Abstract
Major depressive disorder (MDD) presents a substantial health burden with low treatment response rates. Predicting antidepressant efficacy is challenging due to MDD’s complex and varied neuropathology. Identifying biomarkers for antidepressant treatment requires thorough analysis of clinical trial data. Multimodal neuroimaging, combined with advanced data-driven methods, can enhance our understanding of the neurobiological processes influencing treatment outcomes. To address this, we analyzed resting-state fMRI and EEG connectivity data from 130 patients treated with sertraline and 135 patients with placebo from the Establishing Moderators and Biosignatures of Antidepressant Response in Clinical Care (EMBARC) study. A deep learning framework was developed using graph neural networks to integrate data-augmented connectivity and cross-modality correlation, aiming to predict individual symptom changes by revealing multimodal brain network signatures. The results showed that our model demonstrated promising prediction accuracy, with an R2 value of 0.24 for sertraline and 0.20 for placebo. It also exhibited potential in transferring predictions using only EEG. Key brain regions identified for predicting sertraline response included the inferior temporal gyrus (fMRI) and posterior cingulate cortex (EEG), while for placebo response, the precuneus (fMRI) and supplementary motor area (EEG) were critical. Additionally, both modalities identified the superior temporal gyrus and posterior cingulate cortex as significant for sertraline response, while the anterior cingulate cortex and postcentral gyrus were common predictors in the placebo arm. Additionally, variations in the frontoparietal control, ventral attention, dorsal attention, and limbic networks were notably associated with MDD treatment. By integrating fMRI and EEG, our study established novel multimodal brain network signatures to predict individual responses to sertraline and placebo in MDD, providing interpretable neural circuit patterns that may guide future targeted interventions. Trial Registration: Establishing Moderators and Biosignatures of Antidepressant Response for Clinical Care for Depression (EMBARC) ClinicalTrials.gov Identifier: NCT#01407094.
Similar content being viewed by others
Introduction
Major depressive disorder (MDD) remains a major global mental health concern, affecting millions of people. Despite the widespread use of serotonin reuptake inhibitors and other antidepressants, their effectiveness varies substantially, leaving many patients without sufficient relief [1]. This variability underscores the urgent need to understand the biological and psychological mechanisms driving different treatment responses. Personalized treatment strategies, considering the heterogeneity of MDD, are becoming increasingly important [2, 3]. Emerging evidence suggests that brain network (or connectivity) variations may capture pivotal information associated with treatment effects [4, 5]. Understanding these complex variations is vital for developing more effective, personalized therapeutic strategies for MDD.
In neuropsychiatric research, functional magnetic resonance imaging (fMRI) and electroencephalography (EEG) are essential for studying the neurobiology of MDD [6]. By measuring hemodynamic responses to neural activity, fMRI provides spatially high-resolution images of brain function, identifying regions implicated in psychiatric disorders [7]. In contrast, EEG records electrical oscillations from neural activity, offering superior temporal resolution for capturing network dynamics [8]. Together, these modalities enhance our understanding of the complex neural circuits associated with MDD, supporting the advancement of more tailored therapeutic interventions.
The fusion of multimodal brain imaging techniques is gaining popularity in mental disorder analysis, since it allows for a multidimensional analysis, providing comprehensive insights into the interrelationships between various imaging modalities. This integrative technique offers a more reliable foundation for precision diagnosis and prognosis, enhancing the analytical capabilities of each technique. Recent studies have successfully used multimodal fusion to predict and understand various mental health conditions. For instance, utilizing complementary information from MRI, PET, assessment metrics, and demographic data has shown promise for improved diagnoses of Alzheimer’s disease and autism [9]. Another recent study employed multimodal fusion of functional and structural data to provide a comprehensive view of longitudinal changes in brain patterns [10]. Integrating functional and structural connectivity from fMRI and diffusion tensor imaging emphasized dynamic inter-modal dependencies [11]. Furthermore, coupled tensor/matrix factorization was used to extract joint latent components from MEG and fMRI data, revealing fine-grained spatiotemporal brain dynamics and improving understanding of brain function and development in adolescents [12]. A recent study on MDD identified common and unique structural and functional connectivity coupling changes, highlighting the structural and functional connectivity coupling changes, highlighting the vulnerability of these connections to external stressors and adverse behaviors [13]. Despite these advances, there is a lack of research employing the fusion of fMRI and EEG to predict antidepressant outcomes. Combining EEG’s temporal resolution with fMRI’s spatial detail at the brain network level may offer a comprehensive view of neural circuits, enhancing our understanding of the neurobiological basis of MDD and its interaction with antidepressant treatment.
Recent advancements in machine learning and artificial intelligence have demonstrated potential in identifying brain biomarkers for more precise mental health interventions [14, 15]. The human brain can be modeled as a complex network comprising numerous spatially distributed but functionally interconnected brain regions [16]. Graph neural networks (GNNs) [17], a type of deep learning models, are particularly suited for analyzing brain functional architecture due to their ability to handle complex node interactions. This unique advantage has led to their increasing application in studying mental disorders through brain network modeling [11, 18,19,20,21,22]. Nevertheless, very few studies employing GNNs have focused on multi-modality brain network analysis for predicting treatment responses leveraging clinical trial data.
Utilizing data from the Establishing Moderators and Biosignatures of Antidepressant Response in Clinical Care (EMBARC) study, a large neuroimaging-coupled randomized clinical trial, we propose a novel deep learning framework integrating fMRI and EEG connectivity to establish robust biomarkers for predicting antidepressant and placebo treatment responses in MDD. This framework augments the functional connectivity of both modalities and constructs brain graphs for pre-treatment fMRI and EEG data, respectively. It subsequently employs GNNs to capture the spatial dependencies within each modality, thereby optimizing spatial characteristics. By maximizing the correlation between latent features, our approach harnesses complementary information through modality fusion to predict treatment outcomes. An interpretability analysis conducted based on the trained model parameters further identifies critical biomarkers from both brain region and connectivity perspectives, advancing our comprehension of neural circuits that dictate response to antidepressant treatment.
