Abstract
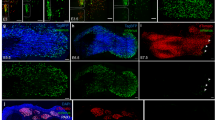
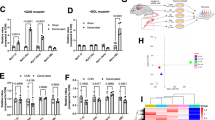
Skeletal muscle is divided into slow- and fast-type muscles, which possess distinct contractile and metabolic properties. Myogenic progenitors associated with each muscle fiber type are known to intrinsically commit to specific muscle fiber lineage during embryonic development. However, it is still unclear whether the functionality of postnatal adult myogenic cells is attributable to the muscle fiber in which they reside, and whether the characteristics of myogenic cells derived from slow- and fast-type fibers can be distinguished at the genetic level. In this study, we isolated adult satellite cells from slow- and fast-type muscle individually and observed that satellite cells from each type of muscle generated myotubes expressing myosin heavy chain isoforms similar to their original muscle, and showed different metabolic features. Notably, we discovered that slow muscle-derived cells had low potential to differentiate but high potential to self-renew compared with fast muscle-derived cells. Additionally, cell transplantation experiments of slow muscle-derived cells into fast-type muscle revealed that slow muscle-derived cells could better contribute to myofiber formation and satellite cell constitution than fast muscle-derived cells, suggesting that the recipient muscle fiber type may not affect the predetermined abilities of myogenic cells. Gene expression analyses identified T-box transcriptional factor Tbx1 as a highly expressed gene in fast muscle-derived myoblasts. Gain- and loss-of-function experiments revealed that Tbx1 modulated muscle fiber types and oxidative metabolism in myotubes, and that Tbx1 stimulated myoblast differentiation, but did not regulate myogenic cell self-renewal. Our data suggest that metabolic and myogenic properties of myogenic progenitor cells vary depending on the type of muscle from which they originate, and that Tbx1 expression partially explains the functional differences of myogenic cells derived from fast-type and slow-type muscles.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5.
Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–38.
Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301.
Day K, Shefer G, Shearer A, Yablonka-Reuveni Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev Biol. 2010;340:330–43.
Verdijk LB, Snijders T, Drost M, Delhaas T, Kadi F, van Loon LJ. Satellite cells in human skeletal muscle; from birth to old age. Age (Dordr). 2013;36:545–7.
Webster C, Blau HM. Accelerated age-related decline in replicative life-span of Duchenne muscular dystrophy myoblasts: implications for cell and gene therapy. Somat Cell Mol Genet. 1990;16:557–65.
Shefer G, Van de Mark DP, Richardson JB, Yablonka-Reuveni Z. Satellite-cell pool size does matter: defining the myogenic potency of aging skeletal muscle. Dev Biol. 2006;294:50–66.
Bernet JD, Doles JD, Hall JK, Kelly-Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–71.
Cosgrove BD, Gilbert PM, Porpiglia E, Mourkioti F, Lee SP, Corbel SY, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–64.
Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev Biol. 2010;337:29–41.
Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–9.
Lagord C, Soulet L, Bonavaud S, Bassaglia Y, Rey C, Barlovatz-Meimon G, et al. Differential myogenicity of satellite cells isolated from extensor digitorum longus (EDL) and soleus rat muscles revealed in vitro. Cell Tissue Res. 1998;291:455–68.
Hughes SM, Blau HM. Muscle fiber pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–71.
Lexell J, Jarvis JC, Currie J, Downham DY, Salmons S. Fibre type composition of rabbit tibialis anterior and extensor digitorum longus muscles. J Anat. 1994;185:95–101.
Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–531.
Nicholls DG, Darley-Usmar VM, Wu M, Jensen PB, Rogers GW, Ferrick DA. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010;46:2511.
Bockman EL, McKenzie JE. Tissue adenosine content in active soleus and gracilis muscles of cats. Am J Physiol. 1983;244:H552–9.
Kushmerick MJ, Meyer RA, Brown TR. Regulation of oxygen consumption in fast- and slow-twitch muscle. Am J Physiol. 1992;263:C598–606.
Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24.
Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801.
Baroffio A, Bochaton-Piallat ML, Gabbiani G, Bader CR. Heterogeneity in the progeny of single human muscle satellite cells. Differentiation. 1995;59:259–68.
Yoshida N, Yoshida S, Koishi K, Masuda K, Nabeshima Y. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J Cell Sci. 1998;111:769–79.
Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–57.
Gayraud-Morel B, Chretien F, Jory A, Sambasivan R, Negroni E, Flamant P, et al. Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. J Cell Sci. 2012;125:1738–49.
Bassaglia Y, Gautron J. Fast and slow rat muscles degenerate and regenerate differently after whole crush injury. J Muscle Res Cell Motil. 1995;16:420–9.
Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 1996;60:39–45.
Salmons S. Exercise, stimulation and type transformation of skeletal muscle. Int J Sports Med. 1994;15:136–41.
Gambke B, Lyons GE, Haselgrove J, Kelly AM, Rubinstein NA. Thyroidal and neural control of myosin transitions during development of rat fast and slow muscles. FEBS Lett. 1983;156:335–9.
