Abstract
Genetic information is generally transferred from RNA to protein according to the classic “Central Dogma”. Here, we made a striking discovery that post-translational modification of a protein specifically regulates the editing of its own mRNA. We show that S-nitrosylation of cathepsin B (CTSB) exclusively alters the adenosine-to-inosine (A-to-I) editing of its own mRNA. Mechanistically, CTSB S-nitrosylation promotes the dephosphorylation and nuclear translocation of ADD1, leading to the recruitment of MATR3 and ADAR1 to CTSB mRNA. ADAR1-mediated A-to-I RNA editing enables the binding of HuR to CTSB mRNA, resulting in increased CTSB mRNA stability and subsequently higher steady-state levels of CTSB protein. Together, we uncovered a unique feedforward mechanism of protein expression regulation mediated by the ADD1/MATR3/ADAR1 regulatory axis. Our study demonstrates a novel reverse flow of information from the post-translational modification of a protein back to the post-transcriptional regulation of its own mRNA precursor. We coined this process as “Protein-directed EDiting of its Own mRNA by ADAR1 (PEDORA)” and suggest that this constitutes an additional layer of protein expression control. “PEDORA” could represent a currently hidden mechanism in eukaryotic gene expression regulation.
Similar content being viewed by others
Introduction
S-nitrosylation is a ubiquitous post-translational modification of proteins by which the thiol group of a cysteine residue is modified by nitric oxide (NO), a molecule generated by NO synthase (NOS).1,2,3 Protein S-nitrosylation is considered as a regulator of diverse cellular functions, including signal transduction, cellular metabolism, membrane trafficking, and differentiation.4,5,6 Hence, dysregulation of S-nitrosylation is closely associated with pathophysiological consequences.7,8,9
The Stamler’s group has identified cathepsin B (CTSB), a papain-family cysteine protease,10 as a target of S-nitrosylation modification.11 While CTSB is mainly located in lysosomes, it has been shown to be secreted extracellularly.12,13 Under physiological conditions, CTSB plays an important role in proteolysis, processing of antigens, autophagy and mitosis.14,15,16,17 However, stress-induced lysosomal destabilization can cause the release of CTSB into the cytoplasm, inducing inflammasome activation, mitochondrial dysfunction, cell death, angiogenesis and tumor metastasis.18,19,20,21,22,23
In this study, we surprisingly found that S-nitrosylation of CTSB enhances its protein expression. This response is independent of gene transcriptional regulation, but rather occurs through a signaling mechanism that leads to an increase exclusively in its own mRNA stability dependently on adenosine-to-inosine (A-to-I) RNA editing. Importantly, this newly identified flow of gene expression information from protein to its own RNA may be a supplement to the classic “Central Dogma”. It is fascinating that a protein could regulate its own mRNA, but few reports have been published on this phenomenon. Recently, Ou’s group reported that hyperactive DYF-5 kinase could induce RNA editing of its own mRNA via adenosine deaminase acting on RNA type 2 (ADAR2).24 However, the underlying mechanism remains unclear. Here, our work uncovered that S-nitrosylated CTSB regulates its own mRNA editing via an ADD1/MATR3/ADAR1 regulatory axis. This provides the first evidence for a novel feedforward mechanism of protein expression control which we named “Protein-directed EDiting of its Own mRNA by ADAR1 (PEDORA)”. “PEDORA” constitutes a novel layer of protein expression regulation linking protein post-translational modification and its own RNA editing.
