Abstract
Psychiatric conditions, known for their hereditary nature, exert significant impacts on various life domains. Leveraging this heritability, we examine the relations between genetic susceptibility to major psychiatric disorders and the multifaceted aspects of quality of life in two population-based cohorts, the Adolescent Brain Cognitive Development (ABCD) study (N = 3909 preadolescent children) and the UK Biobank (N = 269,293 adults). Genetic susceptibility to seven major psychiatric disorders was quantified by polygenic scores derived from extensive genome-wide association studies (N = 21,000–413,000). Pervasive associations were found between genetic risk for all seven major psychiatric disorders investigated and age-relevant real-life quality of life indices, with varied patterns of associations for different life domains. We especially highlight the role of genetic risks for ADHD and major depressive disorders. Our findings emphasize the continuous nature of psychiatric traits, extending their influence on daily life experiences and societal functioning beyond symptomatology and diagnostic classifications.
Similar content being viewed by others
Introduction
The impact of psychiatric disorders transcends the confines of mental wellbeing, exerting influence across a multifaceted spectrum of life domains, encompassing educational, occupational, physical, social, and psychological outcomes [1]. This comprehensive reach is well-documented in extensive epidemiological studies, which consistently associate psychiatric disorders with a decrement in overall quality of life and specific functional impairments [2, 3]. Crucially, these burdens extend not only to individuals who meet the clinical symptom and impairment thresholds for a psychiatric diagnosis but also to those who exhibit subclinical symptomatology. This phenomenon aligns with the liability threshold model in psychiatric genetics [4], which posits that the genetic risk for psychiatric disorders exists on a continuum, and individuals may experience varying degrees of genetic susceptibility, irrespective of clinical diagnosis.
In the pursuit of enhancing citizens’ well-being and satisfaction, many nations have made quality of life improvement a central objective of their policy agendas. However, there exists no universally accepted framework for conceptualizing and measuring quality of life. A commonly endorsed model recognizes quality of life as a construct encompassing dimensions of standard of living that conform to societal expectations. This is quantifiable across domains such as social, health, and economic well-being, in addition to subjective assessments of personal well-being, reflecting the degree to which individual life expectations are met [5]. Quality of life, however, is not a static construct and can vary significantly across different life stages, particularly between childhood and adulthood. In children, physical health and safety are paramount, as are education and social interactions that foster cognitive and emotional development [6, 7]. These elements are critical in shaping a child’s foundational sense of well-being and future prospects. In adulthood, economic stability, career satisfaction, and the quality of personal relationships become more important indicators of quality of life [8]. In addition to these stage-specific factors, core elements like health and social connections remain consistently important throughout the lifespan [1, 6], highlighting both the dynamic and enduring nature of quality of life.
Large-scale genome-wide association studies (GWASs) have enabled the identification of common genetic variation contributing to psychiatric disorders [9]. The polygenic architecture of many prevalent and debilitating psychiatric disorders has been characterized with large GWAS samples, including attention-deficit/hyperactivity disorder (ADHD) [10], autism spectrum disorder (ASD) [11], major depressive disorder (MDD) [12], anxiety disorders (ANX) [13, 14], schizophrenia (SCZ) [15], bipolar disorder (BIP) [16], and cannabis use disorder (CUD) [17]. Combining information across the genome through a weighted sum of the number of the disorder-associated alleles, polygenic scores (PGSs) [18] can provide proxies for individuals’ genetic loading for a psychiatric condition on a continuous scale in the population. Such quantification of genetic susceptibility to different phenotypes has proven useful in risk stratification of common complex diseases [19], and is increasingly powerful as GWAS sample sizes increase. Prior research [20] demonstrated that genetic predisposition to schizophrenia contributes to additional variance in quality of life beyond clinical factors. However, there is still a need for systematic quantification of PGS effects across psychiatric conditions on diverse quality of life domains.
Here, we aimed to assess how genetic susceptibility to seven major psychiatric disorders indexed by PGSs relates to various quality of life-relevant indices at different phases of the lifespan, specifically preadolescent children and middle-aged adults. To this end, we took advantage of two large population cohorts, the Adolescent Brain Cognitive Development (ABCD) study and the UK Biobank, and derived multi-faceted quality of life constructs at these two life stages, capturing general and specific domains of human functioning and experiences.
