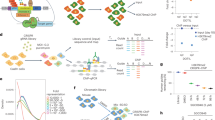
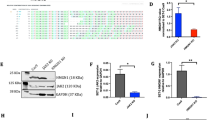
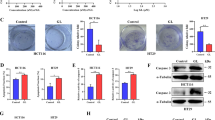
Abstract
DNA damage response (DDR) is a complicated network that responds to DNA lesions to prevent their accumulation; a defective DDR is one hallmark of cancer. Although targeting DDR pathways has been considered as a therapeutic approach, DDR inhibitors have also been reported ineffective for treating some low mutation burden cancers, such as Mixed-lineage leukemia (MLL)-rearranged (MLL-r) leukemia, a clinically fatal and refractory malignancy. Exploring the roles and mechanisms of DDR pathways in these low mutation burden cancers may help understand the chromatin biology and develop therapeutic strategies. Here, we identified a set of DDR-related chromatin-associated circular RNAs (cacircRNAs) that regulate DNA repair via the non-homologous end joining (NHEJ) pathway, which is vital for meeting the high DNA repair demands during the progression of MLL-r leukemia. Among these cacircRNAs, we identified ciCRLF3(2) as a previously unknown component of the NHEJ complex. We showed that ciCRLF3(2) recruits NHEJ regulators to DNA lesions, supporting abundant DNA repair in leukemia cells. ciCRLF3(2) abundance is abnormally upregulated in MLL-r leukemia and indicates a poor prognosis. Targeting ciCRLF3(2) suppressed NHEJ-mediated DNA repair, leading to DNA damage and broad anti-cancer effects in vitro and in vivo. A patient-derived xenograft model of MLL-r leukemia further indicated that ciCRLF3(2) depletion can decrease the leukemic burden. These findings demonstrate the function of cacircRNAs in DDR and chromatin biology and reveal a new avenue for developing strategies to treat low mutation burden cancers, such as MLL-r leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The circRNA RNA-seq data in this study has been uploaded to Sequence Read Archive (SRA) database. The BioProject No. is PRJNA1276304. All other study data are included in the article and/or Supplemental Information. Any information required to reanalyze the data reported in this study is available from the corresponding authors (wangwt8@mail.sysu.edu.cn) upon request.
References
Panier S, Wang S, Schumacher B. Genome instability and DNA repair in somatic and reproductive aging. Annu Rev Pathol. 2024;19:261–290.
Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer. 2023;23:78–94.
Kellner V, Luke B. Molecular and physiological consequences of faulty eukaryotic ribonucleotide excision repair. EMBO J. 2020;39:e102309.
Lanz MC, Dibitetto D, Smolka MB. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019;38:e101801.
Alghoul E, Basbous J, Constantinou A. Compartmentalization of the DNA damage response: Mechanisms and functions. DNA Repair (Amst). 2023;128:103524.
Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33.
Thomas AF, Kelly GL, Strasser A. Of the many cellular responses activated by TP53, which ones are critical for tumour suppression? Cell Death Differ. 2022;29:961–971.
Jessica L, Hopkins LL, Lee Z. DNA repair defects in cancer and therapeutic opportunities. Genes Dev. 2022;36:278–293.
Zhao B, Rothenberg E, Ramsden DA, Lieber MR. The molecular basis and disease relevance of non-homologous DNA end joining. Nat Rev Mol Cell Biol. 2020;21:765–781.
Pilié PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104.
Ngoi NYL, Pilie PG, McGrail DJ, Zimmermann M, Schlacher K, Yap TA. Targeting ATR in patients with cancer. Nat Rev Clin Oncol. 2024;21:278–293.
Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18:610–621.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158.
Martincorena Iñigo. PJC. Somatic mutation in cancer and normal cells. Science. 2015;349:1483–1489.
Conn VM, Gabryelska M, Toubia J, Kirk K, Gantley L, Powell JA, et al. Circular RNAs drive oncogenic chromosomal translocations within the MLL recombinome in leukemia. Cancer Cell. 2023;41:1309–1326.
Esposito MT, Zhao L, Fung TK, Rane JK, Wilson A, Martin N, et al. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med. 2015;21:1481–1490.
Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–542.
Malcikova J, Pavlova S, Kunt Vonkova B, Radova L, Plevova K, Kotaskova J, et al. Low-burden TP53 mutations in CLL: clinical impact and clonal evolution within the context of different treatment options. Blood. 2021;138:2670–2685.
Kwok M, Agathanggelou A, Stankovic T. DNA damage response defects in hematologic malignancies: mechanistic insights and therapeutic strategies. Blood. 2024;143:2123–2144.
Wong N-HM, So CWE. Novel therapeutic strategies for MLL-rearranged leukemias. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194584.
Baxter JS, Zatreanu D, Pettitt SJ, Lord CJ. Resistance to DNA repair inhibitors in cancer. Mol Oncol. 2022;16:3811–3827.
Kalamara V, Garinis GA. The epitranscriptome: reshaping the DNA damage response. Trends Cell Biol. 2024;35:294–304.
Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016–2034.
Papaspyropoulos A, Hazapis O, Lagopati N, Polyzou A, Papanastasiou AD, Liontos M, et al. The role of circular RNAs in DNA damage response and repair. Cancers (Basel). 2021;13:5352.
Chen L, Wang Y, Lin J, Song Z, Wang Q, Zhao W, et al. Exportin 4 depletion leads to nuclear accumulation of a subset of circular RNAs. Nat Commun. 2022;13:5769.
Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206.
Li X, Zhang JL, Lei YN, Liu XQ, Xue W, Zhang Y, et al. Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci China Life Sci. 2021;64:1795–1809.
Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491.
Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer. 2020;19:128.
Abaza Y, Zeidan AM. Immune checkpoint inhibition in acute myeloid leukemia and myelodysplastic syndromes. Cells. 2022;11:2249.
Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301.
Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–346.
Meyer C, Larghero P, Lopes BA, Burmeister T, Gröger D, Sutton R, et al. The KMT2A recombinome of acute leukemias in 2023. Leukemia. 2023;37:988–1005.
Han C, Sun LY, Luo XQ, Pan Q, Sun YM, Zeng ZC, et al. Chromatin-associated orphan snoRNA regulates DNA damage-mediated differentiation via a non-canonical complex. Cell Rep. 2022;38:110421.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.
Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P, et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature. 2020;580:147–150.
Zuckerman B, Ulitsky I. Predictive models of subcellular localization of long RNAs. RNA. 2019;25:557–572.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264.
Chen L-L, Bindereif A, Bozzoni I, Chang HY, Matera AG, Gorospe M, et al. A guide to naming eukaryotic circular RNAs. Nat Cell Biol. 2023;25:1–5.
Padella A, Ghelli Luserna Di Rorà A, Marconi G, Ghetti M, Martinelli G, Simonetti G. Targeting PARP proteins in acute leukemia: DNA damage response inhibition and therapeutic strategies. J Hematol Oncol. 2022;15:10.
Takagi M. DNA damage response and hematological malignancy. Int J Hematol. 2017;106:345–356.
Li S, Li X, Xue W, Zhang L, Yang LZ, Cao SM, et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. 2021;18:51–59.
Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang Z, et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat Methods. 2021;18:499–506.
Gibbons HR, Aune TM. Immunoprecipitation of DNA:RNA Hybrids Using the S9.6 Antibody. Methods Mol Biol. 2020;2161:195–207.
Sun YM, Wang WT, Zeng ZC, Chen TQ, Han C, Pan Q, et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood. 2019;134:1533–1546.
Iioka H, Loiselle D, Haystead TA, Macara IG. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res. 2011;39:e53.
Chen Y, Jiang T, Zhang H, Gou X, Han C, Wang J, et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat Cell Biol. 2020;22:1276–1285.
Head PE, Kapoor-Vazirani P, Nagaraju GP, Zhang H, Rath SK, Luong NC, et al. DNA-PK is activated by SIRT2 deacetylation to promote DNA double-strand break repair by non-homologous end joining. Nucleic acids Res. 2023;51:7972–7987.
Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, Pe’Ery T, et al. The NF90/NF45 Complex Participates in DNA Break Repair via Nonhomologous End Joining. Mol Cell Biol. 2011;31:4832–4843.
Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, et al. METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol Cell. 2020;79:425–442.
Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem Biol. 2011;6:628–635.
Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F, et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J Cell Biol. 2021;220:e202008030.
Bianchini RM, Kurz EU. The analysis of protein recruitment to laser microirradiation-induced DNA damage in live cells: Best practices for data analysis. DNA Repair (Amst). 2023;129:103545.
Tang M, Chen G, Tu B, Hu Z, Huang Y, DuFort CC, et al. SMYD2 inhibition–mediated hypomethylation of Ku70 contributes to impaired nonhomologous end joining repair and antitumor immunity. Sci Adv. 2023;9:eade6624.
Wang PL, Teng L, Feng YC, Yue YM, Han MM, Yan Q, et al. The N-Myc-responsive lncRNA MILIP promotes DNA double-strand break repair through non-homologous end joining. Proc Natl Acad Sci USA. 2022;119:e2208904119.
Walsh KD, Kato TA. Alkaline comet assay to detect DNA damage. Methods Mol Biol. 2023;2519:65–72.
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell. 2017;67:214–227.
Patino C, Haenni AL, Urcuqui-Inchima S. NF90 isoforms, a new family of cellular proteins involved in viral replication? Biochimie. 2015;108:20–24.
Kuwano Y, Pullmann R Jr., Marasa BS, Abdelmohsen K, Lee EK, Yang X, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238.
Heikamp EB, Henrich JA, Perner F, Wong EM, Hatton C, Wen Y, et al. The menin-MLL1 interaction is a molecular dependency in NUP98-rearranged AML. Blood. 2022;139:894–906.
Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7:716–735.
Gray ZH, Chakraborty D, Duttweiler RR, Alekbaeva GD, Murphy SE, Chetal K, et al. Epigenetic balance ensures mechanistic control of MLL amplification and rearrangement. Cell. 2023;186:4528–4545.