Results
S-nitrosylation of CTSB at a phylogenetically conserved cysteine
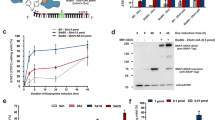
S-nitrosylation of cysteine residues regulates protein function in numerous cellular processes.25 Angiotensin II (Ang II) is implicated in inflammation, endothelial dysfunction, atherosclerosis, hypertension, aortic dissection, and other cardiovascular diseases.26 Studies indicate that Ang II regulates NO production and protein S-nitrosylation modification.27 In this study, we treated human umbilical vein endothelial cells (HUVECs) with Ang II and examined S-nitrosylated proteins using a modification of the biotin switch method in which biotin is irreversibly conjugated to the S-nitrosylated sites and all intermolecular disulfide bonds are broken before purification of biotinylated proteins and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.28 Among the S-nitrosylated proteins identified by mass spectrometry, CTSB was S-nitrosylated at only one cysteine residue Cys319 (Fig. 1a; Supplementary information, Fig. S1a and Table S1).29 Human CTSB possesses 14 cysteine residues, of which Cys108 and Cys319 are unpaired, and 12 other cysteine residues participate in disulfide bond formation. Cys319 is positioned in a slight depression framed by the side chains of five aromatic residues facilitating the removal of its thiol group from the surface29 (Fig. 1b; Supplementary information, Fig. S1b). We speculate that these characteristics of Cys319 of CTSB provide the structural basis for NO recognition and subsequent S-nitrosylation modification. Additionally, Cys319 is highly conserved across phylogeny, including human CTSB and its orthologs from different eukaryotic species (Fig. 1c). Previous studies showed that Cys108 and Cys211 are S-nitrosylated in blood and liver tissues.30 To explore whether Cys319 is a primary locus of NO modification in endothelial cells, we silenced endogenous CTSB by transfecting HUVECs with siRNA targeting CTSB 3′UTR and transfected these cells with either wild-type (WT) CTSB or mutant CTSB plasmids (C108A, C211A, C319A). When treated with Ang II, the WT, C108A and C211A constructs, all showed evidence of S-nitrosylation of CTSB. In stark contrast, the C319A construct abolished the S-nitrosylation of CTSB induced by Ang II (Fig. 1d–g). These data demonstrated that CTSB was capable of being S-nitrosylated in a manner dependent on C319. We next set out to further confirm that this post-translational modification could be induced in endothelial cells under pathophysiological conditions. We targeted cells with three known stressors of endothelial cells: Ang II, oxidized low-density lipoprotein (OxLDL), and high glucose (HG). Once again, we treated HUVECs with siRNA targeting CTSB 3′UTR and transfected WT CTSB or C319A mutant, and exposed them to these three stressors. Importantly, all three stimuli induced S-nitrosylation in WT but not C319A CTSB (Fig. 1h). These data clearly show that CTSB is targeted for S-nitrosylation in endothelial cells under certain stress conditions. Since protein S-nitrosylation plays an important role in virtually all aspects of cardiovascular function and pathophysiology,5 it was thus conceivable that S-nitrosylation of CTSB may be an important regulatory modification.
a The irreversible biotinylation procedures and LC-MS/MS assay of CTSB S-nitrosylation. b 3D structure of CTSB showing the conserved S-nitrosylation (SNO) site Cys319 in red. c Site of S-nitrosylation in human CTSB is conserved. d Western blot analysis of CTSB protein levels in endothelial cells with CTSB 3’UTR-targeted siRNA (siCTSB) transfection. e Western blot analysis of CTSB protein levels in endothelial cells with indicated plasmids transfected. f Western blot analysis of CTSB protein levels in endothelial cells with siCTSB and indicated plasmids transfected. g Biotin switch and western blot analysis for S-nitrosylation of CTSB in endothelial cells with siCTSB and indicated plasmids transfected followed by Ang II treatment. h Biotin switch and western blot analysis for S-nitrosylation of CTSB in endothelial cells with siCTSB and indicated plasmids transfected after the treatment with three known stressors of endothelial cells, including Ang II, OxLDL and HG, respectively. i Schematic diagram for generating an endothelial cell line with the mutation of the cysteine site of NO modification (C319S) by CRISPR/Cas9 genome editing method using immortalized endothelial cell line (HUVEC/TERT2). j, k Biotin switch and western blot analysis of CTSB S-nitrosylation in endothelial cells with stressor treatment. l Western blot analysis of caspase-1 p20 and IL-1β in endothelial cells with stressor treatment. m ELISA analysis of XOR activity in endothelial cells with stressor treatment. n The Griess Reagent System was used to determine relative NO levels in endothelial cells after treatment with XOR inhibitor allopurinol followed by three stressors. o, p Biotin switch and immunoblot analysis of CTSB S-nitrosylation in endothelial cells after treatment with XOR inhibitor allopurinol followed by three stressors. For all graphs, data represent means ± SEM of three independent experiments. Statistical analyses were performed using Student’s t-test for d, one-way ANOVA for the rest. ***P < 0.001.