Methods
Participants
Our study sample consisted of 3909 non-Hispanic white preadolescent children (47% females, age 9.92 ± 0.62 years) recruited across the United States of America as part of the ABCD study cohort (request 11315, data release 4.0) and 269,293 white British, unrelated adults (54% females, age 56.95 ± 7.94 years) from the population-based UK Biobank (application 23668, data release 3.0). Participants and their caregivers in the studies provided written or verbal consent, as appropriate.
Quality of life outcomes
We hypothesized several models capturing different quality of life domains in both cohorts, and assessed their fit through confirmatory factor analyses. Given the great phenotyping depth of both cohorts, more than one well-fitting model structure with sets of reasonable indicators was available. Here, for each cohort, we selected one theoretically sound and statistically best-fit model as the primary model used in the main analyses, as described in the main text, and present a less well-fitting alternative model in eFig. 1.
In the ABCD cohort, we fitted the model with nine observed variables (eTable 1) indicating three latent factors – educational performance and cognition (Edu), physical health (Hea1), and adverse peer experience (Peer). Observed variables are, for Edu, school grades in the past year reported by parents (sag_grade_type) and youths (sag_grades_last_yr), cognition total composite score (abcd_tbss01:nihtbx_totalcomp_uncorrected); for Hea1, ever seen doctors before the past year (excluding regular check-ups) (abcd_mx01:medhx_1b), ever seen doctor for any (severe) diseases (derived from abcd_mx01), emergency room visits before the past year (derived from abcd_mx01), ever been in the hospital overnight or longer (abcd_mx01:medhx_8a); for Peer, ever been cyberbullied (abcd_cb01:cybb_phenx_harm), experienced victimization from peers (derived from abcd_peq01). We used variables from the second-year follow-up, except for medical history reports and cognition total composite score, which were only available as baseline measures and expected to capture the underlying traits that are relatively stable across time. Individuals who reported ‘do not know’, ‘not applicable’, or ‘refuse to answer’ to the abovementioned items were excluded from the analyses. A final list of 3909 participants with phenotypic data available was included in downstream analyses.
In the UK Biobank cohort, we included three latent factors – socioeconomic status (SES), health (Hea2), and social wellbeing (Soc) – which were indicated by eight binary or ordinal variables (eTable 2): average total household income before tax (UK Biobank index 738) and educational qualifications (6138) for the first factor; overall health rating (2178), long-standing illness, disability or infirmity (2188), and whether diagnosed with any serious medical conditions (derived by aggregating item 6150, 6152, 2443, 2453, and 2473) for the second factor; the frequency of being able to confide with someone (2110), whether often feel loneliness or in isolation (2020), and the tendency to worry too long after embarrassment (1930) for the third factor. For educational qualifications, we converted each individual’s highest qualification to an International Standard Classification of Education (ISCED) category and removed other qualifications that were not included in such classification scheme. Individuals who reported ‘do not know’ or ‘prefer not to answer’ to the abovementioned items were excluded from the analyses.
Confirmatory factor analysis
The CFA models were implemented in lavaan package [21] using the method of weighted least squares mean and variance adjusted (WLSMV), which used diagonally weighted least squares to estimate model parameters, but the full weight matrix to compute robust standard errors. Final models contained the abovementioned three first-level factors and one second-level general QoL factor. To ease the interpretation, variables were sign-flipped so that larger values always correspond to higher quality of life levels. The variances of all latent variables (i.e., ‘factor scores’) in the models were fixed to unity, and their estimated values were computed using Empirical Bayes Method (EBM) [21]. The metrics of Comparative Fit Index (CFI), Root Mean Square Error of Approximation (RMSEA) and 90% confidence intervals, Standardized Root Mean Square Residual (SRMR), together with Tucker-Lewis Index (TLI) were employed to assess the model fit. CFI values above 0.95, RMSEA values below 0.06, SRMR values below 0.08, and TLI values above 0.95 were considered as evidence for good model fit [22].