Liang K, Volk AG, Haug JS, Marshall SA, Woodfin AR, Bartom ET, et al. Therapeutic Targeting of MLL degradation pathways in MLL-rearranged Leukemia. Cell. 2017;168:59–72.
Chen TQ, Huang HJ, Zhu SX, Chen XT, Pu KJ, Wang D, et al. Blockade of the lncRNA-DOT1L-LAMP5 axis enhances autophagy and promotes degradation of MLL fusion proteins. Exp Hematol Oncol. 2024;13:18.
Wang WT, Chen TQ, Zeng ZC, Pan Q, Huang W, Han C, et al. The lncRNA LAMP5-AS1 drives leukemia cell stemness by directly modulating DOT1L methyltransferase activity in MLL leukemia. J Hematol Oncol. 2020;13:78.
Feng XY, Zhu SX, Pu KJ, Huang HJ, Chen YQ, Wang WT. New insight into circRNAs: characterization, strategies, and biomedical applications. Exp Hematol Oncol. 2023;12:91.
Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad Sci USA. 2018;115:E7970–E7977.
Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017;549:548–552.
De Bragança S, Aicart-Ramos C, Arribas-Bosacoma R, Rivera-Calzada A, Unfried JP, Prats-Mari L, et al. APLF and long non-coding RNA NIHCOLE promote stable DNA synapsis in non-homologous end joining. Cell Rep. 2023;42:111917.
Feng P, Wang Y, Liu N, Chen Y, Hu Y, Huang Z, et al. High expression of PPP1CC promotes NHEJ-mediated DNA repair leading to radioresistance and poor prognosis in nasopharyngeal carcinoma. Cell Death Differ. 2024;31:683–696.
Ramsden DA, Carvajal-Garcia J, Gupta GP. Mechanism, cellular functions and cancer roles of polymerase-theta-mediated DNA end joining. Nat Rev Mol Cell Biol. 2022;23:125–140.
Castella S, Bernard R, Corno M, Fradin A, Larcher JC. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip Rev RNA. 2015;6:243–256.
Ye J, Jin H, Pankov A, Song JS, Blelloch R. NF45 and NF90/NF110 coordinately regulate ESC pluripotency and differentiation. RNA. 2017;23:1270–1284.
Grasso G, Kiernan R. The Polyvalent Role of NF90 in RNA Biology. Int J Mol Sci. 2022;23:13584.
Barbier J, Chen X, Sanchez G, Cai M, Helsmoortel M, Higuchi T, et al. An NF90/NF110-mediated feedback amplification loop regulates dicer expression and controls ovarian carcinoma progression. Cell Res. 2018;28:556–571.
Li B, Wang J, Cheng X, Liu Y, Yang Y, Yang X, et al. Molecular mechanism underlying the subtype-selectivity of competitive inhibitor NF110 and its distinct potencies in human and rat P2X3 receptors. Sci Bull (Beijing). 2018;63:1616–1625.
Sun L, Wang W, Han C, Huang W, Sun Y, Fang K, et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol Cell. 2021;81:4493–4508.
Zeng ZC, Pan Q, Sun YM, Huang HJ, Chen XT, Chen TQ, et al. METTL3 protects METTL14 from STUB1-mediated degradation to maintain m(6)A homeostasis. EMBO Rep. 2023;24:e55762.
Hu Z, Mi S, Zhao T, Peng C, Peng Y, Chen L, et al. BGL3 lncRNA mediates retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J. 2020;39:e104133.
Funding
This research was supported by National Key R&D Program of China (Nos.2021YFA1300502 and 2022YFA1303302), National Natural Science Foundation of China (Nos. 32170570,32370594, and 32400439), and Guangdong Province (Nos. 2021B1515020002 and 2022A1515140018), and Guangzhou (No. 2024A04J5004).
Author information
Authors and Affiliations
Contributions
K.J.P., X.T.C. and S.X.Z. designed and performed the research, analyzed the data, and wrote the manuscript. Y.A., X.Y.F., J.Y.L., H.J.H., C.L.Z., M.Y.Y., B.R.H., Y.C.W., Y.X.M., C. F., N. Z., D. W., T.Q.C. and Y.M.S. performed the research and analyzed the data. K.J.P., X.T.C. and S.X.Z. collected and analyzed the clinical data. Y.Q.C. and W.T.W. designed the research, analyzed the data, and wrote the manuscript. The authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interest.
Ethics
All patient samples were obtained with informed consent from the first Affiliated Hospital of Sun Yat-sen University. Sample collection was approved by the Hospital’s Protection of Human Subjects Committee. Additionally, all animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Approval No. SYSU-IACUC-2023-B0438) and were performed in accordance with the approved protocols.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pu, KJ., Chen, XT., Zhu, SX. et al. Chromatin-associated circRNA ciCRLF3(2) regulates cell differentiation blockage via activating non-homologous end joining–based DNA repair. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01574-9
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41418-025-01574-9