To examine a potential pathophysiologic role for CTSB S-nitrosylation, we constructed the endothelial cell line with cysteine-to-serine mutation at position 319 (CTSB-MUT) using CRISPR/Cas9 method (Fig. 1i). Treatment of these cells confirmed that Ang II, OxLDL and HG, could all induce the S-nitrosylation of CTSB in CTSB-WT cells (Fig. 1j, k). Surprisingly, while at baseline no difference in CTSB protein levels were observed in the two cell lines, steady-state levels of CTSB protein were increased in WT cells under conditions that induced CTSB S-nitrosylation, but not in the CTSB-MUT cells (Fig. 1j, k). Importantly, while the mutation of S-nitrosylation modification site did not affect CTSB activity (Supplementary information, Fig. S1c), CTSB activity was elevated upon stimulation in a manner correlated with increasing CTSB protein levels at acid pH. Moreover, Ang II, OxLDL and HG treatment all induced lysosome injury and CTSB cytoplasm translocation (Supplementary information, Fig. S1d, e). CTSB is important for NOD-like receptor thermal protein domain associated protein 3 (NLRP3) inflammasome activation.31 Indeed, all three stimuli led to increased levels of caspase-1 p20 and IL-1β suggesting induction of inflammasome activity, but only in WT cells, and not in CTSB-MUT cells (Fig. 1l; Supplementary information, Fig. S1f). These data support a role for S-nitrosylation of CTSB in stress-induced inflammasome pathway activation.
In endothelial cells, NO is produced mainly by endothelial nitric oxide synthase (eNOS), but it can also be released non-enzymatically from S-nitrosothiols or nitrate/nitrite.3 In addition, NO produced by eNOS is impaired under endothelial dysfunction.32 Indeed, treatment with the NOS inhibitor, l-NAME, had no impact on the S-nitrosylation of CTSB in endothelial cells under certain stress conditions (Supplementary information, Fig. S1g). Considering that xanthine oxidoreductase (XOR) catalyzes the nitrite to NO by reduction reaction,33 we measured the XOR activity in WT cells after Ang II, OxLDL and HG treatment. Endothelial XOR activity was elevated upon treatment by all three stressors (Fig. 1m). Remarkably, these three stressors increased NO levels of WT cells, and XOR inhibitor allopurinol abolished the increasing of NO levels (Fig. 1n). Furthermore, allopurinol eliminated S-nitrosylation of CTSB and the subsequent increase in steady-state CTSB protein levels in WT cells under stress stimuli (Fig. 1o, p). Collectively, these data suggest that XOR-mediated CTSB S-nitrosylation promotes endothelial inflammation under conditions of pathologic stress.
S-nitrosylation of CTSB increases the steady-state levels of its own protein via mRNA stabilization
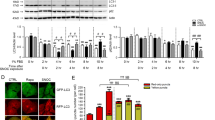
To understand the mechanism of how S-nitrosylation modification impacts on CTSB protein steady-state levels, we confirmed our earlier observation and expanded it by treating the cells with either a proteasome inhibitor (MG132) or a translational inhibitor (cycloheximide, CHX). MG132 had no impact on the steady-state levels of CTSB, suggesting proteasomal turnover does not play a role here (Fig. 2a, b). In contrast, CHX treatment completely blocked the stress-induced increase in CTSB steady-state levels (Fig. 2c). Additionally, neither chloroquine nor leupeptin (inhibitors of lysosomal protein degradation) inhibited the upregulated CTSB protein steady-state levels under stress stimuli (Supplementary information, Fig. S2). These data suggest that the increase in the steady-state CTSB is at the level of protein expression but not protein stability. In support of this, an increase in CTSB mRNA was observed in WT cells treated with stress stimuli, but not in CTSB-MUT cells (Fig. 2d). These results suggested that S-nitrosylation of CTSB could positively regulate its own mRNA levels.
