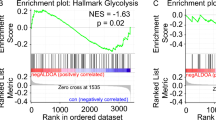
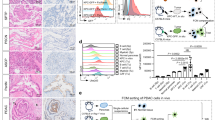
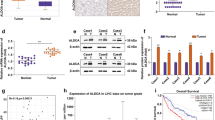
Abstract
The function of cytosolic aldolase A (ALDOA) in glycolysis is well recognized. However, the cytosol-to-nucleus redistribution of ALDOA and its nuclear function is poorly understood. Here, we uncover inflammatory factor-stimulated nuclear function of ALDOA in augmenting pancreatic carcinogenesis by activating NF-κB signaling in a ubiquitination-dependent manner. TNF-α-triggered K11- and K29-linked ubiquitination of ALDOA at Lys200 promotes its interaction with RelA/p65 and facilitates importin-β-dependent nuclear translocation, establishing a positive feedback regulation in the tumor microenvironment by elevating the TNF-α expression in pancreatic ductal adenocarcinoma (PDAC). USP4 is identified as a negative regulator that deubiquitinates ALDOA. Instead of broadly targeting ALDOA, which causes glycolysis impairment, the specific elimination of ALDOA ubiquitination enhances chemosensitivity and the synergistic effect of chemotherapy combined with p65-specific anti-inflammatory therapy by selectively suppressing inflammation-induced proliferation in cancer cells. Collectively, we unveil the multifaceted mechanisms by which ALDOA promotes PDAC carcinogenesis, from metabolic to gene regulatory perspectives, providing potential therapies combatting cancer.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD059292 and 10.6019/PXD059292, PXD059293 and 10.6019/PXD059293.
References
Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol. 2019;20:436–50.
Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46.
Elia I, Haigis MC. Metabolites and the tumour microenvironment: from cellular mechanisms to systemic metabolism. Nat Metab. 2021;3:21–32.
Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21:669–80.
Xiao Z, Dai Z, Locasale JW. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun. 2019;10:3763.
Campbell SL, Wellen KE. Metabolic Signaling to the Nucleus in Cancer. Mol Cell. 2018;71:398–408.
Boon R, Silveira GG, Mostoslavsky R. Nuclear metabolism and the regulation of the epigenome. Nat Metab. 2020;2:1190–203.
Cluntun AA, Huang H, Dai L, Liu X, Zhao Y, Locasale JW. The rate of glycolysis quantitatively mediates specific histone acetylation sites. Cancer Metab. 2015;3:10.
Cai L, Sutter Benjamin M, Li B, Tu Benjamin P. Acetyl-CoA Induces Cell Growth and Proliferation by Promoting the Acetylation of Histones at Growth Genes. Mol Cell. 2011;42:426–37.
Xu D, Shao F, Bian X, Meng Y, Liang T, Lu Z. The Evolving Landscape of Noncanonical Functions of Metabolic Enzymes in Cancer and Other Pathologies. Cell Metab. 2021;33:33–50.
Sivanand S, Rhoades S, Jiang Q, Lee JV, Benci J, Zhang J, et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol Cell. 2017;67:252–65.e6.
Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee J-H, et al. Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell. 2017;66:684–97.e9.
Pardo-Lorente N, Gkanogiannis A, Cozzuto L, Gañez Zapater A, Espinar L, Ghose R, et al. Nuclear localization of MTHFD2 is required for correct mitosis progression. Nat Commun. 2024;15:9529.
Wang Z, Li M, Jiang H, Luo S, Shao F, Xia Y, et al. Fructose-1,6-bisphosphatase 1 functions as a protein phosphatase to dephosphorylate histone H3 and suppresses PPARα-regulated gene transcription and tumour growth. Nat Cell Biol. 2022;24:1655–65.
Snaebjornsson MT, Schulze A. Non-canonical functions of enzymes facilitate cross-talk between cell metabolic and regulatory pathways. Exp Mol Med. 2018;50:1–16.
Pan C, Li B, Simon MC. Moonlighting functions of metabolic enzymes and metabolites in cancer. Mol Cell. 2021;81:3760–74.
Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic Enzymes Moonlighting in the Nucleus: Metabolic Regulation of Gene Transcription. Trends Biochem Sci. 2016;41:712–30.
Chang YC, Yang YC, Tien CP, Yang CJ, Hsiao M. Roles of Aldolase Family Genes in Human Cancers and Diseases. Trends Endocrinol Metab. 2018;29:549–59.
Zhang C, Hawley SA, Zong Y, Li M, Wang Z, Gray A, et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548:112–6.
Mamczur P, Gamian A, Kolodziej J, Dziegiel P, Rakus D. Nuclear localization of aldolase A correlates with cell proliferation. Biochim Biophys Acta Mol Cell Res. 2013;1833:2812–22.
Kusakabe T, Motoki K, Hori K. Mode of Interactions of Human Aldolase Isozymes with Cytoskeletons. Arch Biochem Biophys. 1997;344:184–93.
Fu H, Gao H, Qi X, Zhao L, Wu D, Bai Y, et al. Aldolase A promotes proliferation and G1/S transition via the EGFR/MAPK pathway in non-small cell lung cancer. Cancer Commun. 2018;38:18.
Gizak A, Wiśniewski J, Heron P, Mamczur P, Sygusch J, Rakus D. Targeting a moonlighting function of aldolase induces apoptosis in cancer cells. Cell Death Dis. 2019;10:712.
Niu Y, Lin Z, Wan A, Sun L, Yan S, Liang H, et al. Loss-of-Function Genetic Screening Identifies Aldolase A as an Essential Driver for Liver Cancer Cell Growth Under Hypoxia. Hepatology. 2021;74:1461–79.
Song J, Li H, Liu Y, Li X, Shi Q, Lei Q-Y, et al. Aldolase A Accelerates Cancer Progression by Modulating mRNA Translation and Protein Biosynthesis via Noncanonical Mechanisms. Adv Sci. 2023;10:2302425.
Wan N, Wang N, Yu S, Zhang H, Tang S, Wang D, et al. Cyclic immonium ion of lactyllysine reveals widespread lactylation in the human proteome. Nat Methods. 2022;19:854–64.
Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell. 2019;179:561–77.e22.
Cha Y, Han M-J, Cha H-J, Zoldan J, Burkart A, Jung JH, et al. Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c–SIRT2 axis. Nat Cell Biol. 2017;19:445–56.
Qin W, Qin K, Zhang Y, Jia W, Chen Y, Cheng B, et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat Chem Biol. 2019;15:983–91.
Lin ZP, Gan G, Xu X, Wen C, Ding X, Chen XY, et al. Comprehensive PTM profiling with SCASP-PTM uncovers mechanisms of p62 degradation and ALDOA-mediated tumor progression. Cell Rep. 2025;44:115500.
Park W, Chawla A, O’Reilly EM. Pancreatic Cancer: A Review. JAMA. 2021;326:851–62.
Engle DD, Tiriac H, Rivera KD, Pommier A, Whalen S, Oni TE, et al. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science. 2019;364:1156–62.
Padoan A, Plebani M, Basso D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int J Mol Sci. 2019;20:676.
Del Poggetto E, Ho I-L, Balestrieri C, Yen E-Y, Zhang S, Citron F, et al. Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis. Science. 2021;373:eabj0486.
Yuan C, Morales-Oyarvide V, Khalaf N, Perez K, Tabung FK, Ho GYF, et al. Prediagnostic Inflammation and Pancreatic Cancer Survival. J Natl Cancer Inst. 2021;113:1186–93.
Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012;12:121–32.
Chang YC, Chan YC, Chang WM, Lin YF, Yang CJ, Su CY, et al. Feedback regulation of ALDOA activates the HIF-1α/MMP9 axis to promote lung cancer progression. Cancer Lett. 2017;403:28–36.
Lin J, Xia L, Oyang L, Liang J, Tan S, Wu N, et al. The POU2F1-ALDOA axis promotes the proliferation and chemoresistance of colon cancer cells by enhancing glycolysis and the pentose phosphate pathway activity. Oncogene. 2022;41:1024–39.
