 16 microsatellite markers was used for this purpose. These markers targeted common sites for tumor-suppressor genes associated with pancreaticobiliary cancers and have previously undergone analytic and clinical validation for pancreaticobiliary disease as reported in prior studies,16, 17, 18, 19, 20, 21, 22, 23 and present at the following chromosomal locations: 1p (CMM1, Lmyc), 3p (VHL, OGG1), 5q (MCC, APC), 9p (CDKN2A, CDKN2B), 10q (PTEN, MXI1), 17p (TP53), 17q (NME1, RNF43), 21q, 22q (NF2) using quantitative fluorescent PCR/capillary electrophoresis.
Quantitative allelic imbalance determination was performed as previously described.38 The threshold for determining a significant allelic imbalance for each microsatellite marker of this LOH marker panel was based on a large database of non-neoplastic aspirated pancreatic cyst fluid and microdissected stained cytology samples, all of which had confirmed equally non-neoplastic outcome, as determined by surgical pathology and clinical follow-up. This large data set of over 1000 specimens encompassed the majority of allelic combinations seen in the general patient population. The range for normal allelic balance was defined as two standard deviations from the average allelic ratio in which the fluorescence derived from the shorter allele copy is divided by that of the longer allele copy.38 Allelic ratios falling outside the thresholds were considered as demonstrating a significant imbalance (LOH). When imbalance was shown to be present, and the shorter microsatellite allele copy was found to be the relatively deficient, an LOH clonality (degree of clonal expansion) measurement was approximated using the following formula:

Conversely, when the longer allele copy was relatively deficient, the inverse formula was applied:

KRAS point mutation was assessed by dideoxy chain termination and the ratio of wild-type and mutant peak heights was used as an approximation of mutated versus non-mutated (non-neoplastic) DNA for each individual sample. The range of length differences between allelic pairs for each locus was between 4 and 8 bp. This difference is very small to cause a preferential loss of the longer allele. Expectedly, loss of the longer allele and loss of the shorter allele occurred with equal frequencies among the loci examined. Capillary electrophoresis was used for the determination of clonality for both KRAS point mutations and LOH. It is however widely recognized that the clonality determination for oncogene point mutation, as well as for allelic imbalance is at best an approximation as the fluorescence output, measured in capillary electrophoresis, is not necessarily stoichiometric but representative for the allelic pairing ratio of an individual patient sample.39, 40
Results
A total of 23 cytocentrifugation supernatants and the corresponding microdissected cytology slide specimens were produced from 23 pancreatic FNAs (two of which belonging to the same patient, obtained at different times), were analyzed for DNA content and mutational profiling, and were compared with their respective cytology findings (Table 1). Each case first received a preliminary cytology assessment for adequacy by microscopic evaluation. Molecular analysis was carried out on DNA extracted from microdissected cells judged to be most representative of the cytological diagnosis, meeting the morphological criteria for the greatest degree of cellular atypia present in each individual sample.
DNA levels were compared between each slide microdissection specimen and the corresponding cytocentrifugation supernatant fluid specimens to establish the relative amount of DNA obtainable from each type of specimen. In each case, a greater amount of DNA was obtained from the supernatant fluid, when compared with that extracted from the cytology slide microdissected cells, with an average of 31.04 ng/μl for the former compared with an average of 4.45 ng/μl for the latter (Table 1). With regard to the microdissected cell DNA specimens subgroup, three samples did not yield detectable amounts of DNA, two samples failed to amplify for LOH target detection and one sample yielded low quantities of DNA, but no PCR amplification (for LOH or KRAS). Conversely, all DNA samples obtained from the supernatant were amplifiable for LOH and KRAS analysis.
When comparing the two sources of DNA, the ability to detect the presence of mutational changes was equal to or greater in the supernatant specimens, compared with the corresponding microdissected cytology slide. In all the cases where either a KRAS point mutation or LOH was identified, the mutational clonality was equal to or higher in the supernatant DNA compared with the microdissected cell DNA (Table 2). Furthermore, no mutations detected from the microdissected slide specimens were missed by their supernatant specimens.
Mutational analysis of the supernatant fluid revealed that all cytologically malignant samples were positive upon mutational profiling. Similarly, no samples with benign cytological diagnosis were found to be positive on mutational analysis. In one out of five cases cytologically diagnosed as adenocarcinoma (number 4), no mutations were detected in the mutational analysis of the microdissected slide, while the corresponding supernatant showed the presence of both a KRAS mutation and LOH in two targets (Table 2). Likewise, all specimens diagnosed as mucinous by cytology showed the presence of KRAS mutations and/or LOH.
