Abstract
There is increasing evidence that dysregulated long non-coding RNA (lncRNA) is implicated in tumorigenesis and progression. We aim to explore the role of lncRNA MIR600HG in glycometabolism and cisplatin (DDP) resistance of oral squamous cell carcinoma (OSCC) cells via regulating microRNA-125a-5p (miR-125a-5p) and RING finger 44 (RNF44). Expression of MIR600HG, miR-125a-5p, and RNF44 in OSCC clinical samples, cell lines, and DDP-resistant OSCC cells (SCC-9/DDP) was determined. In SCC-9 cells, proliferation, IC50 value of DDP, migration, invasion, and apoptosis were detected; in SCC-9/DDP cells, proliferation, IC50 value of DDP, apoptosis, glucose consumption, and production of lactic acid and ATP were evaluated. The interaction of MR600HG, miR-125a-5p, and RNF44 was verified. MIR600HG and RNF44 were upregulated while miR-125a-5p was downregulated in OSCC tissues and cell lines, and also in SCC-9/DDP cells. In SCC-9 cells, MIR600HG overexpression improved cell growth, metastasis, and inhibited cell susceptibility to DDP; in SCC-9/DDP cells, silencing of MIR600HG promoted apoptosis, improved DDP sensitivity, and inhibited cell glycolysis. Downregulation of miR-125a-5p showed the opposite effect to downregulation of MIR600HG. MIR600HG bound to miR-125a-5p and miR-125a-5p targeted RNF44. Downregulation of miR-125a-5p reversed the improvement of DDP sensitivity and the inhibition of cell glycolysis by downregulated MIR600HG on SCC-9/DDP cells. Downregulating RNF44 reversed the promotion of DDP resistance and cell glycolysis of SCC-9/DDP cells mediated by downregulation of miR-125a-5p. Collectively, our study addresses that MIR600HG downregulation elevates miR-125a-5p and reduces RNF44 expression, thereby improving DDP sensitivity and inhibiting glycolysis in DDP-resistant OSCC cells.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) is a metastatic and aggressive malignant oral malignancy with an increasing incidence [1]. Currently, although progresses have been made in the administration and diagnosis of OSCC, the survival rate of OSCC patients is still disappointing due to the high recurrence rate, later diagnosis at advanced stages, and a lack of early diagnostic biomarkers [2]. As a gene-related disease, the detailed molecular mechanism of OSCC tumorigenesis still remains to be further explored. Therefore, it is of importance to investigate diagnostic strategies and find therapeutic targets to improve the treatment of OSCC. Cisplatin (DDP) is an effective first-line chemotherapy drug for OSCC treatment, while its therapeutic effect often fails as a result of the drug resistance [3]. Tumor cells can take up more glucose, and glucose undergo aerobic glycolytic synthesis to generate more intermediate substances and eventually to generate lactic acid. These intermediate products such as NADPH and 5-phosphoribose are often the raw materials for the synthesis of nucleic acids and proteins [4], and these products can promote the development of tumor resistance [5]. Related reports have shown that tumor cells have highly active glucose metabolism levels which is related to chemotherapeutic drug resistance [6,7,8]. Thus, it is vital to identify the underlying mechanisms of chemoresistance to promote DDP sensitivity.
Long non-coding RNAs (lncRNAs) are important regulatory transcripts lacking protein-coding functions [9]. Previous studies have reported that dysregulated lncRNAs are involved in the occurrence and development of various tumors, including OSCC [10, 11]. MIR600HG has been reported to suppress metastasis and enhance oxaliplatin chemosensitivity in colorectal cancer (CRC) [12]. Moreover, it has been demonstrated that MIR600HG is related to autophagy in OSCC [13] and acts as a prognostic lncRNA in pancreatic ductal adenocarcinoma (PDAC) [14]. We found through the bioinformatic analysis that there existed binding sites between MIR600HG and microRNA (miR)-125a-5p. miRs are small non-coding RNA molecules that modulate the genetic expression at the post-transcriptional level [15]. Importantly, miR-125a-5p has been revealed to affect the chemosensitivity to DDP in osteosarcoma cells [16], and it has also been unraveled that miR-125a-5p participates in the progression of head and neck squamous cell carcinoma [17] and esophageal squamous cell carcinoma [18]. Nevertheless, the role of miR-125a-5p in OSCC remains unknown. Through the bioinformatic analysis, we also found the binding sites between miR-125a-5p and RING finger 44 (RNF44). RNF44 is a novel E3 ligase that has been reported to be responsible for AMP-activated protein kinase-α1 degradation [19]. Its role in OSCC is still unknown.
We speculated that MIR600HG may sponge miR-125a-5p to affect DDP resistance in OSCC via regulating RNF44.
