Abstract
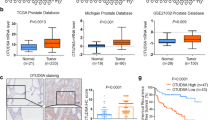
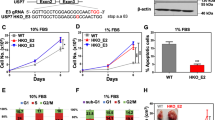
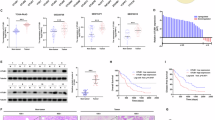
The DNA damage response (DDR) is critical for maintaining cellular homeostasis and genome integrity. Mounting evidence has shown that posttranslational protein modifications play vital roles in the DDR. In this study, we showed that deubiquitinase OTUD6A is involved in the DDR and is important for maintaining genomic stability. Mechanistically, in response to DNA damage, the abundance of OTUD6A was increased; meanwhile, PP2A interacted with OTUD6A and dephosphorylated OTUD6A at sites S70/71/74, which promoted nuclear localization of OTUD6A. Subsequently, OTUD6A was recruited to the damage site, where it interacted with TopBP1 and blocked the interaction between TopBP1 and its ubiquitin E3 ligase UBR5, decreasing K48-linked polyubiquitination and increasing the stability of TopBP1. OTUD6A depletion impaired CHK1 S345 phosphorylation and blocked cell cycle progression under DNA replication stress. Consistently, knockout of OTUD6A rendered mice hypersensitive to irradiation, shortened survival, and inhibited tumor growth by regulating TopBP1 in xenografted nude mice. Moreover, OTUD6A is expressed at high levels in breast cancer, and OTUD6A overexpression promotes cell proliferation, migration and invasion, indicating that dysregulation of OTUD6A expression contributes to genomic instability and is associated with tumor development. In summary, this study demonstrates that OTUD6A plays a critical role in promoting tumor cell resistance to chemoradiotherapy by deubiquitinating and stabilizing TopBP1.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Data availability
All data generated or analyzed during this study are included in this article and its supplementary data files and all original data are available from the corresponding authors upon request.
References
Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl J Med. 2009;361:1475–85.
Pilie PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009;461:1071–8.
Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204.
Sarangi P, Zhao X. SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci. 2015;40:233–42.
Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell. 2013;49:897–907.
Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26.
Kim JJ, Lee SY, Miller KM. Preserving genome integrity and function: the DNA damage response and histone modifications. Crit Rev Biochem Mol Biol. 2019;54:208–41.
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009;136:435–46.
Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, et al. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 2015;10:226–38.
Krais JJ, Wang Y, Bernhardy AJ, Clausen E, Miller JA, Cai KQ, et al. RNF168-mediated ubiquitin signaling inhibits the viability of BRCA1 null cancers. Cancer Res. 2020;80:2848–60.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007;131:887–900.
Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–78.
Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007;316:1198–202.
Cheng YC, Shieh SY. Deubiquitinating enzyme USP3 controls CHK1 chromatin association and activation. Proc Natl Acad Sci USA 2018;115:5546–51.
Li F, Sun Q, Liu K, Han H, Lin N, Cheng Z, et al. The deubiquitinase OTUD5 regulates Ku80 stability and non-homologous end joining. Cell Mol Life Sci. 2019;76:3861–73.
Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, et al. The Deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 2015;13:93–107.
Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9.
Peng Y, Liao Q, Tan W, Peng C, Hu Z, Chen Y, et al. The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat Commun. 2019;10:1224.
Wijnhoven P, Konietzny R, Blackford AN, Travers J, Kessler BM, Nishi R, et al. USP4 Auto-Deubiquitylation promotes homologous recombination. Mol Cell. 2015;60:362–73.
Yang C, Zang W, Tang Z, Ji Y, Xu R, Yang Y, et al. A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. 2018;78:1069–82.
Yang Y, Yang C, Li T, Yu S, Gan T, Hu J, et al. The Deubiquitinase USP38 promotes NHEJ repair through regulation of HDAC1 activity and regulates cancer cell response to genotoxic insults. Cancer Res. 2020;80:719–31.
Fraile JM, Quesada V, Rodriguez D, Freije JM, Lopez-Otin C. Deubiquitinases in cancer: New functions and therapeutic options. Oncogene 2012;31:2373–88.
Komander D, Clague MJ, Urbe S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63.
Antao AM, Tyagi A, Kim KS, Ramakrishna S. Advances in Deubiquitinating enzyme inhibition and applications in cancer therapeutics. Cancers (Basel) 2020;12:1579.
Shi L, Liu J, Peng Y, Zhang J, Dai X, Zhang S, et al. Deubiquitinase OTUD6A promotes proliferation of cancer cells via regulating Drp1 stability and mitochondrial fission. Mol Oncol. 2020;14:3169–83.
Wardlaw CP, Carr AM, Oliver AW. TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst). 2014;22:165–74.
Sokka M, Parkkinen S, Pospiech H, Syvaoja JE. Function of TopBP1 in genome stability. Subcell Biochem. 2010;50:119–41.
Ma S, Cao C, Che SY, Wang YJ, Su DX, Liu S, et al. PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability. Sci Adv 2021;7:eabf7684.no.19
Liu T, Lin YH, Leng W, Jung SY, Zhang H, Deng M, et al. A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint. Mol Cell. 2014;56:681–95.
Leimbacher PA, Jones SE, Shorrocks AK, de Marco Zompit M, Day M, Blaauwendraad J, et al. MDC1 Interacts with TOPBP1 to Maintain Chromosomal Stability during Mitosis. Mol Cell. 2019;74:571–83 e8.
Bigot N, Day M, Baldock RA, Watts FZ, Oliver AW, Pearl LH. Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint. Elife 2019;8:e44353.
Feng H, Lu J, Song X, Thongkum A, Zhang F, Lou L, et al. CK2 kinase-mediated PHF8 phosphorylation controls TopBP1 stability to regulate DNA replication. Nucleic Acids Res. 2021;49:2400–1.