Methods and materials
Clinical trial dataset
Our study utilized data from the EMBARC study [1], a comprehensive, neuroimaging-integrated, placebo-controlled, randomized clinical trial focusing on depression. Participants aged 18 to 65 were enrolled from four study locations: Massachusetts General Hospital, University of Texas Southwestern Medical Center, University of Michigan, and Columbia University. In this double-blind trial, individuals diagnosed with MDD were randomly assigned to receive either an eight-week treatment of sertraline or placebo. Out of 309 patients diagnosed with MDD, 296 met the inclusion criteria and received treatment. For subjects who have both fMRI and EEG data, there are 130 from the sertraline arm (38 male, 92 female) and 135 from the placebo arm (50 male, 85 female) after preprocessing. The primary outcome was measured using the 17-item Hamilton Rating Scale for Depression (HAMD17) [23] at multiple time points (baseline, weeks 1, 2, 3, 4, 6, and 8). Missing HAMD17 scores at week 8 were estimated through Bayesian regression methods [24] using baseline HAMD17, week 1 HAMD17, week 2 HAMD17, week 3 HAMD17, week 4 HAMD17, week 6 HAMD17, baseline Quick Inventory of Depressive Symptoms (QIDS) total score, baseline Mood and Symptom Questionnaire subscale scores for Anxious Arousal, Anhedonic Depression, and General Distress, Snaith-Hamilton Pleasure Scale (SHAPS) total score, age, years of education, gender, and Wechsler Abbreviated Scale of Intelligence (WASI) t-scores for Vocabulary and Matrix Reasoning. Treatment outcome was measured by the change in HAMD17 scores from the baseline to week 8.
fMRI acquisition and preprocessing
Resting-state functional magnetic resonance imaging (rs-fMRI) scans were conducted using T2* weighted images via a single-shot gradient echo-planar pulse sequence. Each session lasted eight minutes using parameter settings: repetition time 2000 ms, echo time 28 ms, matrix size 64 × 64, voxel size 3.2 × 3.2 × 3.1 mm3. More details of the fMRI acquisition can be found in [25].
The rs-fMRI data were preprocessed using the fMRIPrep pipeline [26]. This involved correcting the T1 weighted images for intensity nonuniformity and skull stripping, followed by spatial normalization via nonlinear registration to a T1w reference [27]. Brain tissues, including cerebrospinal fluid, white matter, and grey matter, were segmented using FSL from the brain-extracted T1 weighted images [28]. Fieldmap information was utilized to correct distortions in the echo-planar imaging, improving the co-registration with the anatomical reference. The BOLD signals were aligned to the T1-weighted images using boundary-based registration with nine degrees of freedom to address remaining distortions [29]. Head-motion parameters were estimated using MCFLIRT (FSL), and the BOLD signals were subjected to slice-time correction, susceptibility distortion correction, and were resampled into MNI152NLin2009cAsym space. Motion artifacts were removed using ICA-AROMA [30], and following spatial smoothing with a 6 mm FWHM Gaussian kernel, the data underwent quality control for head motion.
EEG acquisition and preprocessing
Resting-state EEG (rs-EEG) data were recorded from each of the four study sites (Columbia University: 72-channel BioSemi system at 256 Hz sampling rate, McLean Hospital: 129-channel Geodesic Net system at 250 Hz, University of Michigan: 60-channel NeuroScan Synamp system at 250 Hz, University of Texas Southwestern Medical Center: 62-channel NeuroScan Synamp system at 250 Hz). Amplifier calibrations were consistently performed across all sites. The rs-EEG recording protocol consisted of four two-minute segments—two with eyes closed and two with eyes open, executed in a counterbalanced sequence. Participants were advised to minimize blinking and eye movements, maintaining fixation on a central cross during the eyes-open condition.
The rs-EEG data was processed offline using a fully automated artifact rejection pipeline in EEGLAB [2]. Initially, EEG signals were downsampled to a frequency of 250 Hz, followed by the elimination of the 60 Hz AC line noise [31]. Subsequently, a high-pass filter with a cutoff frequency of 0.01 Hz was employed to filter out low-frequency elements that were not physiological. The next stages involved the identification and removal of epochs and channels that did not meet specific threshold criteria for magnitude and spatial correlation, respectively. Participants with an excess of 20% problematic channels were excluded. For channels with compromised signals, a reconstruction was carried out using spherical spline interpolation based on signals from adjacent channels [32]. To further cleanse the data, independent component analysis was applied to remove additional artifacts like those caused by scalp muscles, eye movements, and ECG [33]. After artifact rejection, 54 EEG channels common to all four study sites were identified and extracted for each subject. The EEG signals were then standardized to a common average reference. Lastly, the signals were divided into four key frequency bands: theta (4–7 Hz), alpha (8–12 Hz), beta (13–30 Hz), and low gamma (31–50 Hz) for further analysis.
Source localization using the Brainstorm toolbox [34] with a minimum-norm estimation [35] was then conducted to convert channel-space EEG into source-space signals over 3003 vertices. This conversion was achieved through a three-layer boundary element head model, comprising scalp, skull, and cortical surface, calculated using the OpenMEEG plugin [36], which utilizes the FreeSurfer average brain template [37]. The model facilitated the generation of 3003 dipoles with free orientations. A lead-field matrix, critical for linking the activity of these dipoles to EEG data, was subsequently derived from the boundary element model. Principal component analysis was then applied to condense the three-dimensional estimated source signal at each vertex into a one-dimensional time series of the principal component.
Functional connectivity calculation
For rs-fMRI, regional time series was first computed by averaging the preprocessed voxel-level BOLD signals into time series for 100 regions of interest (ROIs) defined by the Schaefer parcellation [38]. Functional connectivity was then calculated as Pearson correlation coefficient between the time series of every pair of ROIs. The resulting connectivity values were z-score normalized and then subjected to Fisher’s r-to-z transformation to improve normality.
For rs-EEG, power envelope connectivity (PEC) was calculated in preference to other metrics such as coherence or phase lag index, as PEC not only has shown promise in mitigating spurious correlations caused by volume conduction, a common issue in EEG that can produce misleading connectivity patterns between adjacent channels [39, 40], but also demonstrates superior stability in resting-state analyses as power envelopes exhibit more consistent temporal characteristics than instantaneous phase relationships [41]. More importantly, PEC’s focus on power envelope fluctuations provides a more compatible temporal framework for integration with fMRI’s hemodynamic responses, as it captures slow temporal dynamics of neural oscillation amplitudes (typically <1 Hz) [42]. This temporal correspondence has been empirically validated by multiple studies showing significant spatial overlap between PEC-derived networks and fMRI connectivity patterns [43]. PEC addresses the volume conduction problem by focusing on the oscillatory power of signals, rather than their raw phase or amplitude, which are more susceptible to such artifacts. Specifically, Hilbert transform was first used to convert source estimates into analytical time series, followed by orthogonalizing the analytical time series of each pair of brain signals to eliminate zero-phase-lag correlation. Unlike phase lag index which may discard genuine zero-phase-lag connectivity, PEC’s orthogonalization approach maintains sensitivity to physiologically meaningful synchronization while effectively eliminating artifactual correlations [44]. The power envelopes were then obtained by squaring these orthogonalized analytical signals. A logarithmic transformation was subsequently applied to refine the data and enhance the normality of the measurements. PEC was then calculated using Pearson’s correlation coefficient between the log-transformed power envelopes of each brain region pair. Similar to fMRI, EEG connectivity features were extracted based on the Schaefer parcellation. The resulting connectivity values were z-score normalized before undergoing Fisher’s r-to-z transformation to improve normality and ensure comparability across modalities.