GWAS data
In selecting phenotypes for our study, we employed the following criteria: diversity in ages of onset, variety in symptom profiles, robust availability of large and recent GWAS datasets of European ancestry, and the ability to avoid sample overlap with our target cohorts. Guided by these considerations, we included seven major psychiatric disorders—ADHD, ASD, MDD, ANX, SCZ, BIP, and CUD—and ensured that all GWAS summary statistics excluded UK Biobank participants when computing PGSs for the UK Biobank. Notably, we chose CUD to represent substance use disorders because of its substantial sample size (N ≈ 358,000) and strong genetic correlations with other SUDs (e.g., rg = 0.75 with alcohol use disorder, rg = 0.74 with opioid use disorder, and rg = 0.63 with tobacco use disorder) [23]. All GWAS summary statistics were based on cohorts of European ancestry, and were annotated to the Genome Reference Consortium (GRCh) 37/hg19 build. We reviewed the study protocols for each GWAS analysis to ensure good quality of summary statistics and sufficient SNP heritability of traits (h2SNP > 0.05) before including them in the current study. An overview of the discovery GWASs with publication references is provided in Table 1. We removed strand-ambiguous SNPs, duplicated SNPs, multi-allelic SNPs, and SNPs with out-of-bounds values, or with MAF < 0.01, imputation quality INFO < 0.9, or not matching to HapMap3 reference panel.
Polygenic score calculation and statistical analyses
Detailed descriptions of the QC and imputation steps in both cohorts have been provided in previous publications [24, 25]. Subsequent sample and variant filtering was conducted prior to polygenic score analyses based on the protocol described in eMethods. We adopted PRS-CS [26] as the primary approach to compute polygenic scores for the four quality of life factors. PRS-CS used a Bayesian continuous shrinkage method to adjust SNP effect estimates from the original GWAS summary statistics and infer posterior SNP weights. We then used the --score command in PLINK 2.0 to sum over all SNPs weighted by their posterior effect sizes and derive an additive score per individual. Two other PGS approaches were also tested to further validate the results (eMethods), and support power analysis as described in the following section.
Simple linear regression was performed between each polygenic score (as predictor) and quality of life factor (as outcome). Sex, age in years, batch, site, and the first ten ancestry informative genotype PCs for the UK Biobank cohort and batch, site, plate, sex, age in months, and the first ten ancestry informative genotype PCs for the ABCD cohort were included as covariates. R2 was calculated for each PGS by subtracting variance explained by the covariates from variance explained by the full model including both the PGS and covariates. Multiple regression models with all 7 PGSs were further constructed to assess the overall variance explained by the different PGSs altogether on top of the covariates. Bonferroni correction was applied accounting for the number of polygenic scores and outcome variables tested. To assess the effect of differential sample sizes of the base GWAS samples, we performed power calculation for the PGS analyses as described in eMethods.
SNP heritability estimation and genetic correlation analyses
Genome-wide association analyses on the four estimated latent factors were conducted in the UK Biobank cohort using PLINK 2.0 [27], with an assumed additive genetic model. Generalized linear regression model was performed on the imputed data, with sex, age, batch, plate, site, and the first ten PCs as the covariates. SNP-heritability estimates of each latent factor and their genetic correlations with the seven psychiatric disorders were then calculated using linkage disequilibrium score regression (LDSC) v1.0.1 [28] and LD scores precomputed from the European reference samples from the 1000 Genomes Project.
Results
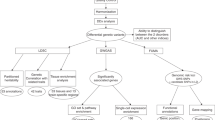
The main data processing and analytic processes were illustrated in Fig. 1. In a final sample of 3909 preadolescent children and 269,293 adults for which both genotype data and quality of life factor scores were available, we modeled the covariances of the quality of life indicators (correlation matrices shown in Fig. 2A and Fig. 2B) and assessed the fit of the hypothesized model structures using confirmatory factor analysis (CFA). In the ABCD cohort, the model consisted of three first-order latent factors, namely educational performance and cognition (Edu), physical health (Hea1), and peer experience (Peer), as well as one second-order general quality of life (QoL) factor. This model had an excellent model fit (CFI = 0.989, RMSEA = 0.021, SRMR = 0.030, TLI = 0.984) (Fig. 2C). A similarly structured second-order model was estimated in the UK Biobank cohort (CFI = 0.971, RMSEA = 0.046, SRMR = 0.043, TLI = 0.952), with socioeconomic status (SES), physical health (Hea2), and social wellbeing (Soc) factors on the first-order level and general QoL factor on the second-order level (Fig. 2D). All model parameters are presented in eTable 3 and eTable 4. In a subset of the UK Biobank sample, in which self-rated life satisfaction measures were available, the three first-level quality of life factors estimated from the model structure were significantly associated with subjective satisfaction in their corresponding life domain (i.e., SES with individuals’ financial situation satisfaction, ρ = 0.25, p < 0.001; physical health with health satisfaction, ρ = 0.58, p < 0.001; social wellbeing with general happiness in life, ρ = 0.41, p < 0.001, family relationship, ρ = 0.32, p < 0.001, and friendship satisfaction, ρ = 0.28, p < 0.001) (eTable 5).
