Abstract
A recent study demonstrated that rodents exposed to early life stress (ELS) showed changes in fat tissue composition and exhibited depression-like phenotype. Decreased Sirt1 expression in the reward-related nucleus accumbens (NAc) was a key component of this alteration. Our aim was to translate these findings into humans using a population genetic approach and brain imaging data. We tested the interaction of genetic risk of NAD + /SIRT1 pathway and ELS on depression and investigated whether body fat mediates this interaction effect. We also investigated the effect of the genetic risk and ELS interaction on functional connectivity of the NAc. Our findings suggest that interaction between NAD + /SIRT1 pathway risk and ELS contributes to depression in males (beta = 2.9275, p = 0.0002). Additionally, this interaction influences sex-dependent functional connectivity of the NAc with the middle frontal gyrus and triangular part of the inferior frontal gyrus (p = 0.0139). The observed interaction effect is independent of body fat percentage in adults, indicating that these depressogenic genetic effects are not mediated through adiposity. Overall, these results pave the way for potential therapeutic interventions in depressed male patients with NAD + /SIRT1 risk variants who experienced ELS, regardless of their body fat percentage.
Similar content being viewed by others
Introduction
Depression is a common mental disorder that contributes to the decline of quality of life and causes significant burdens. Existing antidepressant treatments are often ineffective [1], which may be due to the remarkable heterogeneity of depression, evident in its symptomatic manifestation and in the genetic and neurobiological background underlying its symptoms. One important contributor to depression’s heterogeneity is sex. Studies consistently show differences between males and females in the onset, course, and symptoms of depression, likely due to hormonal, genetic, and developmental factors influencing stress and emotional regulation. Recognizing sex as a biological variable is therefore crucial for understanding its mechanisms and developing more targeted treatments.
Nicotinamide-adenine dinucleotide (NAD + )-Dependent Protein Deacetylase Sirtuin-1 (SIRT1) has become a key target of several research studies [2,3,4]. SIRT1 is involved in several essential physiological processes [2, 5,6,7], and impairment in its deacetylase activity contributes to the formation of disease conditions. Morató et al. [8] demonstrated that rodents exposed to peripubertal stress (henceforth early life stress - ELS) manifested increased adiposity and reduced sociability in adulthood. The study also established ELS-induced altered stress-fat-brain connection as key factor leading to the observed behavioral change in male rodents. In these changes, a decrease in the expression of Sirt1, specifically in the nucleus accumbens (NAc), and impairment of NAD + /SIRT1 pathway played a central role. Furthermore, nicotinamide mononucleotide (NMN) treatment, capable to compensate for impaired SIRT1 function by boosting NAD+ level, could reverse behavioral changes. Since reduced sociability and social withdrawal are indicators of depression, these findings suggest that ELS may lead to depression via the NAD + /SIRT1 pathway. However, the ELS- and sex-dependent connections of this pathway with depression, its relationship with obesity, and its impact on the function of the NAc remained untested in humans, preventing the utilization of components of NAD + /SIRT1 pathway as biomarkers or treatment targets. The aim of our study was to investigate this pathway to determine its contribution to the pathophysiology of human depression.
Based on the above, our aims were:
1) To explore the association between NAD + /SIRT1 pathway genes considering its interaction with ELS in human depression.
Genome-wide association studies (GWAS) have attempted to identify new single nucleotide polymorphisms (SNPs) in depression. Given the polygenic nature of depression [9], rather entire genes and pathways than individual variants, would be worth investigating. In statistical genetic analyses the investigated single nucleotide polymorphisms, located in non-coding or intronic regions, have limited or no evidence for their functional impact on gene expression. As a result, it is currently unknown whether these variants lead to upregulation, downregulation, or broader dysregulation of the genes involved. Therefore, rather than reflecting a clear expression-level change, the calculated genetic risk captures a general vulnerability conferred by this pathway, aggregating small effects of many variants that may influence depression risk through complex and potentially regulatory mechanisms. An effective method to represent a combined risk of a pathway is the estimation of polygenic risk scores (PRS), which can compress GWAS signals of pathways into a single number [10]. Calculation of PRSs enables for testing an aggregated risk score and its association with depression in interaction with environmental factors in regression models. Therefore, we performed PRS estimations and linear regression analyses.
2) To assess whether the hypothesized interacting effect of NAD + /SIRT1 pathway PRS with ELS acts directly or through body fat percentage on depression.
ELS increases the risk for obesity [11, 12], which can lead to depression, anhedonia, and social withdrawal [13], prominent manifestations of obesity-associated diseases [14]. There is a known link between body fat and obesity and depression [15, 16], while SIRT1 has been also linked to energy metabolism and adiposity [6, 17]. It remains unclear if the link between body fat and depression is mediated via SIRT1 or other factors in humans. Therefore, to distinguish direct and indirect effects of NAD + /SIRT1 pathway and adiposity on depression, we performed mediation analyses.
3) To investigate whether interaction of NAD + /SIRT1 pathway PRS and ELS has a role in the alterations of NAc’s functional connectivity with other brain regions.
Functional and structural changes in the brain can be detectable using functional magnetic resonance imaging (fMRI). Studies have previously demonstrated that genetic risk is associated with altered brain connectivity in psychiatric disorders like schizophrenia [18] and major depression [19]. Morató and colleagues [8] identified specific impairment of NAD + /SIRT1 pathway in NAc, which is known to be a key brain region in reward processes and depression [20,21,22]. To further investigate whether genetic risk within NAD + /SIRT1 pathway in interaction with ELS correlates with functional alterations in NAc’s connectivity we used fMRI data and performed regression analyses.