Results
MIR600HG is upregulated in OSCC
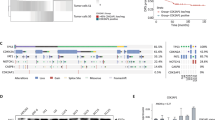
To investigate whether MIR600HG participates in OSCC progression, we detected the expression of MIR600HG in OSCC tissues and normal tissues from 85 patients using quantitative reverse trabscription PCR (RT-qPCR). It was found that versus the normal tissues, MIR600HG was upregulated in OSCC tissues (Fig. 1A). MIR600HG expression in OSCC cell lines and normal human oral keratinocyte (NHOK) was assessed. The results indicated that it was upregulated in OSCC cell lines as well, compared to NHOK (Fig. 1B). MIR600HG expression in SCC-9/DDP cells and the DDP sensitivity were assessed. As expected, MIR600HG expression was increased (Fig. 1C) and DDP sensitivity was reduced in SCC-9/DDP cells (Fig. 1D). These data suggested that MIR600HG may be related to DDP resistance of OSCC cells.
A MIR600HG expression in clinical samples was assessed using RT-qPCR (n = 85); B MIR600HG expression in NHOK and OSCC cell lines was assessed using RT-qPCR; C MIR600HG expression in SCC-9 and SCC-9/DDP cells was assessed using RT-qPCR; D IC50 of SCC-9 and SCC-9/DDP cells was detected using MTT assay; N = 3; *P < 0.05; the results were exhibited as mean ± standard deviation; t-test or ANOVA was used to analyze data.
MIR600HG affects growth, DDP sensitivity, and glycometabolism of OSCC cells
MIR600HG was overexpressed through oe-MIR600HG transfection in SCC-9 cells while it was knocked down in SCC-9/DDP cells through si-MIR600HG transfection (Fig. 2A, B). After transfection, colony formation assay and MTT assay were performed and it was found that overexpression of MIR600HG induced cell growth (Fig. 2C, D) and DDP resistance of SCC-9 cells (Fig. 2E). Migration and invasion were determined using Transwell assay and the results showed that MIR600HG overexpression facilitated migration and invasion of SCC-9 cells (Fig. 2F). Moreover, flow cytometry was utilized to assess cell apoptosis and it was discovered that the apoptosis rate of SCC-9 cells was reduced when MIR600HG was overexpressed (Fig. 2G).
A MIR600HG expression in SCC-9 cells after transfection of oe-NC or oe-MIR600HG was assessed using RT-qPCR. B The knockdown effect after MIR600HG siRNA transfection was confirmed by RT-qPCR in SCC-9/DDP cells. C Colony formation ability of SCC-9 cells was determined using colony formation assay. D viability of SCC-9 cells was determined using MTT assay. E IC50 of SCC-9 cells was determined using MTT assay. F Migration and invasion of SCC-9 cells was detected using Transwell assay. G Apoptosis of SCC-9 cells was detected using flow cytometry. H Viability of SCC-9/DDP cells was determined using MTT assay. I IC50 of SCC-9/DDP cells was determined using MTT assay. J Apoptosis of SCC-9/DDP cells was detected using flow cytometry. K Glycometabolism of SCC-9/DDP cells was analyzed; N = 3; *P < 0.05; the data were expressed as mean ± standard deviation and t-test was used to analyze data.
For SCC-9/DDP cells, reduction of MIR600HG reduced cell viability and elevated apoptosis rate (Fig. 2H–J). Furthermore, metabolism of SCC-9/DDP cells was determined, including production of lactic acid and adenosine triphosphate (ATP), as well as glucose consumption. The results indicated that silencing of MIR600HG reduced glucose consumption and production of lactic acid and ATP (Fig. 2K). Thus, it was concluded that MIR600HG could affect growth, DDP sensitivity, and glycometabolism of OSCC cells.
MIR600HG inhibits miR-125a-5p expression
It was predicted using miRcode and RNA22 that there existed binding sites between MIR600HG and miR-125a-5p (Fig. 3A). We further confirmed using dual-luciferase reporter gene assay that the co-transfection of miR-125a-5p mimic and WT-MIR600HG suppressed the luciferase activity (Fig. 3B). MIR600HG was overexpressed in SCC-9 cells to assess its regulatory role in miR-125a-5p expression. It was found through RT-qPCR that miR-125a-5p expression was inhibited by MIR600HG overexpression (Fig. 3C). These data suggested that MIR600HG may mediate miR-125a-5p expression in OSCC.
A Binding sites between miR-125a-5p and MIR600HG were predicted using bioinformatic websites; B targeting relationship between miR-125a-5p and MIR600HG was confirmed using dual luciferase reporter gene assay; C miR-215a-5p expression in SCC-9 cells after transfection of oe-NC or oe-MIR600HG was assessed using RT-qPCR; D miR-125a-5p expression in clinical samples was assessed using RT-qPCR (n = 85); E miR-125a-5p expression in NHOK and OSCC cell lines was assessed using RT-qPCR; F miR-125a-5p expression in SCC-9 and SCC-9/DDP cells was assessed using RT-qPCR; G miR-215a-5p expression in SCC-9 cells after transfection of mimic NC or miR-125a-5p mimic was assessed using RT-qPCR; H colony formation ability of SCC-9 cells was determined using colony formation assay; I viability of SCC-9 cells was determined using MTT assay; J IC50 of SCC-9 cells was determined using MTT assay; K migration and invasion of SCC-9 cells was detected using Transwell assay; L apoptosis of SCC-9 cells was detected using flow cytometry; M miR-215a-5p expression in SCC-9/DDP cells after transfection of inhibitor NC or miR-125a-5p inhibitor was assessed using RT-qPCR; N viability of SCC-9/DDP cells was determined using MTT assay; O IC50 of SCC-9/DDP cells was determined using MTT assay; P apoptosis of SCC-9/DDP cells was detected using flow cytometry; Q glycometabolism of SCC-9/DDP cells was analyzed; N = 3; *P < 0.05; Data were expressed as mean ± standard deviation; t-test or ANOVA was used to analyze data.
