Abstract
Cancer has become a leading cause of mortality worldwide, with alarming increases in incidence and mortality rates. Emerging evidence suggests that tRNA modification enzymes play a crucial role in cancer development by modulating codon-specific translation. In this review, we focus on 18 tRNA modification enzymes and elucidate their mechanisms of action and roles in disease. We highlight the functions and mechanisms of seven tRNA regulators that mediate favorable tRNA translation in tumorigenesis and cancer progression, providing deeper insights into their clinical potential as cancer-related biomarkers and prognostic indicators. These findings emphasize the need for further investigation into the therapeutic potential of tRNA modification enzymes in cancer management and their potential application in personalized cancer therapy and diagnostics.
Similar content being viewed by others
Facts
-
tRNA modifications mediate codon-specific translation, thereby exerting a pivotal influence on tumor initiation and progression.
-
There are cancer specific differences in codon-specific translation, which not only exist in different types of cancer, but also in different subtypes.
-
The substrate specificity and mechanistic axes of the promising ALKB, TRMT, NSUN and other emerging enzyme families remain to be elucidated.
-
tRNA modifying enzymes have shown great potential as biomarkers and targeted therapeutic drugs.
Introduction
Globally, the incidence of cancer is steadily increasing, representing a growing public health burden. A report released by the International Agency for Research on Cancer (IARC) of the World Health Organization in February 2024 stated that there were 20 million new cancer cases and 9.7 million cancer deaths worldwide in 2022. It is estimated that by 2050, the number of new cancer cases globally will exceed 35 million [1].
Studies have shown that regulating the malignant biological behavior of tumors can effectively inhibit their occurrence and development. The biological behavior of tumors mainly includes self-renewal, proliferation, invasion, and metastasis, which promote the continuous proliferation and spread of tumor cells to other parts of the body. Various factors, such as the tumor microenvironment, cell signaling pathways, and epigenetics, influence malignant behavior.
With increasing research, the influence of epigenetic factors on tumors has become increasingly valued. Epigenetic factors include DNA and RNA methylation modifications, noncoding RNA regulation, chromosome remodeling, and histone modifications. Since the first record of RNA modifications in the 1950s, more than 170 types of RNA modifications have been discovered [2]. Among them, methylation accounts for approximately two-thirds of RNA modifications. RNA methylation sites include most nitrogen atoms, in addition to the oxygen atom of ribose 2’OH, the carbon atom at position 5 of pyrimidine, and the second and eighth carbon atoms of adenosine, which also undergo methylation reactions. The types of RNA methylation also differ and include m6A, m1A, m5C, m1G, m7G, m5U, etc. Research has shown that m6A methylation plays an important role in tumors; for example, METTL16, an enzyme responsible for RNA m6A modification, interacts with EIF3A/B in hepatocellular carcinoma (HCC) cells, mainly through the R1 and R2 regions of METTL16. This interaction promotes mRNA translation and the survival and proliferation of HCC cells [3].
Previous studies have mostly emphasized mRNA modification as a key regulatory factor determining tumor occurrence and development [4]. Recent emerging evidence suggests that some tRNA modifications play crucial roles in regulating tumors [5]. The tRNA modifications found thus far include the base modifications m1A, m3C, m5C, 2’-o-methylribose, Ψ, D, and I and 2’-O-methylated and phosphorylated ribose modifications; tRNA modification may participate in regulating the fate of cancer cells by controlling codon-specific translation [6, 7]. Subsequent research has indeed confirmed these views [8].
However, as a form of epigenetic modification, the mechanism of action of many tRNA modifications in tumors is still unclear [9]. In this review, we provide the latest information on common tRNA modifications by analyzing the structure of tRNAs, indicating that tRNA modifications need to be studied through tRNA enzyme modification-mediated codon-specific translation to influence diseases and cancers, providing more insights into their clinical potential as cancer-related biomarkers, prognostic markers, and more.
Types of tRNA modification
Nucleobase modification
N⁶-methyladenosine (m⁶A)
N⁶-methyladenosine is a modified nucleotide formed by adding a methyl group to the N⁶ position of adenosine (A). This modification is the most common internal posttranscriptional modification in animals, plants, and yeast [10,11,12,13]. It is widely present in various RNA molecules, such as mRNA, tRNA, lncRNA, and hnRNA (heterogeneous nuclear RNA). In hnRNA and mRNA, it occurs mainly on the 6th nitrogen atom of adenine and appears in the clear sequence -N1-(GA)-m6A-C-N2- [14]. There are many types of m6A; for example, METTL3 and METTL14 form stable coordination compounds that methylate m6A [15].
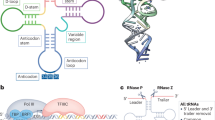
However, m6A modification is very rare in tRNAs, and the common adenosine methylation modifications in tRNAs are m1A and m2A, which occur mainly at the N¹ and N² positions of adenosine. These modifications serve many functions: m1A can promote antitumor immunity in CD8+ T cells by enhancing the translation of ATP citrate lyase [16]. m2A in tRNA enhances protein translation by decoding m2A-tRNA-dependent codons [17]. (Fig. 1)
Several types of tRNA modifications: A Methyladenosine modification, Taking m1A modification as an example. B Methylcytosine modification, Taking m5C modification as an example. C Methylguanosine modification, Taking m1G modification as an example. D Pseudouridine modification. E Dihydrouracil modification. F Hypoxanthine modification. G 2’-O-Methylation modification.
m6A RNA methylation modulates tumorigenesis, inflammation, and aging by orchestrating posttranscriptional circuits that govern oncogenic signaling, DNA repair capacity, and mitochondrial stress responses [18]. Hypomethylation drives endometrial carcinogenesis via derepression of AKT signaling [19]. METTL3-mediated hyper-methylation promotes gastric cancer through the HDGF axis, positioning METTL3 as both prognostic biomarker and therapeutic target [20]. YTHDF2-dependent m6A recognition destabilizes inflammation-related transcripts, thereby tuning the intensity of immune responses [21]. Overall, m6A epitranscriptomic rewiring couples cellular stress adaptation to clinical phenotypes across cancer, immune, and aging.
5-Methylcytosine (m⁵C)
5-Methylcytosine (m5C) is a modified base formed by the addition of a methyl group (-CH3) to the 5th carbon of the pyrimidine ring of cytosine. It is a common RNA modification that occurs in the rRNA, tRNA, noncoding RNA and ribosomal RNA of plants and animals and can help the body cope with changes in internal and external environments [22,23,24,25,26]. In tRNA, m5C is catalyzed by the NOL1/NOP2/sun (Nsun) family and DNA methyltransferase member 2 (DNMT2, TRNA aspartate methyltransferase 1 or TRDMT1) [27,28,29].
m⁵C RNA methylation is involved in diverse human diseases, including cancer, viral infection, cardiovascular disease, and neurological disorders [30]. Aberrant m⁵C modification drives intrinsic resistance to gefitinib in EGFR-mutant NSCLC through the NSUN2/YBX1/QSOX1 axis, providing new insight into lung cancer resistance mechanisms and therapeutic strategies [31]. Targeting NSUN2 or m⁵C modification may suppress HBV replication and prevent related liver disease [32]. m⁵C modification also contributes to cardiovascular pathogenesis by modulating gene expression and mitochondrial dysfunction, highlighting novel molecular mechanisms and potential therapeutic targets in cardiovascular disease [33].
1-Methylguanosine (m¹G)
1-Methylguanosine (m1G) modification involves the addition of a methyl (-CH3) group to the first nitrogen atom (N1) of guanosine (G) to ensure the correct pairing of codons and anticodons during translation. This modification is widely present in various types of RNA and has important effects on the structure, function, and physiological activities of RNA [34]. For example, in yeast cell tRNA, the formation of m1G modifications is closely related to the open reading frame YOL093w (named TRM10). The Trm10p protein encoded by TRM10 is responsible for the methylation of the 9th G of tRNA. Therefore, m1G modification may be crucial for maintaining the normal structure and function of tRNA [35]. In E. coli tRNA, m1G modification is located at position 37 of the tRNA sequence, and m 1G37 is necessary for the terminal methylation of U at position 34 (forming mcmo5U34) in tRNA (Pro) [36].
In cancer, m1G modification has multiple functions. The m1G methylation level of mitochondrial tRNA in tumor tissue is significantly altered; this abnormal modification may be closely related to the occurrence and development of tumors and is expected to serve as an important biomarker or potential therapeutic target in tumor research [37, 38]. This modification can also threaten human health, such as by inducing disease of the human reproductive system [39].
Pseudouridine (Ψ)
Pseudouridine (Ψ) modification is a common RNA epigenetic modification, which refers to the substitution of uracil (U) in RNA molecules with pseudouracil (Ψ). This modification is widely present in various RNA molecules, including mRNA, noncoding RNA, and rRNA, and has a significant effect on the function and stability of RNA [40].
For example, in embryonic stem cells, the Ψ modification of tRNA is mediated by the “writing enzyme” PUS7, which modifies tRNA and its derived small fragments (tRFs) to regulate protein synthesis. In addition, Ψ modification plays a crucial role in the development and functional maintenance of hematopoietic stem cells and is closely related to the occurrence and development of hematological diseases [41]. Through its abnormal modification (mediated by PUSs), PSI affects RNA function, participates in the occurrence and development of cancer, and can serve as a potential biomarker for cancer diagnosis and prognosis. Targeted PUS therapy strategies provide new research ideas for cancer treatment [42, 43].
Dihydrouracil (D)
Dihydrouracil (D) is present mainly in the D loop of tRNA and is catalyzed by Dus. Its modification process involves regions such as the D loop, T loop, and anticodon stem. The presence of D helps to stabilize the overall structure of tRNA, increase its flexibility, and promote accurate recognition of codons and transport of amino acids during translation, ensuring the accuracy and efficiency of translation [44]. D modification is located on the dihydrouracil arm of tRNA and may play an important role in the later stages of tRNA evolution. It helps stabilize specific structural domains of tRNA, affecting its interactions with ribosomes, aminoacyl tRNA synthetases, and mRNA and enhancing the specificity and accuracy of tRNA function, thereby ensuring efficient protein synthesis [45].
Hypoxanthine (I)
The modification of hypoxanthine (I) involves an enzymatic reaction that inserts hypoxanthine into the swing position of the tRNA anticodon, resulting in the formation of inosine. This modification significantly expands the codon recognition range of tRNA, enabling it to more flexibly interpret the genetic code in protein synthesis, thereby improving the efficiency and accuracy of protein synthesis and maintaining normal physiological functions [46].
Ribose modification
2’-O-methylation
2 ‘- O-methylation refers to the substitution of the hydroxyl group (-OH) at the 2’ position of RNA nucleotides with a methyl group (-CH3), thereby altering the chemical structure and functional properties of RNA. This modification is widely present in various RNA molecules, such as mRNA, tRNA, rRNA, and miRNA, and significantly affects gene expression regulation, protein synthesis, and other biological processes [47]. The 2’-O-methylation of tRNA affects mainly its structural stability and function. This modification maintains the secondary and tertiary structure of tRNA, ensuring correct folding and spatial conformation, thereby promoting interactions between tRNA and ribosomes and between aminoacyl tRNA synthetases, thereby increasing codon recognition and amino acid loading efficiency and improving the accuracy and efficiency of protein synthesis [48]. In addition, it plays many roles in immune and other diseases. In T cells, interferon-induced ISG20 impairs the reverse transcription of hypomethylated HIV-1, indicating that 2′-O-methylation directly antagonizes ISG20-mediated antiviral activity, facilitating HIV-1 evasion of host restriction and promoting viral replication [49, 50].
Function of tRNA modification enzymes
ALKBH8
The ALKB (ALKB homologs) family belongs to the 2-oxoglutarate (2OG)- and iron (Fe(II))-dependent dioxygenase superfamily and is widely present in mammals, plants, and viruses. The family members include ALKBH1 to ALKBH8 and FTO [51,52,53]. The AlkB family repairs DNA damage through oxidative demethylation and can repair alkylated damage, such as methyladenine at N1 and methylcytosine at N3, through oxidative dealkylation [54,55,56,57].
ALKBH8 is a tRNA methyltransferase with DNA repair and tRNA modification activities that also functions as an m6A demethylase [58]. ABH8 is the most unique among AlkB homologous genes in other mammals because of its fusion of the RNA recognition motif (RRM) and S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MT) motif to the amino and carboxyl termini of the AlkB oxygenase motif, respectively [59].
The catalytic activity of ALKBH8 is mediated by two key structural domains: the C-terminal methyltransferase (MT) domain and the central AlkB oxygenase (Ox) domain. The MT domain can methylate CM5U in tRNA to mcm5U, while the Ox domain further modifies it to (S)-5-methoxycarbonylhydroxymethyluridine ((S)-mcm5U) and has stereoselectivity for the enzymatic hydroxylation of mcm5U [60,61,62]. ALKBH8 and Trm9 have similar functions and can catalyze the conversion of cm5U to mcm5U. tRNAArg (UCU) and tRNAGlu (UUC) have been shown to be two uridines containing swing tRNAs associated with the ALKBH8 complex. Therefore, ALKBH8 regulates protein translation by modifying the swing uridine of tRNAs [59, 63].
The mechanism of action of ALKBH8 is complex, and its target genes are not fully understood; however, its biochemical characteristics indicate that it is associated with various diseases. For example, the loss of ALKBH8 function may interfere with the tRNA modification process, thereby affecting brain function and leading to intellectual developmental disorders [61, 64, 65]. In addition, ALKBH8 plays important roles in protecting against acetaminophen (APAP) toxicity, regulating selenoprotein levels, aging, stress response gene regulation, mitochondrial weight programming, and maintaining neurological function [66,67,68]. ALKBH8 also promotes the growth of bladder cancer by regulating survivin expression, but its relationship with tRNA levels has not been confirmed [69].