4) To examine potential sex differences, raised by results of Morató et al., by analyzing sex-separated groups.
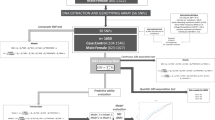
Figure 1 illustrates all the above-described aims, and the methods used to reach these aims.
Methods
Ethical approvals
UK Biobank
Data used in our analyses were part of the UK Biobank (UKB, application number 1602). All participants (N = 334,248, Supplementary Tables S2-S3) provided written informed consent, and procedures were carried out by the Declaration of Helsinki. Details of the data collection were previously published in the paper of Bycroft et al. [23].
Hungarian cohort
The study was conducted in accordance with the Helsinki Declaration for Research and approved by the local ethical committee of the Medical Research Council (23609-1/2011-EKU, 23421-1/2015-EKU).
Genotypes
In the UKB data, genomic quality control steps and filtering were performed as described in the supplementary information of Eszlari et al. [24] Participants were of white British ancestry (as per UKB data field 22006).
Regarding genetic data of the Hungarian cohort, we performed filtering steps as in case of UKB data.
NAD + /SIRT1 pathway
We selected all genes encoded on the nuclear DNA from the NAD + /SIRT1 pathway based on the work of Morató et al. [8] We selected variants within the genes of the pathway and their 10,000 base pair vicinity [25], yielding 20 genes. We annotated our SNPs with biomaRt R package [26, 27], and LDlink databases [28].
Altogether, 3827 and 1885 SNPs remained in the UKB were involved.
In the Hungarian population, altogether 1885 SNPs remained after QC filtering steps (Supplementary Table S1). Details of the genes and variants are shown in Supplementary Table S1 and Supplementary Material, section 1.
Phenotypes
UK biobank
We used current depressive symptoms (henceforth depression score) as a measure of depression in both the discovery and target samples and calculated the score from UKB data fields: 2050, 2060, 2070, and 2080 based on the study of Hullam et al. [29]
ELS score was measured with the Childhood Trauma Screener (CTS), which included the following UKB data fields: 20487, 20488, 20489, 20490, 20491 as described previously by Walker et al. [30].
The used body fat percentage variable, UKB data field: 23099, was measured by body composition estimation through impedance measurement for every participant.
In our analyses, we used the sex and age (UKB data fields 31 and 21003 respectively) variables respectively as covariates (except in sex-separated analyses).
Hungarian cohort
Mental health problems were assessed using the Mini International Neuropsychiatric Interview (MINI) [31] and depression was measured with the Zung Self-Rating Depression Scale (SDS depression scale [32]). ELS was measured with the Childhood Trauma Questionnaire [33]. Descriptive statistics can be found in Supplementary Table 4.
Statistical analyses
Genome-wide association analysis on the discovery sample
PRS estimation requires a discovery (summary statistics of a GWAS) and an independent target dataset, with individual-level data. Thus, we divided the UKB cohort into discovery and target samples. From the 334,248 individuals, we selected 105,654 individuals who had data related to ELS, as a target sample. The other 228,595 individuals formed the discovery sample.
We performed GWAS for depression score in the discovery sample with PLINK2.0 [34]. In our analysis, age, sex, 10 principal components of the genome, and genotype array were added as covariates. Manhattan plot constructed with FUMA v1.5.1 [35] and top significant loci of the GWAS can be found in Supplementary Figures S1-S2. Descriptive statistics of the discovery sample are shown in Supplementary Table S2.
Polygenic risk score (PRS) analyses on the target sample
For the PRS calculations in the target sample (N = 105,654) we used LDpred2 [36] (R package versions: bigsnpr v1.12.2, bigstatsr v1.5.12 [37]). We calculated PRS on the target sample, using beta values of the NAD + /SIRT1 pathway variants from GWAS results of depression score in the total target population, and sex-separated subpopulations: Nfemales = 59,073, Nmales = 46,581.
Details of the methods are described in the Supplementary Material, section 3. Descriptive statistics of the target sample can be found in Supplementary Tables 3-4.
Calculating interaction of PRS and ELS scores
We examined how much phenotypic variance of depression score is explained by the interaction of ELS and the genetic risk of NAD + /SIRT1 pathway genes.
As a null model we performed linear regression separately for depression score, including age, sex (only in the total sample), 10 principal components of the genome, genotype array, main effect of the PRS, and ELS. In the risk model besides the same covariates as in the null model, we added the interaction term of the PRS and ELS to the regression. The change in the explained variance (ΔR2) was calculated from these independent models as in Eq. 1:
Variance change by the interaction of the PRS and ELS.
Where ΔR2 is the change in the explained variance caused by the interaction term of the genetic risk and ELS, R2 PRS x ELS is the variance of the phenotype explained by the genetic risk x ELS interaction term and covariates, and R2 null is the variance in the phenotype explained by the covariates, and main effects of the genetic risk and ELS. We calculated the explained variance change in percentage with the following formula:
Variance change in percentage
Linear regressions were performed with R version 4.1.2. Details of the regression models are described in Supplementary Material, section 3.2.