a Western blot analysis of CTSB in WT and CTSB-MUT endothelial cells with stressor treatment. b Western blot analysis of CTSB in MG132-treated WT and CTSB-MUT endothelial cells with stressor treatment. c Western blot analysis of CTSB in CHX-treated WT and CTSB-MUT endothelial cells with stressor treatment. d Real-time PCR analysis of CTSB mRNA in WT and CTSB-MUT endothelial cells with stressor treatment. e Luciferase analysis of CTSB promoter activity in WT and CTSB-MUT endothelial cells with stressor treatment. f–i Real-time PCR analysis and linear regression of CTSB mRNA levels in WT and CTSB-MUT endothelial cells with ActD treatment followed by stressor treatment. j Real-time PCR analysis of CTSB mRNA in CTSB-MUT endothelial cells with CTSB (WT) transfection followed by stressor treatment. k, l Western blot analysis of CTSB in CTSB-MUT endothelial cells with CTSB (WT) transfection followed by stressor treatment. m–p Real-time PCR analysis and linear regression of endogenous CTSB mRNA levels in CTSB-MUT endothelial cells with CTSB (WT) transfection followed by ActD and stressor treatment. For all graphs, data represent means ± SEM of three independent experiments. Statistical analyses were performed using linear regression for f–i and m–p, one-way ANOVA for the rest. **P < 0.01, ***P < 0.001.
To examine the CTSB S-nitrosylation-induced increase in CTSB mRNA, we next tested CTSB promotor activity. We cloned the CTSB promoter in front of a luciferase reporter and measured the effects of Ang II, OxLDL and HG treatment on luciferase activity in WT or CTSB-MUT cells. To our surprise, none of the stimuli had any impact on CTSB promoter activity (Fig. 2e). Based on this finding, we suggested that CTSB S-nitrosylation might affect its own mRNA stability. To test this hypothesis directly, we treated WT and CTSB-MUT cells under control and S-nitrosylation-inducing conditions with the transcription inhibitor actinomycin D (ActD). RNA was extracted over time and CTSB mRNA levels were quantified by real-time PCR. The decay rate of CTSB mRNA was unchanged at baseline between the WT and CTSB-MUT cell lines; in contrast, the CTSB mRNA half-life was significantly increased under stress stimuli in WT cells, but not in the CTSB-MUT cell lines (Fig. 2f–i). To confirm that this effect was dependent on S-nitrosylation of CTSB, we transfected the CTSB-MUT endothelial cells with CTSB (WT CDS construct) followed by the analysis of endogenous CTSB expression and mRNA stability using CTSB 3′UTR primers. As shown in Fig. 2j–p, exogenous WT CTSB expression in CTSB-MUT endothelial cells significantly increased the expression levels of endogenous CTSB mRNA and protein, which is also coupled with enhanced CTSB mRNA stability. These results suggest that the S-nitrosylation of CTSB promotes its own protein expression via increasing CTSB mRNA stability.
S-nitrosylation of CTSB controls its own RNA editing to promote CTSB mRNA stability via recruitment of HuR
Steady-state protein levels of another cathepsin family member, Cathepsin S (CTSS) has been shown to be increased under hypoxic or inflammatory conditions in endothelial cells through an increase in mRNA stability mediated by A-to-I RNA editing of the 3’UTR.34 A-to-I RNA editing of the 3’UTR enables the recruitment of the RNA-binding protein ELAV-like protein 1 (HuR) to the 3′UTR of the CTSS, thereby controlling CTSS mRNA stability and translation.34 We therefore examined whether a similar A-to-I RNA editing was involved in CTSB S-nitrosylation-mediated increase of its own mRNA stability. First, we predicted several HuR-binding sites and identified three clusters of RNA editing events in the CTSB 3′UTR, which correspond to the AluSx1, AluSg, and AluSq2 regions (Fig. 3a). To unravel whether S-nitrosylation of CTSB promotes the A-to-I editing of its own mRNA, RNA sequencing (RNA-seq) and GIREMI assays were employed to identify all the A-to-I-edited genes in WT and CTSB-MUT cells after treatment with Ang II. To our surprise, compared with Ang II-treated CTSB-MUT cells (with denitrosylated CTSB), only the CTSB mRNA showed an enhanced A-to-I RNA editing level in Ang II-treated WT cells (with S-nitrosylated CTSB) (Fig. 3b, c; Supplementary information, Table S2). Most notably, the A-to-I editing rate of CTSB mRNA increased in WT cells under all three stress conditions, but CTSB mRNA editing did not increase in CTSB-MUT cells (Fig. 3d, e). We speculated that RNA editing of the Alu regions in the CTSB 3′UTR may influence CTSB mRNA stability. To test this directly, we cloned the CTSB 3′UTR into our luciferase reporter construct. Consistent with our hypothesis, luciferase activity increased in WT, but not in CTSB-MUT cells upon treatment with stimuli (Fig. 3f). To further confirm whether ADAR1 was responsible for mediating CTSB mRNA editing, Ang II-treated endothelial cells were transfected with ADAR1- or ADAR2-siRNA, and CTSB mRNA editing was examined. We found that the level of CTSB A-to-I RNA editing rate was specifically reduced by ADAR1 knockdown in endothelial cells under Ang II treatment (Fig. 3g–i). As expected, stress-induced CTSB protein and mRNA expression was also reduced by ADAR1 knockdown in CTSB-WT cells, but not by ADAR2 knockdown (Fig. 3j–l). Finally, we used a cross-linked RNA immunoprecipitation (RIP) assay to determine the interaction between ADAR1 proteins and CTSB mRNA. We found that all three stimuli promoted the binding of ADAR1 proteins and CTSB mRNA in WT cells, but not in CTSB-MUT cells (Fig. 3m). These data suggest that S-nitrosylation of CTSB exclusively enhances the A-to-I editing of its own mRNA by ADAR1.
