Abstract
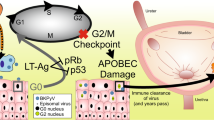
BK polyomavirus (BKPyV)-associated cancer after transplantation has gained increasing attention. However, the role of BKPyV integration on oncogenesis is still unclear. In this study, next-generation virome capture sequencing of primary and metastatic tumors were performed in three patients with BKPyV-associated urothelial carcinoma after renal transplantation. As a result, a total of 332 viral integration sites were identified in the six tumors. Integration of BKPyV in both primary and metastatic tumors followed the mechanism of microhomology-mediated end joining mostly, since microhomologies between human and BKPyV genomes were significantly enriched in flanking regions of 84% of the integration sites. Viral DNA breakpoints were nonrandom and tended to assemble in large T gene, small T gene and viral protein 2 gene. There were three, one and one consensus integration sites between the primary and metastatic tumors, which affected LINC01924, eIF3c, and NEIL2 genes in the three cases respectively. Thus, we concluded that integration of BKPyV was a continuous process occurring in both primary and metastatic tumors, generating heterogenous tumor cell populations. Through this ongoing process, certain cell populations might have gained growth advantage or metastatic potential, as a result of viral integration either affecting the cellular genes where the viral DNA integrated to or altering the expression or function of the viral genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Whole genome sequencing data of the six samples are available at NCBI Sequence ReadArchive with the BioProject accession number of PRJNA643598. Integration sites identified by capture sequencing were summarized in Supplementary Table S1. Virome capture sequencing data and probe sequences are available from the corresponding author on reasonable request.
References
Knowles WA, Pipkin P, Andrews N, Vyse A, Minor P, Brown DW, et al. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J Med Virol. 2003;71:115–23.
Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis. 2009;199:837–46.
Schwarz A, Linnenweber-Held S, Heim A, Brocker V, Rieck D, Framke T, et al. Factors influencing viral clearing and renal function during polyomavirus BK-associated nephropathy after renal transplantation. Transplantation. 2012;94:396–402.
Starrett GJ, Buck CB. The case for BK polyomavirus as a cause of bladder cancer. Curr Opin Virol. 2019;39:8–15.
Delbue S, Ferrante P, Provenzano M. Polyomavirus BK and prostate cancer: an unworthy scientific effort? Oncoscience. 2014;1:296–303.
Brugge JS, Butel JS. Role of simian virus 40 gene A function in maintenance of transformation. J Virol. 1975;15:619–35.
Cicala C, Pompetti F, Carbone M. SV40 induces mesotheliomas in hamsters. Am J Pathol. 1993;142:1524–33.
Kenan DJ, Mieczkowski PA, Burger-Calderon R, Singh HK, Nickeleit V. The oncogenic potential of BK-polyomavirus is linked to viral integration into the human genome. J Pathol. 2015;237:379–89.
Kenan DJ, Mieczkowski PA, Latulippe E, Cote I, Singh HK, Nickeleit V. BK polyomavirus genomic integration and large T antigen expression: evolving paradigms in human oncogenesis. Am J Transpl. 2017;17:1674–80.
Fu F, Deng W, Yu S, Liu Y, Yu L, Liu R, et al. Occurrence and regression of BK polyomavirus associated carcinoma: a clinical and next-generation sequencing study. Clin Sci. 2018;132:1753–63.
Jin Y, Zhou Y, Deng W, Lee R, Liu Y, Blute M, et al. Genome-wide profiling of BK polyomavirus integration in urothelial carcinoma of kidney transplant recipients reveals a potential microhomology or nonhomologous end joining mediated integration mechanism. Am J Transpl. 2019;193:1158.
Borchert S, Czech-Sioli M, Neumann F, Schmidt C, Wimmer P, Dobner T, et al. High-affinity Rb binding, p53 inhibition, subcellular localization, and transformation by wild-type or tumor-derived shortened Merkel cell polyomavirus large T antigens. J Virol. 2014;88:3144–60.
Houben R, Dreher C, Angermeyer S, Borst A, Utikal J, Haferkamp S, et al. Mechanisms of p53 restriction in Merkel cell carcinoma cells are independent of the Merkel cell polyoma virus T antigens. J Investig Dermatol. 2013;133:2453–60.
Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8.
Merid SK, Goranskaya D, Alexeyenko A. Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis. BMC Bioinforma. 2014;15:308.
Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24.
Ferber MJ, Thorland EC, Brink AA, Rapp AK, Phillips LA, McGovern R, et al. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–42.
Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–83.
Hu J, Luo H, Xu Y, Luo G, Xu S, Zhu J, et al. The prognostic significance of EIF3C gene during the tumorigenesis of prostate cancer. Cancer Investig. 2019;37:199–208.
Wei H, Kamat A, Chen M, Ke HL, Chang DW, Yin J, et al. Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guerin. PLos ONE. 2012;7:e38533.
Jin L, Gibson PE, Booth JC, Clewley JP. Genomic typing of BK virus in clinical specimens by direct sequencing of polymerase chain reaction products. J Med Virol. 1993;41:11–17.
Moens U, Van Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 2005;331:209–31.
Starrett GJ, Thakuria M, Chen T, Marcelus C, Cheng J, Nomburg J, et al. Clinical and molecular characterization of virus-positive and virus-negative Merkel cell carcinoma. Genome Med. 2020;12:30.
Muller DC, Ramo M, Naegele K, Ribi S, Wetterauer C, Perrina V, et al. Donor-derived, metastatic urothelial cancer after kidney transplantation associated with a potentially oncogenic BK polyomavirus. J Pathol. 2018;244:265–70.
Anzivino E, Zingaropoli MA, Iannetta M, Pietropaolo VA, Oliva A, Iori F, et al. Archetype and rearranged non-coding control regions in urothelial bladder carcinoma of immunocompetent individuals. Cancer Genom Proteom. 2016;13:499–509.
Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992.
Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675–93.
Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300.
Javorka P, Raxwal VK, Najvarek J, Riha K. artMAP: a user-friendly tool for mapping ethyl methanesulfonate-induced mutations in Arabidopsis. Plant Direct. 2019;3:e146.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-né P, Nicolas A, et al. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics. 2010;26:1895–6.
Acknowledgements
The work was funded by Natural Science Foundation of Guangdong Province [grant number 2020A1515010674], National Natural Science Foundation of China [grant number 81500573], Science and Technology Planning Project of Guangzhou [grant number 201803010109] and President Funding of Nanfang Hospital [grant number 2018B009, 2018C003]. And we thanked Dr. Min-Hua Luo (Wuhan Institute of Virology, Wuhan, China) for her assisstant in the study design and critical revision of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Liu, Y., Deng, W. et al. Viral integration in BK polyomavirus-associated urothelial carcinoma in renal transplant recipients: multistage carcinogenesis revealed by next-generation virome capture sequencing. Oncogene 39, 5734–5742 (2020). https://doi.org/10.1038/s41388-020-01398-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-020-01398-6
This article is cited by
-
Report on post-transplantation cancer in southeast Asia from the Thai kidney transplantation cohort
Scientific Reports (2024)
-
Unraveling the role of hepatitis B virus DNA integration in B-cell lymphomagenesis
British Journal of Cancer (2024)
-
Fatal BK polyomavirus-associated pneumonia: report of two cases with literature review
BMC Infectious Diseases (2023)
-
Genome-wide profiling of BK polyomavirus integration in bladder cancer of kidney transplant recipients reveals mechanisms of the integration at the nucleotide level
Oncogene (2021)