We detected miR-125a-5p expression in OSCC clinical samples and cell lines, and found that it was downregulated in OSCC tissues (Fig. 3D) and cell lines (Fig. 3E), and also in DDP-resistant cells (Fig. 3F). MiR-125a-5p was upregulated in SCC-9 cells using miR-125a-5p mimic transfection (Fig. 3G), which subsequently reduced cell viability (Fig. 3H, I), promoted drug sensitivity (Fig. 3J), suppressed migration and invasion (Fig. 3K), and facilitated apoptosis of SCC-9 cells (Fig. 3L). The above data indicated that miR-125a-5p inhibits SCC-9 cell growth.
SCC-9/DDP cells were transfected with miR-125a-5p inhibitor, and the transfection efficacy was confirmed using RT-qPCR (Fig. 3M). Then, we found through a series of assays that inhibition of miR-125a-5p promoted cell viability (Fig. 3N), inhibited DDP sensitivity (Fig. 3O), repressed apoptosis (Fig. 3P), increased the production of lactic acid and ATP, and the amount of glucose consumption (Fig. 3Q).
Therefore, MIR600HG inhibits miR-125a-5p and miR-125a-5p affects the functions of DDP-resistant OSCC cells.
MIR600HG/miR-125a-5p axis regulates the proliferation, apoptosis, DDP sensitivity, and glycometabolism of DDP-resistant OSCC cells
To verify whether the functions of MIR600HG on DDP-resistant OSCC cells were correlated with miR-125a-5p, si-MIR600HG and miR-125a-5p inhibitor were transfected into SCC-9/DDP cells, and the transfection efficiency was determined using RT-qPCR (Fig. 4A). Then, experimental results suggested that inhibition of miR-125a-5p could reverse the effects of MIR600HG knockdown on the viability (Fig. 4B), DDP sensitivity (Fig. 4C), apoptosis (Fig. 4D, E), and glycometabolism (Fig. 4F) of SCC-9/DDP cells. These findings indicated that the functions of MIR600HG on OSCC cells are mediated by miR-125a-5p.
A miR-215a-5p expression in SCC-9/DDP cells after transfection of si-MIR600HG+miR-125a-5p inhibitor or si-MIR600HG+inhibitor NC was assessed using RT-qPCR; B viability of SCC-9/DDP cells was determined using MTT assay; C IC50 of SCC-9/DDP cells was determined using MTT assay; D, E apoptosis of SCC-9/DDP cells was detected using flow cytometry; F glycometabolism of SCC-9/DDP cells was analyzed; N = 3; *P < 0.05; data were expressed as mean ± standard deviation; and ANOVA was used to analyze data.
RNF44 is upregulated in OSCC, and RNF44 affects growth, DDP sensitivity, and glycometabolism of OSCC cells
RNF44 expression in OSCC clinical samples and cell lines was determined and we found that it was upregulated in OSCC tissues and cell lines (Fig. 5A, B), especially in SCC-9/DDP cells (Fig. 5C).
A RNF44 mRNA expression in clinical samples was assessed using RT-qPCR (n = 85); B RNF44 mRNA expression in NHOK and OSCC cell lines was assessed using RT-qPCR; C RNF44 mRNA expression in SCC-9 and SCC-9/DDP cells was assessed using RT-qPCR; D two effective si-RNF44 were designed to knockdown the expression of RNF44 and its interference efficiency was detected by RT-qPCR and western blot analysis in SCC-9 cells; E colony formation ability of SCC-9 cells was determined using colony formation assay after transfection of si-RNF44; F viability of SCC-9 cells was determined using MTT assay after transfection of si-RNF44; G IC50 of SCC-9 cells was determined using MTT assay after transfection of si-RNF44; H migration and invasion of SCC-9 cells was detected using Transwell assay after transfection of si-RNF44; I apoptosis of SCC-9 cells was detected using flow cytometry; J glycometabolism of SCC-9/DDP cells was analyzed after transfection of si-RNF44; N = 3; *P < 0.05; Data were expressed as mean ± standard deviation; t-test or ANOVA was used to analyze data.
To further explore the effect of RNF44 on OSCC cells, we first designed two si-RNAs specific to RNF44 (si-RNF44-1 and si-RNF44-2), and transfected them into SCC-9 cells and SCC-9/DDP cells. RT-qPCR and western blot showed that the mRNA and protein levels of RNF44 were effectively interfered by si-RNF44-1 and si-RNF44-2 in SCC-9 cells (Fig. 5D) and SCC-9/DDP cells (Supplementary Fig. 1A). Furthermore, a series of experiments indicated that silencing of RNF44 by si-RNF44-1 and si-RNF44-2 inhibited colony formation ability (Fig. 5E) and cell viability (Fig. 5F), enhanced DDP sensitivity (Fig. 5G), repressed migration and invasion (Fig. 5H), and increased apoptosis of SCC-9 cells (Fig. 5I). These findings suggested that RNF44 could affect the growth of SCC-9 cells.