a Distribution and pattern of A-to-I RNA-editing sites and HuR-binding sites in CTSB 3’UTR. b RNA-seq analysis of RNA editing in CTSB-WT and CTSB-MUT endothelial cells with Ang II treatment. c Numbers of CTSB RNA-editing sites in CTSB-WT and CTSB-MUT endothelial cells with Ang II treatment. d, e RNA editing rate and site distribution in CTSB 3’UTR in endothelial cells with stressor treatment (n = 10). f Luciferase analysis of CTSB 3’UTR in endothelial cells with stressor treatment. g, h Western blot analysis of ADAR1 (g) or ADAR2 (h) levels in endothelial cells with ADAR1 or ADAR2 knockdown. i RNA editing rate analysis of CTSB 3’UTR in endothelial cells with ADAR1 or ADAR2 knockdown followed by Ang II treatment (n = 10). j, k Western blot analysis of CTSB in endothelial cells with ADAR1 (j) or ADAR2 (k) knockdown followed by stressor treatment. l Real-time PCR analysis of CTSB mRNA in endothelial cells with ADAR1 knockdown followed by stressor treatment. m RIP and Real-time PCR analyses of CTSB 3’UTR enrichment in endothelial cells with stressor treatment. The CTSB mRNA was pulled down by anti-ADAR1 antibody. n RNA editing rate analysis of CTSB 3’UTR in endothelial cells with stressor treatment. The CTSB mRNA was pulled down by anti-HuR antibody (n = 5). o Western blot analysis of HuR level in endothelial cells with HuR knockdown. p Luciferase analysis of CTSB 3’UTR in endothelial cells with ADAR1 or HuR knockdown followed by Ang II treatment. q Western blot analysis of CTSB in endothelial cells with HuR knockdown followed by stressor treatment. r Real-time PCR analysis of CTSB mRNA in endothelial cells with HuR knockdown followed by stressor treatment. s RIP and real-time PCR analyses of CTSB 3’UTR enrichment in endothelial cells with stressor treatment. The CTSB mRNA was pulled down by anti-HuR antibody. t RNA editing rate analysis of CTSB 3’UTR in CTSB-MUT endothelial cells with WT CTSB transfection followed by stressor treatment (n = 10). u, v RIP and real-time PCR analyses of CTSB 3’UTR enrichment in CTSB-MUT endothelial cells with WT CTSB transfection followed by stressor treatment. The CTSB mRNA was pulled down by anti-ADAR1 (u) or anti-HuR (v) antibody. For d, i, n, t, data were shown as violin plot of indicated independent experiments. For other panels, data represent means ± SEM of three independent experiments. Statistical analyses were performed using Student’s t-test for g, h, o, one-way ANOVA for the rest. *P < 0.05, **P < 0.01, ***P < 0.001.