Kim W, Zhao F, Gao H, Qin S, Hou J, Deng M, et al. USP13 regulates the replication stress response by deubiquitinating TopBP1. DNA Repair (Amst). 2021;100:103063.
Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, et al. Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem. 2002;277:3599–605.
Forma E, Krzeslak A, Bernaciak M, Romanowicz-Makowska H, Brys M. Expression of TopBP1 in hereditary breast cancer. Mol Biol Rep. 2012;39:7795–804.
Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, et al. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol Cell Biol. 2009;29:2673–93.
Liu K, Ling S, Lin WC. TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73. Mol Cell Biol. 2011;31:4464–81.
Peng B, Wang J, Hu Y, Zhao H, Hou W, Zhao H, et al. Modulation of LSD1 phosphorylation by CK2/WIP1 regulates RNF168-dependent 53BP1 recruitment in response to DNA damage. Nucleic Acids Res. 2015;43:5936–47.
Hu B, Li S, Zhang X, Zheng X. HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res. 2014;42:5582–93.
Lim KS, Li H, Roberts EA, Gaudiano EF, Clairmont C, Sambel LA, et al. USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol Cell. 2018;72:925–41.e4.
Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–89.
Herhaus L, Perez-Oliva AB, Cozza G, Gourlay R, Weidlich S, Campbell DG, et al. Casein kinase 2 (CK2) phosphorylates the deubiquitylase OTUB1 at Ser16 to trigger its nuclear localization. Sci Signal. 2015;8:ra35.
Chen C, Wei X, Wang S, Jiao Q, Zhang Y, Du G, et al. Compression regulates gene expression of chondrocytes through HDAC4 nuclear relocation via PP2A-dependent HDAC4 dephosphorylation. Biochim Biophys Acta. 2016;1863(7 Pt A):1633–42.
Mehta S, McKinney C, Algie M, Verma CS, Kannan S, Harfoot R, et al. Dephosphorylation of YB-1 is required for nuclear localisation during G2 phase of the cell cycle. Cancers (Basel). 2020;12:315.
Peng Y, Liu J, Wang Z, Cui C, Zhang T, Zhang S, et al. Prostate-specific oncogene OTUD6A promotes prostatic tumorigenesis via deubiquitinating and stabilizing c-Myc. Cell Death Differ. 2022. https://doi.org/10.1038/s41418-022-00960-x.
Fu X, Zhao J, Yu G, Zhang X, Sun J, Li L, et al. OTUD6A promotes prostate tumorigenesis via deubiquitinating Brg1 and AR. Commun Biol. 2022;5:182.
Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell 2006;124:943–55.
Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37.
Wang J, Gong Z, Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol. 2011;193:267–73.
Yoshida K, Inoue I. Expression of MCM10 and TopBP1 is regulated by cell proliferation and UV irradiation via the E2F transcription factor. Oncogene 2004;23:6250–60.
Liu K, Graves JD, Lee YJ, Lin FT, Lin WC. Cell Cycle-Dependent Switch of TopBP1 Functions by Cdk2 and Akt. Mol Cell Biol. 2020;40:e00599–19.
Acknowledgements
We sincerely thank Prof. Lingqiang Zhang for providing OTUD6A cDNA, Prof. Jiadong Wang for providing the Myc-TopBP1 plasmid, Prof. Shimin Zhao for providing Myc-UBR5 plasmids, and Prof. Daochun Kong for providing materials used in EMSA experiment. We thank the National Center for Protein Sciences at Peking University, particularly Liying Du, Huan Yang, Dong Liu, Qi Zhang, Hongxia Lv, LiQin Fu, and Guilan Li, for technical help. We also appreciate the assistance of Xiaochen Li and Siying Qin from the Core Facilities of Life Sciences at Peking University for their assistance with microscopic imaging. This work was supported by the National Natural Science Foundation of China (82130081 and 81730080) and the Natural Science Foundation of Beijing Municipality (5212008).
Author information
Authors and Affiliations
Contributions
YZ designed and performed the experiments, analyzed the data, and wrote the manuscript. XH, DZ, and JL performed the experiments. MW analyzed the pathway data and performed experiments. SY provided technical support for metaphase spread assays. YT performed the cancer cell injection into mice. XZ designed and supervised this study and wrote the manuscript.
Corresponding author
Ethics declarations
COMPETING INTERESTS
The authors declare no competing interests.
Ethical approval
All animal procedures were performed under the guidelines of the Ethics Committee of Peking University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by K. Engeland
Rights and permissions
About this article
Cite this article
Zhao, Y., Huang, X., Zhu, D. et al. Deubiquitinase OTUD6A promotes breast cancer progression by increasing TopBP1 stability and rendering tumor cells resistant to DNA-damaging therapy. Cell Death Differ 29, 2531–2544 (2022). https://doi.org/10.1038/s41418-022-01036-6
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41418-022-01036-6
This article is cited by
-
A comprehensive molecular characterization of a claudin-low luminal B breast tumor
Biology Direct (2024)
-
Deubiquitination of CDC6 by OTUD6A promotes tumour progression and chemoresistance
Molecular Cancer (2024)
-
E3 ubiquitin ligase UBR5 promotes gemcitabine resistance in pancreatic cancer by inducing O-GlcNAcylation-mediated EMT via destabilization of OGA
Cell Death & Disease (2024)
-
Genomic and transcriptomic profiling of hepatocellular carcinoma reveals a rare molecular subtype
Discover Oncology (2024)
-
Deubiquitylase YOD1 regulates CDK1 stability and drives triple-negative breast cancer tumorigenesis
Journal of Experimental & Clinical Cancer Research (2023)