The key analytic steps performed in the study were described in a modular fashion, with the arrows illustrating the input and output data involved in different steps. Dashed arrows and boxes denote filters applied to obtain the final analytic sample. QC quality control, PCA principal component analysis, PCs principal components, ADHD attention-deficit/hyperactivity disorder, ASD autism spectrum disorder, MDD major depressive disorder, ANX anxiety disorder, SCZ schizophrenia, BIP bipolar disorder, CUD cannabis use disorder.
Correlation matrices of quality of life indicators included in either primary or alternative models for ABCD (A) and UK Biobank (B) cohorts. Primary confirmatory factor analysis model structures for ABCD (C) and UK Biobank (D) cohorts. Observed variables (indicators) are represented in rectangular boxes. Unobserved (latent) variables derived from the models are shown in circles. Single-headed arrows denote the influence of latent variables on indicators (or another latent variable), with the values on the arrows represent factor loading coefficients. The double-headed arrow connecting the variable to itself denotes residual variances. Higher scores of the indicators correspond to higher levels of quality of life when constructing the models. A pair of alternative models is presented in eFigure 1. QoL quality of life, Edu educational performance and cognition, Hea1 physical health modeled in ABCD cohort, Peer peer experience, SES socioeconomic status, Hea2 physical health modeled in UK biobank cohort, Soc social wellbeing.
We examined patterns of associations between PGSs for psychiatric disorders and four latent quality of life factors estimated from the fitted model. In the ABCD cohort, the PGS for ADHD significantly explained 1.67% of Edu (β = −0.133, SE = 0.016, p = 1.53 × 10−16), 0.84% of Peer (β = −0.094, SE = 0.016, p = 7.81 × 10−9), and 1.85% of general QoL factors (β = −0.140, SE = 0.016, p = 3.37 × 10−18), respectively. The PGSs for other disorders were not significantly associated with any of the latent factors (Fig. 3A, eTable 6). In the UK Biobank cohort, PGSs based on all seven psychiatric disorders were associated with the general QoL factor and at least one first-order subdomain (Fig. 3B, eTable 6). Among them, ADHD-PGS showed the largest effect on the general QoL factor (β = −0.096, SE = 0.002, p < 2.23 × 10−308, R2 = 0.009), Hea2 (β = −0.083, SE = 0.002, p < 2.23 × 10−308, R2 = 0.007), and SES (β = −0.081, SE = 0.002, p < 2.23 × 10−308, R2 = 0.007). The largest effect on the Soc subdomain was seen with the MDD-PGS (β = −0.060, SE = 0.002, p = 9.96 × 10−214, R2 = 0.004), which was also associated with the domains of Hea2 (β = −0.059, SE = 0.002, p = 4.48 × 10−211, R2 = 0.003) and SES (β = −0.042, SE = 0.002, p = 3.63 × 10−125, R2 = 0.002). Genetic risk for CUD was most strongly associated with SES (β = −0.037, SE = 0.002, p = 2.05 × 10−99, R2 = 0.001), compared to the other two subdomains, whereas genetic risk for ASD was linked most strongly to the Soc subdomain (β = −0.024, SE = 0.002, p = 2.06 × 10−36, R2 = 0.001), and ANX to Hea2 (β = −0.028, SE = 0.002, p = 4.57 × 10−51, R2 = 0.001).
