Abstract
The miR17~92 cluster plays important roles in haematopoiesis. However, it is not clear at what stage of differentiation and through which targets miR17~92 exerts this function. Therefore, we generated miR17~92fl/fl; RosaCreERT2 mice for inducible deletion of miR17~92 in haematopoietic cells. Bone marrow reconstitution experiments revealed that miR17~92-deleted cells were not capable to contribute to mature haematopoietic lineages, which was due to defects in haematopoietic stem/progenitor cells (HSPCs). To identify the critical factor targeted by miR17~92 we performed gene expression analysis in HSPCs, demonstrating that mRNA levels of pro-apoptotic Bim inversely correlated with the expression of the miR17~92 cluster. Strikingly, loss of pro-apoptotic BIM completely prevented the loss of HSPCs caused by deletion of miR17~92. The BIM/miR17~92 interaction is conserved in human CD34+ HSPCs, as miR17~92 inhibition or blockade of its binding to the BIM 3′UTR reduced the survival and growth of these cells. Despite the prediction that miR17~92 functions by impacting a plethora of different targets, the absence of BIM alone is sufficient to prevent all defects caused by deletion of miR17~92 in haematopoietic cells.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98.
Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–35.
Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53.
Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86.
Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14.
Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–97.
Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–8.
Chamorro-Jorganes A, Lee MY, Araldi E, Landskroner-Eiger S, Fernandez-Fuertes M, Sahraei M, et al. VEGF-induced expression of miR-17-92 cluster in endothelial cells is mediated by ERK/ELK1 activation and regulates angiogenesis. Circ Res. 2016;118:38–47.
Landskroner-Eiger S, Qiu C, Perrotta P, Siragusa M, Lee MY, Ulrich V, et al. Endothelial miR-17 approximately 92 cluster negatively regulates arteriogenesis via miRNA-19 repression of WNT signaling. Proc Natl Acad Sci USA. 2015;112:12812–7.
Lau KW, Stiffel VM, Rundle CH, Amoui M, Tapia J, White TD, et al. Conditional disruption of miR17~92 in osteoclasts led to activation of osteoclasts and loss of trabecular bone in part through suppression of the miR17-mediated downregulation of protein-tyrosine phosphatase-oc in mice. JBMR. 2017;1:73–85.
Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3:1398–406.
de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–30.
Regelin M, Blume J, Pommerencke J, Vakilzadeh R, Witzlau K, Lyszkiewicz M, et al. Responsiveness of developing T cells to IL-7 signals is sustained by miR-17 approximately 92. J Immunol. 2015;195:4832–40.
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–8.
Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, et al. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA. 1991;88:8661–5.
McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88.
Han YC, Vidigal JA, Mu P, Yao E, Singh I, Gonzalez AJ, et al. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet. 2015;47:766–75.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11.
Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–49.
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–72.
Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12.
Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8.
Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol. 2010;477:109–23.
Brinkmann K, Grabow S, Hyland CD, Teh CE, Alexander WS, Herold MJ, et al. The combination of reduced MCL-1 and standard chemotherapeutics is tolerable in mice. Cell Death Differ. 2017;24:2032–43.
Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–42.
Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4.
Morelli E, Biamonte L, Federico C, Amodio N, Di Martino MT, Gallo Cantafio ME, et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-mir-17-92. Blood. 2018;132:1050–63.
Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci USA. 2010;107:3710–5.
de Graaf CA, Choi J, Baldwin TM, Bolden JE, Fairfax KA, Robinson AJ, et al. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Rep. 2016;7:571–82.
Choi J, Baldwin TM, Wong M, Bolden JE, Fairfax KA, Lucas EC, et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 2019;47(D1):D780–D785.
Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, et al. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–96.
Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–29.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:05005.
Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci USA. 2001;98:14541–6.
Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–16.
Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94.
Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82.
Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106:4131–8.