Investigating the role of body fat percentage
Our objective was to investigate whether the interaction effect of ELS and the NAD + /SIRT1 pathway PRS on depression score is mediated by body fat percentage. To test these hypotheses, we performed mediation analyses with the “lavaan” R package (version 0.6-12) [38] for structural equation modeling on the UKB target sample.
In these analyses, we tested the direct effect of the interaction of the PRS and ELS on depressive symptoms score and its indirect effect through body fat percentage, in the total population and male/female subpopulations. The covariates in models were age, sex (only in the total population), 10 principal components of the genome, genotype array, and main effects of the PRS and ELS. The same models without PRS and the interaction term were run, to examine the effect of ELS on body fat percentage and depression without the genetic contribution as well. In our analyses, we calculated the direct effect (c), indirect effect (a x b), and the total effect (c + (a x b)), with c being the direct effect, and a and b the two direct effects (Fig. 1). Details of the equations are written in Supplementary material, section 3.3.
Based on the work of Baron & Kenny [39], mediation models, like lavaan, use separate regression equations to test three hypotheses. The first one is that the investigated X variable (in our analyses, PRS x ELS) significantly influences the observed Y outcome variable (depression score). Second is that X significantly impacts M (body fat percentage), and third, M significantly predicts Y.
Resting-state functional connectivity analysis
We used data from the Hungarian dataset comprising 102 healthy participants (Nfemales=58, Nmales = 44) who underwent a resting-state functional magnetic resonance imaging (fMRI) session and provided a genetic sample. Resting-state functional magnetic resonance imaging (fMRI) measures changes in blood oxygen level-dependent (BOLD) signals in the brain, reflecting neural activity. Synchronous BOLD signal time series between brain regions indicate possible functional connectivity, which can be quantified by converting the correlations into Z-scores. 23 subjects performed a special pain task before the MRI scan [40]. Parameters of the scanners can be found in Supplementary Material, section 3.4, and imaging data preprocessing steps were the same for both scanners as detailed in our previous publication [41]. Seed-based resting-state functional connectivity analysis was performed with the NAc serving as seed region (MNI coordinates: x = 10; y = 12; z = -8, radius 4 mm) [42]. The individual connectivity maps were used in second-level analysis of Statistical Parametric Mapping (SPM12) software package implemented in Matlab (version 2016). All the analyses were corrected for the possible effect of the scanners, sex, age, and motion parameters. For data visualization, MRIcroGL was used (http://www.mccauslandcenter.sc.edu/mricrogl/).
Descriptive statistics, functional annotation, multiple testing correction
Descriptive statistical analyses were performed with R (version 4.1.2) [43]. We performed 3 regression analyses (total sample, and two sex subpopulations) to test the interaction of PRS and ELS on depression using the UKB data. Main effect and mediation analyses can be considered independent post-hoc analyses. Thus, the Bonferroni corrected p-value is 0.05/3 = 0.0167, since we ran 3 related analyses (total population, and sex separated sub-populations).
Results
Descriptives
Detailed descriptive statistics for the UKB and the Hungarian cohorts used for functional connectivity analyses are provided in Supplementary Tables S2-S4.
Discovery GWAS on depression score
Results and significant risk loci are presented in Supplementary Figures S1-S2.
NAD + /SIRT1 pathway PRS: main effect on depression score
P-value distribution of the pathway-level variants and their functional properties are shown in Supplementary Figure S3 and Supplementary Table S1.
We conducted main effect analyses within the target sample with linear regression for depression score, using covariates and NAD + /SIRT1 pathway PRS as predictors. The main effects of ELS and age were significant across the total population and the sex-separated subpopulations. The main effect of the PRS was non-significant indicating a lack of association without interaction. (Supplementary Table S5).
NAD + /SIRT1 pathway PRS: effect of interaction with ELS on depression score
We investigated the effect of the interaction between NAD + /SIRT1 pathway PRS and ELS on depression score. The interaction is mathematically the product of ELS and PRS scores, which can model non-linear relationships between the variables, as detailed in the Methods section.
P-values corresponding to the interaction effect were significant (p ≤ 0.016) in total population and in the male subpopulation, in which explained variance change in percentage (ΔR2perc) was the highest (Table 1).
The role of body fat percentage in mediating the effect of the interaction between genetic risk of the NAD + /SIRT1 pathway and ELS
In this model, similarly to regression analyses in section 2.4 (Table 1), genetic risk of pathway in interaction with ELS had significant direct effect (p ≤ 0.05) on depression in total population and in male subpopulation. Body fat percentage showed a significant (p ≤ 0.05) direct effect on depression score in all populations. However, we did not identify significant indirect effects of interaction through body fat percentage (Table 2) showing that body fat ratio does not mediate this effect. Visualization of the mediation analyses is shown in Fig. 1.
Association between resting state functional connectivity of the NAc and interaction of genetic risk of NAD + /SIRT1 pathway with ELS in a sex-specific manner in an independent cohort
In our present study the resting-state functional connectivity map of NAc with cortical and limbic regions showed similarities with findings from previous studies [44] (Supplementary Figure S4 and Supplementary Table S6). There was no difference between males and females in baseline NAc connectivity or in the effect of ELS on NAc connectivity in the whole population. ELS, however, showed significant effects on NAc functional connectivity in males, but not in females (for detailed results, see Supplementary Section 2.5.1).