Regression estimates between polygenic scores for different major psychiatric disorders and quality of life latent factors for ABCD study (A) and UK Biobank (B) cohorts. ADHD attention-deficit/hyperactivity disorder, ASD autism spectrum disorder, MDD major depressive disorder, ANX anxiety disorder, SCZ schizophrenia, BIP bipolar disorder, CUD cannabis use disorder, QoL quality of life, Edu educational performance and cognition, Hea1 physical health modeled in ABCD cohort, Peer peer experience, SES socioeconomic status, Hea2 physical health modeled in UK Biobank cohort; Soc social wellbeing. Dark blue indicates statistical significance after correction. Error bars represent standard errors.
To evaluate the influence of statistical power (including differences in discovery GWAS sample size) on identifying PGS effects, the lower bound of the power estimates was obtained in both cohorts based on the sample sizes of both the base and target cohorts, SNP heritability, and trait prevalence. The power estimates in the UK Biobank cohort for all disorders and target phenotypes were above 0.990 across genetic correlation (rg) thresholds of 0.2, 0.4, 0.6, 0.8, and 1. In the ABCD cohort, all disorders except ASD and CUD were sufficiently powered for rg = 0.4 and above. Assuming the same degree of genetic correlations with quality of life domains across disorders, the ranking of the seven disorders based on their power estimates would be as follows: ANX, SCZ, BIP, ADHD, MDD, CUD, and ASD. Full results of the power analyses are shown in eTables 7 and eTable 8. Further, the results were consistent across three polygenic scoring methods examined (eFigure 2 and eFigure 3).
Using the estimated latent quality of life factors as the target phenotype in the UK Biobank cohort, we further conducted GWAS and estimated the SNP heritability (h2SNP) of the latent factors using LD score regression. The estimated h2SNP of the latent factors are in line with estimates for the individual indicators (SES: h2SNP = 0.113 (0.005), Hea2: h2SNP = 0.094 (0.004), Soc: h2SNP = 0.063 (0.003), QoL: h2SNP = 0.115 (0.005)). In the multiple regression model, seven PGSs together explained R2 = 0.011, 0.010, 0.006, 0.014 variances, which is 9.7%, 10.4%, 9.7%, and 12.2% of the SNP heritability of SES, Hea2, Soc, and general QoL, respectively. Variances explained proportional to SNP heritability by single PGSs are presented in eTable 9. To explore the genetic relationships between psychiatric disorders and quality of life domains, we estimated their pairwise genetic correlations. Consistent with PGS analysis results, a widespread pattern of negative associations was observed across quality of life domains (eTable 10). MDD (largest rg = −0.688 for Soc factor), ANX (largest rg = −0.682 for Soc factor), and ADHD (largest rg = −0.633 for general QoL factor) were highly negatively genetically correlated with all the quality of life factors. CUD was most strongly correlated with SES (rg = −0.400), and ASD with Soc (rg = −0.298). The genetic correlations for SCZ and BIP were weaker, with BIP exclusively associated with the Hea2 subdomain (rg = −0.133); no significant genetic correlation was seen between ASD and SES.
Discussion
In this study, we set out to systematically evaluate the impact of psychiatric genetic liabilities on the quality of life in individuals during two distinct phases of life. We utilized PGSs that encompassed seven major psychiatric disorders and characterized their relations with various aspects of quality of life, with a specific focus on domains of academic performance, socio-economic factors, physical health, and social wellbeing. Our results unveiled a pervasive yet varied pattern of associations with quality of life across different psychiatric disorders, such that ADHD was more strongly associated with educational performance and cognition in children and with socioeconomic status in adults; while MDD was most strongly associated with social well-being in adults only. Notably, we accentuated the prominent role played by genetic burden for ADHD during childhood, while also highlighting the impact of genetic predisposition associated with ADHD, MDD, and CUD in adulthood.
Our study revealed that the impact of genetic susceptibility, as measured by PGSs, on quality of life exists across the lifespan, even in individuals without psychiatric diagnoses within the general population. This aligns with the observations of attenuated impairments in unaffected family members in twin or family studies. For instance, lower cognitive functioning has been observed in unaffected first-degree relatives of patients with schizophrenia [29], bipolar disorder [30], and ADHD [31], compared to healthy controls. While there is little research, and no evidence in identifying reductions in quality of life-related traits in high-risk family studies [32], our results revealed the overarching trend of decreased everyday functioning relating to genetic risks across disorders, encouraging further efforts exploiting the continuous nature of psychiatric traits.