Data augmentation
Data augmentation is widely adopted in deep learning as a strategy to enhance performance and prevent overfitting. To increase the sample size for more robust multimodal fusion and prevent overfitting, we employed Common Orthogonal Basis Extraction (COBE) [45], a well-established algorithm in group component analysis, to augment functional connectivity data for both fMRI and EEG. COBE is adept at extracting a common basis from multi-block datasets. By removing the common components from the original functional connectivity of fMRI or EEG, individualized features are isolated. These individualized features, as newly generated data, retain essential discriminatory information related to their original functional connectivity, thereby forming the augmented data in this study. Additional details about the augmentation process are provided in Supplementary S1.
Deep learning of multimodal brain network signatures
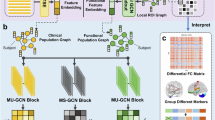
We developed a GNN-based deep learning model to integrate multimodal brain networks from fMRI and EEG, identifying robust and powerful brain signatures indicative of antidepressant response. Our model consists of two main components: 1) utilizing parallel GNNs to encode the connectivity features of fMRI and EEG, performing spatial optimization; 2) training pairs of fMRI and EEG weights to ensure that the linear combination of node features maximizes the correlation in resulting latent features between these two modalities to achieve effective modality fusion. The overview of the framework is illustrated in Fig. 1. GNNs are particularly effective for processing graph-structured data due to their capacity to encapsulate the intricate relationships among nodes [46, 47]. Methods and implementation details are provided in Supplementary S2 and S3.
Functional connectivity (FC) features derived from fMRI and EEG are firstly obtained based on parcellation and subsequently encoded via two parallel GNNs. The correlation between these multimodal representations is then maximized to enhance their compatibility for modality fusion. Subsequently, these highly correlated latent variables are concatenated and fed into a multilayer perceptron (MLP) to predict changes in HAMD17 score. More detailed implementation aspects of the architecture are available in Supplementary Figure S2.
Brain region and connectivity importance evaluation
Besides the novel multimodal graph fusion strategy, our model also offers interpretable brain network patterns, advancing the understanding of neurobiological mechanisms underlying treatment effects. We used two learnable weight matrices (see Supplementary S2) in our model function as mappings for maximizing the correlation between fMRI and EEG latent features. These matrices quantify the contributions of different ROIs to the fMRI-EEG association as self-learned model parameters during end-to-end training, thus providing an intuitive reflection of the brain regional importance in modality fusion. We integrated these pairs of vectors into a singular pair using the weights in the aggregation block, followed by absolute value computation and normalization. An average was then computed over ten folds in the optimal run as the evaluation of the brain region importance.
In addition, the adjacency matrix of the graphs was constructed by calculating similarities between node pairs in real-time, reflecting the inherent associations among ROIs. However, it may not represent the optimal topology for subsequent modality fusion and final predictions. To address this, we introduced scaling matrices for fMRI and EEG, enabling dynamic estimation and adjustment of connection strength between ROI pairs during end-to-end training for each modality. This approach captures and refines crucial connectivity patterns tailored for modality fusion and final prediction, rather than solely relying on similarity measures. We assessed the significance of connections by averaging the matrix values from the most predictive runs for each modality in both treatment arms. It is important to note that the learnable weight matrices and the scaling matrices serve distinct but complementary roles in our model. The learnable weight matrices are used for selecting salient brain regions for modality fusion, while the scaling matrices focus on interpreting optimized dependencies within graph-structured unimodal data.
Results
Multimodal prediction of treatment outcome
We hypothesize that different EEG frequency bands contain distinct discriminative information relevant to treatment prediction. Therefore, instead of using a single unified model, we combined EEG data from each frequency band with fMRI to train separate models. Figure 2a shows the predictive results of two treatment arms across four EEG bands combined with fMRI, derived from ten independent runs of 10-fold cross-validation using various seeds for model parameter initialization. Combining fMRI with EEG in the theta, alpha, and beta bands show notable predictiveness. Given the alpha band’s superior predictive accuracy relative to the other bands (Paired t-test: pfdr < 0.01, pfdr < 0.001, pfdr < 0.01 for theta, beta, gamma in sertraline arm, and pfdr < 0.05, pfdr < 0.01 for theta, beta in placebo arm based on R2), all subsequent analyses in this study focused exclusively on the alpha band. Figure 2b illustrates the outcomes of the optimal predictive performance runs for the sertraline and placebo arms respectively. A substantial correlation was observed between the actual and predicted changes in HAMD17 scores in both arms (For sertraline: R2 = 0.31, Pearson’s r = 0.58, p = 5.36 × 10−13; For placebo, R2 = 0.28, Pearson’s r = 0.56, p = 1.66 × 10−12). The statistical significance of predictive accuracy was further validated through 1000 random permutation tests (Ppermutation<0.001 for both sertraline and placebo). Results from the other nine runs are presented in Supplementary Figure S3. To account for the influence of gender, we also regressed out gender covariate before evaluating model performance, and the results are presented in Supplementary Figure S4.
a Prediction results across four EEG bands under our deep learning-based multimodal analytical framework. b The optimal run of predictive outcomes using alpha band EEG in conjunction with fMRI. The statistical significance of predictive accuracy was validated through 1000 random permutation tests with Ppermutation<0.001 for both sertraline and placebo. Each point represents a patient in the test set.
Multimodal versus unimodal prediction
To validate the advantage of multimodal prediction over unimodal prediction, we evaluated the performance of using fMRI and EEG independently, i.e., using unimodality for both training and testing. The model maintains its core structure, excluding only the unilateral GNN and the modality fusion component, aligning it with the original model’s design. The results shown in Fig. 3 indicate that the multimodal fusion model outperformed models trained with a single modality, whether fMRI or EEG. This finding suggests that, despite the higher cost of collecting both EEG and fMRI data in a clinical setting, it remains practically valuable for enhancing the accuracy of treatment outcome predictions and identifying more informative biomarkers.
Blue and orange represent sertraline and placebo arms respectively. fMRI, training and testing using fMRI only. EEG, training and testing using alpha EEG only. Multimodal→fMRI, training with multimodal data and testing with fMRI. Multimodal\({{{\boldsymbol{\to }}}}\)EEG, training with multimodal data and testing with alpha EEG. Multimodal, training and testing using multimodal data.
Considering the challenges and costs of collecting both fMRI and EEG data in real-world clinical settings, we also aim to adapt our multimodal model for patients who only have unimodal neuroimaging. Our model maximizes correlation, suggesting that the latent features derived from fMRI and EEG reflect similar brain patterns while providing complementary information for treatment prediction. We trained a standalone multilayer perceptron (MLP) regressor, half the size of the original multimodal model’s MLP, using the latent features from both modalities in the training set. This allows us to use the corresponding latent features from one side of the GNN pipeline for patients with only unimodal neuroimaging, inputting them directly into the reduced MLP to generate predictions, without needing to concatenate features from the other modality. Figure 3 shows the results of training with both modalities and testing with either fMRI or alpha EEG. The data splitting and random seeds for model parameter initialization were kept identical to previous settings to ensure a fair comparison and avoid data leakage. Although predictive performance was lower with fMRI, alpha EEG yielded results only slightly inferior to multimodal prediction. This suggests that alpha EEG played a dominant role during training, with fMRI providing supplementary information. In other words, multimodal fusion provided the best predictive performance for new patients. Unimodal prediction, such as using only alpha EEG, could be improved when informed by multimodal signatures, indicating its promising clinical application value due to its much lower cost. However, it is important to note that this phenomenon may be case-specific; different training data could result in EEG not always being dominant.