It was also discovered that downregulation of RNF44 decreased cell viability (Supplementary Fig. 1B), increased DDP sensitivity (Supplementary Fig. 1C), enhanced apoptosis rate (Supplementary Fig. 1D), and restrained glycometabolism of SCC-9/DDP cells (Fig. 5J). These results showed that RNF44 is needed for the development of SCC-9/DDP cells.
MiR-125a-5p/RNF44 axis affects proliferation, apoptosis, DDP sensitivity, and glycometabolism of DDP-resistant OSCC cells
Binding sites between hsa-miR-125a-5p and RNF44 were found through Targetscan website (Fig. 6A) and their targeting relationship was confirmed using dual luciferase reporter gene assay (Fig. 6B). To further investigate the relationship between miR-125a-5p and RNF44, miR-125a-5p inhibitor and si-RNF44 were transfected into SCC-9/DDP cells, and the success of the transfection was confirmed using RT-qPCR (Fig. 6C). Moreover, the results of our experiments indicated that silencing of RNF44 could abolish the effects of miR-125a-5p inhibition on cell viability (Fig. 6D), DDP sensitivity (Fig. 6E), apoptosis (Fig. 6F), and glycometabolism of SCC-9/DDP cells (Fig. 6G). The above data implied that miR-125a-5p regulates RNF44 to affect DDP resistance in OSCC.
A Binding sites between miR-125a-5p and RNF44 were predicted using Targetscan website; B Targeting relationship between miR-125a-5p and RNF44 was confirmed using dual luciferase reporter gene assay; C RNF44 expression in SCC-9/DDP cells after transfection of miR-125a-5p inhibitor+si-NC or miR-125a-5p inhibitor+si-RNF44 was detected using RT-qPCR; D Viability of SCC-9/DDP cells was determined using MTT assay; E IC50 of SCC-9/DDP cells was determined using MTT assay; F apoptosis of SCC-9/DDP cells was detected using flow cytometry; G glycometabolism of SCC-9/DDP cells was analyzed; N = 3; *P < 0.05; Data were expressed as mean ± standard deviation; t-test or ANOVA was used to analyze data.
MIR600HG knockdown reduces the growth of DDP-resistant OSCC cells in vivo
The in vivo assay was performed to observe the role of MIR600HG in tumor growth. SCC-9/DDP cells that had been transfected with sh-MIR600HG were injected into the nude mice, and then the tumor growth on days 7, 14, 21, and 28 was assessed. The results suggested that DDP reduced the tumor weigh and volume, which were further inhibited by MIR600HG knockdown (Fig. 7A–C). Thus, we demonstrated that MIR600HG inhibition strengthens DDP sensitivity of OSCC cells in vivo.
Discussion
OSCC is the sixthh most prevalent malignancy globally. It occurs due to the alteration in gene expression affected by specific genetic characteristics and environmental conditions [20]. We found that the MIR600HG/miR-125a-5p/RNF44 axis regulated glycometabolism and DDP resistance of OSCC cells.
MIR600HG is a lncRNA that has been reported to be related to autophagy in OSCC [13]. Here we found that MIR600HG was upregulated in OSCC tissues and cells. Similar to our result, Song et al. [14] have demonstrated that MIR600HG acted as a prognostic target for predicting PDCA and was significantly associated with the overall survival of PDCA patients. In this study, it was found that in SCC-9 cells, MIR600HG overexpression promoted proliferation, migration, invasion and DDP resistance, and inhibited apoptosis of cells. In SCC-9/DDP cells, silencing of MIR600HG suppressed proliferation and glycometabolism, and enhanced apoptosis and DDP sensitivity of cells. These findings indicated an oncogenic role of MIR600HG in the development of OSCC. Yao et al. [12] have reported that MIR600HG suppressed metastasis and enhanced oxaliplatin chemosensitivity by targeting ALDH1A3 in CRC. Due to the limited literature, the accurate mechanism of MIR600HG needs further explorations in human cancers.
LncRNAs are known to regulate the biological function of miRNAs by acting as a competitive endogenous RNA (ceRNA) [21]. In the present study, the interaction between MIR600HG and miR-125a-5p was confirmed. Furthermore, our results suggested that miR-125a-5p was expressed at a low level in OSCC cell lines and tissues, as well as in DDP-resistant OSCC cells. In accordance with our finding, Cao et al. [22] have clarified that miR-125a-5p was downregulated in both cervical cancer tissues and cell lines, and miR-125a-5p levels shows a marked reduction in patients with prostate cancer in comparison to healthy controls [23]. Also, our results implied that miR-125a-5p elevation inhibited malignant phenotypes and DDP resistance of OSCC cells, and reduction of miR-125a-5p promoted proliferation and glycometabolism, and reduced apoptosis and DDP sensitivity of DDP-resistant OSCC cells. Consistently, it has been reported that miR-125a-5p inhibition attenuated ANRIL knockdown-induced chemosensitivity to DDP in osteosarcoma cells [16]. Moreover, a recent study has indicated that miR-125a-5p inhibited cell proliferation, cell cycle progression, and migration whereas promoted apoptosis of head and neck cancer cells [24]. It has also been revealed that miR-125a-5p reduced glycolysis to improve pulmonary arterial hypertension [25] and ameliorated hepatic glycolipid metabolism disorder in type 2 diabetes mellitus [26].