To our knowledge, this is the first study to quantify the contribution of genetic risks across major psychiatric disorders to different aspects of quality of life. The quality of life domains we found associated with the genetic risk for each disorder were consistent with previous findings from case-control group contrasts based on clinical diagnosis [33,34,35,36]. For instance, a meta-analysis of over 6000 youths and their parents found that children with ADHD were most strongly impacted in aspects of academic performance and social functioning. Their physical functioning was only slightly lower, but the discrepancy compared to typically developing peers became larger as they grew older [36], consistent with our finding that higher genetic liability for ADHD is more strongly associated with compromised health status in adults than in children, whereas the academic and social domains were impacted both in children and adults. Notably, the lack of associations for other disorders in the pre-adolescent children’s cohort is likely not fully explained by statistical power, highlighting a potential temporal distinction in the manifestation of genetic risks for different psychiatric disorders. ADHD genetic risk is more likely to manifest early in life, significantly influencing brain development processes such as cortical maturation, particularly in the prefrontal cortex—which is crucial for executive functions including attention and motor planning [37]. Genetic risks for disorders such as MDD and SCZ may be linked to later developmental events, coinciding with neurobiological changes during adolescence and early adulthood, and often additionally influenced by environmental stressors that occur at later stages [38, 39]. The consistent strong effect of the ADHD-PGS in both childhood and adulthood challenges the prevailing perception of ADHD as a relatively mild childhood condition that individuals tend to outgrow during development and a condition less likely to require treatment [40, 41]. Our work highlights the importance of research to understand the etiology, and long-term impact of ADHD, along with other psychiatric disorders, which can have far-reaching consequences on various aspects of individuals’ lives, extending beyond those directly affected by the disorders themselves.
In assessing quality of life, we employed a comprehensive approach by integrating self-perceived [5] and objectively quantified impairment in adaptive functioning measures across diverse domains, informants, and scales, and extended beyond the health-related quality of life as often examined in the context of psychiatric disorders [42]. The derived constructs align with individuals’ subjective satisfaction in corresponding life domains. Our findings advocate for an increased emphasis on researching and incorporating the concept of quality of life into clinical practices, thus complementing the conventional focus on reducing psychiatric symptoms as the primary measure of intervention effectiveness.
The utilization of PGSs in this study has allowed for estimation of genetic risk proxies in cohorts without the requirement for specific symptom or trait measures, providing insights into the contribution of different psychiatric disorders to the variables of interest. PGSs offer a cost-effective and efficient means to approximate genetic risk burden and capture the cumulative effects of multiple genetic variants associated with a particular disorder in large general population datasets [18]. By leveraging PGSs, we were able to assess the relative contributions of various disorders to the overall quality of life and its subdomains. The negative relationships we identified between genetic liability to major psychiatric disorders and quality of life related outcomes were consistent across different PGS approaches, despite their small effect sizes. While PGSs offer advantages such as scalability and broad applicability, the common variants captured only reflect a fraction of the total heritability of the disorders and can thus explain a small portion of variance in their primary phenotypes (i.e., diagnostic status) [43]. Joint efforts to increase the sample sizes and ethnic diversity of GWASs, as well as exploiting data for rare and structural genetic variants are essential to provide a more complete individual genetic risk profiles for psychiatric disorders.
Caution should be exercised when interpreting our study due to certain limitations. First, prior research [44] suggested a ‘healthy volunteer selection bias’ in the UK Biobank cohort, where the sample was enriched in wealthier and healthier individuals. This may limit the generalizability of the current results, especially for disorders such as SCZ and ASD, where the debilitating genetic effect might be more pronounced in the samples at the higher end of the liability spectrum. Second, this study only provided snapshots of childhood (around 10 years old) and a part of adulthood (40–70 years old). Longitudinal data are needed to further elucidate how the genetic risks manifest along the trajectory of human development and aging.
In summary, combining newly available GWAS results with genotyped and richly phenotyped cohorts of children and adults, our study highlights the inverse relationship between psychiatric liability and different domains of quality of life. PGSs provided a means to evaluate the contributions of genetic liability for different psychiatric disorders to different aspects of life in the general population.