Treatment outcome-predictive brain regions
The ROI’s importance is presented in Fig. 4 through circular barplots. The Schaefer atlas [38] parcellated the 100 ROIs into seven spatially distributed functional networks, including visual network (VN), somatomotor network (SMN), dorsal attention network (DAN), ventral attention network (VAN), limbic network (LN), frontoparietal control network (FPCN), and default mode network (DMN). The radar chart in the center of the figure calculates the average ROI importance in each functional network, reflecting the importance at the network level. The most salient 20 ROIs with the highest weights are visualized on the cerebral cortex in Fig. 5a.
The color reflects the importance of brain regions, with higher values indicating greater significance. a Top 20 salient ROIs derived from multimodal model. Blue circles identify the ROIs that are prominent in multimodal analysis but not in unimodal analysis. b Top 20 salient ROIs derived from unimodal model.
In the sertraline arm, fMRI primarily highlighted the superior and inferior temporal gyri within the temporal lobe, in addition to the middle occipital and cuneus regions in the occipital lobe, the cingulate cortex, and associative areas including the supramarginal gyrus, precuneus, and anterior insula. On the other hand, EEG pinpointed key areas in the frontal lobe, particularly in the Brodmann area 8 and inferior frontal gyrus, while also emphasizing the precuneus, posterior cingulate, calcarine, and precentral areas. In the placebo arm, fMRI predominantly revealed the precuneus, precentral, and postcentral gyrus in the parietal lobe, along with the anterior cingulate cortex, middle temporal gyrus, and posterior insula. Concurrently, EEG detected distinct regions in the frontal lobe, encompassing the supplementary motor area and anterior cingulate, as well as the precuneus, inferior parietal gyri, middle temporal, and posterior cingulate cortices. At the network level, VAN was prominently displayed in the fMRI findings, whereas FPCN was distinctly observed in the EEG data for sertraline. Moreover, FPCN was significantly represented in the fMRI, while DAN was clearly apparent in the EEG for the placebo arm.
To elucidate the advantages of multimodal approaches over unimodal ones in predicting treatment outcomes, we further explored the distinctions in salient ROIs identified by models trained separately on fMRI and EEG alone. Figure 5b displays the top 20 salient ROIs derived from a single modality. Analysis of this figure reveals that while the multimodal and unimodal models generally demonstrate similar brain patterns, notable differences exist in specific regions. In the sertraline arm, the multimodal model accentuates the inferior parietal and superior temporal regions more than the fMRI-based model, and the precentral and calcarine regions more than the EEG-based model. In the placebo arm, the multimodal model places increased emphasis on the posterior insula and rostral middle frontal regions compared to the fMRI model, and on the middle cingulate and middle occipital regions compared to the EEG model.
Critical brain connections
The 20 most significant connections are visualized in edge bundling figures and brain connectomes in Fig. 6. In the sertraline arm, notable connections include strong negative connections between the middle frontal/superior orbitofrontal and inferior frontal/temporal pole in the fMRI connectome, and a strong positive connection between the precuneus/inferior parietal in the EEG connectome. In the placebo arm, the most prominent positive and negative connections are between the inferior frontal/middle occipital and superior temporal/superior frontal in the fMRI connectome, and between the precuneus/precentral and fusiform/supplementary motor area in the EEG connectome.
To further analyze network-level connectivity patterns, we evaluated the connection intensity by calculating the average of the absolute edge scaling values between each network pair. The results for each condition are presented using heatmaps and chord charts in Fig. 7. In the sertraline arm, LN exhibits notably high within-network connection intensity for both fMRI and EEG, with LN-DAN displaying significant inter-network connection intensity in fMRI. In the placebo arm, VN shows high within-network connection intensity for both fMRI and EEG, and VN-DAN exhibits significant inter-network connection intensity in fMRI. For EEG, LN-SMN and LN-VAN demonstrated stronger inter-network connection intensity. The bar plots in Fig. 7 represent the averaged between-network and within-network connection intensities for each condition. The between-network intensity was calculated by averaging the connection intensities across each network and the other six networks.
Associations of brain network signatures with MDD clinical symptoms
In the EMBARC dataset, patients with MDD were additionally evaluated using the Mood and Anxiety Symptom Questionnaire (MASQ) and the Quick Inventory of Depressive Symptomatology (QIDS). The MASQ, designed to assess mood and anxiety symptoms, categorizes conditions such as ‘General Distress’, ‘Anhedonic Depression’, and ‘Anxious Arousal’. The QIDS quantifies the severity of depressive symptoms, featuring metrics like the ‘QIDS Total Score’. We applied previously trained multimodal models to derive latent representations of test samples, which served as the inputs to the MLP regressor, subsequently using Generalized Additive Models (GAMs) [48] and conducting 10-fold cross-validation to predict these scales. As depicted in Fig. 8, the results show correlations between the clinical scales predicted by the GAMs and the actual clinical scales. In the sertraline arm, the predicted results are significantly correlated with the true scores for General Distress (r = 0.23, p = 0.026) and Anhedonic Depression (r = 0.23, p = 0.024). In the placebo arm, the predicted results are significantly correlated with the trues scores for Anxious Arousal (r = 0.19, p = 0.049) and QIDS Total Score (r = 0.26, p = 0.013). Additionally, we examined the predictability of brain signatures for baseline HAMD17 score. Although some correlations are observed (r = 0.18, p = 0.048 for sertraline; r = 0.28, p = 0.008 for placebo), they are notably weaker than the prediction of HAMD17 change (Fisher’s z-test p < 0.001 for sertraline and p = 0.003 for placebo). This implies that the treatment outcome prediction was not primarily driven by the baseline HAMD17 variation despite its correlation with brain signatures.
Brain signatures of test samples were generated through previously trained multimodal models in the optimal run of 10-fold cross-validation. GAMs were then employed to predict these clinical scales using these brain signatures. P values are false discovery rate (FDR) corrected. N.S. means non-significant.
Clinical implications of identified signatures
To enhance the clinical applicability of the identified predictive signatures in both treatment arms, disregarding the actual treatments received and their outcomes, we stratified all patients into either a sertraline preferred, or a placebo preferred group based on predicted responses derived from well-trained sertraline and placebo prediction models. The predictive analysis indicated 99 sertraline preferred and 31 placebo preferred patients in the sertraline arm, and 84 sertraline preferred and 51 placebo preferred patients in the placebo arm. Responder rates (defined as more than a 50% reduction in HAMD17 score) were significantly higher among patients whose predicted preferences aligned with their assigned treatments compared to those whose preferences did not align (Chi-square test for responder rates: sertraline arm: 57.6% [57/99] vs. 3.2% [1/31], \({\chi }^{2}\) = 26.1, p < 0.0001; placebo arm: 58.8% [30/51] vs. 14.3% [12/84], \({\chi }^{2}\) = 27.3, p < 0.0001). Similarly, actual HAMD17 reduction rates were significantly higher among patients whose assigned treatments matched their preferences compared to those whose treatments did not (two-sample t-test for treatment preference: sertraline arm: t = 9.5, p < 0.0001; placebo arm: t = 6.6, p < 0.0001). These findings provide robust clinical context for our predictions and underscore their practical relevance for treatment selection. Further details and visualizations are provided in Supplementary Figure S5.