Guo Z, Mu X, Ouyang H, Cheng Z, Wang Z, Xu B. Operative treatment of huge adult frontal nasoethmoid meningoencephalocele. J Craniofac Surg. 2013;24:1669–70.
Xu Z, Sharp PP, Yao Y, Segal D, Ang CH, Khaw SL, et al. BET inhibition represses miR17-92 to drive BIM-initiated apoptosis of normal and transformed hematopoietic cells. Leukemia. 2016;30:1531–41.
Clybouw C, Merino D, Nebl T, Masson F, Robati M, O’Reilly L, et al. Alternative splicing of Bim and Erk-mediated Bim(EL) phosphorylation are dispensable for hematopoietic homeostasis in vivo. Cell Death Differ. 2012;19:1060–8.
Herold MJ, Rohrbeck L, Lang MJ, Grumont R, Gerondakis S, Tai L, et al. Foxo-mediated Bim transcription is dispensable for the apoptosis of hematopoietic cells that is mediated by this BH3-only protein. EMBO Rep. 2013;14:992–8.
Acknowledgements
We thank C Stivala, S Russo, J Mansheim, T Baldinger, M Patsis and G Siciliano for expert animal care; B Helbert and K Mackwell for genotyping; J Corbin and J McManus for automated blood analysis; S Monard and his team for help with flow cytometry; S Mifsud for assistance with colony assays and Dr P Bouillet for providing Bim−/− mice. This work was supported by grants and fellowships from the Deutsche Krebshilfe (Dr Mildred Scheel post-doctoral fellowship to KB) the Australian National Health and Medical Research Council (NHMRC) (Project Grants 1145728 to MJH, 1143105 to MJH and AS, 1122783 to APN, Program Grants 1016701 to AS and 1113577 to WSA and Fellowships 1020363 to AS, 1058344 to WSA, 1156095 to MJH, GNT1035229 to CAdG and 1124788 to NDH), the Leukemia and Lymphoma Society of America (LLS SCOR 7001-13 to AS and MJH), the Cancer Council of Victoria (project grant 1052309 to AS and Venture Grant to MJH and AS), a Victorian Cancer Agency Fellowship (ECSG18020 to JR) as well as by operational infrastructure grants through the Australian Government Independent Research Institute Infrastructure Support Scheme (361646 and 9000220) and the Victorian State Government Operational Infrastructure Support Program.
Author information
Authors and Affiliations
Contributions
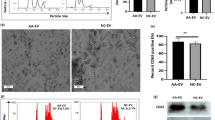
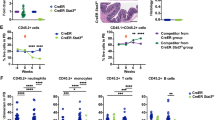
KB, MJH and AS designed and conceived the study, planned experiments and prepared the paper. KB conducted and analysed the experiments. CAdG helped with the analysis of the RNA expression data (Fig. 1b) CH and WSA provided advice for the analysis of the LT-HSC compartment, helped with these experiments and data analysis and provided the antibodies for staining cells (Figs. 3–5). APN and LDR performed the colony formation assays (Fig. 6a). JR and NDH gave advice on experiments with human CD34+ cord blood cells and provided the cells and media (Fig. 6). EM provided MIR17PTi and helped with planning the colony formation assays (Fig. 6a).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Brinkmann, K., Ng, A.P., de Graaf, C.A. et al. miR17~92 restrains pro-apoptotic BIM to ensure survival of haematopoietic stem and progenitor cells. Cell Death Differ 27, 1475–1488 (2020). https://doi.org/10.1038/s41418-019-0430-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-019-0430-6
This article is cited by
-
The nucleobase guanine at the 3’-terminus of oligonucleotide RGLS4326 drives off-target AMPAR inhibition and CNS toxicity
Nature Communications (2025)
-
Regulation of programmed cell death by Brd4
Cell Death & Disease (2022)
-
The transcription factor TAL1 and miR-17-92 create a regulatory loop in hematopoiesis
Scientific Reports (2020)