Data availability
This research has been conducted using data from UK Biobank (http://www.ukbiobank.ac.uk/), under application 23668, and the ABCD Study (https://abcdstudy.org/), under request 11315.
Code availability
Analysis code is available from the corresponding author on request.
References
The World Health Organization quality of life assessment (WHOQOL): Position paper from the World Health Organization. Soc Sci Med. 1995;41:1403–9.
GBD 2016 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1260–344.
Alonso J, Angermeyer MC, Bernert S, Bruffaerts R, Brugha TS, Bryson H, et al. Disability and quality of life impact of mental disorders in Europe: results from the European Study of the Epidemiology of Mental Disorders (ESEMeD) project. Acta Psychiatr Scand. 2004;109:38–46.
Falconer DS. The inheritance of liability to diseases with variable age of onset, with particular reference to diabetes mellitus. Ann Hum Genet. 1967;31:1–20.
Costanza R, Fisher B, Ali S, Beer C, Bond L, Boumans R, et al. An Integrative Approach to Quality of Life Measurement, Research, and Policy. SAPIENS Surveys and Perspectives Integrating Environment and Society [Internet]. 2008 Nov 26 [cited 2022 Oct 20];(1.1). Available from: https://journals.openedition.org/sapiens/169.
Eiser C, Morse R. The measurement of quality of life in children: past and future perspectives. J Dev Behav Pediatr. 2001;22:248.
Wallander JL, Koot HM. Quality of life in children: A critical examination of concepts, approaches, issues, and future directions. Clin Psychol Rev. 2016;45:131–43.
Felce D, Perry J. Quality of life: Its definition and measurement. Res. Dev Disabil. 1995;16:51–74.
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101:5–22.
Demontis D, Walters GB, Athanasiadis G, Walters R, Therrien K, Nielsen TT, et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet. 2023;55:198–208.
Matoba N, Liang D, Sun H, Aygün N, McAfee JC, Davis JE, et al. Common genetic risk variants identified in the SPARK cohort support DDHD2 as a candidate risk gene for autism. Transl Psychiatry. 2020;10:1–14.
Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81.
Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2020;25:3292–303.
Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry. 2016;21:1391–9.
Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8.
Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29.
Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry. 2020;7:1032–45.
Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–8.
Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24.
Pazoki R, Lin BD, van Eijk KR, Schijven D, de Zwarte S, Investigators G. et al. Phenome-wide and genome-wide analyses of quality of life in schizophrenia. BJPsych Open. 2021;7:e13.
Rosseel Y. lavaan: an R package for structural equation modeling. J Stat Softw. 2012;48:1–36.
Hu L, Bentler PM. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Struct Equ Modeling: A Multidiscip J. 1999;6:1–55.
Gerring ZF, Thorp JG, Treur JL, Verweij KJH, Derks EM. The genetic landscape of substance use disorders. Mol Psychiatry. 2024;29:3694–705.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9.
Jernigan TL, Brown SA, Dowling GJ. The adolescent brain cognitive development study. J Res Adolesc. 2018;28:154–6.
Ge T, Chen CY, Ni Y, Feng YCA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7.
Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–48.
Glahn DC, Almasy L, Blangero J, Burk GM, Estrada J, Peralta JM, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet Part B: Neuropsychiatric Genet. 2007;144B:242–9.
Bora E, Özerdem A. A meta-analysis of neurocognition in youth with familial high risk for bipolar disorder. Eur Psychiatry. 2017;44:17–23.
Rommelse NNJ, Altink ME, Oosterlaan J, Buschgens CJM, Buitelaar J, Sergeant JA. Support for an independent familial segregation of executive and intelligence endophenotypes in ADHD families. Psychological Med. 2008;38:1595–606.
Tempelaar WM, Termorshuizen F, MacCabe JH, Boks MPM, Kahn RS. Educational achievement in psychiatric patients and their siblings: a register-based study in 30 000 individuals in The Netherlands. Psychol Med. 2017;47:776–84.
Sivertsen H, Bjørkløf GH, Engedal K, Selbæk G, Helvik AS. Depression and quality of life in older persons: a review. Dement Geriatric Cognit Disord. 2015;40:311–39.