DNMT2
DNMT2 (DNA methyltransferase 2) belongs to the DNA methyltransferase family and has an evolutionary history older than that of DNMT1 and DNMT3. It is widely present in various organisms, including plants, fungi, insects, fish, amphibians, birds, and mammals [70]. DNMT2 has multiple domains, including a target recognition domain (TRD) and a target recognition extension domain (TRED). Its domain is similar to that of prokaryotic m5C MTase and is involved in DNA damage recognition, recombination, and mutation repair [71,72,73].
DNMT2 plays an important role in tRNA modification, is capable of methylating tRNA (Asp GTC), tRNA (Val AAC), and tRNA (Gly GCC), and plays a role in the biosynthesis of tRNA-derived small RNAs [74,75,76]. During the formation of tRNA m5C38, quinine (Q) can increase DNMT2 activity [77,78,79]. DNMT2 recognizes the C32U33 (G/I) 34N35 (C/U) 36A37C38 motif in the tRNA anticodon loop, U11 in the D stem A24, and the correct size variable loop, selectively modifying tRNA [80, 81].
DNMT2 also participates in the regulation of protein translation. For example, DNMT2-mediated tRNA Asp methylation achieves posttranscriptional regulation by controlling the synthesis of target proteins containing polyaspartic acid sequences [82]. The DNMT2-mediated loss of tRNA Asp (GTC) C38 methylation leads to the downregulation of proteins with an Asp GAC codon bias [83]. In addition, DNMT2 and NSUN2 jointly promote the stability of tRNA and protein synthesis [84]. At the cellular level, DNMT2 silencing can induce oxidative stress and DNA damage in human fibroblasts and regulate cell proliferation and lifespan, and its regulation may become a potential anticancer strategy [85, 86]. In addition, DNMT2 also participates in antiviral defense mechanisms [83].
Mettl family
METTL1
METTL1 is a methyltransferase with an S-adenosylmethionine (SAM)-binding domain. Its characteristic 7-β chain catalytic domain enables it to modify DNA, RNA, and proteins [87, 88].
METTL1 works synergistically with WDR4 to introduce m7G modification at position 46 of tRNA, which is crucial for the normal function of tRNA and is the most common methylation site in prokaryotic and eukaryotic organisms [89, 90]. m7G modification not only affects overall methylation levels but also dynamically regulates internal methylation sites, especially in the GA/GG enrichment region of the 5’-untranslated region (UTR), as well as significantly increasing and increasing the accumulation of m7G modifications in coding sequences (CDSs) and 3’-UTRs under oxidative and heat stress [91,92,93,94].
WDR4 regulates the activity of METTL1 through two mechanisms, the first of which is by enhancing the binding affinity between METTL1 and SAM. The N-terminus of METTL1 regulates the methylation activity of m7G46 by coordinating SAM binding, RNA binding, and conformational changes in tRNA and the METTL1-WDR4 complex [95]. The second is by providing a scaffold for RNA binding, promoting the correct orientation of substrate tRNA and thereby achieving efficient catalysis [96]. In addition, METTL1 can be phosphorylated and inactivated by PKB and RSK both in vitro and in cells [97].
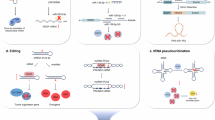
METTL1 plays important roles in various biological processes. METTL1-mediated m7G modification can maintain normal mRNA translation, self-renewal and differentiation in embryonic stem cells and, furthermore, can affect fertility by regulating tRNA homeostasis levels and the translation efficiency of genes required for spermatogenesis [98, 99]. In addition, METTL1-WDR4-catalyzed tRNA m7G46 modification can regulate translation, thereby affecting aging [100]. The conditional deletion of METTL1 or missense mutation of WDR4 can impair endochondral bone formation and accumulation, the mechanism of which is related to METTL1 knockout, which reduces the abundance of m7G-modified tRNA and inhibits the translation of mRNA related to the cytoskeleton and Rho GTPase signaling [101, 102]. METTL1 silencing can reduce the translation efficiency of hiPSC marker genes in stem cells and provides new ideas for the treatment of neurological and vascular diseases via the inhibition of mesodermal differentiation and angiogenesis [103]. METTL1 is also a potential therapeutic target, and its silencing can inhibit the growth of tumor cells [104] (Fig. 2).
A METTL1 silencing can reduce the translation efficiency of hiPSC marker genes in stem cells. B Knockout of METTL1 leads to a decrease in m7G modification, resulting in damage to the Rho GTPase signaling pathway. C The m7G modification of METTL1 can activate the mTORC1 pathway and promote tumor development. D METTL1 affects the EGFR signaling pathway through m7G modification, regulates codon translation efficiency, and influences lenvatinib resistance.
METTL1 enhances the translation of GADD45A and RB1 through an m⁷G dependent, codon-specific mechanism that increases tRNA-m⁷G levels, thereby inducing G₂/M arrest and suppressing breast cancer cell proliferation while increasing the antitumor efficacy of CDK4/6 inhibitors such as abemaciclib. Given that CDK4/6 inhibitors are primarily employed in the luminal A/B (ER⁺) breast cancer subtype, METTL1 appears to be closely associated with this subtype. In contrast, the mRNA expression levels of METTL1 and WDR4 are lower in the HER2⁺ and triple-negative subtypes than in luminal A/B tumors and normal breast tissue. Collectively, these findings indicate that METTL1 may exert distinct effects across different breast cancer subtypes [105, 106]. Whether tRNA modifications at specific codons are correlated with tumor subtypes and the precise molecular mechanisms through which they participate in tumorigenesis and progression of these subtypes warrant further investigation.
METTL6
METTL6 is an S-adenosylmethionine-dependent methyltransferase that can specifically modify tRNA (Ser) to resist N(3)-methylcytosine (m3C) at the 32nd nucleotide of the copper ring [107]. METTL6 shares homology with Trm141 in Streptococcus pyogenes and targets serine tRNA [108]. METTL6 can modify tRNA through its unique structural domain, m3C-RBD (m3C-specific RNA binding domain). This domain consists of an N-terminal region, an internal insertion, and an extended hairpin with a central β fold, forming a positively charged groove that accommodates the anticodon stem through nonspecific electrostatic contact with the base while recognizing the modified C32 through base-specific contact [109].
The modification function of METTL6 involves codon bias, which can improve the translation efficiency of tRNA Ser GCT decoding AGU codons, thereby regulating the cell cycle and DNA damage response [110]. In addition, METTL6 plays important roles in various cellular functions. For example, METTL6 knockout significantly reduces the sensitivity of lung cancer cells to cisplatin, indicating its potential role in chemotherapy resistance [111]. In hepatocellular carcinoma (HCC), METTL6 depletion inhibits cell growth, colony formation, migration, invasion, and cell adhesion and reduces the expression levels of cell adhesion proteins such as ITGA1, SPON1, and CLDN14 [112]. The expression of METTL6, a tumor-associated gene (TRG), is strongly correlated with the prognosis of patients with HCC and can serve as a potential prognostic indicator, providing new directions for the treatment strategies of patients with HCC [113] (Fig. 3).
tRNA modifying enzymes affect codon-specific translation: A TRMT10C promotes cancer progression via m1A modification. B Trm1 modifies tRNA SerUGA and tRNA LeuAAG. C TRMT12 inactivation reduces tRNA modifications, causing ribosomal -1 frameshifts. D NSUN6 upregulation increases tRNA m5C modification, enhancing translation of cancer-related proteins. E METTL6-mediated m3C modification boosts translation efficiency of tRNA SerGCT decoding AGU codons.
PUS1
Pseudouridylate synthase (PUS) catalyzes pseudouridine (Ψ) modification, which is an RNA modification that plays an important role in cellular function [114]. Pseudouridylate synthase 1 (PUS1) is a key member of this family, and its functions vary significantly among different organisms. Yeast Pus1 is a multisite-specific enzyme that synthesizes Ψ 34 and Ψ 36 in tRNAIle (UAU). In contrast, Pus1 (cmPus1) in Cyanidioschyzon merolae only catalyzes the Ψ modification at positions 34, 36, and/or 55 in certain specific introns containing precursor tRNA (UAU) variants [115].
PUS1 plays important roles in various biological processes. Pseudouridine modification is critical for the regulation of tRNA homeostasis, cytoplasmic translation, and erythropoiesis [116]. PUS1 gene mutation can lead to the loss of pseudouridine modification in mitochondrial tRNA, resulting in mitochondrial dysfunction and impaired protein synthesis [117]. In addition, PUS1 may participate in DNA repair, E2F targeting, MYC targeting, and the G2M checkpoint and promote malignant transformation in non-small cell lung cancer (NSCLC) through mcm5 or XPO1 [118]. PUS1 silencing can significantly inhibit the proliferation and invasion of breast tumors and may serve as a potential diagnostic biomarker for various cancers, including renal cell carcinoma (RCC) [119, 120]. PUS1 may also be involved in metabolic pathways, mitochondrial function, nonalcoholic fatty liver disease (NAFLD), and important oncogenic pathways and can be used as a clinical diagnostic biomarker for sepsis [121, 122]. In addition, the homozygous C656T mutation in the PUS1 gene is one of the pathogenic mutations in MLASA syndrome [123].
TRMT family
TRM1
TRM1 (tRNA methyltransferase 1) is a zinc ion binding, S-adenosylmethionine-dependent methyltransferase involved in tRNA processing, with RNA binding and tRNA (guanine N2) methyltransferase activity, and is mainly responsible for modifying m2G in tRNA. TRM1 exhibits substrate specificity and multisite modification ability in different organisms. For example, TRM1 in A. aeolicus not only catalyzes the methylation of G26 to generate m2G but also catalyzes the methylation of G27 to generate m22G [124]. In addition, endogenous and overexpressed Trm1 significantly modifies tRNA SerUGA and tRNA LeuAAG, indicating a preference for specific tRNA substrates. Mechanistically, Trm1 recognizes the secondary and tertiary structures of tRNA through domain specificity and targets the precursor tRNA that has been terminally processed and contains introns. This combination does not rely on catalytic activity. The binding of precursor tRNA to La inhibits the methylation of Trm1, while the substrate binds weakly to the RNA chaperone protein La because of its 3’ end sequence characteristics, resulting in reduced inhibition. At the same time, the RNA annealing partner activity of Trm1 promotes the formation of a preferred substrate conformation suitable for G26 modification, thereby efficiently completing the modification [125]. Among Archaeans, the TRM1 enzyme of Pyrococcus abyssi is a homologous tetramer with site specificity that can catalyze m1A modifications at positions 57 and 58 of certain tRNA T loops, whereas the TRM1 enzyme of Pyrococcus furiosus can be expressed and function in Escherichia coli [126,127,128].
TRM1 plays important roles in various biological processes. It may participate in the regulation of neuronal function after birth and maintain cell proliferation and cell survival under oxidative stress [129, 130]. These functions indicate that TRM1 plays important roles in cellular physiology and stress responses.
TRM1 generally exists in the human body in the form of its homolog, which is TRMT1. Its function and function are similar to those of TRM1. TRMT1 can methylate all tRNAs known to contain guanosine at position 26, catalyzing the formation of N2,N2-dimethylguanosine (m2,2G). Mutations in TRMT1 may lead to tRNA modification defects, which in turn can cause related diseases [131].
TRM4/NSUN2
TRM4 is a tRNA methyltransferase that uses S-adenosylmethionine (AdoMet) as a methyl donor to catalyze cytosine methylation on tRNA molecules to generate m5C modifications [132, 133]. m5C modification is widely present in RNA and affects mainly biological processes, such as RNA stability, splicing, and nuclear cytoplasmic transport. TRM4 acts as a tRNA m5C methyltransferase in brewing yeast and Methanococcus jannaschii specifically by modifying the C34 site in yeast pre-tRNA Leu (CAA) [132,133,134].
TRM4 plays important roles in various biological processes. In yeast cells, TRM4 participates in codon bias-based gene expression regulation by modulating tRNA modifications. Under oxidative stress conditions in yeast cells, TRM4 regulates tRNA modification, selectively translates mRNAs containing more TTG genes, and can also alter ribosome composition [135]. In addition, TRM4 is associated with sensitivity to 5-fluorouracil and has genetic interactions with other tRNA-modified genes, such as PUS1, PUS4, PUS7, and TRM2 [136]. The absence of TRM4 triggers the RTD (ribosome rescue) pathway, affecting the normal lifecycle of tRNA. TRM4 also synergizes with TRM8 to regulate cell growth and predict the accumulation of protein–RNA complexes after homocysteine mutations [137,138,139] (Fig. 4).
A ELP3 affects cancer development by influencing the tumor microenvironment and specific codon translations. B FTSJ1 mediated methylation modification enhances translation of related selenoproteins and inhibits cancer cell metastasis. C NSUN2 regulates m⁵C modification, selectively translating mRNAs with TTG codons and promoting cancer-related protein synthesis. D Elp1 mutation leads to codon specific translation, activates the PTCH signaling pathway, and affects tumor susceptibility. E Elp5 deficiency affects U34 modification and reduces sensitivity to gemcitabine.
NSUN2 is a mammalian homolog of yeast TRM4, belongs to the RNA methyltransferase family and is encoded by the human NSUN2 gene. It adds a methyl group to the C5 position of the RNA cytosine base through the catalytic mechanism of homocysteine, resulting in m5C modification. This modification is widely present in tRNA, rRNA, mRNA, and various types of noncoding RNA and is highly important for the structural stability, metabolic regulation, and functional performance of RNA [84, 140, 141].