One specimen diagnosed as pancreatitis (number 16) failed to amplify for LOH analysis on the microdissected slide, while the supernatant did amplify, with no LOH detected. One specimen, that was called suboptimal on cytology (number 22), failed to yield amplifiable DNA from the microdissected slide, while the supernatant produced amplifiable DNA, with no mutations identified.
Specimens 7 and 8 came from the same patient and were obtained at two different times. The mutational profiles were identical, when comparing between the first and second microdissected cytology slide specimens and when comparing between the first and second supernatant specimens, demonstrating consistency of test findings.
Interestingly, one of the cases that was interpreted as suboptimal on cytological evaluation (number 23) showed low clonality KRAS mutation and failed PCR for LOH on the microdissected slide. The corresponding supernatant DNA showed two, low clonality KRAS mutations and produced amplifiable DNA for LOH analysis, with no LOH detected (Table 2).
Discussion
Although FNA has contributed significantly in the minimally invasive diagnosis of pancreatic cancer, the cytomorphological analysis of these FNA samples is occasionally limited by inconclusive and false negative diagnoses.9, 10, 41 FNA diagnosis of pancreatic samples is occasionally limited by low sample volumes, low cellularity, poor cell preservation, and non-diagnostic cytomorphological findings on cytological examination.10, 41, 42 The integration of molecular studies with cytological analysis can increase the diagnostic yield available for the patient and provide a more accurate diagnostic approach for pancreatic lesions, especially in cases of suboptimal specimens.16, 17, 18, 19, 20, 21
When comparing between biological material to be used for DNA extraction and mutational analysis, the extraction of DNA from the supernatant has advantages compared with analysis of microdissected cytology cells. Microdissection of selected targets of cell clusters from the cytology slide will give information pertinent solely to the selected cells, whereas sampling DNA from supernatant will provide DNA that is more reflective of the neoplastic process, as this cell-free DNA is presumably derived mainly from the actively cycling neoplastic cells. Furthermore, in vitro DNA degradation effects related to fixative exposure and staining could be responsible, although minimally, for diminished amplifiability of microdissected stained cytology cells (especially for the detection of LOH, which uses longer amplicons), a problem not incurring with the cytocentrifugation supernatant that is unfixed.
Using this small group of pancreatic solid mass FNA specimens, this study demonstrated that the cytocentrifugation supernatant component of the specimens, that is discarded in normal cytology practice, contained abundant cell-free DNA to generate mutational profiles that outperformed the corresponding DNA of the cells present in the samples. The molecular panel examined in the supernatant fluids was detectable genetic alterations in all five cases diagnosed as adenocarcinomas while the same panel examined in the microdissected cytology slides failed to show genetic alterations in one of the five cases (sample 4). Furthermore, out of the three cases where cytology was inconclusive (samples 21–23, Table 2), the cytocentrifugation supernatant showed the presence of mutations. In this case (sample 23), a repeat FNA obtained on follow-up showed a diagnostic material for a mucinous neoplasm.
Because of the different processing steps and type of material sampled factors, the amounts of DNA between the two subgroups are not directly comparable, but from a practical standpoint, the supernatant fluid still yielded distinctly higher amounts and more intact, amplifiable DNA compared with the microdissected cells thus supporting the concept that supernatant is enriched with cell-free DNA, shed from the most active, potentially neoplastic, cells. The use of Hank’s Balanced Salt Solution provided an optimal source for unfixed supernatant DNA, eliminating the risk of DNA degradation due to fixation. The specimens were analyzed after ∼48 h, reflecting the time necessary for the cytology samples to be prepared and evaluated morphologically for the eventual necessity of molecular testing. By respecting the true processing times of the specimens in actual clinical practice, it was shown that this could be a reliable procedure in clinical cytology practice. The available literature supports the validity of similar testing with the use of fixative-based supernatant specimens with comparable results.42 In any case, the microdissected cells underwent fixation with Shandon Cytospin® Collection Fluid, thus validating the use of this particular fixative for this type of molecular studies.
Since this was not a large base study, these results will need further confirmation with a larger study set. Moreover, this study was limited to the use of Shandon Cytospin® Collection Fluid. While data on the use of Saccomanno’s fixative, another commonly used cytology fixation medium are available,42 more information with regard to the different commonly used fixatives needs to become available regarding their capacity to yield adequate supernatant DNA, followed by comparison with the fixative-free supernatant method used in this study. Notwithstanding, available data on cytology specimens have consistently demonstrated that microdissected stained cytology cells are especially suitable for mutational analysis and applicable to common cytology practice.17, 18, 20, 23, 43 Furthermore, as most of the commonly used fixatives in cytology are alcohol based, and are not expected to induce significant DNA degradation, it would be reasonable to expect favorable results with these other methods of sample preparation of the supernatant fluid as well.