Xu M, Xi S, Li H, Xia Y, Mei G, Cheng Z. Prognosis significance and potential association between ALDOA and AKT expression in colorectal cancer. Sci Rep. 2024;14:6488.
Snaebjornsson MT, Poeller P, Komkova D, Röhrig F, Schlicker L, Winkelkotte AM, et al. Targeting aldolase A in hepatocellular carcinoma leads to imbalanced glycolysis and energy stress due to uncontrolled FBP accumulation. Nat Metab. 2025;7:348–66.
Ronai ZE, Robinson RC, Rutberg SE, Lazarus P, Sardana MK. Aldolase-DNA interactions in a SEWA cell system. Biochim Biophys Acta. 1992;1130:20–8.
Adesso L, Calabretta S, Barbagallo F, Capurso G, Pilozzi E, Geremia R, et al. Gemcitabine triggers a pro-survival response in pancreatic cancer cells through activation of the MNK2/eIF4E pathway. Oncogene. 2013;32:2848–57.
Genkinger JM, Spiegelman D, Anderson KE, Bergkvist L, Bernstein L, van den Brandt PA, et al. Alcohol Intake and Pancreatic Cancer Risk: A Pooled Analysis of Fourteen Cohort Studies. Cancer Epidemiol Biomark Prev. 2009;18:765–76.
Ho WJ, Jaffee EM, Zheng L. The tumour microenvironment in pancreatic cancer — clinical challenges and opportunities. Nat Rev Clin Oncol. 2020;17:527–40.
Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18:309–24.
Huber M, Brehm CU, Gress TM, Buchholz M, Alashkar Alhamwe B, von Strandmann EP, et al. The Immune Microenvironment in Pancreatic Cancer. Int J Mol Sci. 2020;21:7307.
Zhang T, Sun L, Hao Y, Suo C, Shen S, Wei H, et al. ENO1 suppresses cancer cell ferroptosis by degrading the mRNA of iron regulatory protein 1. Nat Cancer. 2022;3:75–89.
Zhang Y, Sampathkumar A, Kerber SM-L, Swart C, Hille C, Seerangan K, et al. A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat Commun. 2020;11:4509.
Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–70.
Thomas GE, Egan G, García-Prat L, Botham A, Voisin V, Patel PS, et al. The metabolic enzyme hexokinase 2 localizes to the nucleus in AML and normal haematopoietic stem and progenitor cells to maintain stemness. Nat Cell Biol. 2022;24:872–84.
Liang P, Zhang H, Wang G, Li S, Cong S, Luo Y, et al. KPNB1, XPO7 and IPO8 Mediate the Translocation of NF-κB/p65 into the Nucleus. Traffic. 2013;14:1132–43.
Li Y, Dammer EB, Gao Y, Lan Q, Villamil MA, Duong DM, et al. Proteomics Links Ubiquitin Chain Topology Change to Transcription Factor Activation. Mol Cell. 2019;76:126–37.e7.
Boughton AJ, Krueger S, Fushman D. Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1. Structure. 2020;28:29–43.e6.
Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell. 2017;171:918–33.e20.
Paul A, Wang B. RNF8- and Ube2S-Dependent Ubiquitin Lysine 11-Linkage Modification in Response to DNA Damage. Mol Cell. 2017;66:458–72.e5.
Zhao J, Cai B, Shao Z, Zhang L, Zheng Y, Ma C, et al. TRIM26 positively regulates the inflammatory immune response through K11-linked ubiquitination of TAB1. Cell Death Differ. 2021;28:3077–91.
Kaiho-Soma A, Akizuki Y, Igarashi K, Endo A, Shoda T, Kawase Y, et al. TRIP12 promotes small-molecule-induced degradation through K29/K48-branched ubiquitin chains. Mol Cell. 2021;81:1411–24.e7.
Nucifora FC, Nucifora LG, Ng C-H, Arbez N, Guo Y, Roby E, et al. Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nat Commun. 2016;7:11792.
