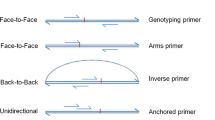
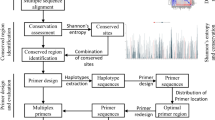
Abstract
Here we describe protocols for designing, optimizing and implementing conserved anchor primers for use in genome mapping or phylogenetic applications, with particular emphasis on homologous gene sequences among mammals. The increasing number of whole genome sequences in public databases makes this approach applicable across a wide range of taxa. Genome sequences from representatives of two or more divergent subclades within a taxonomic group of interest are used to identify candidate local alignments (i.e., exons, exons spanning introns or conserved 5′- or 3′-untranslated regions) that contain sequences with appropriate variability for the chosen downstream application. PCR primers are designed to maximize amplification success across a broad range of taxa, and are optimized under a touchdown thermocycling protocol. Based on the initial optimization results, primers are selected for application in a diverse sampling of species, or for mapping the genome of a target species of interest. We discuss factors that have to be considered for experimental design of broad-scope phylogenetic studies. With this protocol, primers can be designed, optimized and implemented within as little as 1–2 weeks.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
O'Brien, S.J. et al. Anchored reference loci for comparative genome mapping in mammals. Nat. Genet. 3, 103–112 (1993).
Venta, P.J., Brouillette, J.A., Yuzbasiyan-Gurkan, V. & Brewer, G.J. Gene-specific universal mammalian sequence-tagged sites: application to the canine genome. Biochem. Genet. 34, 321–341 (1996).
Lyons, L.A. et al. Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat. Genet. 15, 47–56 (1997).
Jiang, Z., Priat, C. & Galibert, F. Traced orthologous amplified sequence tags (TOASTS) and mammalian comparative maps. Mamm. Genome 9, 577–587 (1998).
Lyons, L.A., Kehler, J.S. & O'Brien, S.J. Development of comparative anchor tagged sequences (CATS) for canine genome mapping. J. Hered. 90, 15–26 (1999).
Murphy, W.J., Shan, S., Pecon-Slattery, J., Chen, Z.Q. & O'Brien, S.J. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 9, 1223–1230 (1999).
Chen, Z.Q., Lautenberger, J.A., Lyons, L.A., McKenzie, L. & O'Brien, S.J. A human genome map of comparative anchor tagged sequences. J. Hered. 90, 477–484 (1999).
Murphy, W.J. et al. A radiation hybrid map of the cat genome: implications for comparative mapping. Genome Res. 10, 691–702 (2000).
Menotti-Raymond, M. et al. Second generation integrated linkage and radiation hybrid maps of the domestic cat. J. Hered. 94, 95–106 (2003).
Hitte, C. et al. Facilitating genome navigation: survey sequencing and dense radiation-hybrid gene mapping. Nat. Rev. Genet. 6, 643–648 (2005).
Rosenbloom, K. et al. Phylogenomic resources at the UCSC genome browser. in Methods in Molecular Biology: Phylogenomics (ed. Murphy, W.J.) (Humana Press, Totowa, NJ, 2007).
Kohn, M., Murphy, W.J., Ostrander, E.A. & Wayne, R.K. Genomics and conservation genetics. Trends Ecol. Evol. 21, 629–637 (2006).
Murphy, W.J. et al. Molecular phylogenetics and the origin of placental mammals. Nature 409, 614–618 (2001).
Roca, A.L., Georgiadis, N. & O'Brien, S.J. Cytonuclear genomic dissociation in African elephant species. Nat. Genet. 37, 96–100 (2005).
Teeling, E.C. et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584 (2005).
Johnson, W.E. et al. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311, 73–77 (2006).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 (2000).
Palumbi, S. Nucleic acids II: The polymerase chain reaction. in: Molecular Systematics 2nd edn. (eds. Hillis, D.M., Moritz, C., Mable, B.K.) (Sinauer, Sundlerland, MA, 1996).
Acknowledgements
We thank Jan Janečka and Alison Wilkerson for helpful comments on this manuscript. This work was funded by the National Institutes of Health and the National Science Foundation.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Table 1
Conserved PCR primers optimized and applied in mammalian phylogenetic studies (PDF 32 kb)
Rights and permissions
About this article
Cite this article
Murphy, W., O'Brien, S. Designing and optimizing comparative anchor primers for comparative gene mapping and phylogenetic inference. Nat Protoc 2, 3022–3030 (2007). https://doi.org/10.1038/nprot.2007.429
Published:
Issue date:
DOI: https://doi.org/10.1038/nprot.2007.429
This article is cited by
-
Regional Comparison of Snow Leopard (Panthera uncia) Diet using DNA Metabarcoding
Biodiversity and Conservation (2021)
-
The Prevalence of Trypanosoma cruzi, the Causal Agent of Chagas Disease, in Texas Rodent Populations
EcoHealth (2017)
-
Application of dissociation curve analysis to radiation hybrid panel marker scoring: generation of a map of river buffalo (B. bubalis) chromosome 20
BMC Genomics (2008)