NSUN2 plays crucial roles in various biological processes. Research has shown that NSUN2 deficiency can lead to reduced efficiency of glycine codon-specific translation, which in turn affects protein synthesis, synaptic transmission in mature neurons, and other complex behaviors. In addition, NSUN2 may affect intellectual development and disease occurrence through genetic mutation or abnormal methylation [141,142,143,144]. In cancer, abnormal NSUN2 expression is associated with the occurrence and development of various cancers, such as oral cancer and rectal cancer [145].
TRM9
TRM9 is a product of the yeast YML014w gene, which can catalyze the esterification reaction of uridine nucleotides and form specific modifications in tRNA [146]. TRM9 is mainly responsible for catalyzing the modification of mcm5 and mcm5s2 in tRNAArg (UCU) and tRNAGlu (UUC), which can regulate the translation selectivity of proteins. For example, Trm9 deficiency leads to the loss of the mcm5 and mcm5s2 modification of tRNAArg (UCU) and tRNAGlu (UUC), resulting in a decrease in their pairing efficiency with AGA and GAA codons. It is difficult for unmodified tRNA to quickly match the AGA and GAA codons on mRNA during translation, causing ribosome arrest, translation deceleration, and reduced protein synthesis [147]. In addition, TRM9-mediated specific tRNA modification can enhance codon-specific translation extension and increase the levels of key damage response proteins such as Yef3, Rnr1, and Rnr3 [146].
In terms of tumor biology, the expression of the human homolog hTRM9L of TRM9 is significantly downregulated in a variety of cancers, including testicular cancer, cervical cancer and bladder cancer. The absence of hTRM9L may increase the sensitivity of tumor cells to drugs that induce translation errors, making hTRM9L a potential target for treating hTRM9L-deficient tumors. hTRM9L also exerts antitumor effects by inhibiting tumor growth, regulating the cell cycle, and affecting the hypoxic response [148]. However, research on the specific mechanism through which TRM9 affects tumors is still insufficient and is worth exploring.
TRMT2A
Human tRNA methyltransferase 2 homolog A (hTRMT2A) is an enzyme that can bind and methylate RNA with low specificity. hTRMT2A contains multiple domains, including an RNA-binding domain (RBD), a central domain, and a fusion domain, which play synergistic roles in binding to tRNA and jointly participate in the interaction between hTRMT2A and tRNA. The uridine (U) at position 54 of tRNA is crucial for the binding and methylation activity of hTRMT2A, and its specific recognition mechanism enables hTRMT2A to accurately recognize and modify its target tRNA molecule. hTRMT2A plays an important role in maintaining translational fidelity, and loss or impairment of its function may lead to an increase in protein synthesis errors, thereby affecting the normal physiological functions of cells [149].
In addition, hTRMT2A has multiple functions in cell biology. It can inhibit cell proliferation and cell cycle progression and is considered a potential drug target for treating polyQ disease (a neurodegenerative disease) [150, 151].
TRMT10A
The TRM10 family of methyltransferases is responsible for the N1 methylation of the 9th purine in the tRNA of Archaeans and eukaryotes. The human genome encodes three types of TRM10 enzymes, namely, TRM10A, TRM10B, and TRM10C [152]. Among them, TRMT10A can catalyze the methylation of adenine (A) or guanine (G) at the 9th position of tRNA, resulting in m1G modification, and enhance the m6A demethylase activity of FTO through interactions with the mRNA demethylase FTO, stabilizing unmodified mRNA [153, 154].
TRMT10A has specific substrates in different species. In humans, its substrates include tRNAGln (UUG/CUG) and tRNAiniMeth (CAU), whereas in yeast, its substrates are tRNAGly (GCC) and tRNAVal (UAC). tRNAGln is responsible for decoding CAA and CAG codons (corresponding to glutamine) in mRNA. The deletion of TRMT10A leads to functional defects in tRNAGln, which may reduce its recognition efficiency of CAA/CAG codons and affect the translation of genes containing these codons [155]. TRMT10A deficiency can lead to translational distortion, decreased postsynaptic density, and impaired synaptic plasticity and can affect learning and memory function in mice, indicating its crucial role in brain function [156].
In addition, TRMT10A gene mutation is associated with diabetes and primary microcephaly in young people. Its defects can lead to a decrease in tRNA methylation levels, induce beta-cell apoptosis, and thus affect insulin secretion [155, 157,158,159,160].
TRMT10C
TRMT10C is a tRNA methyltransferase and a subunit of mitochondrial ribonuclease P (RNase P). When it binds to MRPP2, it forms a stable m1R9 tRNA methyltransferase complex, and when it binds to MRPP3, it forms an RNase P complex responsible for the 5’ end cleavage of mitochondrial tRNA [161, 162]. TRMT10C is expressed in mitochondria and participates in the positive regulation of RNA metabolism and mitochondrial translation, which is of significance for the maturation and functional expression of tRNA [163,164,165].
In addition, TRMT10C is involved in various biological functions. It indirectly inhibits lung cancer growth by mediating m7G modification of circFAM126A [166]. TRMT10C also participates in N1 methyladenosine (m1A) modification, which plays a key role in various cancers, such as glioma, hepatocellular carcinoma, and renal clear cell carcinoma [167,168,169,170,171]. However, the specific mechanism is still unclear and worthy of further study.
TRMT12
TRMT12 (tRNA methyltransferase 12) is an enzyme involved in tRNA modification, and its gene is also known as TYW2 or TRM12 in yeast. TRMT12 is essential for the synthesis of wybutosine (yW) in yeast, which is an important tRNA modification that regulates mRNA translation efficiency and maintains the correct reading framework.
TRMT12 exhibits significant changes in expression in various cancers. For example, in head and neck squamous cell carcinoma (HNSCC) tissue, the expression of TRMT12 is significantly upregulated, which may be related to genetic and epigenetic changes [172, 173]. In addition, through whole-exome sequencing of tumor and nonmalignant samples from 12 patients with special peripheral T-cell lymphoma (PTCL), researchers identified TRMT12 as one of 70 genes with somatic mutations [174]. However, the specific mechanism of action of TRMT12 in cancer is still unclear and deserves further investigation, which may provide a new perspective for the study of tumor development.
Trmt61A
TRMT61A belongs to the tRNA methyltransferase family and methylates adenine (A) to generate N1 methyladenosine (m1A) [175]. This modification occurs in the nucleus, where TRMT61A and TRMT6 work together to modify specific tRNAs with m1A58. The m1A58 modification may enhance the interaction between tRNA and ribosomes and between aminoacyl tRNA synthetases, accelerate the translation process, and rapidly synthesize MYC and other key functional proteins by changing the local conformation of tRNA. MYC, as a core transcription factor that drives cell proliferation, contains high-frequency codons in its mRNA that match the modified tRNA. The m1A58 modification enables tRNAs to quickly decode these codons and ensure the rapid accumulation of MYC protein after T-cell activation, thereby promoting the rapid expansion of activated T cells and guaranteeing the timeliness of the immune response [176]. (Fig. 5)
Three examples of m1A modification of TRMT61A: A TRMT61A knockout reduces tRNA m¹A modification, inhibits ACL translation, and impairs CD8⁺ T cell function, promoting tumor immune evasion. B TRMT61A overexpression enhances tRNA m¹A modification, boosts MYC synthesis, and upregulates PD-L1, aiding tumor immune escape. C The TRMT61A-TRMT6 complex increases m¹A methylation, particularly on tRNAAla(AGC) and tRNAGlu(CTC), thereby boosting translation efficiency and PPARδ synthesis.
In addition, TRMT6-TRMT61A complex-mediated tRNA-m1A58 modification plays an important role in the homeostasis of hematopoietic stem cells (HSCs) and performs nonclassical functions during HSC aging. These findings indicate that TRMT61A plays multifaceted regulatory roles in cell proliferation and stem cell aging [177,178,179].
TRMT61B
TRMT61B is a tRNA methyltransferase. In human mitochondria, it is responsible for 1-methyladenosine (m1A) modification at position 58 of mitochondrial tRNA (Leu (UUR)), tRNA (Lys), and tRNA (Ser (UCN)). This modification is crucial for mitochondrial protein synthesis and cellular function and plays a key role in various cancers [168].
TRMT61B regulates mitochondrial function through methylation, which may have significant implications for the pathogenesis of Alzheimer’s disease [180]. In addition, the role of TRMT61B in hepatoblastoma and melanoma has been preliminarily revealed: in the SK-MEL-103 melanoma cell line, TRMT61B knockout significantly inhibited tumor growth and metastasis and prolonged the survival of mice. Similarly, in zebrafish embryos and nude mouse models, TRMT61B inhibition slowed tumor cell proliferation. These findings suggest that targeting TRMT61B may provide new strategies for treating certain tumors [181, 182]. In addition, TRMT61B may affect protein translation through tRNA modification, thereby regulating cellular biological behavior and influencing the occurrence and development of cancer. For example, the TRMT61B gene may be related to susceptibility to breast cancer, but its specific mechanism of action in breast cancer still needs to be clarified through further research [177, 183, 184].
TRMU
TRMU is a highly conserved mitochondrial tRNA modification enzyme that is primarily responsible for the 2-thiouridine (s2U) modification of three types of mitochondrial tRNA (mt tRNA) at the U34 position. This modification is crucial for maintaining the structural stability and normal function of tRNA. TRMU plays a crucial role in vertebrates, participating in the processing and modification of tRNA [185].
It has several functions; for example, TRMU variants can cause liver failure in infants [186, 187]. The functional abnormalities of TRMU are associated with various diseases. For example, mutations in the TRMU gene may lead to structural and functional abnormalities in the enzyme, especially homozygous mutations in its N-terminal conserved region (such as A10S), which can severely affect mitochondrial tRNA metabolism and ultimately result in hearing loss [188, 189]In a zebrafish model, deletion of the MTU1 gene resulted in complete disappearance of the 2-thiouridine modification of mitochondrial tRNA (Lys), tRNA (Glu), and tRNA (Gln) at position U34, indicating that TRMU plays an important role in maintaining mitochondrial tRNA modification and hearing development [190].
TYW2
TYW2 is a tRNA modification enzyme that is involved in the synthesis of wybutosine (yW) modification in tRNA. This modification is crucial for maintaining the normal biological function of tRNA [191, 192].
In Archaeans, TYW2 plays a key role in the biosynthesis of guanosine derivatives, and its function is similar to that of homologous enzymes in eukaryotes, as it is involved in the construction of the basic skeleton of guanosine derivatives. TYW2 works synergistically with other related enzymes, such as TRM5, TYW1, TYW3, and TYW4, to determine the type and content of guanosine derivatives in Archaeans [193].
NSUN6
The NSUN family belongs to the methyltransferase family and is the main RNA m5C-modifying enzyme family in eukaryotes, consisting of seven members, from NSUN1 to NSUN7. Among them, NSUN6 is mainly responsible for catalyzing the m5C modification of tRNA, specifically by acting on the 72nd cytosine of the tRNA amino acid arm [194]. NSUN6 recognizes the CCA end and D-stem region of tRNA through its PUA domain and precisely identifies the target base using its MTase domain, inducing conformational changes in tRNA and exposing and modifying the target base [195].
NSUN6 plays important roles in various biological processes. When NSUN6 expression is upregulated, the m5C modification level of tRNA increases, promoting the translation of proteins related to cancer cell proliferation and invasion. Its functional impairment can lead to abnormal m5C modification of tRNA and mRNA, which in turn affects the accuracy and efficiency of protein synthesis, interferes with neurological development, and leads to cognitive dysfunction [196, 197]. In addition, NSUN6 affects the synthesis of cell proliferation-related proteins by regulating tRNA methylation, thereby regulating the proliferation of pancreatic cancer cells [198]. In glioblastoma, NSUN6-mediated m5C modification regulates transcriptional pauses and controls the tumor response to alkylating agents through the accumulation of NELFB and transcription factor complexes (POLR2A, TBP, TFIIA, and TFIIE) at the TATA binding site [199]. NSUN6 is also involved in the development of lung cancer and other cancers and can serve as a potential therapeutic target. Its role in cognitive impairment also deserves further research [200, 201].
Elongator complex
The elongator complex is a highly conserved multisubunit protein complex in eukaryotes that consists of six subunits, ELP1–ELP6. Among them, ELP1-ELP3 constitute the core subunit, and ELP4-ELP6 interact with the core subunit to maintain the stability and function of the complex. This complex plays important roles in gene transcription extension, tRNA modification, and other cellular processes.
Its catalytic subunit Elp3 contains an S-adenosylmethionine binding (rSAM) domain and a lysine acetyltransferase (KAT) domain, which are crucial for modification of the tRNA swing position (U34) and can promote the formation of modified nucleoside side chains such as 5-aminoformylmethyluridine (ncm⁵U₃₄), 5-methoxycarbonylmethyluridine (mcm⁵U₃₄), and 5-methoxycarbonylmethyl-2-thiouridine (mcm⁵s²U₃₄). These modified nucleosides are of significance for efficient decoding in the translation process, helping to maintain the accuracy and efficiency of protein synthesis and affecting both telomere gene silencing and the DNA damage response [202].
The elongator complex serves multiple functions. In vivo, Elp3 can promote polarization of M2 macrophages. During this process, Elp3 expression is upregulated by IL-4 and IL-13 induction. By modifying tRNA, it promotes the translation of mRNAs containing specific codons, such as LysAAA, GlnCAA, and GluGAA. These mRNA-encoded proteins participate in M2 macrophage polarization [203]. In addition, Elp3 plays an important role in the differentiation of intestinal cluster cells by regulating tRNA modification [204]. Notably, dysregulation of the tRNA modification activity of Elp3 is associated with various human diseases, including various cancers and neurodegenerative diseases, and Elp2 also participates in the regulation of these diseases [205, 206].