Chen Y-H, Huang T-Y, Lin Y-T, Lin S-Y, Li W-H, Hsiao H-J, et al. VPS34 K29/K48 branched ubiquitination governed by UBE3C and TRABID regulates autophagy, proteostasis and liver metabolism. Nat Commun. 2021;12:1322.
Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–209.
Gao P, Ma X, Yuan M, Yi Y, Liu G, Wen M, et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat Commun. 2021;12:1194.
Zhang ZD, Li HX, Gan H, Tang Z, Guo YY, Yao SQ, et al. RNF115 Inhibits the Post-ER Trafficking of TLRs and TLRs-Mediated Immune Responses by Catalyzing K11-Linked Ubiquitination of RAB1A and RAB13. Adv Sci. 2022;9:e2105391.
Feng X, Jia Y, Zhang Y, Ma F, Zhu Y, Hong X, et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy. 2019;15:1130–49.
Xie W, Jin S, Wu Y, Xian H, Tian S, Liu D-A, et al. Auto-ubiquitination of NEDD4-1 Recruits USP13 to Facilitate Autophagy through Deubiquitinating VPS34. Cell Rep. 2020;30:2807–19.e4.
Hu H, Sun SC. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–83.
Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–89.
Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure. 2020;28:1071–81.e3.
Ianevski A, Giri AK, Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50:W739–W43.
Acknowledgements
We sincerely thank Prof. Daming Gao from Shanghai Institute of Biochemistry and Cell Biology for providing Flag-ALDOA plasmid, Prof. Chensong Zhang from Xiamen University for providing sg-ALDOA plasmids, Prof. Lingqiang Zhang from Academy of Military Science for providing HA-ubiquitin-mutated plasmids, Prof. Danying Chen from Peking University for providing luciferase reporter plasmids, Prof. Jinfang Zhang from Wuhan University for providing the Myc-tagged MARCH1-11 plasmids, Prof. Chengqi Yi from Peking University for providing ALDOB expressing plasmid, Prof. Jianguo Ji from Peking University for providing MiaPaCa-2 cell line, Prof. Jihui Hao from Tianjin Medical University for providing SW1990 cell line, Prof. Peng Du from Peking University for providing PANC-1 cell line, Prof. Xianghong Yang from Shengjing Hospital of China Medical University for providing HPDE6C7 cell line, Prof. Deng Pan from Tsinghua University for providing Panc02 cell line and Prof. Zhe Zhang from Peking University for providing HEK293F cell line. We thank Dr. Shichen Geng from Peking University for instructing the construction of site-mutation cell lines. We thank the National Center for Protein Sciences and the Core Facilities of Life Sciences at Peking University, particularly Qi Zhang, Wenling Gao, Siying Qin, Liying Du, Huan Yang, Jia Luo, Feifei Cheng, and Guilan Li for technical assistance. We also appreciate Biorender for its easy-to-use platform that helped illustrate our research findings (agreement no. ON28RU2H3S).
Funding
This work was supported by National Key R&D Program of China (2024YFA1306200 and 2022YFA1302803), and the National Natural Science Foundation of China (82130081 and 32270756).
Author information
Authors and Affiliations
Contributions
S Zhou and X Zheng designed this study. S Zhou conducted the experiments. S Zhou analyzed the data and wrote the manuscript. Y Li, C Wang and Y Zhao helped the mice xenograft assay. X Zheng supervised this study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experiments were approved by the Ethics Committee of Peking University (IACUC No. LSC-ZhengXF-1). All animal handling and experimental protocols strictly adhered to the ethical principles outlined in the Declaration of Helsinki, as well as relevant national guidelines and regulations. The human tissue arrays were purchased from Shanghai Superchip Company Ltd. with the ethics approval number (SHXC2021YF01).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, S., Li, Y., Wang, C. et al. K11- and K29-ubiquitination-mediated nuclear translocation of glycolytic enzyme aldolase A promotes pancreatic cancer progression by NF-κB activation. Cell Death Differ (2025). https://doi.org/10.1038/s41418-025-01592-7
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41418-025-01592-7