Functional connectivity of the NAc did not show an association with the NAD + /SIRT1 pathway PRS x ELS interaction in the total population. A sex-dependent association was found between the NAc functional connectivity and PRS x ELS interaction. More precisely, compared to females, males displayed stronger functional connectivity of the NAc with middle frontal (Peak T-value = 3967, Peak MNI (x,y,z)=44; 30; 30; and triangular part of inferior frontal gyri (Peak T-value = 4142, Peak MNI coordinates (x,y,z) = 42; 22; 30) (Fig. 2) (pFWE=0.0139, k = 159). In males, we observed that a higher interaction score between PRS and ELS led to stronger connection between the NAc, inferior frontalis gyrus pars triangularis and middle frontal gyrus (beta=0.3859, SE = 0.1513, R2 = 0.1135). Conversely, in females, we found the inverse pattern, where higher interaction score resulted in diminished connectivity (beta = -0.3850, SE = 0.1104, R2 = 0.1637), (Table 3 and Fig. 3).
Significantly associated brain regions with NAc in males with respect to the genetic risk of the NAD + /SIRT1 pathway and ELS (PRS x ELS). Green: Nucleus accumbens (NAc) as seed region (MNI: x = 10; y = 12; z = -8, radius 4 mm) Red: Middle frontal and triangular part of inferior frontal gyri. Secondary cluster-level threshold pFWE < 0.05. MNI: Montreal Neurological Institute. MRIcroGL was used for visualization of significant clusters.
Discussion
Our study provides novel evidence that genetic risk of the NAD + /SIRT1 pathway interacts with ELS in influencing current depressive symptom score, in males, and this effect in humans is not mediated by body fat percentage. Furthermore, we also found genetic risk of the NAD + /SIRT1 pathway in interaction with ELS influences the NAc functional connectivity network, and NAc connectivity changes in males are opposite to those in females. Overall, our results successfully translated and extended animal findings into humans and provide compelling genetic evidence for the NAD + /SIRT1 pathway-mediated risk of ELS on depression in humans.
NAD + /SIRT1 pathway risk on depression score is dependent on early life stress exposure
The significant interaction between NAD + /SIRT1 pathway PRS and ELS supports the key role of ELS to manifest genetic liability in depression and is in line with the known environmental dependence of depression [1, 45,46,47]. Findings are consistent with previous studies. For example, in humans, childhood adversity decreased the expression of SIRT1 and reduced SIRT1 levels correlated with the severity of depressive symptoms [48]. Another study, in a rodent model, investigated the link between adverse childhood experiences and adult depression risk and found that isolation stress in early life reduced levels of Sirt1 in the brain and blood cells, correlating with depression-like behavior in adulthood [49]. These findings, and the study of Morató et al., [8] also suggested SIRT1’s role in the long-term effects of adverse childhood experiences on depression risk. The interaction of the genetic risk of the NAD + /SIRT1 pathway and ELS on depression in humans in the present study, thus, provides key findings suggesting a potential mechanism, how effects of ELS may be translated into depression beyond SIRT1. PRSs derived from the whole genome range between 2-12% on the liability scale [50, 51], thus, 0.5037% explained variance change in males caused by PRS x ELS can be considered large.
Findings support a pathway-level effect, beyond SIRT1. Most of the components of NAD + /SIRT1 pathway (TFAM, MFN1, MFN2, NDUFS4, UQCRB, COX6A, ACO2, CS, BNIP3, CLPP) are mitochondrial protein-coding genes. Our findings support previous reports of genes encoded in nuclear DNA and involved in mitochondrial functions playing a role in depression [52]. The connection between mitochondrial functions and depression, for example in cognitive disturbances, suggests that evaluating cognitive symptoms of depression in association with the NAD + /SIRT1 pathway risk may be an interesting future direction of research.
Taken together our findings provide substantial evidence about how ELS may be translated into depression via the NAD + /SIRT1 pathway and implicate mitochondrial energy homeostasis as a potential mediator of this process on a functional level.
Genetic risk in NAD + /SIRT1 pathway on depression score is not mediated by body fat
In spite of the animal experiment connecting ELS, decreased sociability, increased adiposity, and SIRT1 [8], the stress-dependent effects of NAD + /SIRT1 pathway genetic risk in our analyses were not mediated by body fat percentage, and potentially obesity. This indicates that variants of the NAD + /SIRT1 pathway are independent risk factors for ELS-associated depression. These suggest a different relationship between ELS, adiposity, NAD + /SIRT1 pathway, and mood disorders in animal models of depression and human disease. We must mention that the mean age of the investigated UK Biobank cohort was ~57 years (Supplementary Tables 2-3), and ELS may have occurred over a longer time interval than in the animal study, thus, this might have influenced the results on the role of body fat percentage. To understand the role of body fat percentage, we tested whether body fat percentage mediates the main effect of ELS on depression score without genetic contribution. The results showed that ELS has a significant effect on depression score independently of sex, both directly and through body fat percentage (Supplementary Table S7). Thus, although body fat percentage significantly contributes to depression score directly, and partially mediates the effect of ELS, this effect does not seem to involve the genetic risk related to the NAD + /SIRT1 pathway. Several genes from the NAD + /SIRT1 pathway (PPARGC1A, CAT, GPX4, HSPD1) encode proteins involved in metabolic and stress response processes [53, 54]. For example, NPY, which encodes neuropeptide Y, was previously implicated in major depressive disorder in interaction with ELS [55], as well as in obesity and energy metabolism [56]. Despite: 1) all these lines of evidence and the original findings of Morató et al., [8] 2) evidence that a high body mass index may be a trigger-like risk factor for depression [57], and 3) the known genetic overlap between depression and obesity [57], we could not confirm the role of body fat percentage in ELS-associated human depression in relation with genetic risk of NAD + /SIRT1 pathway.