Role of codon-specific translational reprogramming mediated by tRNA modification in cancer
ALKBH8 mediates tRNA codon-specific translation in cancer
ALKBH8 is a methyltransferase that can affect codon-specific translation by modulating tRNA modifications, thereby influencing the occurrence and development of cancer. Under oxidative stress conditions, ALKBH8 can increase the 5-methoxycarbonylmethyl-2’-methyluridine (mc5Um) modification of tRNA, thereby driving the expression of selenocysteine-containing ROS-detoxifying enzymes such as Gpx1, Gpx3, Gpx6, and TrxR1. This modification is crucial for maintaining the redox balance of cells. In ALKBH8-deficient (Alkbh8-/-) cells, tRNA modification is impaired, leading to translational reprogramming and codon reprogramming abnormalities, which in turn affect cell function [207]. In addition, ALKBH8 is abnormally expressed in various cancers, and its function is closely related to tumor progression. For example, in colorectal cancer (CRC), by regulating the translation of selenoproteins and affecting the oxidative stress response, ALKBH8 may become a new target for CRC treatment. ALKBH8 expression is also significantly altered in other cancers, such as glioma, suggesting its potential role in tumorigenesis and drug resistance [208,209,210,211,212]. (Fig. 6) (Table 1)
Modifying enzymes influence cancer occurrence and progression by affecting tRNA codon-specific translation. Multiple enzymes can impact the same cancer type. Using Springer Nature Author Services, compare the increase or decrease in the expression levels of different modifying enzymes relative to normal tissues in different tumor types.
In summary, ALKBH8 affects the ROS detoxification network and protein translation by regulating tRNA modification, thereby influencing the occurrence and development of cancer. This suggests its potential application value in cancer treatment, which deserves further exploration.
DNMT2 mediates tRNA codon-specific translation in cancer
DNMT2 (TRDMT1) is an RNA cytosine methyltransferase that converts cytosine (C) in tRNA to 5-methylcytosine (m5C), primarily targeting the C38 site. This modification can affect the complementary pairing between tRNAs and codons, thereby regulating the accuracy and efficiency of translation. When the modification function of DNMT2 is abnormal, it may lead to codon translation errors, thereby regulating the occurrence and development of cancer; thus, DNMT2 is a potential target for cancer treatment [73, 213]. DNMT2 is a highly conserved tRNA Asp methyltransferase that maintains cellular function. In cancer, mutations in the DNMT2 gene can affect its catalytic activity. For example, the E63K mutation doubles enzyme activity, whereas mutations such as G155S, L257V, R371H, and G155V significantly reduce or completely decrease enzyme activity. These mutations affect protein synthesis by altering tRNA binding ability or methylation, which may promote tumorigenesis. These findings provide important information for studying the mechanism of action of DNMT2 in tumorigenesis and offer potential targets for cancer treatment and diagnosis [214].
In cancer cells, DNMT2 affects codon-specific translation by modifying tRNA. For example, 5-azacytidine-mediated RNA immunoprecipitation (Aza IP) technology can be used to identify C > G translocations at cytosine residues targeted by DNMT2, thereby recognizing specific methylated cytosines in target RNA. In addition, 5-azacytidine can inhibit RNA methylation at the DNMT2 target site, thereby affecting the function of tRNA [215]. When DNMT2 is absent, FTO expression increases, leading to a decrease in the m6A methylation level of TNFSF10, which in turn upregulates TNFSF10 expression and significantly inhibits the proliferation and metastasis of DNMT2-deficient liver cancer cells. These findings suggest that DNMT2 may indirectly affect the biological behavior of liver cancer cells. Moreover, the absence of DNMT2 may also activate the intracellular apoptotic signaling pathway, allowing bortezomib to more effectively induce apoptosis in liver cancer cells [216]. In addition, DNMT2 affects the development of breast cancer, cervical cancer, osteosarcoma, glioblastoma and other cancers [217].
These findings suggest that DNMT2 affects protein synthesis and cellular function by regulating tRNA modifications and codon-specific translation, thereby influencing the development of cancer. Therefore, DNMT2 and its regulated tRNA modification pathway may become new targets for cancer therapy.
METTL1 mediates tRNA m7G in cancer
METTL1 is a key tRNA methyltransferase located in the region of chromosome 12 (12q13-14), which is frequently amplified in various cancers and closely associated with tumorigenesis [90, 218]. METTL1 modifies the m7G site of tRNA to affect the translation efficiency of specific codons, thereby regulating the occurrence and development of cancer.
During the occurrence and development of tumors, the METTL1/WDR4 complex modifies the tRNA subpopulation that decodes m7G-dependent codons, promotes stable ribosome extension, and increases the cell cycle and translation efficiency of oncogenic mRNA, thereby promoting cancer progression [219, 220]. For example, in HCC, METTL1-mediated tRNA m7G modification significantly increases the translation of target mRNA with high-frequency m7G-related codons. METTL1 knockdown can reduce the level of m7G-modified tRNA in HCC cells and inhibit cell proliferation, migration, and invasion [221, 222].
From the perspective of drug resistance, the EGFR gene is a key target of METTL1, and its mRNA has a high m7G-related codon frequency. METTL1 enhances the function of tRNA corresponding to these codons, thereby promoting the translation efficiency of EGFR mRNA, increasing EGFR protein expression, and activating downstream signaling pathways such as the Akt and p44/42 MAPK pathways [222]. Activation of the EGFR pathway enhances the proliferation ability of liver cancer cells, inhibits apoptosis, and allows cells to survive and proliferate under treatment with lenvatinib, ultimately leading to the development of drug resistance [223, 224]. Moreover, METTL1/WDR4 selectively upregulates the translation of ECM remodeling-related genes through m7G tRNA modification, driving osteosarcoma resistance to doxorubicin. This mechanism suggests that inhibiting METTL1 may reverse drug resistance and provide a new strategy for combination chemotherapy [225]. In addition, METTL1 provides a new therapeutic intervention strategy against oral squamous cell carcinoma (OSCC) resistance by enhancing overall mRNA translation and stimulating oxidative phosphorylation (OXPHOS) [102, 226]. From a clinical feasibility perspective, drug resistance risk can be predicted, and treatment plans can be guided by detecting METTL1 expression or m7G modification levels; METTL1 inhibition can reverse drug resistance and be used as a targeted development inhibitor to enhance efficacy in combination with chemotherapy/targeted drugs. However, it is necessary to address issues such as specificity, standardization of detection, and response to multichannel drug resistance, which is expected to promote the precise treatment of tumors.
In various cancers, METTL1 affects tumor occurrence and progression by regulating tRNA codon-specific translation. For example, in esophageal squamous cell carcinoma (ESCC), METTL1 promotes tumor progression through the regulation of RPTOR gene translation. In ESCC cells METTL1 knockdown, the expression of RPTOR protein is reduced, leading to a decrease in mTORC1 activity, which specifically manifests as a decrease in the phosphorylation levels of the downstream target proteins p4EBP1 and pS6K1 of mTORC1 [227]. In cholangiocarcinoma (ICC), METTL1 affects the expression of cell cycle- and EGFR pathway-related genes by regulating the translation of oncogenic mRNA. Mechanistically, genes whose translation efficiency is reduced after METTL1 is knocked out have more m7G tRNA decoding codons [228, 229]. In prostate cancer, METTL1 promotes tumorigenesis through the biogenesis of tRNA-derived fragments, while its deletion leads to the loss of m7G modification, inhibiting tumor growth [230]. In breast cancer, METTL1-mediated m7G tRNA modification is necessary for codon recognition during mRNA translation [231]. For example, METTL1-mediated m7G tRNA modification can drive the progression and metastasis of thyroid carcinoma by regulating the codon-specific translation of TNF-α [232]. METTL1-mediated m7G modification affects codon translation and is also associated with various other cancers, such as lung cancer, leukemia, nasopharyngeal carcinoma, neuroblastoma, and gastric cancer [233,234,235,236,237].
TRMT12 modifies the G37 site on tRNA in cancer
TRMT12 (also known as TYW2) is a tRNA modification enzyme that affects codon-specific translation by modulating the G37 site of tRNA, thereby regulating the occurrence and development of cancer. Research has shown that the transcriptional silencing of TRMT12 is associated with promoter CpG island hypermethylation in colorectal cancer (CRC). TRMT12 inactivation leads to the depletion of wybutosine derivatives (oHyW and o2yW), resulting in the loss of G37 site modification, which is crucial for maintaining the accuracy of ribosome reading frames. Therefore, the inactivation of TRMT12 triggers an increase in ribosomal -1 frameshift events, leading to the incorrect reading of codons and a decrease in translation efficiency [238].
Changes in the modification at the G37 site may affect the interaction between tRNAPhe and codons, increase the stability of codon‒codon interactions, and prevent code shift errors during translation. In terms of drug resistance, the downregulation of TRMT12 leads to an increase in imG-14 levels, which becomes the main modification of tRNAPhe in paclitaxel-resistant strains. Knocking out TRMT12 in HeLa cells leads to the accumulation of imG-14 and reduces the efficacy of paclitaxel. Therefore, low TRMT12 expression not only promotes cancer survival but also enhances resistance to paclitaxel treatment [239]. From a clinical feasibility perspective, detecting the mRNA or protein levels of TRMT12 in tumor tissues can be used to screen for patients at high risk for drug resistance and guide treatment plan adjustments (such as combining other drugs to reverse drug resistance). Targeted drugs (such as TRMT12 activators) can be developed to address the modification abnormalities caused by low TRMT12 expression, restore its normal expression to correct tRNA modification imbalance, and enhance the killing effect of paclitaxel on tumor cells. In addition, the combined inhibition of abnormal modifications such as imG-14 synthesis (such as by interfering with its upstream metabolic pathways) may further increase chemotherapy sensitivity.
In early colorectal cancer patients, high methylation of the TRMT12 promoter is significantly associated with poor overall survival and is an independent prognostic predictor. Cell model experiments have shown that colorectal cancer cells lacking TRMT12 exhibit enhanced migration ability and epithelial mesenchymal transition (EMT) characteristics, which are associated with the susceptibility of tumor cells to metastasis [238]. From a clinical feasibility perspective, TRMT12 detection relies on mature DNA methylation technology, which can be achieved through tumor tissue samples. TRMT12 detection can assist in early patient risk stratification and guide personalized treatment (such as enhanced follow-up or adjuvant chemotherapy); however, it is necessary to address the limitations of relying on tissue samples (such as exploring liquid biopsy) and validate methods in multicenter cohorts to improve specificity. In the future, TRMT12 is expected to be combined with other biomarkers to construct predictive models or develop targeted therapies, thereby promoting precise diagnosis and treatment. In addition, TRMT12 affects the occurrence and development of breast cancer and head and neck squamous cell carcinoma [173].
These results indicate that TRMT12 affects codon-specific translation by modulating the G37 site modification of tRNA, thereby regulating cancer progression and drug resistance. The epigenetic regulatory mechanism of its expression provides a new target for cancer treatment.
TRMT61A mediates tRNA m1A in cancer
TRMT61A affects codon-specific translation by modulating the m1A modification of tRNA, thereby regulating the occurrence and development of cancer. In hepatocellular carcinoma (HCC), the methyltransferase complex formed by TRMT61A and TRMT6 is highly expressed, leading to a significant increase in the m1A methylation of tRNA, especially on tRNAAla (AGC) and tRNAGlu (CTC). This modification improves the translation efficiency of corresponding codons, increases the translation of peroxisome proliferator activated receptor δ (PPARδ), and is associated with a poor prognosis in HCC patients. Therefore, targeting the TRMT6/TRMT61A complex may be an effective strategy for treating HCC [9]. In addition, the depletion of TRMT6/TRMT61A in C6 glioma cells reduced cell proliferation and increased cell death, which could be partially rescued by the overexpression of tRNAi (Met) [240]. Moreover, the knockout or downregulation of TRMT61A resulted in a significant decrease in the m1A modification of tRNA. In the absence of the TRMT61A gene, the m1A modification of tRNA is reduced, thereby inhibiting ACL translation, blocking cholesterol biosynthesis, and weakening the tumor-killing function and proliferation ability of CD8⁺ T cells. CD8⁺ T cells play crucial roles in the immune defense against solid tumors such as melanoma and lung cancer, as well as hematological tumors such as leukemia. Abnormal expression of TRMT61A may lead tumor cells to escape immune surveillance and promote tumor progression [16]. High expression of TRMT61A can increase the m1A modification of tRNA, improve the recognition efficiency of corresponding codons in MYC mRNA, and promote MYC protein synthesis. MYC, as a transcription factor, activates the transcription of PD-L1, thereby upregulating its expression. As an immune checkpoint protein, highly expressed PD-L1 can bind to PD-1 on the surface of T cells, inhibit T-cell activity, help tumor cells evade immune surveillance, and promote tumor immune escape [241]. In head and neck squamous cell carcinoma (HNSCC), TRMT61A-mediated tRNA m1A58 modification directly regulates the translation of MYC mRNA through codon decoding, thereby affecting tumor occurrence and development [241].
TRMT6/TRMT61A plays a carcinogenic role in bladder cancer (BLCA) and participates in the regulation of the cell stress response. However, its specific mechanism in bladder cancer needs further study [242]. In addition, TRMT61A may play a role in various cancers, such as testicular germ cell tumors, but its specific mechanism is still unclear and deserves further exploration. These studies will help reveal the role of TRMT61A in cancer and provide new targets for cancer treatment [243,244,245,246].