As the supernatant fluid is routinely discarded, analysis of this residual specimen offers an additional, optimal way to characterize the molecular changes, without compromising the cytology slides used for morphological evaluation, or the cytology cell block material that can also be utilized for immunohistochemical or other studies. This analysis can provide valuable, additive information in the differential diagnosis of a reactive versus a neoplastic pancreatic cell proliferation. Should this type of analysis be sought for any pancreatic FNA cytology case, it does not require any additional efforts from a laboratory perspective, with the exception of storing the cytocentrifugation supernatant at 4°C. The findings of this study suggest how the supernatant fluid can be utilized as a source of molecular information and could become a powerful addition to standard cytology evaluation. Mutational profiling of DNA in normally discarded supernatant fluid may help resolve occasional diagnostic challenges and may serve as a useful, complementary tool for cytopathologists when microscopic examination yields no conclusive diagnosis or when a specimen is suboptimal.
References
American Cancer Society. American Cancer Society Facts and Figures 2007. American Cancer Society: Atlanta, GA, 2007, accessed 18 June 2013 athttp://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2007/index.
Ferrone CR, Pieretti-Vanmarcke R, Bloom JP et al. Pancreatic ductal adenocarcinoma: Long-term survival does not equal cure. Surgery 2012;153:S43–S49.
Howlader N, Noone AM, Krapcho M et al SEER Cancer Statistics Review, 1975-2009 (Vintage 2009 Populations). National Cancer Institute: Bethesda, MD, 2012, accessed 18 June 2013 athttp://seer.cancer.gov/csr/1975_2009_pops09).
Jemal A, Siegel R, Xu J et al. Cancer statistics. 2010 CA Cancer J Clin 2010;60:277–300.
Carpizo DR, Allen PJ, Brennan MF . Current management of cystic neoplasms of the pancreas. Surgeon 2008;6:298–307.
Fasanella KE, McGrath K . Cystic lesions and intraductal neoplasms of the pancreas. Best Pract Res Clin Gastroenterol 2009;23:35–48.
Rogart JN, Loren DE, Singu BS et al. Cyst wall puncture and aspiration during EUS-guided fine needle aspiration may increase the diagnostic yield of mucinous cysts of the pancreas. J Clin Gastroenterol 2011;45:164–169.
Siddiqui AA, Brown LJ, Hong SK et al. Relationship of pancreatic mass size and diagnostic yield of endoscopic ultrasound-guided fine needle aspiration. Dig Dis Sci 2011;56:3370–3375.
Hewitt MJ, McPhail MJ, Possamai L et al. EUS-guided FNA for diagnosis of solid pancreatic neoplasms: a meta-analysis. Gastrointest Endosc 2012;75:319–331.
Layfield LJ, Jarboe EA . Cytopathology of the pancreas: neoplastic and nonneoplastic entities. Ann Diagn Pathol 2010;14:140–151.
Siddiqui AA, Kowalski TE, Shahid H et al. False-positive EUS-guided FNA cytology for solid pancreatic lesions. Gastrointest Endosc 2011;74:535–540.
Wood MD, Horst JA, Bibbo M . Weeding atypical glandular cell look-alikes from the true atypical lesions in liquid-based Pap tests: a review. Diagn Cytopathol 2007;35:12–17.
Gleeson FC, Kipp BR, Caudill JL et al. False positive endoscopic ultrasound fine needle aspiration cytology: incidence and risk factors. Gut 2010;59:586–593.
Khalid A, Nodit L, Zahid M et al. Endoscopic ultrasound fine needle aspirate DNA analysis to differentiate malignant and benign pancreatic masses. Am J Gastroenterol 2006;101:2493–2500.
Wu J, Dal Molin M, Maitra A et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA 2011;108:21188–21193.
Khalid A, Zahid M, Finkelstein SD et al. Pancreatic cyst fluid DNA analysis in evaluating pancreatic cysts: a report of the PANDA study. Gastrointest Endosc 2009;69:1095–1102.
Krishnamurti U, Sasatomi E, Swalsky PA et al. Analysis of loss of heterozygosity in atypical and negative bile duct brushing cytology specimens with malignant outcome: are "false-negative" cytologic findings a representation of morphologically subtle molecular alterations? Arch Pathol Lab Med 2007;131:74–80.
Lapkus O, Gologan O, Liu Y et al. Determination of sequential mutation accumulation in pancreas and bile duct brushing cytology. Mod Pathol 2006;19:907–913.
Mertz H . K-ras mutations correlate with atypical cytology and elevated CEA levels in pancreatic cystic neoplasms. Dig Dis Sci 2011;56:2197–2201.
Schoedel KE, Finkelstein SD, Ohori NP . K-Ras and microsatellite marker analysis of fine-needle aspirates from intraductal papillary mucinous neoplasms of the pancreas. Diagn Cytopathol 2006;34:605–608.