Furthermore, RNF44 was confirmed as a target gene of miR-125a-5p in this study, and RNF44 was found to be upregulated in OSCC. For OSCC cells, RNF44 silencing reduced the growth and enhanced DDP sensitivity; for DDP-resistant OSCC cells, RNF44 reduction could reverse the role of miR-125a-5p knockdown. As previously reported, RNF44 expression in BRAF inhibitors (BRAFi)-resistant melanoma (BR) cells was increased, and high levels of RNF44 also appeared in BR xenografts and melanoma tumor tissues from BR and BRAFi/MEK inhibitor-resistant melanoma patients [19].
Taken together, this research indicated that MIR600HG depletion reduces glycometabolism and DDP resistance of OSCC cells through acting as a ceRNA of miR-125a-5p to regulate RNF44. Our study may provide novel biomarkers for OSCC diagnosis and treatment. However, the effects of the MIR600HG/miR-125a-5p/RNF44 are scarcely studied; thus, more efforts are needed for further explorations.
Materials and methods
Ethics statement
Written informed consents were acquired from all patients before this study. The protocol of this study was confirmed by the Ethic Committee of School and Hospital of Stomatology, Cheeloo College of Medicine, Shandong University (ethical number: KY2015-020) and based on the ethical principles for medical research involving human subjects of the Helsinki Declaration. Animal experiments were strictly in accordance with the Guide to the Management and Use of Laboratory Animals issued by the National Institutes of Health. The protocol of animal experiments was approved by the Institutional Animal Care and Use Committee of School and Hospital of Stomatology, Cheeloo College of Medicine, Shandong University (ethical number: SYDW2015-322). The maximum diameter of the tumor allowed by the ethical permit was 1.5 cm.
Study subjects
Eighty-five cases of OSCC tissues and the matched normal tissues were harvested from OSCC patients accepted treatment in School and Hospital of Stomatology, Cheeloo College of Medicine, Shandong University. These tissues were immediately frozen in liquid nitrogen and then preserved at −80 °C. Patients’ information is detailed in Supplementary Table 1.
Cell culture
NHOK, OSCC cell lines (CAL-27, Tca8113, SCC-9, and TSCCA) and DDP-resistant OSCC cell line SCC-9/DDP (all from ATCC, VA, USA) were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (Invitrogen, CA, USA) [27].
Cell transfection and grouping
The synthesized full-length of human MIR600HG cDNA was amplified using RT-qPCR and molecularly cloned into pcDNA3.1 (−) eukaryotic expression vector. MiR-125a-5p mimic, miR-125a-5p inhibitor, mimic negative control (NC), and inhibitor NC were purchased from Shanghai GenePharma Co., Ltd (Shanghai, China). MIR600HG small-interfering (si) RNA (si-MIR600HG), RNF44 siRNA (si-RNF44), Scrambled/nontargeting siRNA were obtained from Guangzhou RiboBio Co., Ltd (Guangzhou, China). siRNA knockout procedure was performed based on the manufacturer’s instructions. siRNA sequence was not provided due to commercial factors.
The target plasmid (2 μg), siRNA, miRNA mimic or inhibitor (50 nM) was transfected into cells using Lipofectamine 3000 (Invitrogen) [22]. After the OSCC cells were transfected onto six-well plates for 48 h, the transfection efficiency was evaluated and subsequent experiments were performed.
SCC-9 cells were classified into the control pcDNA3.1 (oe-NC), pcDNA3.1-MIR600HG overexpression (oe-MIR600HG), mimic NC, miR-125a-5p mimic, si-con, and si-RNF44 groups.
SCC-9/DDP cells were divided into the si-NC, si-MIR600HG, inhibitor NC, miR-125a-5p inhibitor, si-NC + inhibitor NC, si-MIR600HG + inhibitor NC, si-NC + miR-125a-5p inhibitor, si-MIR600HG + miR-125a-5p inhibitor, si-con, si-RNF44, inhibitor NC + si-con, miR-125a-5p inhibitor + si-con, inhibitor NC + si-RNF44 and miR-125a-5p inhibitor + si-RNF44 groupss.
Colony formation assay
Proliferation and colony formation of transfected SCC-9 cells were measured by colony formation assay. At 48 h post-transfection, cells were seeded into six-well plates at a density of 400 cells/well, cultured for 15 days, and then were fixed and stained with crystal violet (Sigma-Aldrich, CA, USA) for 5 min. The colonies were counted under a microscope (Olympus, Tokyo, Japan) [28].