NSUN2 mediates tRNA codon-specific translation in cancer
NSUN2 is a tRNA modification enzyme that affects codon-specific translation by regulating the m5C modification of tRNA, thereby regulating the occurrence and development of cancer. Research has shown that the expression level of NSUN2 is elevated in various cancers and that its mediated tRNA m5C modification can significantly affect the efficiency and accuracy of protein translation.
The expression of NSUN2 gradually increases in normal thyroid, poorly differentiated thyroid cancer (PDTC), and undifferentiated thyroid cancer (ATC) tissues. Knocking out NSUN2 leads to a decrease in m5C methylation levels of cytoplasmic tRNAs, which have species and secondary structure preferences; this decrease manifests as a reduction in m5C modifications of certain specific tRNAs (such as tRNA Leu CAA and CAG). A decrease in the methylation rate of m5C modification sites can affect the secondary structural stability and function of tRNA, thereby affecting codon recognition and protein translation. For example, m5C modification of stable tRNA mediated by NSUN2 can more effectively recognize genes rich in TTG codons, thereby promoting the protein translation of these genes. This mechanism is crucial for the synthesis of oncogene-related proteins such as c-Myc, BCL2, RAB31, JUNB, and TRAF2, which play critical roles in tumor cell growth and proliferation. Therefore, NSUN2 regulates the m5C modification of tRNA, affects codon-specific translation, and thereby regulates cancer progression and drug resistance [247]. In addition, NSUN2 plays important roles in various cancers, such as esophageal cancer, liver cancer, gastric cancer, ovarian cancer, and head and neck squamous cell carcinoma, by affecting tRNA modification and may become a valuable target for cancer treatment and a diagnostic marker for cancer [145, 248,249,250].
Modification of tRNA U34 position in cancer
The elongator complex plays an important role in cancer. From the perspective of the interaction between microorganisms and human immunity, Elp3 is the catalytic subunit of the Elongator complex, is responsible for modifying the U34 site of cytoplasmic tRNA, and is crucial for maintaining the mcms²U modification of tRNA. In Aspergillus niger, knocking out the elp3 gene leads to the loss of mcms²U in tRNA, which in turn affects hyphal growth, the production of biofilm-associated extracellular polysaccharides (GAGs), adhesion ability, and virulence. When human immune function is low, Aspergillus fumigatus infection may trigger an inflammatory response, and a persistent inflammatory microenvironment is associated with the occurrence and development of cancer. In addition, in Aspergillus niger, Elp3 deficiency upregulates the expression of genes related to amino acid metabolism and overexpression of the transcription factor CpcA. If this abnormal regulation of amino acid metabolism occurs in the tumor microenvironment, it may affect the proliferation and survival of tumor cells. Therefore, Elp3 may play an important role in the development of the tumor microenvironment by regulating amino acid metabolism through affecting tRNA modification [251].
In addition, dysfunction of the Elongator complex is closely related to the progression of various tumors. In intestinal epithelial cells, the Wnt signaling pathway can upregulate Elp3 expression, promote Sox9 protein expression, and maintain cancer stem cell subpopulations. In breast cancer, the oncogenic expression of T protein in multiple oncoviruses can increase the expression of Elp3 and Ctu1/2, promote the translation of DEK oncoproteins, and upregulate the characteristics of the invasion-promoting transcriptome. In melanoma, Elp3 modifies tRNA to affect the decoding of specific codons in HIF1A mRNA, thereby promoting the translation of HIF1A protein. The increase in HIF1A protein expression enables melanoma cells to acquire invasive features and drug resistance, leading to resistance of tumor cells to targeted therapeutic drugs such as BRAF V600E inhibitors. In breast cancer, Elp3 is associated with tRNA modification and the IRES-dependent translation of LEF1 to maintain metastasis in breast cancer. In pediatric medulloblastoma, germline mutations in Elp1 and sustained activation of the Sonic Hedgehog signaling pathway increase patient susceptibility to disease [203, 206, 252]. In gallbladder cancer, patients with low expression of Elp5 respond poorly to gemcitabine treatment. In medulloblastoma, the loss of mutations in Elp1 can cause instability of the Elongator complex, loss of tRNA modifications, and trigger codon-dependent translational reprogramming and unfolded protein responses. This, in conjunction with PTCH1 somatic changes and activation of the SHH signaling pathway, increases the susceptibility of individuals to tumors and affects their molecular characteristics [253,254,255].
In addition to the Elongator complex, FTSJ1-mediated U34 methylation (forming mcm5Um, i.e., Um34) can increase the efficiency of selenocysteine (Sec) insertion into protein sequences, thereby increasing the translation level of redox-regulated selenoproteins such as GPX4. However, this process may reduce the oxidative stress tolerance of cancer cells and inhibit tumor metastasis. Therefore, FTSJ1 and its mediated Um34 modification can serve as potential targets for antitumor metastasis drugs. In melanoma cells, exposure to selenium and reactive oxygen species (ROS) can increase FTSJ1-mediated Um34 formation, promote stress-responsive selenoprotein synthesis, alleviate ROS damage, and support cell survival under oxidative conditions [256].
Role of codon-specific translation in cancer
Codon-specific translation exhibits both cancer type specificity and dynamic changes with tumor progression stage. In terms of cancer-specific differences, studies have shown that different types of cancer affect the same tissue in terms of different patterns of synonymous codon usage changes. For example, in breast cancer, invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), and mixed invasive ductal and lobular carcinoma (IDLC) are associated with a significant increase in the use of the glycine codon GGT. However, in IDC and IDLC, but not in ILC, the change in GGT use preference is related to the change in GGC use preference. These findings indicate that different types of breast cancer have different codon usage preferences [257]. In various cancers, the expression of multiple codons of tRNAArg (such as CGT, AGA, CGG, and CGA) is generally upregulated, with tRNAArg (CGT) and tRNAArg (AGA) being highly expressed in renal clear cell carcinoma (KIRC) and associated with a poor prognosis. Other codons, such as tRNAThr (ACA) and tRNAPro (CCA), are downregulated in KIRC and are associated with a poor prognosis. TRNAVal is upregulated in 9 types of cancer and downregulated in 5 types of cancer, reflecting the cancer specificity of its function. Moreover, the upregulated tRNA in prostate cancer corresponds mainly to the AGC codon of serine (Ser) and the ACC codon of threonine (Thr), with little difference in codon usage compared with that in normal prostate tissue (low MSE value). The upregulated tRNA coverage in cholangiocarcinoma is wider, including not only Ser AGC and Thr ACC, which are involved in prostate cancer, but also the AAA codon of lysine (Lys) and the CTT codon of leucine (Leu). There is a significant difference in codon usage patterns between cholangiocarcinoma tissue and normal bile duct tissue (high MSE value). This reflects the heterogeneity of codon-specific translation among different cancer types [258].
There are also some examples in terms of tumor progression stages. For example, through survival analysis, it was found that the degree of change in codon usage (MSE value) between normal and tumor tissues of patients is correlated with prognosis; the greater the change is, the higher the mortality rate of patients. This finding indirectly suggests that as the tumor progresses (such as with increasing malignancy), abnormal changes in codon preference may intensify, but their specific stage-specific patterns still need further research for verification [257]. Moreover, TRMT6 is highly expressed in colorectal cancer (CRC), stabilizing the TRMT6-TRMT61A complex, enhancing the m1A modification of tRNA-Lys-TTT-1-1, and promoting histone translation with an AAA/AAG codon preference, thereby driving cell cycle and tumor progression. High expression of TRMT6 is associated with a poor prognosis in CRC patients, and its translational regulation is key for CRC cell proliferation, indirectly suggesting that a preference for AAA/AAG codons may continue to play a role in CRC progression. The specific correlation mechanism is worthy of further exploration [179]. These findings provide a basis for considering codon preference features in personalized cancer treatment and exploring their potential research value.
Discussion and outlook
In this review, we explore the structure, modification enzymes, and mechanisms of tRNA, as well as their roles in tumor development. tRNA modifications, such as m6A, m5C, m1G, Ψ, D, I, and 2’-O, can influence tumor progression by modulating codon preference during translation. This is often mediated by tRNA modification enzymes, which can induce stop codon rewriting and ribosomal frameshifting or alter base status and translation efficiency. Common tumor-associated modifications include m7G and m5G.
From the perspective of tumor diagnosis, treatment, and prognosis, tRNA modification enzymes can serve as biomarkers and regulate drug resistance. For example, modulating the codon frequency to regulate EGFR translation can maintain resistance to lenvatinib in hepatocellular carcinoma. However, challenges remain in fully understanding the mechanisms underlying tRNA-mediated codon translation in tumor development. Many promising tRNA modification enzymes, such as PUS1, the Trm family, the TRM10 family, TRMT61A-C, and the NSUN family, require further investigation. Therefore, in terms of future clinical relevance and basic research, standardized detection reagents for tRNA modification and related molecules can be developed for early diagnosis, prognostic stratification, and drug resistance risk prediction in cancer; moreover, specific tRNA modification enzyme inhibitors can be developed and combined with existing chemotherapy/targeted drugs to reverse drug resistance, or drug combinations can be adjusted based on tumor cell tRNA modification profiles and codon preferences. Future basic research requires systematic mapping of the correlation between tRNA modification profiles and codon usage frequency in different cancers to clarify the specific axis of action for various tRNA modification enzymes and tRNA codon target genes; explore the effect of tumor microenvironment regulation on tRNA function; explore the cross-interactions between tRNA modifications and other forms of epigenetic regulation; explore the role of noncoding RNA; and develop tRNA modification editing tools and high-throughput screening platforms. By combining clinical translation with basic research, this field is expected to offer breakthroughs in the precision oncology by providing new strategies for overcoming drug resistance and improving prognosis.
In addition to affecting tumors, tRNA modification enzymes also play significant roles in neurological, reproductive, and hematological diseases. Thus, studying tRNA and its modifications is of great importance.
In conclusion, tRNA modifications are closely related to tumor development and progression. The mechanisms discussed here highlight the potential of tRNA-mediated codon-specific translation in influencing disease outcomes. This field holds great potential for further exploration, which will contribute to advancing our understanding of tumor biology and ultimately benefit human health.
Conclusion
In conclusion, our review highlights the critical role of tRNA modifications and their enzymes in tumor development and progression. These modifications, including m6A, m5C, m1G, and others, can influence tumor behavior by modulating codon preference and translation efficiency. Despite emerging insights into the underlying mechanisms, such as ribosomal frameshifting and stop codon rewriting, many questions remain unanswered. The potential of tRNA modifications as biomarkers and therapeutic targets in cancer, as well as their broader implications in other diseases, underscores the importance of further research in this field. Understanding these mechanisms will not only advance our knowledge of tumor biology but also pave the way for novel diagnostic and therapeutic strategies, ultimately benefiting human health.
References
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63.
Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7.
Xue M, Dong L, Zhang H, Li Y, Qiu K, Zhao Z, et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17:7.
Liang W, Lin Z, Du C, Qiu D, Zhang Q. mRNA modification orchestrates cancer stem cell fate decisions. Mol Cancer. 2020;19:38.
Wang Y, Tao EW, Tan J, Gao QY, Chen YX, Fang JY. tRNA modifications: insights into their role in human cancers. Trends Cell Biol. 2023;33:1035–48.
Orellana EA, Siegal E, Gregory RI. tRNA dysregulation and disease. Nat Rev Genet. 2022;23:651–64
Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19:20–30.
Ren D, Mo Y, Yang M, Wang D, Wang Y, Yan Q, et al. Emerging roles of tRNA in cancer. Cancer Lett. 2023;563:216170.
Wang Y, Wang J, Li X, Xiong X, Wang J, Zhou Z, et al. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat Commun. 2021;12:6314.
Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30:4509–18.
Adams JM, Cory S. Modified nucleosides and bizarre 5’-termini in mouse myeloma mRNA. Nature. 1975;255:28–33.
Wang G, Li H, Ye C, He K, Liu S, Jiang B, et al. Quantitative profiling of m6A at single base resolution across the life cycle of rice and Arabidopsis. Nat Commun. 2024;15:4881.
Kan L, Ott S, Joseph B, Park ES, Dai W, Kleiner RE, et al. A neural m6A/Ythdf pathway is required for learning and memory in Drosophila. Nat Commun. 2021;12:1458.
Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977;115:695–714.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5.
Miao S, Li H, Song X, Liu Y, Wang G, Kan C, et al. tRNA m1A modification regulates cholesterol biosynthesis to promote antitumor immunity of CD8+ T cells. J Exp Med. 2025;222:e20240559.
Duan HC, Zhang C, Song P, Yang J, Wang Y, Jia G. C2-methyladenosine in tRNA promotes protein translation by facilitating the decoding of tandem m2A-tRNA-dependent codons. Nat Commun. 2024;15:1025.
Ozkurede U, Kala R, Johnson C, Shen Z, Miller RA, Garcia GG. Cap-independent mRNA translation is upregulated in long-lived endocrine mutant mice. J Mol Endocrinol. 2019;63:123–38.
Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–83.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–205.
Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int J Mol Sci. 2019;20:1323.
Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. 2015;217:337–44.
Liang Z, Riaz A, Chachar S, Ding Y, Du H, Gu X. Epigenetic modifications of mRNA and DNA in plants. Mol Plant. 2020;13:14–30.
Delaunay S, Pascual G, Feng B, Klann K, Behm M, Hotz-Wagenblatt A, et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature. 2022;607:593–603.
Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, et al. Biological roles of RNA m5C modification and its implications in cancer immunotherapy. Biomark Res. 2022;10:15.
Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m5C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639.
Gonskikh Y, Tirrito C, Bommisetti P, Mendoza-Figueroa MS, Stoute J, Kim J, et al. Spatial regulation of NSUN2-mediated tRNA m5C installation in cognitive function. Nucleic Acids Res. 2025;53:gkae1169.
Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–30.
Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–63.