Larsson L, Biral D, Campione M, Schiaffino S. An age-related type IIB to IIX myosin heavy chain switching in rat skeletal muscle. Acta Physiol Scand. 1993;147:227–34.
Holloszy JO, Chen M, Cartee GD, Young JC. Skeletal muscle atrophy in old rats: differential changes in the three fiber types. Mech Ageing Dev. 1991;60:199–213.
Lexell J. Human aging, muscle mass, and fiber type composition. J Gerontol A Biol Sci Med Sci. 1995;50:11–6.
Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol. 2013;45:2191–9.
Launay T, Noirez P, Butler-Browne G, Agbulut O. Expression of slow myosin heavy chain during muscle regeneration is not always dependent on muscle innervation and calcineurin phosphatase activity. Am J Physiol Regul Integr Comp Physiol. 2006;290:R1508–14.
Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–60.
Motohashi N, Asakura A. Muscle satellite cell heterogeneity and self-renewal. Front Cell Dev Biol. 2014;2:1.
Scambler PJ. The 22q11 deletion syndromes. Hum Mol Genet. 2000;9:2421–6.
Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101.
Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–73.
Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, et al. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics. 1997;43:267–77.
de Wilde J, Hulshof MF, Boekschoten MV, de Groot P, Smit E, Mariman EC. The embryonic genes Dkk3, Hoxd8, Hoxd9 and Tbx1 identify muscle types in a diet-independent and fiber-type unrelated way. BMC Genomics. 2010;11:176.
Chemello F, Bean C, Cancellara P, Laveder P, Reggiani C, Lanfranchi G. Microgenomic analysis in skeletal muscle: expression signatures of individual fast and slow myofibers. PLoS ONE. 2011;6:e16807.
Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–76.
Lee KY, Singh MK, Ussar S, Wetzel P, Hirshman MF, Goodyear LJ, et al. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat Commun. 2015;6:8054.
Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38.
Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–36.
Baar K, Song Z, Semenkovich CF, Jones TE, Han DH, Nolte LA, et al. Skeletal muscle overexpression of nuclear respiratory factor 1 increases glucose transport capacity. FASEB J. 2003;17:1666–73.
Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13:2829–40.
Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91.
Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, et al. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell. 2009;16:810–21.
Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–9.
Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–13.
Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–25.
Yamamoto M, Legendre NP, Biswas AA, Lawton A, Yamamoto S, Tajbakhsh S, et al. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Rep. 2018;10:956–69.
Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS ONE. 2009;4:e6049.
Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105:842–51.
van Bueren KL, Papangeli I, Rochais F, Pearce K, Roberts C, Calmont A, et al. Hes1 expression is reduced in Tbx1 null cells and is required for the development of structures affected in 22q11 deletion syndrome. Dev Biol. 2010;340:369–80.
Pane LS, Zhang Z, Ferrentino R, Huynh T, Cutillo L, Baldini A. Tbx1 is a negative modulator of Mef2c. Hum Mol Genet. 2012;21:2485–96.
Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–73.
Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–9.
Motohashi N, Asakura Y, Asakura A. Isolation, culture, and transplantation of muscle satellite cells. J Vis Exp. 2014. https://doi.org/10.3791/50846.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Acknowledgements
This work was supported by a Grant-in-Aid for a Japan Society for the Promotion of Science (JSPS) Fellowship for Research Abroad (NM), JSPS KAKENHI (NM: 15H05364), the Nakatomi Foundation (NM), the Uehara Memorial Foundation (NM), a General Insurance Association of Japan medical research grant on traffic accidents (AU), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS; AA: R01 AR062142), and the Muscular Dystrophy Association (MDA; AA).
Author contributions
NM designed and performed the experiments, analyzed the data, and wrote the manuscript. AU designed the experiments and edited the manuscript. YS and ST contributed the mouse line. AA, MU, SM, YM, RT, and KS analyzed and interpreted the data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by E. Baehrecke
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Motohashi, N., Uezumi, A., Asakura, A. et al. Tbx1 regulates inherited metabolic and myogenic abilities of progenitor cells derived from slow- and fast-type muscle. Cell Death Differ 26, 1024–1036 (2019). https://doi.org/10.1038/s41418-018-0186-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-018-0186-4
This article is cited by
-
Transcriptome analysis of muscle atrophy in Leizhou black goats: identification of key genes and insights into limb-girdle muscular dystrophy
BMC Genomics (2025)
-
Reduced myogenic differentiation capacity of satellite cell-derived myoblasts in male ICR mice compared with male C57BL/6 and BALB/c mice
In Vitro Cellular & Developmental Biology - Animal (2025)
-
Decreased number of satellite cells-derived myonuclei in both fast- and slow-twitch muscles in HeyL-KO mice during voluntary running exercise
Skeletal Muscle (2024)
-
Myokine BDNF highly expressed in Type I fibers inhibits the differentiation of myotubes into Type II fibers
Molecular Biology Reports (2024)
-
DNA methylation of insulin signaling pathways is associated with HOMA2-IR in primary myoblasts from older adults
Skeletal Muscle (2023)