Although adiposity did not mediate the observed effects in our study, its interaction with the NAD + /SIRT1 pathway may still play a significant role in specific subgroups, such as individuals with metabolic or inflammatory depression. Exploring this interplay may further explain the links between metabolism, stress, and depression. For example, individuals with atypical depression, a subtype characterized by increased appetite, weight gain, and fatigue, may exhibit disrupted SIRT1 activity due to altered NAD+ availability and increased adiposity. Similarly, patients with immunometabolic profiles, such as those with elevated inflammatory markers (CRP, IL-6) or metabolic syndrome, could be particularly affected by SIRT1-related pathways. Other subgroups that may be sensitive to this interaction include individuals with insulin resistance or type 2 diabetes, and those with chronic fatigue symptoms, where impaired energy metabolism may be a contributor to mood dysregulation. These populations may show greater vulnerability to depression through reduced NAD + /SIRT1 function, metabolic dysregulation, and inflammation.
Functional connectivity of the nucleus accumbens is influenced by the NAD + /SIRT1 pathway and ELS interaction
The NAc plays an important role in the reward system by modulating the cortico-striatal circuit. ELS interfering with heightened striatal-medial prefrontal cortex (mPFC) connectivity is related to higher levels of internalizing symptomatology [58], such as self-reported depression and anxiety. In our study, a positive correlation was detected between ELS and NAc-mPFC connectivity in males, which is in line with previous results. The prefrontal cortex exerts a top-down inhibition on striatal response to reward [59], thus, increased connectivity between the middle frontal gyrus, corresponding to the dorsolateral prefrontal cortex and NAc indicates a decreased hedonic function in association with ELS exposure. In a recent study, Chen et al. [60] demonstrated that increased functional connectivity of the NAc with middle frontal gyrus is associated with melancholic depression. In another study, Zhao et al. [20] showed that functional connectivity between NAc and middle frontal gyrus correlates with anhedonia in depressed patients. Our results suggest that ELS in interaction with genetic risk in the NAD + /SIRT1 pathway is associated with increased functional connectivity of the NAc and middle frontal gyrus in males, but not in females. Therefore, the NAD + /SIRT1 pathway in the presence of ELS could interfere with hedonic function regulation in males.
Sex-specific effect of the interaction of NAD + /SIRT1 pathway PRS and ELS
Our findings demonstrated significant effect of PRS x ELS interaction on depression probably only in males. This is supported by the whole population significance, where explained variance values seemed to be a combination of the highly significant male and non-significant female values. Nonetheless, a general effect cannot be completely excluded based on our linear regression results.
In further support of a male-specific effect, however, we also identified sex differences in the association of functional connectivity PRS x ELS. This aligns with the sex-specific results of Morató et al. [8], who also found significant effects of ELS on adiposity and reduced sociability only in male mice. Our study strengthens the assumption that there is a sex-specific biological background of depression. It is known that depression is more prevalent in females during the reproductive age [61], and there are sex differences in depression both in involved neural circuits and in molecular mechanisms [62]. Our primary goal was not to compare prevalence between sexes but to investigate the interaction between genetic risk (NAD + /SIRT1 pathway PRS) and early life stress (ELS) in predicting depressive symptom scores. We chose symptom scores instead of categorical diagnosis to better capture individual-level variation in depression severity, which is crucial for identifying subtle gene-environment interactions. This dimensional framework may also uncover sex-specific pathways contributing to depression risk that are not evident when using binary diagnostic categories.
In rodent studies, it is common for research protocols to focus solely on male animals, due to female reproductive cycles that are hard to control. When studies include females, they often fail to detect the same effects observed in males. In fact, this is already the second study in our lab [63], that successfully translates findings in male rodents into findings in males. These results suggest that despite criticism of rodent study capabilities to capture human depression [64], such criticism is too general and findings may be translated into humans given sex is taken into account. This also emphasizes the need to conduct animal studies with females and underline the importance of sex in cross-species translational studies in depression in general. However, recent literature also highlights that male rodents can exhibit considerable variability, particularly due to social hierarchy and dominance dynamics. These emerging findings emphasize that variability is not exclusive to females and that both sexes bring distinct biological and behavioral complexities to experimental models.
Potential clinical and psychopharmacological consequences of the findings
In the study of Morató et al., [8] nicotinamide mononucleotide (NMN) treatment reversed ELS-induced behavioral changes in adult mice. Moreover, resveratrol, which directly activates SIRT1 [65], was found effective in the treatment of depression in animal experiments [66].
Our study suggests the potential role of NMN, resveratrol, and additional potential therapeutic candidates by proving the importance of NAD + /SIRT1 pathway in ELS-associated probably male-specific, depression, and defines a well-defined human sample, where such treatment strategies promise the largest benefit. These results could open the door to sex-informed, personalized treatment strategies. For example, individuals with high early life stress exposure and elevated NAD + /SIRT1 pathway genetic risk, particularly males, may benefit from interventions that target mitochondrial metabolism or NAD+ homeostasis. This includes NAD+ precursors (e.g., NMN or nicotinamide riboside), SIRT1 activators (e.g., resveratrol), or broader strategies aimed at enhancing mitochondrial function and stress resilience.