Ablation study
We investigated the impact of varying data augmentation rates on predictive performance, as shown in Supplementary Figure S6. Additionally, we conducted an ablation study to evaluate the effectiveness of the components within our proposed framework. The experimental results indicated that the GNNs, scaling matrices, and weight matrices were all crucial to the model. Detailed results are provided in Supplementary Table S1.
Comparison of predictive performance with baseline methods
To demonstrate the superior predictive performance of our GNN-based framework, we also report the results of baseline methods, including SVR, Random Forest, and Elastic Net, in Supplementary Table S2. The computational complexities are summarized in Supplementary Table S3. The experimental results indicated that the baseline methods struggled to effectively predict treatment responses, even with the application of data augmentation.
Discussion
In this study, we developed a novel GNN-based deep learning framework to integrate the functional connectivity of fMRI and EEG for identifying multimodal biomarkers of antidepressant treatment outcomes. Our model demonstrated significant efficacy in predicting responses to both sertraline and placebo. The GNNs effectively mapped interactions between ROIs, yielding spatially optimized representations for fMRI and EEG connectivity. By leveraging adaptive edge scaling matrices that dynamically refine connectivity strength, the model improved modality fusion and prediction accuracy. Integrating correlations between GNN-encoded latent features allowed the extraction of informative multimodal brain patterns, revealing synergistic information from both fMRI and EEG. Importantly, we adapted our model for situations where patients have only EEG data by training a standalone MLP, which yielded more accurate predictions than models trained with a single modality. This adaptation greatly enhances the model’s practical utility in real clinical settings. The findings indicate that multimodal fusion provided the best predictive performance, underscoring the value of combining EEG and fMRI data despite the higher costs. The adaptation for unimodal prediction, using only alpha EEG, showed promise by leveraging multimodal signatures. This approach maintains clinical application value due to its lower cost, although it is important to recognize that the dominance of EEG in predictive performance may be case-specific. Different training data or prediction tasks could result in scenarios where EEG is not always the dominant modality. Further research should explore these dynamics to optimize the use of unimodal in clinical settings. On the other hand, predicting placebo treatment, while not directly relevant to clinical care, holds considerable potential in drug development. A placebo-predictive signature could enhance phase 2 studies by selecting comparator groups with low predicted placebo responses rates, potentially reducing costs by allowing for smaller sample sizes and more robust drug efficacy assessments.
Our model not only delivers impressive predictive performance but also offers valuable insights through interpretative analysis. By examining the learnable parameters, including edge scaling matrices for spatial pattern optimization and weight matrices for modality fusion, we identified common brain patterns across modalities and biomarkers specific to antidepressant sertraline or placebo. This analysis revealed distinct ROIs and connectivity patterns associated with each treatment, enhancing our understanding of the neural circuits involved. Our findings align with previous research, indicating that connectivity across theta, alpha, and gamma bands in EEG provides essential insights into treatment responses [4, 49,50,51]. We observed that the prefrontal cortex, cingulate cortex, and specific areas of the temporal and parietal lobes provide signals crucial for predicting antidepressant response, which are consistent with findings reported in previous studies [52,53,54,55]. Notably, the superior temporal gyrus and posterior cingulate cortex substantially contributed to sertraline response in both fMRI and EEG modalities (Fig. 4). These regions are linked to emotional regulation and social cognition, critical factors in depression [56,57,58,59,60,61,62]. For predicting placebo response, the anterior cingulate cortex and the postcentral gyrus played consistent roles across modalities. Elevated resting activity in this region may indicate better clinical outcomes, as it is linked to adaptive self-referential processing and enhanced cognitive control [63]. Furthermore, numerous studies have demonstrated the association between the anterior cingulate cortex-involved functional connectivity changes and placebo response [49, 64,65,66]. Additionally, the postcentral gyrus, associated with emotional regulation and sensory processing, has also been implicated in various forms of depression [67, 68]. Moreover, our multimodal approach uniquely identifies specific brain regions compared to the unimodal approach, such as the superior temporal gyrus on fMRI and the posterior insula on EEG. These findings underscore the advantages of multimodal fusion, which uncovers associations that are not detectable through single-modality analysis. The posterior insula, in particular, is crucial for emotion processing [69] and has been extensively implicated in the pathophysiology of depression [70,71,72,73].
At the network level, our findings (Fig. 4) indicate that FPCN, VAN, and DAN are essential for predicting treatment responses from a multimodal integration perspective. FPCN, involved in problem-solving, working memory, and emotion regulation [74, 75], was prominent in the EEG modality for the sertraline arm and the fMRI modality for placebo. VAN, responsible for attention switching and integrating emotional and sensory stimuli [76,77,78], along with DAN, involved in goal-directed external attention [79], suggests that the extent to which individuals with MDD respond to antidepressants or placebo may depend on externally oriented attention and processing salience. Figure 7 shows that FPCN typically exhibits weaker within network connections while DMN, associated with internally oriented attention and self-referential thought, shower stronger within-network connections compared to those of DAN. Strong inter-network connections between FPCN and DMN may underlie cognitive control deficits observed in MDD [70], such as difficulties in concentration and emotion regulation. Although LN comprises fewer brain regions, it features multiple high-intensity interconnections and exhibits the strongest within-network connections in the sertraline arm. The LN system has consistently been associated with the control of emotions and mood disorders, autonomic regulation, and cognitive deficits [80], and shown sensitivity to stress correlates with the severity of depression [81,82,83]. These specific network patterns may contribute to core deficits in cognitive and affective functioning, thereby serving as brain signatures to inform treatment outcomes. Our framework provides additional validation for the importance of specific brain regions, functional networks, and connections as biomarkers in predicting treatment outcomes for MDD, offering a new perspective on neurobiological mechanisms and underscoring their potential utility in real-world diagnostic scenarios.
Our results also highlight significant correlations between signature-predicted baseline clinical scores from GAMs and the actual ones in both the sertraline and placebo arms, providing supplementary evidence to confirm the clinical relevance of the identified brain network signatures. The use of the MASQ and QIDS scales in this study underscores their importance in capturing the complex symptomatology of MDD. The correlations found with these scales provide a nuanced understanding of how symptoms, such as anhedonia and anxiety, are reflected in brain network function. This insight could enhance diagnostic criteria and aid in the creation of targeted interventions for these symptoms. Future studies might investigate the stability of these correlations through longitudinal research and across different treatment arms. Moreover, extending the analysis to other forms of depression and anxiety disorders could reveal broader applications of these brain network signatures.