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
Transfected SCC-9 cells or DDP-resistant SCC-9 (SCC-9/DDP) cells were seeded into 96-well plates. At different times 0, 24, 48, and 72 h, respectively, cell viability was determined [29]. To observe drug sensitivity to DDP, transfected SCC-9 cells or SCC-9/DDP cells were incubated with DDP at concentrations ranging from 0 to 160 μmol/L for 24 h [27]. After treatment, 10 mL CCK-8 solution was added to each well, and the 96-well plate was incubated at 37 °C for 2 h. Optical density (OD450 nm) value was measured on a microplate reader (Thermo Fisher Scientific, IL, USA)
Flow cytometry
Transfected SCC-9 cells or SCC-9/DDP cells were seeded in a six-well plate at 2 × 105 cells/well. Cell apoptosis was determined using Annexin V-FITC/PI Apoptosis Detection kit (BD Biosciences, San Diego, CA, USA) and analyzed on a flow cytometer (FACSCalibur; BD Biosciences) [30].
Transwell assay
The migration and invasion of transfected SCC-9 cells were determined using Transwell chambers (Costar, NY, USA) as previously described [31]. Matrigel (BD Bioscience, CA, USA) was used in the invasion assay but not migration assay.
Metabolism parameters of glucose utilization and lactic acid production
The culture medium was collected after SCC-9/DDP cells were incubated for 48 h in the six-well plate. Using a Glucose Assay Kit (Sigma-Aldrich) and a L-Lactate Assay Kit (Eton Bioscience, CA, USA), the concentration of glucose and lactate in spent medium was quantitatively measured. Glucose consumption of SCC-9/DDP cells was calculated as the glucose concentration of the unused cell culture medium minus the glucose concentration in the culture medium [32].
ATP level evaluation
ATP level in cells was assessed using the luminometer (Promega, WI, USA) and CellTiter-Glo 2.0 Assay kits. The total protein in relative cell lysate was used to normalize the relative cellular ATP level [33].
Dual-luciferase reporter gene assay
The amplified DNA sequence of MIR600HG containing wild type (WT) or mutant (MUT) miR-125a-5p-binding sites were amplified by PCR and then cloned into a pmirGLO dual-luciferase plasmid (Promega, WI, USA) to produce MIR600HG-WT and MIR600HG-MUT reporter plasmids. SCC-9 cells were co-transfected with MIR600HG-WT or MIR600HG-MUT and miR-125a-5p mimic or its NC using Lipofectamine 3000 (Invitrogen). At 48 h post-transfection, dual-luciferase reporter assay was performed using the Dual-Glo® Luciferase Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity of the pmirGLO vector was measured to normalize Renilla luciferase activity. The targeting relationship between miR-125a-5p and RNF44 were verified in the same way [34, 35].
RNA immunoprecipitation (RIP) assay
RIP kits (Millipore, MA, USA) were used for this assay. In brief, 50 μL magnetic beads were suspended in 100 μL RIP buffer, and incubated with 5 μg anti-RNF44 (1:1000; Abcam, MA, USA). The bead-antibody complex was incubated with 100 μL lysis buffer at 4 °C overnight, and the purified RNA was analyzed using RT-qPCR [36].
Subcutaneous tumorigenesis in nude mice
Twenty-four BALB/c nude mice (Laboratory Animal Center of Shandong University, Shandong, China) were fed under specific pathogen-free conditions. SCC-9/DDP cells with sh-MIR600HG or sh-NC were suspended in 100 μL medium, and cells (2 × 105) were resuspended in 50 μL normal saline, added with 50 μL Matrigel Matrix and subcutaneously injected into the mouse right abdomen. Seven days after the injection, mice were injected with DDP (4 mg/kg) or phosphate-buffered saline (PBS) every 4 days. The tumor volume was measured at the day 7, 14, 21, and 28. (a × b2)/2 (a: tumor length; b: tumor width). The allowed maximum diameter of the tumors was 1.5 cm. After the experiment, mice were euthanized with their xenografts resected and weighed [37].
RT-qPCR
Total RNA in tissues and cells was extracted using Trizol kits (Invitrogen) and reversely transcribed into cDNA using reverse transcriptase (Epicentre, WI, USA) or the miS-cript Reverse Transcription Kit (Qiagen). cDNA amplification was performed using the SYBR Premix Ex Taq (Takara, Dalian, China) and the PCR was conducted using Applied Biosystems 7900HT PCR system with β-actin as the internal reference. Primers are shown in Supplementary Table 2 and data were analyzed using the 2–ΔΔCt method.
Western blot analysis
Total protein was extracted using extraction kits (KeyGen) with the concentration determined using bicinchoninic acid protein detection kits (Thermo Fisher). Proteins were performed with gel electrophoresis, transferred onto membranes, blocked with 5% skim milk powder, and incubated with primary antibodies RNF44 (1:1000; Proteintech, Chicago, USA), and β-actin (1:10,000; Abcam) at 4 °C overnight. Then, the proteins were incubated with relative secondary antibody for 1 h and the bands were analyzed using the Multi Gauge ver 3.0 software (Fuji Photo Film Co., Ltd, Tokyo, Japan) with β-actin as the internal reference.