Jonsson U, Alaie I, Löfgren Wilteus A, Zander E, Marschik PB, Coghill D, et al. Annual Research Review: Quality of life and childhood mental and behavioural disorders – a critical review of the research. J Child Psychol Psychiatry. 2017;58:439–69.
Bastiaansen D, Koot HM, Ferdinand RF, Verhulst FC. Quality of Life in Children With Psychiatric Disorders: Self-, Parent, and Clinician Report. J Am Acad Child Adolesc Psychiatry. 2004;43:221–30.
Lee YC, Yang HJ, Chen VC, Lee WT, Teng MJ, Lin CH, et al. Meta-analysis of quality of life in children and adolescents with ADHD: By both parent proxy-report and child self-report using PedsQLTM. Res Dev Disabil. 2016;51–52:160–72.
Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch JP, Greenstein D, et al. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci USA. 2007;104:19649–54.
Ross CA, Margolis RL, Reading SAJ, Pletnikov M, Coyle JT. Neurobiology of Schizophrenia. Neuron. 2006;52:139–53.
Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorderresults from the national comorbidity survey replication (NCS-R). JAMA. 2003;289:3095–105.
Lebowitz MS. Stigmatization of ADHD: a developmental review. J Atten Disord. 2016;20:199–205.
Mueller AK, Fuermaier ABM, Koerts J, Tucha L. Stigma in attention deficit hyperactivity disorder. Atten Defic Hyperact Disord. 2012;4:101–14.
Revicki DA, Kleinman L, Cella D. A history of health-related quality of life outcomes in psychiatry. Dialogues Clin Neurosci. 2014;16:127–35.
Wray NR, Lin T, Austin J, McGrath JJ, Hickie IB, Murray GK, et al. From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry. 2021;78:101–9.
Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34.
Acknowledgements
UK Biobank is supported by its founding funders the Wellcome Trust and UK Medical Research Council, as well as the Department of Health, Scottish Government, the Northwest Regional Development Agency, British Heart Foundation and Cancer Research UK. The ABCD study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners/. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not participate in the analysis or writing of this report. This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative (Grant no. EINF1824). YS is funded by a junior researcher PhD grant from the Donders Centre for Medical Neuroscience at Radboudumc. ES is funded by HORIZON Europe (HORIZON-HLTH-2021-STAYHLTH-01-02, project: 101057529) and a Hypatia Tenure Track Grant from Radboudumc. NRM and BF have received funding from the EU Horizon 2020 programme under grant agreement no. 847879 (PRIME) and from the National Institute of Mental Health of the National Institutes of Health under Award Number R01MH124851. BF also received funding from the Netherlands Organization for Scientific Research (NWO) for the GUTS project (grant 024.005.011). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and/or the European Commission. We thank Prof. Frank Dudbridge for the helpful discussions regarding the power analyses.
Author information
Authors and Affiliations
Contributions
YS contributed to conceptualisation, methodology, data analysis, data visualisation, and writing of the original draft. NRM contributed to conceptualisation, methodology, supervision, and the review and editing of the manuscript. ES contributed to conceptualisation, methodology, supervision, and the review and editing of the manuscript. BF contributed to conceptualisation, methodology, supervision, the review and editing of the manuscript, and funding acquisition.
Corresponding author
Ethics declarations
Competing interests
BF has received educational speaking fees from Medice GmbH. All other authors report no potential conflicts of interest.
Ethics approval and consent to participate
All methods described in the manuscript were performed in accordance with the relevant guidelines and regulations. UK Biobank has approval from the North West Multi-centre Research Ethics Committee (MREC) as a Research Tissue Bank (RTB) approval. An electronic signed consent was obtained from the participants. The overall protocol of the ABCD Study was approved by the centralized Institutional Review Board at the University of California, San Diego. Participants and their caregivers in the studies provided written or verbal informed consent, as appropriate.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, Y., Mota, N.R., Franke, B. et al. Genetic liability to major psychiatric disorders contributes to multi-faceted quality of life outcomes in children and adults. Transl Psychiatry 15, 232 (2025). https://doi.org/10.1038/s41398-025-03443-y
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03443-y