Abstract
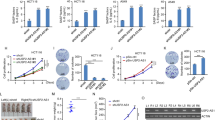
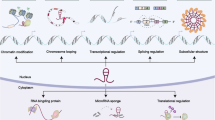
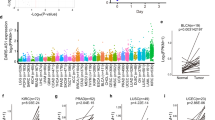
Long noncoding RNAs (lncRNAs) regulating diverse cellular processes implicate in many diseases. However, the function of lncRNAs in cellular senescence remains largely unknown. Here we identify a novel long intergenic noncoding RNA Linc-ASEN expresses in prematurely senescent cells. We find that Linc-ASEN associates with UPF1 by RNA pulldown mass spectrometry analysis, and represses cellular senescence by reducing p21 production transcriptionally and posttranscriptionally. Mechanistically, the Linc-ASEN-UPF1 complex suppressed p21 transcription by recruiting Polycomb Repressive Complex 1 (PRC1) and PRC2 to the p21 locus, and thereby preventing binding of the transcriptional activator p53 on the p21 promoter through histone modification. In addition, the Linc-ASEN-UPF1 complex repressed p21 expression posttranscriptionally by enhancing p21 mRNA decay in association with DCP1A. Accordingly, Linc-ASEN levels were found to correlate inversely with p21 mRNA levels in tumors from patient-derived mouse xenograft, in various human cancer tissues, and in aged mice tissues. Our results reveal that Linc-ASEN prevents cellular senescence by reducing the transcription and stability of p21 mRNA in concert with UPF1, and suggest that Linc-ASEN might be a potential therapeutic target in processes influenced by senescence, including cancer.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217.
Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–53.
Jung SH, Lee HC, Yu DM, Kim BC, Park SM, Lee YS, et al. Heparan sulfation is essential for the prevention of cellular senescence. Cell Death Differ. 2016;23:417–29.
Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–40.
Krtolica A, Campisi J. Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int J Biochem Cell Biol. 2002;34:1401–14.
Prieur A, Peeper DS. Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–5.
Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2011;11:S27–31.
Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci USA. 2001;98:12072–77.
Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–26.
Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Godstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–68.
Muñoz-Espín D, Cañamero M, Maraver A, Gómez-López G, Contreras J, Murillo-Cuesta S, et al. Programmed cell senescence during mammalian embryonic development. Cell. 2013;155:1104–18.
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89.
Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15:423–37.
Kim C, Kang D, Lee EK, Lee JS. Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxid Med Cell Longev. 2017. https://doi.org/10.1155/2017/2062384.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–63.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2018;9:e1471.
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, et al. Jarid2 is implicated in the initial xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell. 2014;53:301–16.
Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708.
Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9.
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69.
Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8.
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55.
Abdelmohsen K, Gorospe M, Noncoding RNA. control of cellular senescence. Wiley Interdiscip Rev RNA. 2015;6:615–29.
Montes M, Lund AH. Emerging roles of lncRNAs in senescence. FEBS J. 2016;283:2414–26.
Kent WJ. BLAT—The BLAST-Like alignment tool. Genome Res. 2003;12:656–64.
Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78.
LeCuyer KA, Behien LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its coorperative binding to RNA. Biochemistry. 1995;34:10600–06.
Sun XL, Perlick HA, Dietz HC, Maquat LE. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc Natl Acad Sci USA. 1998;95:10009–14.
Popp MW, Maquat LE. Leveraging rulges of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016;165:1319–22.
Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712.
Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16:665–77.
Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–35.
Mendell JT, AP Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–22.
Behm-Ansmant I, Kashima I, Rehwinkel J, Saulière J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–53.
Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27.
Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–9.
Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307.
Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–69.
Kim BC, Lee HC, Lee JJ, Choi CM, Kim DK, Lee JC, et al. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J. 2012;31:4289–303.
Lee HC, Jung SH, Hwang HJ, Kang D, De S, Dudekula DB, et al. WIG1 is crucial for AGO2-mediated ACOT7 mRNA silencing via miRNA-dependent and -independent mechanisms. Nucleic Acids Res. 2017;45:6894–910.
Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, et al. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet. 2011;43:745–52.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78.
Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Natl Acad Sci USA. 2012;2:28–36.
Acknowledgements
We are grateful to Prof. Lynne E. Maquat at University of Rochester for pCI-neo-EV, pCI-FLAG-UPF1-WT, and pCI-FLAG-UPF1 R844C plasmids, and Dr Keetae Kim at Daegu Gyeongbuk Institute of Science and Technology (DGIST) for tissues from young and old mouse. This work was supported by grants to JSL from MRC (2014009392), Nuclear Research and Development program (2017M2A2A7A01070591), and Basic Research Science Program (2017R1A2B2007542) through the National Research Foundation (NRF) from Korean government (MSIT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The biospecimens used for this study were provided by the Biobank of Inha University Hospital, and the Ajou University Human Bio-Resource Bank (AHBB) and the Biobank of Korea University Guro Hospital which are members of Korea Biobank Network supported by the Korean Ministry of Health and Welfare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by C. Borner
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, H.C., Kang, D., Han, N. et al. A novel long noncoding RNA Linc-ASEN represses cellular senescence through multileveled reduction of p21 expression. Cell Death Differ 27, 1844–1861 (2020). https://doi.org/10.1038/s41418-019-0467-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-019-0467-6
This article is cited by
-
The chromatin-associated RNAs in gene regulation and cancer
Molecular Cancer (2023)
-
Long non-coding RNAs affecting cell metabolism in cancer
Biology Direct (2022)
-
LncRNA LYPLAL1-AS1 rejuvenates human adipose-derived mesenchymal stem cell senescence via transcriptional MIRLET7B inactivation
Cell & Bioscience (2022)
-
A noncoding regulatory RNA Gm31932 induces cell cycle arrest and differentiation in melanoma via the miR-344d-3-5p/Prc1 (and Nuf2) axis
Cell Death & Disease (2022)
-
The p53 family member p73 in the regulation of cell stress response
Biology Direct (2021)