This study has several limitations and potential areas for expansion that merit consideration. Despite our study having a larger sample size compared to most existing clinical trial studies on antidepressant treatment, the data remains modest for training deep learning models. This limitation was highlighted by our data augmentation ablation analysis. Thus, to validate our biomarker findings, larger sample sizes and replication studies with independent cohorts are essential [84]. Currently, our prediction model is based on baseline data; future research should consider longitudinal data to better understand the relationship between functional connectivity changes and treatment outcomes. Expanding our modality fusion approach to integrate more data types will also provide richer neurophysiological information. Additionally, with larger sample sizes, advanced deep learning techniques could be explored to handle partial modality missing, which is common in clinical settings. Promising approaches include variational autoencoders for learning shared latent representations across modalities [85], self-attention mechanisms for flexible modality fusion [86], and contrastive learning strategies to align representations from different modalities even with incomplete data [87]. These approaches hold promise for improving model robustness and generalizability when faced with real-world data heterogeneity.
Conclusion
This study aimed to predict treatment outcomes in MDD and quantify brain signatures using a GNN-based deep learning method that integrates fMRI and EEG data. Our approach identified key brain regions and connections related to antidepressant response, highlighting the utility of multimodal approach in understanding complex brain interactions. Leveraging data from the EMBARC clinical trial, along with advanced data augmentation and modality fusion techniques, the framework enhanced prediction performance and revealed neurobiological underpinnings of MDD. Our interpretability analysis underscored crucial regions: the inferior temporal gyrus and posterior cingulate for sertraline, and the precuneus and supplementary motor area for placebo responses. The superior temporal gyrus and posterior cingulate cortex were consistently significant across both modalities in the sertraline arm, while the anterior cingulate cortex and postcentral gyrus were notably significant in the placebo arm, underscoring their fundamental role in predicting antidepressant and placebo efficacy. Our findings emphasize the importance of large-scale brain networks, such as the frontoparietal control, ventral and dorsal attention, and limbic networks, in MDD treatment dynamics. Overall, our research enriches the understanding of antidepressant medications in psychiatry through multimodal neuroimaging, offering novel insights on mental health treatment.
Code availability
The COBE augmentation was implemented in MATLAB (v.R2024a). The deep learning framework was implemented in Python (v.3.11.10) and PyTorch (v.2.1.1). The statistical analyses were conducted using the SciPy package (v.1.11.4). The code used in this study is available at https://github.com/YongJiao10/MultimodalGraph4MDD.
References
Trivedi MH, McGrath PJ, Fava M, Parsey RV, Kurian BT, Phillips ML, et al. Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): Rationale and design. J Psychiatr Res. 2016;78:11–23.
Wu W, Zhang Y, Jiang J, Lucas MV, Fonzo GA, Rolle CE, et al. An electroencephalographic signature predicts antidepressant response in major depression. Nat Biotechnol. 2020;38:439–47.
Zhao K, Xie H, Fonzo GA, Tong X, Carlisle N, Chidharom M, et al. Individualized fMRI connectivity defines signatures of antidepressant and placebo responses in major depression. Mol Psychiatry. 2023;28:2490–9.
Rolle CE, Fonzo GA, Wu W, Toll R, Jha MK, Cooper C, et al. Cortical connectivity moderators of antidepressant vs placebo treatment response in major depressive disorder: secondary analysis of a randomized clinical trial. JAMA Psychiatry. 2020;77:397–408.
Zhang Y, Wu W, Toll RT, Naparstek S, Maron-Katz A, Watts M, et al. Identification of psychiatric disorder subtypes from functional connectivity patterns in resting-state electroencephalography. Nat Biomed Eng. 2020;5:309–23.
Klooster D, Voetterl H, Baeken C, Arns M. Evaluating robustness of brain stimulation biomarkers for depression: a systematic review of MRI and EEG studies. Biol Psychiatry. 2024;95:553–63. Published online September 2023:S000632232301569X
Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–78.
Cohen MX. Where does EEG come from and what does it mean? Trends Neurosci. 2017;40:208–18.
Zheng S, Zhu Z, Liu Z, Guo Z, Liu Y, Yang Y, et al. Multi-modal graph learning for disease prediction. IEEE Trans Med Imaging. 2022;41:2207–16.
Saha R, Saha DK, Fu Z, Silva RF, Calhoun VD Multimodal Fusion of Functional and Structural Data to Recognize Longitudinal Change Patterns in the Adolescent Brain. In: 2023 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI). Pittsburgh, USA: IEEE; 2023:1–5.
Yang Y, Ye C, Guo X, Wu T, Xiang Y, Ma T. Mapping multi-modal brain connectome for brain disorder diagnosis via cross-modal mutual learning. IEEE Trans Med Imaging. 2024;43:108–21. Published online 2023:1-1
Belyaeva I, Gabrielson B, Wang Y-P, Wilson TW, Calhoun VD, Stephen JM, et al. Learning spatiotemporal brain dynamics in adolescents via multimodal MEG and fMRI data fusion using joint tensor/matrix decomposition. IEEE Trans Biomed Eng. 2024;71:2189–2200.
Xu M, Li X, Teng T, Huang Y, Liu M, Long Y, et al. Reconfiguration of structural and functional connectivity coupling in patient subgroups with adolescent depression. JAMA Netw Open. 2024;7:e241933.
Durstewitz D, Koppe G, Meyer-Lindenberg A. Deep neural networks in psychiatry. Mol Psychiatry. 2019;24:1583–98.
Squires M, Tao X, Elangovan S, Gururajan R, Zhou X, Acharya UR, et al. Deep learning and machine learning in psychiatry: a survey of current progress in depression detection, diagnosis and treatment. Brain Inf. 2023;10:10.
Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–98.
Kipf TN, Welling M Semi-Supervised Classification with Graph Convolutional Networks. arXiv:160902907. [Preprint] [2017 Feb 22], https://arxiv.org/abs/1609.02907.
Li X, Zhou Y, Dvornek N, Zhang M, Gao S, Zhuang J, et al. BrainGNN: interpretable brain graph neural network for fMRI analysis. Med Image Anal. 2021;74:102233.
Tang H, Ma G, Guo L, Fu X, Huang H, Zhan L. Contrastive brain network learning via hierarchical signed graph pooling model. IEEE Trans Neural Netw Learning Syst. 2022:1–13.
Zhao K, Duka B, Xie H, Oathes DJ, Calhoun V, Zhang Y. A dynamic graph convolutional neural network framework reveals new insights into connectome dysfunctions in ADHD. Neuroimage. 2022;246:118774.
Zhou H, He L, Zhang Y, Shen L, Chen B. Interpretable Graph Convolutional Network Of Multi-Modality Brain Imaging For Alzheimer’s Disease Diagnosis. In: 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI). Kolkata, India: IEEE; 2022. p. 1–5.