In clinical practice, our findings could inform the stratification of patients with depression based on early environmental exposure and genetic profiles, particularly for those with atypical symptoms, anhedonia, or reward-processing impairments. Functional imaging biomarkers, such as altered NAc-mPFC connectivity, may further assist in identifying those who are most likely to respond to NAD + /SIRT1-targeted treatments.
Ultimately, our study supports a precision psychiatry framework, where pathway-specific genetic and environmental interactions help guide therapeutic decision-making, monitor treatment response, and predict prognosis. These insights encourage the integration of genetic screening and ELS history into future clinical trials testing NAD + -related compounds in depression.
Limitations
Our results should be interpreted considering the following limitations and future directions:
1) We divided UKB data into discovery and target sample based on the availability of ELS variable. The data-driven sample splitting is a limitation of our method, while it’s important to consider that the participation in the online follow-up data collection and the completion of the Childhood Trauma Screener (CTS) questionnaire was entirely random, ensuring no bias in the sample.
2) Genes from the NAD + /SIRT1 pathway were not completely covered during our analyses, due to genomic quality control steps and the lack of mitochondrial DNA. However, mitochondrial function depends on nuclear genes as well, and common variants from these nuclear genes influence depressive phenotypes with a minor effect size but a less compensable effect [52]. Therefore, it is unlikely that our study would substantially change if mitochondrial genes would be included. Rather, mitochondrial alterations seem to have a large impact in comparison to nuclearly encoded genes, thus, we would expect the strengthening of our results. Nonetheless, future studies should address such questions.
Future studies on larger populations may focus on compensating for this and use larger coverage.
3) Our results provide no direction of change of the functions of NAD + /SIRT1 pathway as a result of ELS. Therefore, the next step in the development of this direction of research in depression would be to test the pathway, e.g., with transcriptomic and proteomic studies in depressed individuals exposed to ELS. Childhood trauma subtypes can have different effects on the development and psychopathology of psychiatric problems in adulthood [67]. Future studies should investigate the effect of the interaction of different ELS subtypes and NAD + /SIRT1 pathway PRS on depression.
In conclusion, our analyses revealed that 1) males with higher genetic risk in the NAD + /SIRT1 pathway who experienced early life stress are more vulnerable to depression, and 2) higher genetic risk in the NAD + /SIRT1 in the presence of ELS can lead to changes in reward processes in a sex-dependent manner. These findings translate previous animal findings and pave the way for targeted, functional testing of the NAD + /SIRT1 pathway and ELS in depression in humans aiming to reveal novel biomarkers and discover new therapies.
Data availability
UKB data is available for further research upon application to the data owners: UK Biobank, application number:1602.
References
Gonda X, Petschner P, Eszlari N, Baksa D, Edes A, Antal P, et al. Genetic variants in major depressive disorder: from pathophysiology to therapy. Pharmacol Ther. 2019;194:22–43.
Schiedel M, Robaa D, Rumpf T, Sippl W, Jung M. The current state of NAD+-dependent histone deacetylases (sirtuins) as novel therapeutic targets. Med Res Rev. 2018;38:147–200.
Song Y, Wu Z, Zhao P. The protective effects of activating Sirt1/NF-κB pathway for neurological disorders. Rev Neurosci. 2022;33:427–38.
Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, et al. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev. 2012;92:1479–514.
Zhang M, Zhang Q, Hu Y, Xu L, Jiang Y, Zhang C, et al. miR-181a increases FoxO1 acetylation and promotes granulosa cell apoptosis via SIRT1 downregulation. Cell Death Dis. 2017;8:e3088.
Cantó C, Auwerx J. Targeting SIRT1 to improve metabolism: all you need is NAD+? Pharmacol Rev. 2012;64:166–87.
Alam F, Syed H, Amjad S, Baig M, Khan TA, Rehman R. Interplay between oxidative stress, SIRT1, reproductive and metabolic functions. Curr Res Physiol. 2021;4:119–24.
Morató L, Astori S, Zalachoras I, Rodrigues J, Ghosal S, Huang W, et al. eNAMPT actions through nucleus accumbens NAD+/SIRT1 link increased adiposity with sociability deficits programmed by peripuberty stress. Sci Adv. 2022;8:eabj9109.
Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, Middeldorp CM. Research Review: Polygenic methods and their application to psychiatric traits. J Child Psychol Psychiatry. 2014;55:1068–87.
Choi SW, García-González J, Ruan Y, Wu HM, Porras C, Johnson J, et al. PRSet: pathway-based polygenic risk score analyses and software. PLOS Genet. 2023;19:e1010624.
Li L, Chassan RA, Bruer EH, Gower BA, Shelton RC. Childhood maltreatment increases the risk for visceral obesity. Obes Silver Spring Md. 2015;23:1625–32.
Danese A, Tan M. Childhood maltreatment and obesity: systematic review and meta-analysis. Mol Psychiatry. 2014;19:544–54.
Rotenberg KJ, Bharathi C, Davies H, Finch T. Obesity and the social withdrawal syndrome. Eat Behav. 2017;26:167–70.
Pardo PS, Boriek AM. SIRT1 Regulation in Ageing and Obesity. Mech Ageing Dev. 2020;188:111249.
Milaneschi Y, Simmons WK, van Rossum EFC, Penninx BW. Depression and obesity: evidence of shared biological mechanisms. Mol Psychiatry. 2019;24:18–33.