Statistical analysis
All data analyses were conducted using GraphPad Prism 6.0 software (GraphPadSoftware, CA, USA) and SPSS 20.0 software (IBM, NY, USA). Data were presented as mean ± standard deviation. t-test was performed for comparisons between two groups and analysis of variance was used for comparisons among multiple groups, followed by Tukey’s post hoc test. P value <0.05 was indicative of statistically significant difference.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Change history
22 November 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41420-023-01711-8
References
Shi D, Li H, Zhang J, Li Y. CircGDI2 regulates the proliferation, migration, invasion and apoptosis of OSCC via miR-454-3p/FOXF2 axis. Cancer Manag Res. 2021;13:1371–82.
Jiang S, Dong Y. Human papillomavirus and oral squamous cell carcinoma: a review of HPV-positive oral squamous cell carcinoma and possible strategies for future. Curr Probl Cancer. 2017;41:323–7.
Xiao Z, Li J, Jin Q, Liu D. Long non-coding RNA OIP5-AS1 contributes to cisplatin resistance of oral squamous cell carcinoma through the miR-27b-3p/TRIM14 axis. Exp Ther Med. 2021;21:408.
Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta Biochim Biophys Sin. 2013;45:27–35.
Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532.
Zhou Y, Tozzi F, Chen J, Fan F, Xia L, Wang J, et al. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012;72:304–14.
Cui Q, Wen S, Huang P. Targeting cancer cell mitochondria as a therapeutic approach: recent updates. Future Med Chem. 2017;9:929–49.
Motamedian E, Ghavami G, Sardari S. Investigation on metabolism of cisplatin resistant ovarian cancer using a genome scale metabolic model and microarray data. Iran J Basic Med Sci. 2015;18:267–76.
Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–33.
Yuan SJ, Li SY, Wang YH, Zhang HF, Hua Y, Wang T. The clinical significance, prognostic value and biological role of lncRNA LINC01793 in oral squamous cell carcinoma. Arch Oral Biol. 2021;125:105105.
Zhang D, Song Y, Li D, Liu X, Pan Y, Ding L, et al. Cancer-associated fibroblasts promote tumor progression by lncRNA-mediated RUNX2/GDF10 signaling in oral squamous cell carcinoma. Mol Oncol. 2021;16:780–94.
Yao Y, Li N. MIR600HG suppresses metastasis and enhances oxaliplatin chemosensitivity by targeting ALDH1A3 in colorectal cancer. Biosci Rep. 2020;40:BSR20200390.
Jiang Q, Xue D, Shi F, Qiu J. Prognostic significance of an autophagy-related long non-coding RNA signature in patients with oral and oropharyngeal squamous cell carcinoma. Oncol Lett. 2021;21:29.
Song J, Xu Q, Zhang H, Yin X, Zhu C, Zhao K, et al. Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma. J Cell Biochem. 2018;119:4559–69.
Pereira-da-Silva T, Napoleao P, Costa MC, Gabriel AF, Selas M, Silva F, et al. Cigarette smoking, miR-27b downregulation, and peripheral artery disease: insights into the mechanisms of smoking toxicity. J Clin Med. 2021;10:890.
Li G, Zhu Y. Effect of lncRNA ANRIL knockdown on proliferation and cisplatin chemoresistance of osteosarcoma cells in vitro. Pathol Res. Pract. 2019;215:931–8.
Jin S, Liu MD, Wu H, Pang P, Wang S, Li ZN, et al. Overexpression of hsa-miR-125a-5p enhances proliferation, migration and invasion of head and neck squamous cell carcinoma cell lines by upregulating C-C chemokine receptor type 7. Oncol Lett. 2018;15:9703–10.
Lindner K, Eichelmann AK, Matuszcak C, Hussey DJ, Haier J, Hummel R. Complex epigenetic regulation of chemotherapy resistance and biology in esophageal squamous cell carcinoma via microRNAs. Int. J. Mol. Sci. 2018;19:499.
Li YY, Wu C, Shah SS, Chen SM, Wangpaichitr M, Kuo MT, et al. Degradation of AMPK-alpha1 sensitizes BRAF inhibitor-resistant melanoma cells to arginine deprivation. Mol Oncol. 2017;11:1806–25.
Sannam Khan R, Khurshid Z, Akhbar S, Faraz Moin S. Advances of salivary proteomics in oral squamous cell carcinoma (OSCC) detection: an update. Proteomes 2016;4:41.
Nan CC, Zhang N, Cheung KCP, Zhang HD, Li W, Hong CY, et al. Knockdown of lncRNA MALAT1 alleviates LPS-induced acute lung injury via inhibiting apoptosis through the miR-194-5p/FOXP2 axis. Front Cell Dev Biol. 2020;8:586869.
Cao Q, Wang N, Ren L, Tian J, Yang S, Cheng H. miR-125a-5p post-transcriptionally suppresses GALNT7 to inhibit proliferation and invasion in cervical cancer cells via the EGFR/PI3K/AKT pathway. Cancer Cell Int. 2020;20:117.
Li W, Dong Y, Wang KJ, Deng Z, Zhang W, Shen HF. Plasma exosomal miR-125a-5p and miR-141-5p as non-invasive biomarkers for prostate cancer. Neoplasma. 2020;67:1314–8.