Zhang S, Chen X, Shen X, Ren B, Yu Z, Yang H, et al. A-GCL: Adversarial graph contrastive learning for fMRI analysis to diagnose neurodevelopmental disorders. Med Image Anal. 2023;90:102932.
Hamilton M The Hamilton Rating Scale for Depression. In: Sartorius N, Ban TA, editors. Assessment of Depression. Berlin, Heidelberg: Springer Berlin Heidelberg; 1986. p. 143–52.
Fonzo GA, Etkin A, Zhang Y, Wu W, Cooper C, Chin-Fatt C, et al. Brain regulation of emotional conflict predicts antidepressant treatment response for depression. Nat Hum Behav. 2019;3:1319–31.
Chin Fatt CR, Jha MK, Cooper CM, Fonzo G, South C, Grannemann B, et al. Effect of intrinsic patterns of functional brain connectivity in moderating antidepressant treatment response in major depression. AJP. 2020;177:143–54.
Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat Methods. 2019;16:111–6.
Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41.
Zhang Y, Brady JM, Smith S. Hidden Markov random field model for segmentation of brain MR image. In: Medical Imaging 2000: Image Processing. San Diego, USA: SPIE; 2000. p. 1126–37.
Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48:63–72.
Pruim RHR, Mennes M, van Rooij D, Llera A, Buitelaar JK, Beckmann CF. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage. 2015;112:267–77.
NITRC MT. CleanLine: Tool/Resource Info. 2012. 2012.
Perrin F, Pernier J, Bertrand O, Echallier JF. Spherical splines for scalp potential and current density mapping. Electroencephalogr Clin Neurophysiol. 1989;72:184–7.
Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–59.
Tadel F, Baillet S, Mosher JC, Pantazis D, Leahy RM. Brainstorm: a user-friendly application for MEG/EEG analysis. Intell Neurosci. 2011;2011:879716.
Hauk O. Keep it simple: a case for using classical minimum norm estimation in the analysis of EEG and MEG data. Neuroimage. 2004;21:1612–21.
Gramfort A, Papadopoulo T, Olivi E, Clerc M. OpenMEEG: opensource software for quasistatic bioelectromagnetics. Biomed Eng Online. 2010;9:45.
Fischl B. FreeSurfer. Neuroimage. 2012;62:774–81.
Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo X-N, Holmes AJ, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex. 2018;28:3095–114.
Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci. 2012;15:884–90.
Siems M, Pape A-A, Hipp JF, Siegel M. Measuring the cortical correlation structure of spontaneous oscillatory activity with EEG and MEG. Neuroimage. 2016;129:345–55.
Brookes MJ, Hale JR, Zumer JM, Stevenson CM, Francis ST, Barnes GR, et al. Measuring functional connectivity using MEG: methodology and comparison with fMRI. Neuroimage. 2011;56:1082–104.
Tewarie P, Abeysuriya R, Byrne Á, O’Neill GC, Sotiropoulos SN, Brookes MJ, et al. How do spatially distinct frequency specific MEG networks emerge from one underlying structural connectome? The role of the structural eigenmodes. Neuroimage. 2019;186:211–20.
Tewarie P, Bright MG, Hillebrand A, Robson SE, Gascoyne LE, Morris PG, et al. Predicting haemodynamic networks using electrophysiology: the role of non-linear and cross-frequency interactions. Neuroimage. 2016;130:273–92.
O’Neill GC, Barratt EL, Hunt BAE, Tewarie PK, Brookes MJ. Measuring electrophysiological connectivity by power envelope correlation: a technical review on MEG methods. Phys Med Biol. 2015;60:R271.
Zhou G, Cichocki A, Zhang Y, Mandic DP. Group component analysis for multiblock data: common and individual feature extraction. IEEE Trans Neural Netw Learn Syst. 2016;27:2426–39.
Wu Z, Pan S, Chen F, Long G, Zhang C, Philip SY. A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst. 2020;32:4–24.
Jiang W, Luo J. Graph neural network for traffic forecasting: a survey. Expert Syst Appl. 2022;207:117921.
Hastie TJ. Generalized Additive Models. Statistical Models in S. Pacific Grove, USA: Routledge; 1992.
Pizzagalli DA, Webb CA, Dillon DG, Tenke CE, Kayser J, Goer F, et al. Pretreatment rostral anterior cingulate cortex theta activity in relation to symptom improvement in depression: a randomized clinical trial. JAMA Psychiatry. 2018;75:547–54.
Lee PF, Kan DPX, Croarkin P, Phang CK, Doruk D. Neurophysiological correlates of depressive symptoms in young adults: a quantitative EEG study. J Clin Neurosci. 2018;47:315–22.
Fitzgerald PJ, Watson BO. Gamma oscillations as a biomarker for major depression: an emerging topic. Transl Psychiatry. 2018;8:1–7.
Fonseka TM, MacQueen GM, Kennedy SH. Neuroimaging biomarkers as predictors of treatment outcome in Major Depressive Disorder. J Affect Disord. 2018;233:21–35.
Lai C-H. Promising neuroimaging biomarkers in depression. Psychiatry Investig. 2019;16:662–70.
Piani MC, Maggioni E, Delvecchio G, Brambilla P. Sustained attention alterations in major depressive disorder: a review of fMRI studies employing Go/No-Go and CPT tasks. J Affect Disord. 2022;303:98–113.
Pilmeyer J, Huijbers W, Lamerichs R, Jansen JFA, Breeuwer M, Zinger S. Functional MRI in major depressive disorder: a review of findings, limitations, and future prospects. J Neuroimaging. 2022;32:582–95.
Yang X, Huang J, Lan Y, Zhu C, Liu X, Wang Y, et al. Diminished caudate and superior temporal gyrus responses to effort-based decision making in patients with first-episode major depressive disorder. Prog Neuro-psychopharmacol Biol Psychiatry. 2016;64:52–9.
Yang X, Tian K, Wang D, Wang Y, Cheung EFC, Xie G, et al. Anhedonia correlates with abnormal functional connectivity of the superior temporal gyrus and the caudate nucleus in patients with first-episode drug-naive major depressive disorder. J Affect Disord. 2017;218:284–90.
Sui J, Qi S, Van Erp TGM, Bustillo J, Jiang R, Lin D, et al. Multimodal neuromarkers in schizophrenia via cognition-guided MRI fusion. Nat Commun. 2018;9:3028.
McLellan Q, Wilkes TC, Swansburg R, Jaworska N, Langevin LM, MacMaster FP. History of suicide attempt and right superior temporal gyrus volume in youth with treatment-resistant major depressive disorder. J Affect Disord. 2018;239:291–4.
Kang L, Wang W, Zhang N, Nie Z, Gong Q, Yao L, et al. Superior temporal gyrus and cerebellar loops predict nonsuicidal self-injury in major depressive disorder patients by multimodal neuroimaging. Transl Psychiatry. 2022;12:1–8.
Zhu X, Wang X, Xiao J, Liao J, Zhong M, Wang W, et al. Evidence of a dissociation pattern in resting-state default mode network connectivity in first-episode, treatment-naive major depression patients. Biol Psychiatry. 2012;71:611–7.