Marx P, Antal P, Bolgar B, Bagdy G, Deakin B, Juhasz G. Comorbidities in the diseasome are more apparent than real: what bayesian filtering reveals about the comorbidities of depression. PLoS Comput Biol. 2017;13:e1005487. https://doi.org/10.1371/journal.pcbi.1005487.
Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, et al. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008;124:431.
Jameei H, Rakesh D, Zalesky A, Cairns MJ, Reay WR, Wray NR, et al. Linking polygenic risk of schizophrenia to variation in magnetic resonance imaging brain measures: a comprehensive systematic review. Schizophr Bull. 2023;50:32–46.
Cattarinussi G, Delvecchio G, Sambataro F, Brambilla P. The effect of polygenic risk scores for major depressive disorder, bipolar disorder and schizophrenia on morphological brain measures: A systematic review of the evidence. J Affect Disord. 2022;310:213–22.
Hu Y, Zhao C, Zhao H, Qiao J. Abnormal functional connectivity of the nucleus accumbens subregions mediates the association between anhedonia and major depressive disorder. BMC Psychiatry. 2023;23:282.
Ma Y, Kochunov P, Kvarta MD, LeGates T, Adhikari BM, Chiappelli J, et al. Reciprocal relationships between stress and depressive symptoms: the essential role of the nucleus accumbens. Psychol Med. 2024;54:1045–56.
Jiang Y, Zou M, Wang Y, Wang Y. Nucleus accumbens in the pathogenesis of major depressive disorder: A brief review. Brain Res Bull. 2023;196:68–75.
Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9.
Eszlari N, Bruncsics B, Millinghoffer A, Hullam G, Petschner P, Gonda X, et al. Biology of perseverative negative thinking: the role of timing and folate intake. Nutrients. 2021;13:4396.
Schork AJ, Thompson WK, Pham P, Torkamani A, Roddey JC, Sullivan PF, et al. All SNPs are not created equal: genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLOS Genet. 2013;9:e1003449.
Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91.
Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, et al. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinforma Oxf Engl. 2005;21:3439–40.
Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinforma Oxf Engl. 2015;31:3555–7.
Hullam G, Antal P, Petschner P, Gonda X, Bagdy G, Deakin B, et al. The UKB envirome of depression: from interactions to synergistic effects. Sci Rep. 2019;9:1–19.
Walker EA, Gelfand A, Katon WJ, Koss MP, Von Korff M, Bernstein D, et al. Adult health status of women with histories of childhood abuse and neglect. Am J Med. 1999;107:332–9.
Dv S, Y L, Kh S, P A, J J, E W, et al. The mini-international neuropsychiatric interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59:22–33. https://pubmed.ncbi.nlm.nih.gov/9881538/ (accessed 24 Mar2024).
Zung WW. A self-rating depression scale. Arch Gen Psychiatry. 1965;12:63–70.
Bernstein DP, Fink L, Handelsman L, Foote J, Lovejoy M, Wenzel K, et al. Initial reliability and validity of a new retrospective measure of child abuse and neglect. Am J Psychiatry. 1994;151:1132–6.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7.
Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826.
Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics. 2020;36:5424–31.
Privé F, Aschard H, Ziyatdinov A, Blum MGB. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34:2781–7.
Rosseel Y. lavaan: An R package for structural equation modeling. J Stat Softw. 2012;48:1–36.
Baron RM, Kenny DA. The moderator–mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–82.
Kökönyei G, Galambos A, Kocsel N, Szabó E, Édes AE, Gecse K, et al. Inter-individual differences in pain anticipation and pain perception in migraine: Neural correlates of migraine frequency and cortisol-to-dehydroepiandrosterone sulfate (DHEA-S) ratio. PLOS ONE. 2021;16:e0261570.
Gecse K, Baksa D, Dobos D, Aranyi CS, Galambos A, Kocsel N, et al. Sex differences of periaqueductal grey matter functional connectivity in migraine. Front Pain Res. 2021;2:767162. https://doi.org/10.3389/fpain.2021.767162.
Haynos A, Hall L, Lavender J, Peterson C, Crow S, Klimes-Dougan B, et al. Resting state functional connectivity of networks associated with reward and habit in anorexia nervosa. Hum Brain Mapp. 2018;40:652–62. https://doi.org/10.1002/hbm.24402.
R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Nucleus accumbens: a systematic review of neural circuitry and clinical studies in healthy and pathological states in: Journal of Neurosurgery 138(2022).https://thejns.org/configurable/content/journals$002fj-neurosurg$002f138$002f2$002farticle-p337.xml?t:ac=journals%24002fj-neurosurg%24002f138%24002f2%24002farticle-p337.xml (accessed 22 Apr2024).
Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol. 2012;233:102–11.
Sandi C, Haller J. Stress and the social brain: behavioural effects and neurobiological mechanisms. Nat Rev Neurosci. 2015;16:290–304.
Zhao L, Han G, Zhao Y, Jin Y, Ge T, Yang W, et al. Gender differences in depression: evidence from genetics. Front Genet. 2020;11:562316. https://doi.org/10.3389/fgene.2020.562316.
Lo Iacono L, Bussone S, Andolina D, Tambelli R, Troisi A, Carola V. Dissecting major depression: The role of blood biomarkers and adverse childhood experiences in distinguishing clinical subgroups. J Affect Disord. 2020;276:351–60.