Wu P, Gao J, Lan G, Wang Y. The role of RNA m5C modification in central nervous system diseases. Discov Med. 2024;36:1555–71.
Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S, et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. 2023;22:81.
Feng J, Xu T, He M, Li J, Yao P, Ma C, et al. NSUN2-mediated m5C modification of HBV RNA positively regulates HBV replication. PLoS Pathog. 2023;19:e1011808.
Wang YY, Tian Y, Li YZ, Liu YF, Zhao YY, Chen LH, et al. The role of m5C methyltransferases in cardiovascular diseases. Front Cardiovasc Med. 2023;10:1225014.
Björk GR, Wikström PM, Byström AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–9.
Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–85.
Masuda I, Takase R, Matsubara R, Paulines MJ, Gamper H, Limbach PA, et al. Selective terminal methylation of a tRNA wobble base. Nucleic Acids Res. 2018;46:e37.
Monoe Y, Miyamoto S, Jingushi K, Tanimoto M, Tanaka T, Taniguchi K, et al. Hypoxia regulates tumour characteristic RNA modifications in ovarian cancers. FEBS J. 2023;290:2085–96.
Idaghdour Y, Hodgkinson A. Integrated genomic analysis of mitochondrial RNA processing in human cancers. Genome Med. 2017;9:36.
Guo H, Shen X, Hu H, Zhou P, He T, Xia L, et al. Alteration of RNA modification signature in human sperm correlates with sperm motility. Mol Hum Reprod. 2022;28:gaac031.
Begik O, Lucas MC, Pryszcz LP, Ramirez JM, Medina R, Milenkovic I, et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat Biotechnol. 2021;39:1278–91.
Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–16.e26.
Ding H, Liu N, Wang Y, Adam SA, Jin J, Feng W, et al. Implications of RNA pseudouridylation for cancer biology and therapeutics: a narrative review. J Transl Med. 2024;22:906.
Chang Y, Jin H, Cui Y, Yang F, Chen K, Kuang W, et al. PUS7-dependent pseudouridylation of ALKBH3 mRNA inhibits gastric cancer progression. Clin Transl Med. 2024;14:e1811.
Yu F, Tanaka Y, Yamashita K, Suzuki T, Nakamura A, Hirano N, et al. Molecular basis of dihydrouridine formation on tRNA. Proc Natl Acad Sci USA. 2011;108:19593–8.
Maizels N, Weiner AM. Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci USA. 1994;91:6729–34.
Elliott MS, Trewyn RW. Inosine biosynthesis in transfer RNA by an enzymatic insertion of hypoxanthine. J Biol Chem. 1984;259:2407–10.
Häfner SJ, Jansson MD, Altinel K, Andersen KL, Abay-Nørgaard Z, Ménard P, et al. Ribosomal RNA 2’-O-methylation dynamics impact cell fate decisions. Dev Cell. 2023;58:1593–1609.e9.
Zhang M, Lu Z. tRNA modifications: greasing the wheels of translation and beyond. RNA Biol. 2025;22:1–25.
Decombe A, Peersen O, Sutto-Ortiz P, Chamontin C, Piorkowski G, Canard B, et al. Internal RNA 2’-O-methylation on the HIV-1 genome impairs reverse transcription. Nucleic Acids Res. 2024;52:1359–73.
El Kazzi P, Rabah N, Chamontin C, Poulain L, Ferron F, Debart F, et al. Internal RNA 2’O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res. 2023;51:2501–15.
Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2:RESEARCH0007.
Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–8.
Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–82.
Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, et al. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–16.
Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem Rev. 2006;106:215–32.
Ringvoll J, Nordstrand LM, Vågbø CB, Talstad V, Reite K, Aas PA, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–98.
Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–63.
Zdżalik D, Vågbø CB, Kirpekar F, Davydova E, Puścian A, Maciejewska AM, et al. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PLoS ONE. 2014;9:e98729.
Fu D, Brophy JAN, Chan CTY, Atmore KA, Begley U, Paules RS, et al. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol. 2010;30:2449–59.
van den Born E, Vågbø CB, Songe-Møller L, Leihne V, Lien GF, Leszczynska G, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172.
Maddirevula S, Alameer S, Ewida N, de Sousa MML, Bjørås M, Vågbø CB, et al. Insight into ALKBH8-related intellectual developmental disability based on the first pathogenic missense variant. Hum Genet. 2022;141:209–15.
Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl. 2010;49:8885–8.
Leihne V, Kirpekar F, Vågbø CB, van den Born E, Krokan HE, Grini PE, et al. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in arabidopsis tRNA. Nucleic Acids Res. 2011;39:7688–701.
Monies D, Vågbø CB, Al-Owain M, Alhomaidi S, Alkuraya FS. Recessive truncating mutations in ALKBH8 cause intellectual disability and severe impairment of wobble uridine modification. Am J Hum Genet. 2019;104:1202–9.
Waqas A, Nayab A, Shaheen S, Abbas S, Latif M, Rafeeq MM, et al. Case report: biallelic variant in the tRNA methyltransferase domain of the AlkB homolog 8 causes syndromic intellectual disability. Front Genet. 2022;13:878274.
Evke S, Lin Q, Melendez JA, Begley TJ. Epitranscriptomic reprogramming is required to prevent stress and damage from acetaminophen. Genes. 2022;13:421.
Lee MY, Leonardi A, Begley TJ, Melendez JA. Loss of epitranscriptomic control of selenocysteine utilization engages senescence and mitochondrial reprogramming☆. Redox Biol. 2020;28:101375.
Honda K, Hase H, Tanikawa S, Okawa K, Chen L, Yamaguchi T, et al. ALKBH8 contributes to neurological function through oxidative stress regulation. PNAS Nexus. 2024;3:pgae115.
Ohshio I, Kawakami R, Tsukada Y, Nakajima K, Kitae K, Shimanoe T, et al. ALKBH8 promotes bladder cancer growth and progression through regulating the expression of survivin. Biochem Biophys Res Commun. 2016;477:413–8.
Wang FL, Yan LX, Shi HJ, Liu XY, Zheng QY, Sun LN, et al. Genome-wide identification, evolution of DNA methyltransferases and their expression during gonadal development in Nile tilapia. Comp Biochem Physiol B Biochem Mol Biol. 2018;226:73–84.
Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA. 2008;14:1663–70.
Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29:439–48.
Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–32.
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–5.
Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–8.
Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 2007;21:261–6.
Müller M, Legrand C, Tuorto F, Kelly VP, Atlasi Y, Lyko F, et al. Queuine links translational control in eukaryotes to a micronutrient from bacteria. Nucleic Acids Res. 2019;47:3711–27.
Ehrenhofer-Murray AE. Cross-talk between Dnmt2-dependent tRNA methylation and queuosine modification. Biomolecules. 2017;7:14.
Müller M, Hartmann M, Schuster I, Bender S, Thüring KL, Helm M, et al. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015;43:10952–62.
Huang ZX, Li J, Xiong QP, Li H, Wang ED, Liu RJ. Position 34 of tRNA is a discriminative element for m5C38 modification by human DNMT2. Nucleic Acids Res. 2021;49:13045–61.
Shanmugam R, Aklujkar M, Schäfer M, Reinhardt R, Nickel O, Reuter G, et al. The Dnmt2 RNA methyltransferase homolog of geobacter sulfurreducens specifically methylates tRNA-glu. Nucleic Acids Res. 2014;42:6487–96.
Shanmugam R, Fierer J, Kaiser S, Helm M, Jurkowski TP, Jeltsch A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015;1:15010.
Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep. 2013;14:269–75.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–5.
Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Wnuk M. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J Cell Physiol. 2017;232:3714–26.
Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Deregowska A, Semik E, Zabek T, et al. Reduced levels of methyltransferase DNMT2 sensitize human fibroblasts to oxidative stress and DNA damage that is accompanied by changes in proliferation-related miRNA expression. Redox Biol. 2018;14:20–34.
Tooley JG, Catlin JP, Tooley CES. METTLing in stem cell and cancer biology. Stem Cell Rev Rep. 2023;19:76–91.
Wong JM, Eirin-Lopez JM. Evolution of Methyltransferase-Like (METTL) proteins in metazoa: a complex gene family involved in epitranscriptomic regulation and other epigenetic processes. Mol Biol Evol. 2021;38:5309–27.
Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G, et al. Transcriptome-wide mapping of Internal N7-methylguanosine methylome in mammalian mRNA. Mol Cell. 2019;74:1304–16.e8.
Bahr A, Hankeln T, Fiedler T, Hegemann J, Schmidt ER. Molecular analysis of METTL1, a novel human methyltransferase-like gene with a high degree of phylogenetic conservation. Genomics. 1999;57:424–8.
Thongdee N, Jaroensuk J, Atichartpongkul S, Chittrakanwong J, Chooyoung K, Srimahaeak T, et al. TrmB, a tRNA m7G46 methyltransferase, plays a role in hydrogen peroxide resistance and positively modulates the translation of katA and katB mRNAs in pseudomonas aeruginosa. Nucleic Acids Res. 2019;47:9271–81.
Malbec L, Zhang T, Chen YS, Zhang Y, Sun BF, Shi BY, et al. Dynamic methylome of internal mRNA N7-methylguanosine and its regulatory role in translation. Cell Res. 2019;29:927–41.
Ramanathan A, Robb GB, Chan SH. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–26.
Cai M, Yang C, Wang Z. N7-methylguanosine modification: from regulatory roles to therapeutic implications in cancer. Am J Cancer Res. 2023;13:1640–55.
Ruiz-Arroyo VM, Raj R, Babu K, Onolbaatar O, Roberts PH, Nam Y. Structures and mechanisms of tRNA methylation by METTL1-WDR4. Nature. 2023;613:383–90.
Li J, Wang L, Hahn Q, Nowak RP, Viennet T, Orellana EA, et al. Structural basis of regulated m7G tRNA modification by METTL1-WDR4. Nature. 2023;613:391–7.
Cartlidge RA, Knebel A, Peggie M, Alexandrov A, Phizicky EM, Cohen P. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J. 2005;24:1696–705.
Lin S, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m7G tRNA methylome is required for normal mrna translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244–55.e5.
Kaneko S, Miyoshi K, Tomuro K, Terauchi M, Tanaka R, Kondo S, et al. Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster. Nat Commun. 2024;15:8147.
Fu Y, Jiang F, Zhang X, Pan Y, Xu R, Liang X, et al. Perturbation of METTL1-mediated tRNA N7- methylguanosine modification induces senescence and aging. Nat Commun. 2024;15:5713.
Li Q, Jiang S, Lei K, Han H, Chen Y, Lin W, et al. Metabolic rewiring during bone development underlies tRNA m7G-associated primordial dwarfism. J Clin Investig. 2024;134:e177220.
Chen J, Zhou Q, Li S, Ling R, Zhao Y, Chen D, et al. Metabolic reprogramming driven by METTL1-mediated tRNA m7G modification promotes acquired anlotinib resistance in oral squamous cell carcinoma. Transl Res. 2024;268:28–39.
Deng Y, Zhou Z, Ji W, Lin S, Wang M. METTL1-mediated m7G methylation maintains pluripotency in human stem cells and limits mesoderm differentiation and vascular development. Stem Cell Res Ther. 2020;11:306.
Zeng Z, Zhang X, Jiang CQ, Zhang YG, Wu X, Li J, et al. Identifying novel therapeutic targets in gastric cancer using genome-wide CRISPR-Cas9 screening. Oncogene. 2022;41:2069–78.
Du D, Zhou M, Ju C, Yin J, Wang C, Xu X, et al. METTL1-mediated tRNA m7G methylation and translational dysfunction restricts breast cancer tumorigenesis by fueling cell cycle blockade. J Exp Clin Cancer Res. 2024;43:154.
Yang T, Chen C, Wang F, Yue L. N7-methylguanosine (m7G) modification in breast cancer: clinical significances and molecular mechanisms. Cancer Cell Int. 2025;25:303.
Chen R, Zhou J, Liu L, Mao XL, Zhou X, Xie W. Crystal structure of human METTL6, the m3C methyltransferase. Commun Biol. 2021;4:1361.
Ignatova VV, Kaiser S, Ho JSY, Bing X, Stolz P, Tan YX, et al. METTL6 is a tRNA m3C methyltransferase that regulates pluripotency and tumor cell growth. Sci Adv. 2020;6:eaaz4551.
Throll P, G Dolce L, Rico-Lastres P, Arnold K, Tengo L, Basu S, et al. Structural basis of tRNA recognition by the m3C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase. Nat Struct Mol Biol. 2024;31:1614–24.
Cui J, Sendinc E, Liu Q, Kim S, Fang JY, Gregory RI. m3C32 tRNA modification controls serine codon-biased mRNA translation, cell cycle, and DNA-damage response. Nat Commun. 2024;15:5775.
Tan XL, Moyer AM, Fridley BL, Schaid DJ, Niu N, Batzler AJ, et al. Genetic variation predicting cisplatin cytotoxicity associated with overall survival in lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res. 2011;17:5801–11.
Bolatkan A, Asada K, Kaneko S, Suvarna K, Ikawa N, Machino H, et al. Downregulation of METTL6 mitigates cell progression, migration, invasion and adhesion in hepatocellular carcinoma by inhibiting cell adhesion molecules. Int J Oncol. 2022;60:4.
Zhou Y, Li X, Long G, Tao Y, Zhou L, Tang J. Identification and validation of a tyrosine metabolism-related prognostic prediction model and characterization of the tumor microenvironment infiltration in hepatocellular carcinoma. Front Immunol. 2022;13:994259.
Li YH, Zhang G, Cui Q. PPUS: a web server to predict PUS-specific pseudouridine sites. Bioinformatics. 2015;31:3362–4.