Cheng W, Rolls ET, Qiu J, Xie X, Wei D, Huang C-C, et al. Increased functional connectivity of the posterior cingulate cortex with the lateral orbitofrontal cortex in depression. Transl Psychiatry. 2018;8:90.
Pizzagalli DA. Frontocingulate dysfunction in depression: toward biomarkers of treatment response. Neuropsychopharmacology. 2011;36:183–206.
Korb AS, Hunter AM, Cook IA, Leuchter AF. Rostral anterior cingulate cortex activity and early symptom improvement during treatment for major depressive disorder. Psychiatry Research: Neuroimaging. 2011;192:188–94.
Peciña M, Bohnert ASB, Sikora M, Avery ET, Langenecker SA, Mickey BJ, et al. Association between placebo-activated neural systems and antidepressant responses: neurochemistry of placebo effects in major depression. JAMA Psychiatry. 2015;72:1087–94.
Sikora M, Heffernan J, Avery ET, Mickey BJ, Zubieta J-K, Peciña M. Salience network functional connectivity predicts placebo effects in major depression. Biol Psychiatry: Cognit Neurosci Neuroimaging. 2016;1:68–76.
Peng W, Jia Z, Huang X, Lui S, Kuang W, Sweeney JA, et al. Brain structural abnormalities in emotional regulation and sensory processing regions associated with anxious depression. Prog Neuro-psychopharmacol Biol Psychiatry. 2019;94:109676.
Shan X, Cui X, Liu F, Li H, Huang R, Tang Y, et al. Shared and distinct homotopic connectivity changes in melancholic and non-melancholic depression. J Affect Disord. 2021;287:268–75.
Phan KL, Wager T, Taylor SF, Liberzon I. Functional neuroanatomy of emotion: a meta-analysis of emotion activation studies in PET and fMRI. Neuroimage. 2002;16:331–48.
Avery JA, Drevets WC, Moseman SE, Bodurka J, Barcalow JC, Simmons WK. Major depressive disorder is associated with abnormal interoceptive activity and functional connectivity in the insula. Biol Psychiatry. 2014;76:258–66.
Ambrosi E, Arciniegas DB, Madan A, Curtis KN, Patriquin MA, Jorge RE, et al. Insula and amygdala resting‐state functional connectivity differentiate bipolar from unipolar depression. Acta Psychiatr Scand. 2017;136:129–39.
Hu L, Xiao M, Ai M, Wang W, Chen J, Tan Z, et al. Disruption of resting-state functional connectivity of right posterior insula in adolescents and young adults with major depressive disorder. J Affect Disord. 2019;257:23–30.
Guo Z-P, Chen L, Tang L-R, Gao Y, Chand T, Sen ZD, et al. Association between decreased interhemispheric functional connectivity of the insula and duration of illness in recurrent depression. J Affect Disord. 2023;329:88–95.
Koechlin E, Summerfield C. An information theoretical approach to prefrontal executive function. Trends Cognit Sci. 2007;11:229–35.
Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cognit Sci. 2011;15:483–506.
Downar J, Crawley AP, Mikulis DJ, Davis KD. A multimodal cortical network for the detection of changes in the sensory environment. Nat Neurosci. 2000;3:277–83.
Menon V, Uddin LQ. Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct. 2010;214:655–67.
Thomas Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–65.
Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci. 2002;3:201–15.
Adams RD, Victor M, Ropper AH, Daroff RB. Principles of Neurology. Cognit Behav Neurol. 1997;10:220.
Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci. 1996;93:3908–13.
Mingtian Z, Shuqiao Y, Xiongzhao Z, Jinyao Y, Xueling Z, Xiang W, et al. Elevated amygdala activity to negative faces in young adults with early onset major depressive disorder. Psychiatry Research: Neuroimaging. 2012;201:107–12.
Redlich R, Opel N, Bürger C, Dohm K, Grotegerd D, Förster K, et al. The limbic system in youth depression: brain structural and functional alterations in adolescent in-patients with severe depression. Neuropsychopharmacology. 2018;43:546–54.
Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–60.
Kingma DP. Auto-encoding variational bayes. arXiv [Preprint] arXiv:13126114. 2013. 2013 https://arxiv.org/abs/1312.6114.
Tsai YHH, Liang PP, Zadeh A, Morency LP, Salakhutdinov R. Learning Factorized Multimodal Representations. In: International Conference on Learning Representations. 2019. p. 1–11.
Yuan X, Lin Z, Kuen J, Zhang J, Wang Y, Maire M, et al. Multimodal contrastive training for visual representation learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021. p. 6995–7004.
Acknowledgements
This work was supported by NIH grant nos. R01MH129694, R21MH130956, R21AG080425, Alzheimer’s Association Grant (AARG-22-972541), and Lehigh University FIG (FIGAWD35), CORE, and Accelerator grants. Portions of this research were conducted on Lehigh University’s Research Computing infrastructure partially supported by NSF Award 2019035. G.A.F. was also supported by philanthropic funding and NIH grant nos. R01MH132784 and R01MH125886, and grants from the One Mind - Baszucki Brain Research Fund, the SEAL Future Foundation, and the Brain and Behavior Research Foundation.
Author information
Authors and Affiliations
Contributions
YJ conceptualized and designed the work, performed data analysis and result interpretation and drafted and revised the manuscript. KZ and XW performed data analysis and result interpretation and drafted the manuscript. NBC, CJK, DJO and GAF interpreted the data, refined the design of the work and revised the manuscript. YZ conceptualized and designed the work, oversaw the analysis and interpretation of data and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
G.A.F. received monetary compensation for consulting work for SynapseBio AI and owns equity in Alto Neuroscience. C.J.K reports equity from Alto Neuroscience. The remaining authors declare no competing interests.
Ethics approval and consent to participate
The EMBARC data are publicly available through the National Institute of Mental Health Data Archive (NDA) (https://nda.nih.gov/edit_collection.html?id=2199). For collecting the data, written informed consent was obtained from each participant under the Institutional Review Board (IRB) approved protocols at each of the four study sites, including University of Texas Southwestern Medical Center, Columbia University/Stony Brook, Massachusetts General Hospital, University of Michigan, University of Pittsburgh, and McLean Hospital. All analysis methods performed in this study were in accordance with the relevant guidelines and regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiao, Y., Zhao, K., Wei, X. et al. Deep graph learning of multimodal brain networks defines treatment-predictive signatures in major depression. Mol Psychiatry 30, 3963–3974 (2025). https://doi.org/10.1038/s41380-025-02974-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-02974-6
This article is cited by
-
From symptom-based heterogeneity to mechanism-based profiling in youth ADHD: the promise of computational psychiatry
Neuropsychopharmacology (2026)
-
Evaluating the impact of machine learning models on adult major depressive disorder using conventional treatment strategies: a systematic review approach
Discover Public Health (2025)