Xu M, Zhan J, Xie J, Zhu L, Chen L, Luo X, et al. MiR-125a-5p inhibits cell proliferation, cell cycle progression, and migration while promoting apoptosis in head and neck cancers by targeting ERBB3. Auris Nasus Larynx 2020;48:477–86.
Luo L, Xiao L, Lian G, Wang H, Xie L. miR-125a-5p inhibits glycolysis by targeting hexokinase-II to improve pulmonary arterial hypertension. Aging. 2020;12:9014–30.
Xu L, Li Y, Yin L, Qi Y, Sun H, Sun P, et al. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593–609.
Wang X, Hao R, Wang F, Wang F. ZFAS1 promotes cisplatin resistance via suppressing miR-421 expression in oral squamous cell carcinoma. Cancer Manag Res. 2020;12:7251–62.
Liu S, Wu M, Peng M. Circ_0000260 regulates the development and deterioration of gastric adenocarcinoma with cisplatin resistance by upregulating MMP11 via rtargeting MiR-129-5p. Cancer Manag Res. 2020;12:10505–19.
Luo X, Wang GH, Bian ZL, Li XW, Zhu BY, Jin CJ, et al. Long non-coding RNA CCAL/miR-149/FOXM1 axis promotes metastasis in gastric cancer. Cell Death Dis. 2018;9:993.
Yang D, Xu T, Fan L, Liu K, Li G. microRNA-216b enhances cisplatin-induced apoptosis in osteosarcoma MG63 and SaOS-2 cells by binding to JMJD2C and regulating the HIF1alpha/HES1 signaling axis. J Exp Clin Cancer Res. 2020;39:201.
Wang Y, Wang X, Han L, Hu D. LncRNA MALAT1 regulates the progression and cisplatin resistance of ovarian cancer cells via modulating miR-1271-5p/E2F5 axis. Cancer Manag Res. 2020;12:9999–10010.
Jin X, Pan Y, Wang L, Zhang L, Ravichandran R, Potts PR, et al. MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogenesis. 2017;6:e312.
Zhang S, Liao W, Wu Q, Huang X, Pan Z, Chen W, et al. LINC00152 upregulates ZEB1 expression and enhances epithelial-mesenchymal transition and oxaliplatin resistance in esophageal cancer by interacting with EZH2. Cancer Cell Int. 2020;20:569.
Yang G, Zhao Y. Overexpression of miR-146b-5p ameliorates neonatal hypoxic ischemic encephalopathy by inhibiting IRAK1/TRAF6/TAK1/NF-alphaB signaling. Yonsei Med J. 2020;61:660–9.
Zhou M, Mao Y, Yu S, Li Y, Yin R, Zhang Q, et al. LINC00673 represses CDKN2C and promotes the proliferation of esophageal squamous cell carcinoma cells by EZH2-mediated H3K27 trimethylation. Front Oncol. 2020;10:1546.
Yang P, Li J, Peng C, Tan Y, Chen R, Peng W, et al. TCONS_00012883 promotes proliferation and metastasis via DDX3/YY1/MMP1/PI3K-AKT axis in colorectal cancer. Clin Transl Med. 2020;10:e211.
Han YY, Liu K, Xie J, Li F, Wang Y, Yan B. LINC00114 promoted nasopharyngeal carcinoma progression and radioresistance in vitro and in vivo through regulating ERK/JNK signaling pathway via targeting miR-203. Eur Rev Med Pharmacol Sci. 2020;24:2491–504.
Acknowledgements
This work was supported by Shandong Provincial Natural Science Foundation (Grant No. ZR2018PH027), Shandong Provincial Key Research and Development Plan (Grant No. 2019GSF108200) and Shandong Provincial Natural Science Foundation (Grant No. ZR2021MH195).
Funding
This work was supported by Shandong Provincial Natural Science Foundation (Grant No. ZR2018PH027), Shandong Provincial Key Research and Development Plan (Grant No. 2019GSF108200) and Shandong Provincial Natural Science Foundation (Grant No.ZR2021MH195).
Author information
Authors and Affiliations
Contributions
S.G. contributed to study design; X.L. contributed to manuscript editing; T.Z. contributed to experimental studies; Z.Y. contributed to data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41420-023-01711-8"
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Zhao, T., Yuan, Z. et al. RETRACTED ARTICLE: MIR600HG sponges miR-125a-5p to regulate glycometabolism and cisplatin resistance of oral squamous cell carcinoma cells via mediating RNF44. Cell Death Discov. 8, 216 (2022). https://doi.org/10.1038/s41420-022-01000-w
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41420-022-01000-w
This article is cited by
-
A combination of cuproptosis and lncRNAs predicts the prognosis and tumor immune microenvironment in cervical cancer
Discover Oncology (2024)
-
Identification and validation of m5c-related lncRNA risk model for ovarian cancer
Journal of Ovarian Research (2023)
-
Integrative analysis of long noncoding RNAs dysregulation and synapse-associated ceRNA regulatory axes in autism
Translational Psychiatry (2023)