Lo Iacono L, Visco-Comandini F, Valzania A, Viscomi MT, Coviello M, Giampà A, et al. Adversity in childhood and depression: linked through SIRT1. Transl Psychiatry. 2015;5:e629.
Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9.
Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803.
Petschner P, Gonda X, Baksa D, Eszlari N, Trivaks M, Juhasz G, et al. Genes linking mitochondrial function, cognitive impairment and depression are associated with endophenotypes serving precision medicine. Neuroscience. 2018;370:207–17.
Safran M, Rosen N, Twik M, BarShir R, Stein TI, Dahary D et al. The genecards suite. In: Abugessaisa I, Kasukawa T (eds). Practical Guide to Life Science Databases. Springer Nature: Singapore, 2021, pp 27–56.
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma. 2016;54:1.30.1–1.30.33.
Maul S, Giegling I, Fabbri C, Corponi F, Serretti A, Rujescu D. Genetics of resilience: Implications from genome-wide association studies and candidate genes of the stress response system in posttraumatic stress disorder and depression. Am J Med Genet B Neuropsychiatr Genet. 2020;183:77–94.
Zhang W, Cline MA, Gilbert ER. Hypothalamus-adipose tissue crosstalk: neuropeptide Y and the regulation of energy metabolism. Nutr Metab. 2014;11:27.
Anguita-Ruiz A, Zarza-Rebollo JA, Pérez-Gutiérrez AM, Molina E, Gutiérrez B, Bellón JÁ, et al. Body mass index interacts with a genetic-risk score for depression increasing the risk of the disease in high-susceptibility individuals. Transl Psychiatry. 2022;12:1–10.
Hanson JL, Knodt AR, Brigidi BD, Hariri AR. Heightened connectivity between the ventral striatum and medial prefrontal cortex as a biomarker for stress-related psychopathology: understanding interactive effects of early and more recent stress. Psychol Med. 2018;48:1835–43.
Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, Amatya D, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:aac9698.
Chen Z, Ou Y, Liu F, Li H, Li P, Xie G, et al. Increased brain nucleus accumbens functional connectivity in melancholic depression. Neuropharmacology. 2024;243:109798.
Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, et al. Major depressive disorder. Nat Rev Dis Primer. 2016;2:1–20.
Bangasser DA, Cuarenta A. Sex differences in anxiety and depression: circuits and mechanisms. Nat Rev Neurosci. 2021;22:674–84.
Gal Z, Torok D, Gonda X, Eszlari N, Anderson IM, Deakin B, et al. Inflammation and blood-brain barrier in depression: interaction of CLDN5 and IL6 gene variants in stress-induced depression. Int J Neuropsychopharmacol. 2023;26:189–97.
Gencturk S, Unal G. Rodent tests of depression and anxiety: construct validity and translational relevance. Cogn Affect Behav Neurosci. 2024;24:191–224.
Alageel A, Tomasi J, Tersigni C, Brietzke E, Zuckerman H, Subramaniapillai M, et al. Evidence supporting a mechanistic role of sirtuins in mood and metabolic disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:95–101.
Moore A, Beidler J, Hong M. Resveratrol and depression in animal models: a systematic review of the biological mechanisms. Molecules. 2018;23:2197.
Carr CP, Martins CMS, Stingel AM, Lemgruber VB, Juruena MF. The role of early life stress in adult psychiatric disorders: a systematic review according to childhood trauma subtypes. J Nerv Ment Dis. 2013;201:1007.
Acknowledgements
This project was supported by the National Research, Development and Innovation Office, Hungary (2019-2.1.7-ERA-NET-2020-00005), under the frame of ERA PerMed (ERAPERMED2019-108); by the Hungarian Brain Research Program (Grant: 2017-1.2.1-NKP-2017-00002; NAP2022-I-4/2022); KTIA_13_NAPA-II/14; KTIA_NAP_13-1-2013- 0001; KTIA_NAP_13-2- 2015-0001; NAP2022-I-4/2022; by the Ministry of Innovation and Technology of Hungary, Development and Innovation Fund, under TKP2021-EGA-25; by the Hungarian National Research, Development, and Innovation Office Grant: K 143391 and PD 146014. Peter Petschner was an international research fellow of the Japan Society for the Promotion of Science (P20809). Nora Eszlari is supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. Kinga Gecse, Nora Eszlari, and Sandor Krause were supported by the ÚNKP-23-4-I-SE-31, EKÖP-2025-624, and ÚNKP-22-4-II-SE-1 and ÚNKP-23-3-I-SE-73 grants. Dora Torok is supported by EKÖP-2024-68.
Author information
Authors and Affiliations
Contributions
DT - conceptualization, analysis, methodology, writing – original draft, writing – review and editing, SK - analysis, methodology, writing – original draft, KG - analysis, methodology, writing – original draft, ZsG - analysis, methodology, writing – original draft, NE – conceptualization, analysis, methodology, writing – original draft, MCs - analysis, methodology, writing – original draft, GyB - funding acquisition, writing – review and editing, XG conceptualization, writing – original draft, writing – review and editing, *GJ - funding acquisition, conceptualization, supervision, writing – original draft, writing – review and editing, PP - conceptualization, supervision, writing – original draft, writing – review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Torok, D., Krause, S., Gecse, K. et al. Interaction of early life stress and NAD + /SIRT1 pathway genetic risk promotes depression. Transl Psychiatry 15, 509 (2025). https://doi.org/10.1038/s41398-025-03733-5
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03733-5