Toll AD, Kowalski T, Loren D et al. The added value of molecular testing in small pancreatic cysts. JOP 2010;11:582–586.
Wu J, Jiao Y, Dal Molin M et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA 2011;108:21188–21193.
Finkelstein SD, Bibbo M, Loren DE et al. Molecular analysis of centrifugation supernatant fluid from pancreaticobiliary duct samples can improve cancer detection. Acta Cytol 2012;56:439–447.
Anker P, Mulcahy H, Chen XQ et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999;18:65–73.
Aung KL, Board RE, Ellison G et al. Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J 2010;4:11–21.
Bremnes RM, Sirera R, Camps C . Circulating tumour-derived DNA and RNA markers in blood: a tool for early detection, diagnostics, and follow-up? Lung Cancer 2005;49:1–12.
Cabral R, Neto J, Carvalho MG . Circulating DNA as a biomarker for early detection of cancer: a brief update with an emphasis on lung cancer. Open Lung Cancer J 2010;3:38–44.
Mulcahy H, Farthing MJ . Diagnosis of pancreatico-biliary malignancy: detection of gene mutations in plasma and stool. Ann Oncol 1999;10 (Suppl 4):114–117.
Narayan A, Carriero NJ, Gettinger SN et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep-sequencing. Cancer Res 2012;72:3492–3498.
No HJ, Kim K, Park KH et al. level as a prognostic biomarker for epithelial ovarian cancer. Anticancer Res 2012;32:3467–3471.
Schwarzenbach H, Eichelser C, Kropidlowski J et al. Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell-free tumor DNA as indicator of breast cancer progression. Clin Cancer Res 2012;18:5719–5730.
Zhou J, Shi YH, Fan J . Circulating cell-free nucleic acids: promising biomarkers of hepatocellular carcinoma. Semin Oncol 2012;39:440–448.
Choi JJ, Reich CF III, Pisetsky DS . The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 2005;115:55–62.
Brouckaert G, Kalai M, Krysko DV et al. Phagocytosis of necrotic cells by macrophages is phosphatidylserine dependent and does not include inflammatory cytokine production. Mol Biol Cell 2004;15:1089–1100.
Jahr S, Hentze H, Englisch S et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–1665.
Diehl F, Li M, Dressman D et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005;102:16368–16373.
Bibbo M . How technology is reshaping the practice of nongynecologic cytology: frontiers of cytology symposium. Acta Cytol 2007;51:123–152.
Ostrovnaya I, Seshan VE, Olshen AB et al. Clonality: an R package for testing clonal relatedness of two tumors from the same patient based on their genomic profiles. Bioinformatics 2011;27:1698–1699.
Slebos RJ, Umbach DM, Sommer CA et al. Analytical and statistical methods to evaluate microsatellite allelic imbalance in small amounts of DNA. Lab Invest 2004;84:649–657.
Skotheim RI, Diep CB, Kraggerud SM et al. Evaluation of loss of heterozygosity/allelic imbalance scoring in tumor DNA. Cancer Genet Cytogenet 2001;127:64–70.
Lin F, Staerkel G . Cytologic criteria for well differentiated adenocarcinoma of the pancreas in fine-needle aspiration biopsy specimens. Cancer 2003;99:44–50.
Xiao GQ . Fine-needle aspiration of cystic pancreatic mucinous tumor: oncotic cell as an aiding diagnostic feature in paucicellular specimens. Diagn Cytopathol 2009;37:111–116.
Lin X, Finkelstein SD, Zhu B et al. Loss of heterozygosities in Barrett esophagus, dysplasia, and adenocarcinoma detected by esophageal brushing cytology and gastroesophageal biopsy. Cancer 2009;117:57–66.
Acknowledgements
An abstract relative to this study was submitted to the United States & Canadian Academy of Pathology 101st Annual Meeting in Vancouver, BC, Canada in March 2012 and received the First Place, Resident Research Award by the Papanicolaou Society of Cytopathology for Georgios Deftereos, MD. We would like to thank the Papanicolaou Society of Cytopathology for the honor of having received this recognition.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Sydney D Finkelstein, Sara A Jackson, and Eric MG Ellsworth are employees of RedPath Integrated Pathology, Inc. Pittsburgh, PA, USA.
Rights and permissions
About this article
Cite this article
Deftereos, G., Finkelstein, S., Jackson, S. et al. The value of mutational profiling of the cytocentrifugation supernatant fluid from fine-needle aspiration of pancreatic solid mass lesions. Mod Pathol 27, 594–601 (2014). https://doi.org/10.1038/modpathol.2013.147
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/modpathol.2013.147
Keywords
This article is cited by
-
Salvaging the supernatant: next generation cytopathology for solid tumor mutation profiling
Modern Pathology (2018)