Nagato Y, Tomikawa C, Yamaji H, Soma A, Takai K. Intron-dependent or independent pseudouridylation of precursor tRNA containing atypical introns in cyanidioschyzon merolae. Int J Mol Sci. 2022;23:12058.
Shi D, Wang B, Li H, Lian Y, Ma Q, Liu T, et al. Pseudouridine synthase 1 regulates erythropoiesis via transfer RNAs pseudouridylation and cytoplasmic translation. iScience. 2024;27:109265.
Wang B, Shi D, Yang S, Lian Y, Li H, Cao M, et al. Mitochondrial tRNA pseudouridylation governs erythropoiesis. Blood. 2024;144:657–71.
Tan Y, Wang Z, Wang Y, Tian X, Huang Y, Wu G, et al. Multi-omics analysis reveals PUS1 triggered malignancy and correlated with immune infiltrates in NSCLC. Aging. 2023;15:12136–54.
Fang Z, Shen HY, Xu Q, Zhou HL, Li L, Yang SY, et al. PUS1 is a novel biomarker for predicting poor outcomes and triple-negative status in breast cancer. Front Oncol. 2022;12:1030571.
Li L, Zhu C, Xu S, Xu Q, Xu D, Gan S, et al. PUS1 is a novel biomarker for evaluating malignancy of human renal cell carcinoma. Aging. 2023;15:5215–27.
Lan C, Huang X, Liao X, Zhou X, Peng K, Wei Y, et al. PUS1 may be a potential prognostic biomarker and therapeutic target for hepatocellular carcinoma. Pharmgenomics Pers Med. 2023;16:337–55.
Zhang Q, Bao X, Cui M, Wang C, Ji J, Jing J, et al. Identification and validation of key biomarkers based on RNA methylation genes in sepsis. Front Immunol. 2023;14:1231898.
Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet. 2004;74:1303–8.
Awai, Kimura T, Tomikawa S, Ochi C, Ihsanawati null A, Bessho Y, et al. Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J Biol Chem. 2009;284:20467–78.
Porat J, Vakiloroayaei A, Remnant BM, Talebi M, Cargill T, Bayfield MA. Crosstalk between the tRNA methyltransferase Trm1 and RNA chaperone La influences eukaryotic tRNA maturation. J Biol Chem. 2023;299:105326.
Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, et al. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–76.
Constantinesco F, Motorin Y, Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J Mol Biol. 1999;291:375–92.
Constantinesco F, Benachenhou N, Motorin Y, Grosjean H. The tRNA(guanine-26,N2-N2) methyltransferase (Trm1) from the hyperthermophilic archaeon Pyrococcus furiosus: cloning, sequencing of the gene and its expression in Escherichia coli. Nucleic Acids Res. 1998;26:3753–61.
Vauti F, Goller T, Beine R, Becker L, Klopstock T, Hölter SM, et al. The mouse Trm1-like gene is expressed in neural tissues and plays a role in motor coordination and exploratory behaviour. Gene. 2007;389:174–85.
Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol. 2017;37:e00214-17.
Zhang K, Manning AC, Lentini JM, Howard J, Dalwigk F, Maroofian R, et al. Human TRMT1 and TRMT1L paralogs ensure the proper modification state, stability, and function of tRNAs. Cell Rep. 2025;44:115092.
Moon HJ, Redman KL. Trm4 and Nsun2 RNA:m5C methyltransferases form metabolite-dependent, covalent adducts with previously methylated RNA. Biochemistry. 2014;53:7132–44.
Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006;34:6034–43.
Kuratani M, Hirano M, Goto-Ito S, Itoh Y, Hikida Y, Nishimoto M, et al. Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin. J Mol Biol. 2010;401:323–33.
Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun. 2012;3:937.
Görlitz K, Bessler L, Helm M, Schaffrath R, Klassen R. Fluoropyrimidines trigger decay of hypomodified tRNA in yeast. Nucleic Acids Res. 2024;52:5841–51.
Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5’-3’ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–80.
Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96.
King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–25.
Li P, Huang D. NSUN2-mediated RNA methylation: molecular mechanisms and clinical relevance in cancer. Cell Signal. 2024;123:111375.
Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017;8:112–24.
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39.
Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–63.
Blaze J, Navickas A, Phillips HL, Heissel S, Plaza-Jennings A, Miglani S, et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior. Nat Commun. 2021;12:4913.
Okamoto M, Hirata S, Sato S, Koga S, Fujii M, Qi G, et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–71.
Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–70.
Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet. 2015;11:e1005706.
Begley U, Sosa MS, Avivar-Valderas A, Patil A, Endres L, Estrada Y, et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol Med. 2013;5:366–83.
Witzenberger M, Burczyk S, Settele D, Mayer W, Welp LM, Heiss M. Human TRMT2A methylates tRNA and contributes to translation fidelity. Nucleic Acids Res. 2023;51:8691–710.
Chang YH, Nishimura S, Oishi H, Kelly VP, Kuno A, Takahashi S. TRMT2A is a novel cell cycle regulator that suppresses cell proliferation. Biochem Biophys Res Commun. 2019;508:410–5.
Margreiter MA, Witzenberger M, Wasser Y, Davydova E, Janowski R, Metz J, et al. Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death. Comput Struct Biotechnol J. 2022;20:443–58.
Vilardo E, Amman F, Toth U, Kotter A, Helm M, Rossmanith W. Functional characterization of the human tRNA methyltransferases TRMT10A and TRMT10B. Nucleic Acids Res. 2020;48:6157–69.
Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith WA. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–93.
Ontiveros RJ, Shen H, Stoute J, Yanas A, Cui Y, Zhang Y, et al. Coordination of mRNA and tRNA methylations by TRMT10A. Proc Natl Acad Sci USA. 2020;117:7782–91.
Cosentino C, Toivonen S, Diaz Villamil E, Atta M, Ravanat JL, Demine S, et al. Pancreatic β-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46:10302–18.
Tresky R, Miyamoto Y, Nagayoshi Y, Yabuki Y, Araki K, Takahashi Y, et al. TRMT10A dysfunction perturbs codon translation of initiator methionine and glutamine and impairs brain functions in mice. Nucleic Acids Res. 2024;52:9230–46.
Lin H, Zhou X, Chen X, Huang K, Wu W, Fu J, et al. tRNA methyltransferase 10 homologue A (TRMT10A) mutation in a Chinese patient with diabetes, insulin resistance, intellectual deficiency and microcephaly. BMJ Open Diabetes Res Care. 2020;8:e001601.
Stern E, Vivante A, Barel O, Levy-Shraga Y. TRMT10A mutation in a child with diabetes, short stature, microcephaly and hypoplastic kidneys. J Clin Res Pediatr Endocrinol. 2022;14:227–32.
Samhani C, Guerci B, Larose C. Discovery of a TRMT10A mutation in a case of atypical diabetes: case report. Diabetes Metab. 2024;50:101572.
Igoillo-Esteve M, Genin A, Lambert N, Désir J, Pirson I, Abdulkarim B, et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013;9:e1003888.
Oerum S, Roovers M, Leichsenring M, Acquaviva-Bourdain C, Beermann F, Gemperle-Britschgi C, et al. Novel patient missense mutations in the HSD17B10 gene affect dehydrogenase and mitochondrial tRNA modification functions of the encoded protein. Biochim Biophys Acta Mol Basis Dis. 2017;1863:3294–302.
Reinhard L, Sridhara S, Hällberg BM. The MRPP1/MRPP2 complex is a tRNA-maturation platform in human mitochondria. Nucleic Acids Res. 2017;45:12469–80.
Bhatta A, Kuhle B, Yu RD, Spanaus L, Ditter K, Bohnsack KE, et al. Molecular basis of human nuclear and mitochondrial tRNA 3’ processing. Nat Struct Mol Biol. 2025;32:613–24.
Valentín Gesé G, Hällberg BM. Structural basis of 3'-tRNA maturation by the human mitochondrial RNase Z complex. EMBO J. 2024;43:6573–90.
Metodiev MD, Thompson K, Alston CL, Morris AAM, He L, Assouline Z, et al. Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies. Am J Hum Genet. 2016;98:993–1000.
Zhao Q, Li X, Wu J, Zhang R, Chen S, Cai D, et al. TRMT10C-mediated m7G modification of circFAM126A inhibits lung cancer growth by regulating cellular glycolysis. Cell Biol Toxicol. 2024;40:78.
Li L, Tan H, Zhou J, Hu F. Predicting response of immunotherapy and targeted therapy and prognosis characteristics for renal clear cell carcinoma based on m1A methylation regulators. Sci Rep. 2023;13:12645.
Liu Y, Zhang S, Gao X, Ru Y, Gu X, Hu X. Research progress of N1-methyladenosine RNA modification in cancer. Cell Commun Signal. 2024;22:79.
Li D, Li K, Zhang W, Yang KW, Mu DA, Jiang GJ, et al. The m6A/m5C/m1A regulated gene signature predicts the prognosis and correlates with the immune status of hepatocellular carcinoma. Front Immunol. 2022;13:918140.
Liu Y, Zhu J, Wang X, Zhang W, Li Y, Yang Z, et al. TRMT10C gene polymorphisms confer hepatoblastoma susceptibility: evidence from a seven-center case-control study. J Cancer. 2024;15:5396–402.
Oerum S, Dégut C, Barraud P, Tisné C. m1A post-transcriptional modification in tRNAs. Biomolecules. 2017;7:20.
Rodriguez V, Chen Y, Elkahloun A, Dutra A, Pak E, Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer. 2007;46:694–707.
Wang K, Zheng M, Ren Y. Overexpression of TRMT12 may independently predict poor overall survival in patients with head and neck squamous cell carcinoma. Onco Targets Ther. 2019;12:7269–79.
Simpson HM, Khan RZ, Song C, Sharma D, Sadashivaiah K, Furusawa A, et al. Concurrent mutations in ATM and genes associated with common γ chain signaling in peripheral T cell lymphoma. PLoS ONE. 2015;10:e0141906.
Pajdzik K, Lyu R, Dou X, Ye C, Zhang LS, Dai Q, et al. Chemical manipulation of m1A mediates its detection in human tRNA. RNA. 2024;30:548–59.
Liu Y, Zhou J, Li X, Zhang X, Shi J, Wang X, et al. tRNA-m1A modification promotes T cell expansion via efficient MYC protein synthesis. Nat Immunol. 2022;23:1433–44.
Zuo H, Wu A, Wang M, Hong L, Wang H. tRNA m1A modification regulate HSC maintenance and self-renewal via mTORC1 signaling. Nat Commun. 2024;15:5706.
He H, Wang Y, Zhang X, Li X, Liu C, Yan D, et al. Age-related noncanonical TRMT6-TRMT61A signaling impairs hematopoietic stem cells. Nat Aging. 2024;4:213–30.
Tao EW, Wang Y, Tan J, Chen Y, Sun TY, Hao Y, et al. TRMT6-mediated tRNA m1A modification acts as a translational checkpoint of histone synthesis and facilitates colorectal cancer progression. Nat Cancer. 2025;6:1458–76.
Sekar S, McDonald J, Cuyugan L, Aldrich J, Kurdoglu A, Adkins J, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36:583–91.
Zeng D, Zhu J, Li J, Liao F, Yang Z, Li Y, et al. TRMT61B rs4563180 G>C variant reduces hepatoblastoma risk: a case-control study of seven medical centers. Aging (Albany NY). 2023;15:7583–92.
Martín A, Epifano C, Vilaplana-Marti B, Hernández I, Macías RIR, Martínez-Ramírez Á, et al. Mitochondrial RNA methyltransferase TRMT61B is a new, potential biomarker and therapeutic target for highly aneuploid cancers. Cell Death Differ. 2023;30:37–53.
Ali AT, Idaghdour Y, Hodgkinson A. Analysis of mitochondrial m1A/G RNA modification reveals links to nuclear genetic variants and associated disease processes. Commun Biol. 2020;3:147.
Couch FJ, Kuchenbaecker KB, Michailidou K, Mendoza-Fandino GA, Nord S, Lilyquist J. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nat Commun. 2016;7:11375.
Hagervall TG, Pomerantz SC, McCloskey JA. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J Mol Biol. 1998;284:33–42.
Zeharia A, Shaag A, Pappo O, Mager-Heckel AM, Saada A, Beinat M, et al. Acute infantile liver failure due to mutations in the TRMU gene. Am J Hum Genet. 2009;85:401–7.
Vogel GF, Mozer-Glassberg Y, Landau YE, Schlieben LD, Prokisch H, Feichtinger RG, et al. Genotypic and phenotypic spectrum of infantile liver failure due to pathogenic TRMU variants. Genet Med. 2023;25:100828.
Meng F, Cang X, Peng Y, Li R, Zhang Z, Li F, et al. Biochemical evidence for a nuclear modifier allele (A10S) in TRMU (Methylaminomethyl-2-thiouridylate-methyltransferase) related to mitochondrial tRNA modification in the phenotypic manifestation of deafness-associated 12S rRNA mutation. J Biol Chem. 2017;292:2881–92.
Guan MX, Yan Q, Li X, Bykhovskaya Y, Gallo-Teran J, Hajek P, et al. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am J Hum Genet. 2006;79:291–302.
Zhang Q, Zhang L, Chen D, He X, Yao S, Zhang Z, et al. Deletion of Mtu1 (Trmu) in zebrafish revealed the essential role of tRNA modification in mitochondrial biogenesis and hearing function. Nucleic Acids Res. 2018;46:10930–45.
Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–54.
Umitsu M, Nishimasu H, Noma A, Suzuki T, Ishitani R, Nureki O. Structural basis of AdoMet-dependent aminocarboxypropyl transfer reaction catalyzed by tRNA-wybutosine synthesizing enzyme, TYW2. Proc Natl Acad Sci USA. 2009;106:15616–21.
de Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, et al. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol. 2010;27:2062–77.
Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47:2041–55.
Long T, Li J, Li H, Zhou M, Zhou XL, Liu RJ, et al. Sequence-specific and shape-selective RNA recognition by the human RNA 5-methylcytosine methyltransferase NSun6. J Biol Chem. 2016;291:24293–303.
Knight HM, Demirbugen Öz M, PerezGrovas-Saltijeral A. Dysregulation of RNA modification systems in clinical populations with neurocognitive disorders. Neural Regen Res. 2024;19:1256–61.
Mattioli F, Worpenberg L, Li CT, Ibrahim N, Naz S, Sharif S, et al. Biallelic variants in NSUN6 cause an autosomal recessive neurodevelopmental disorder. Genet Med. 2023;25:100900.
Yang R, Liang X, Wang H, Guo M, Shen H, Shi Y, et al. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine. 2021;63:103195.
Awah CU, Winter J, Mazdoom CM, Ogunwobi OO. NSUN6, an RNA methyltransferase of 5-mC controls glioblastoma response to temozolomide (TMZ) via NELFB and RPS6KB2 interaction. Cancer Biol Ther. 2021;22:587–97.
Yu G, Bao J, Zhan M, Wang J, Li X, Gu X, et al. Comprehensive analysis of m5C methylation regulatory genes and tumor microenvironment in prostate cancer. Front Immunol. 2022;13:914577.
Lu Z, Liu B, Kong D, Zhou X, Pei D, Liu D. NSUN6 regulates NM23-H1 expression in an m5C manner to affect epithelial-mesenchymal transition in lung cancer. Med Princ Pr. 2024;33:56–65.
Chen C, Huang B, Eliasson M, Rydén P, Byström AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7:e1002258.
Chen D, Nemazanyy I, Peulen O, Shostak K, Xu X, Tang SC, et al. Elp3-mediated codon-dependent translation promotes mTORC2 activation and regulates macrophage polarization. EMBO J. 2022;41:e109353.
Wathieu C, Lavergne A, Xu X, Rolot M, Nemazanyy I, Shostak K, et al. Loss of Elp3 blocks intestinal tuft cell differentiation via an mTORC1-Atf4 axis. EMBO J. 2024;43:3916–47.
Russo A, Forest C, Leone GJ, Iascone M, Tenconi R, Maffei M, et al. ELP2 compound heterozygous variants associated with cortico-cerebellar atrophy, nodular heterotopia and epilepsy: phenotype expansion and review of the literature. Eur J Med Genet. 2021;64:104361.
Abbassi NEH, Biela A, Glatt S, Lin TY. How elongator acetylates tRNA bases. Int J Mol Sci. 2020;21:8209.
Endres L, Begley U, Clark R, Gu C, Dziergowska A, Małkiewicz A, et al. Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage. PLoS ONE. 2015;10:e0131335.
Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49.
Singhal R, Mitta SR, Das NK, Kerk SA, Sajjakulnukit P, Solanki S, et al. HIF-2α activation potentiates oxidative cell death in colorectal cancers by increasing cellular iron. J Clin Investig. 2021;131:e143691.
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72.
Schwartz AJ, Goyert JW, Solanki S, Kerk SA, Chen B, Castillo C, et al. Hepcidin sequesters iron to sustain nucleotide metabolism and mitochondrial function in colorectal cancer epithelial cells. Nat Metab. 2021;3:969–82.
DeAngelo SL, Zhao L, Dziechciarz S, Shin M, Solanki S, Balia A, et al. Recharacterization of the Tumor Suppressive Mechanism of RSL3 Identifies the Selenoproteome as a Druggable Pathway in Colorectal Cancer. Cancer Res. 2025;85:2788–2804.
Schwickert M, Fischer TR, Zimmermann RA, Hoba SN, Meidner JL, Weber M, et al. Discovery of inhibitors of DNA methyltransferase 2, an epitranscriptomic modulator and potential target for cancer treatment. J Med Chem. 2022;65:9750–88.
Elhardt W, Shanmugam R, Jurkowski TP, Jeltsch A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015;112:66–72.
Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–64.
Lai J, Chen L, Li Q, Zhao G, Li X, Guo D, et al. tRNA methyltransferase DNMT2 promotes hepatocellular carcinoma progression and enhances bortezomib resistance through inhibiting TNFSF10. Cell Signal. 2025;127:111533.
Adamczyk-Grochala J, Bloniarz D, Zielinska K, Lewinska A, Wnuk M. DNMT2/TRDMT1 gene knockout compromises doxorubicin-induced unfolded protein response and sensitizes cancer cells to ER stress-induced apoptosis. Apoptosis: Int J Program Cell Death. 2023;28:166–85.
Wikman H, Nymark P, Väyrynen A, Jarmalaite S, Kallioniemi A, Salmenkivi K, et al. CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes Chromosomes Cancer. 2005;42:193–9.
Orellana EA, Liu Q, Yankova E, Pirouz M, De Braekeleer E, Zhang W, et al. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol Cell. 2021;81:3323–38.e14.
Katsara O, Schneider RJ. m7G tRNA modification reveals new secrets in the translational regulation of cancer development. Mol Cell. 2021;81:3243–5.
Zhu S, Wu Y, Zhang X, Peng S, Xiao H, Chen S, et al. Targeting N7-methylguanosine tRNA modification blocks hepatocellular carcinoma metastasis after insufficient radiofrequency ablation. Mol Ther. 2023;31:1596–614.
Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu H, et al. METTL1 promotes hepatocarcinogenesis via m7 G tRNA modification-dependent translation control. Clin Transl Med. 2021;11:e661.
Liao J, Yi Y, Yue X, Wu X, Zhu M, Chen Y, et al. Methyltransferase 1 is required for nonhomologous end-joining repair and renders hepatocellular carcinoma resistant to radiotherapy. Hepatology. 2023;77:1896–910.
Huang M, Long J, Yao Z, Zhao Y, Zhao Y, Liao J, et al. METTL1-Mediated m7G tRNA modification promotes lenvatinib resistance in hepatocellular carcinoma. Cancer Res. 2023;83:89–102.
Wang Z, Yu P, Zou Y, Ma J, Han H, Wei W. METTL1/WDR4-mediated tRNA m7G modification and mRNA translation control promote oncogenesis and doxorubicin resistance. Oncogene. 2023;42:1900–12.
Li T, Chen Z, Wang Z, Lu J, Chen D. Combined signature of N7-methylguanosine regulators with their related genes and the tumor microenvironment: a prognostic and therapeutic biomarker for breast cancer. Front Immunol. 2023;14:1260195.
Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R, et al. N7-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022;13:1478.
Dai Z, Liu H, Liao J, Huang C, Ren X, Zhu W, et al. N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol Cell. 2021;81:3339–55.e8.
Liu H, Zeng X, Ren X, Zhang Y, Huang M, Tan L, et al. Targeting tumour-intrinsic N7-methylguanosine tRNA modification inhibits MDSC recruitment and improves anti-PD-1 efficacy. Gut. 2023;72:1555–67.
García-Vílchez R, Añazco-Guenkova AM, Dietmann S, López J, Morón-Calvente V, D’Ambrosi S, et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. 2023;22:119.
Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X, et al. METTL1-m7 G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. 2021;11:e675.
Li W, Xie R, Chen H, Lin J, Zhong M, Zhang J, et al. METTL1-mediated m7G tRNA modification drives papillary thyroid cancer progression and metastasis by regulating the codon-specific translation of TNF-α. Cell Death Dis. 2025;16:378.
Huang Y, Ma J, Yang C, Wei P, Yang M, Han H, et al. METTL1 promotes neuroblastoma development through m7G tRNA modification and selective oncogenic gene translation. Biomark Res. 2022;10:68.
Ma J, Han H, Huang Y, Yang C, Zheng S, Cai T, et al. METTL1/WDR4-mediated m7G tRNA modifications and m7G codon usage promote mRNA translation and lung cancer progression. Mol Ther. 2021;29:3422–35.
Zhao P, Xia L, Chen D, Xu W, Guo H, Xu Y, et al. METTL1 mediated tRNA m7G modification promotes leukaemogenesis of AML via tRNA regulated translational control. Exp Hematol Oncol. 2024;13:8.
Chen B, Jiang W, Huang Y, Zhang J, Yu P, Wu L, et al. N7-methylguanosine tRNA modification promotes tumorigenesis and chemoresistance through WNT/β-catenin pathway in nasopharyngeal carcinoma. Oncogene. 2022;41:2239–53.
Xu X, Huang Z, Han H, Yu Z, Ye L, Zhao Z, et al. N7-methylguanosine tRNA modification promotes gastric cancer progression by activating SDHAF4-dependent mitochondrial oxidative phosphorylation. Cancer Lett. 2025;615:217566.
Rosselló-Tortella M, Llinàs-Arias P, Sakaguchi Y, Miyauchi K, Davalos V, Setien F, et al. Epigenetic loss of the transfer RNA-modifying enzyme TYW2 induces ribosome frameshifts in colon cancer. Proc Natl Acad Sci USA. 2020;117:20785–93.
Pan Y, Yan TM, Wang JR, Jiang ZH. The nature of the modification at position 37 of tRNAPhe correlates with acquired taxol resistance. Nucleic Acids Res. 2021;49:38–52.
Li J, Zhang H, Wang H. N1-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J. 2022;20:6578–85.
Li S, Feng T, Liu Y, Yang Q, Song A, Wang S, et al. m1A inhibition fuels oncolytic virus-elicited antitumor immunity via downregulating MYC/PD-L1 signaling. Int J Oral Sci. 2024;16:36.
Monshaugen I, Luna L, Rhodes J, Kristiansen FIS, Lång A, Bøe SO, et al. Depletion of the m1A writer TRMT6/TRMT61A reduces proliferation and resistance against cellular stress in bladder cancer. Front Oncol. 2023;13:1334112.
Zaragoza-Huesca D, Garrido-Rodríguez P, Jiménez-Fonseca P, Martínez de Castro E, Sánchez-Cánovas M, Visa L, et al. Identification of thrombosis-related genes in patients with advanced gastric cancer: data from AGAMENON-SEOM registry. Biomedicines. 2022;10:148.
Yin JJ, Song YL, Guo YF, Dai YH, Chang Q, Wang T, et al. Transcriptome-wide 1-methyladenosine functional profiling of messenger RNA and long non-coding RNA in bladder cancer. Front Genet. 2024;15:1333931.
Cai Z, Jiang Z, Li S, Mo S, Wang S, Liang M, et al. RNA modification Regulators’ Co-Expression Score (RMRCoeS) predicts biochemical recurrence and therapy response in prostate cancer: a multi-omics and experimental validation study. Int Immunopharmacol. 2024;139:112723.
Yao L, Cong R, Ji C, Zhou X, Luan J, Meng X, et al. RNA-binding proteins play an important role in the prognosis of patients with testicular germ cell tumor. Front Genet. 2021;12:610291.
Li P, Wang W, Zhou R, Ding Y, Li X. The m5 C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin Transl Med. 2023;13:e1466.
Yang JC, Risch E, Zhang M, Huang C, Huang H, Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981–90.
Lu L, Zhu G, Zeng H, Xu Q, Holzmann K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Investig. 2018;36:246–53.
Xiang S, Ma Y, Shen J, Zhao Y, Wu X, Li M, et al. m5C RNA methylation primarily affects the ErbB and PI3K-akt signaling pathways in gastrointestinal cancer. Front Mol Biosci. 2020;7:599340.
Zhang Y, Wang Y, Fan J, Zhu G, Lu L. Aspergillus fumigatus Elongator complex subunit 3 affects hyphal growth, adhesion and virulence through wobble uridine tRNA modification. PLoS Pathog. 2022;18:e1010976 .
Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med. 2016;213:2503–23.
Waszak SM, Robinson GW, Gudenas BL, Smith KS, Forget A, Kojic M, et al. Germline elongator mutations in Sonic Hedgehog medulloblastoma. Nature. 2020;580:396–401.
Xu S, Zhan M, Jiang C, He M, Yang L, Shen H, et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat Commun. 2019;10:5492.
Wang XW, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly JM, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–95.
Lee N, Kim D. Adapt or perish: efficient selenocysteine insertion is critical for metastasizing cancer cells. Cancer Res. 2025;85:410–2.
Meyer D, Kames J, Bar H, Komar AA, Alexaki A, Ibla J, et al. Distinct signatures of codon and codon pair usage in 32 primary tumor types in the novel database CancerCoCoPUTs for cancer-specific codon usage. Genome Med. 2021;13:122.
Zhang Z, Ye Y, Gong J, Ruan H, Liu CJ, Xiang Y, et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun Biol. 2018;1:234.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82273367, 82303383), the Excellent Youth Training Program of the First Affiliated Hospital of Fujian Medical University (2023FY-YXQN-2), the Natural Science Foundation of Fujian Province (2024J09033), the Fujian Provincial Health Technology Project (2024GGB07), Joint Funds for the Innovation of Science and Technology, Fujian Province (2024Y9193).
Author information
Authors and Affiliations
Contributions
XS and JC contributed to conception and design of the manuscript; WH and ZJ drafted the manuscript; WH and ZJ conducted table and image processing; JC critically reviewed and edited the manuscript; XS given the final approval of the version to be published. All authors have read and approved the final version of the manuscript for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr Francesca Bernassola
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, H., Zhang, J., Jiang, C. et al. Emerging roles of tRNA modification-mediated codon-specific translational reprogramming in cancer biology. Cell Death Dis 17, 4 (2026). https://doi.org/10.1038/s41419-025-08234-3
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41419-025-08234-3