To further investigate whether edited RNA facilitates the recruitment of RNA-binding protein HuR to the 3′UTR of CTSB, we included HuR in the RIP assay as above. We found that A-to-I RNA editing events of CTSB 3′UTR were enriched in HuR immunoprecipitates (Fig. 3n). We reasoned that ADAR1-mediated RNA editing of Alu regions in CTSB 3′UTR may alter dsRNA formation and enable the recruitment of the mRNA-stabilizing protein HuR to CTSB 3′UTR, thereby influencing CTSB mRNA stability. Luciferase reporter assays with CTSB 3′UTR demonstrated that the silencing of ADAR1 or HuR could effectively reduce the increase of luciferase level induced by Ang II (Fig. 3o, p). Furthermore, HuR knockdown also led to decreased levels of CTSB protein and mRNA expression in WT endothelial cells under all three stimuli (Fig. 3q, r). Consistent with these findings, the enrichment of CTSB mRNA was higher from HuR immunoprecipitates in WT cells after all three stimuli, but unchanged in CTSB-MUT cells (Fig. 3s). Moreover, CTSB mRNA could not bind with HuR upon ADAR1 silence in WT endothelial cells under all three stimuli (Supplementary information, Fig. S3a). Further, restoration of exogenous WT CTSB, but not mutant CTSB expression (C319A), in CTSB-MUT cells effectively increased the CTSB mRNA editing rate, and the binding of ADAR1 and HuR with CTSB mRNA (Supplementary information, Fig. S3b–e, t–v). Collectively, these data establish that S-nitrosylation of CTSB promotes CTSB mRNA stability in a manner dependent on ADAR1 A-to-I RNA editing of the CTSB 3’UTR and the recruitment of HuR.
Matrin-3 binds to CTSB mRNA and recruits ADAR1 for CTSB mRNA editing
ADAR1-mediated A-to-I RNA editing events are essential in mRNA regulation, but few trans-regulators of ADAR1 are known. In order to unravel the underlying mechanism by which S-nitrosylation modification promotes ADAR1-mediated CTSB mRNA editing, we employed chromatin isolation by RNA purification-mass spectrometry (ChIRP-MS) assay35 to identify the RNA-binding proteins (RBPs) associated with CTSB mRNA. Biotinylated oligonucleotides were used to enrich CTSB mRNA from WT and CTSB-MUT endothelial cells, respectively, after treatment with Ang II; and the purified proteins were subjected to mass spectrometry analysis (Fig. 4a; Supplementary information, Fig. S4a). U1 or U2 probe was used as a positive control (Supplementary information, Fig. S4b and Table S3). A real-time PCR assay confirmed efficient enrichment of CTSB mRNA from cells (Supplementary information, Fig. S4c). Using CTSB mRNA probes and ChIRP-MS, we identified different proteins associated with CTSB mRNA in WT, but not in CTSB-MUT cells when treated with Ang II (Supplementary information, Table S4). A gene ontology analysis of these candidate genes revealed significant enrichment of proteins functionally associated with RNA processing (Fig. 4b). Previous studies have identified a number of RBPs as key regulators of A-to-I RNA editing.36 Here, we found that X-ray repair cross-complementing protein 5 (XRCC5), XRCC6, splicing factor proline- and glutamine-rich (SFPQ) and matrin-3 (MATR3) are significantly enriched in our dataset. To determine which RBP interacts with CTSB mRNA and regulates ADAR1-mediated A-to-I RNA editing in Ang II-treated WT cells, the RBPs screened above were further tested by western blot and RIP assays. Notably, Ang II increased the binding of SFPQ and MATR3 with CTSB mRNA (Fig. 4c–e). Furthermore, only MATR3 knockdown could effectively inhibit the Ang II-induced CTSB A-to-I RNA editing (Fig. 4f–h). MATR3 has been previously shown to bind to RNA to control RNA processing.37 Here, we found several predicted MATR3-binding elements in CTSB mRNA (Fig. 4i). Thus, we asked whether MATR3 binds to CTSB mRNA and recruits ADAR1 for CTSB A-to-I RNA editing. Indeed, the binding of MATR3 to CTSB mRNA increased in WT cells but not in CTSB-MUT cells upon treatment with any of the three stimuli (Fig. 4j). These data correlate with the dynamic changes of RNA editing in CTSB-S-nitrosylated endothelial cells. Co-immunoprecipitation (Co-IP) assays showed that conditions promoting CTSB S-nitrosylation also enhanced the interaction of MATR3 and ADAR1 (Fig. 4k). Furthermore, we performed RIP assay to detect the enrichment of CTSB mRNA in MATR3 immunoprecipitates upon ADAR1 knockdown in WT endothelial cells under all three stimuli. The results showed that CTSB mRNA still bound with MATR3 after ADAR1 knockdown (Supplementary information, Fig. S4d). Collectively, our data suggest that MATR3 binds to CTSB mRNA and recruits ADAR1, thus promoting CTSB A-to-I RNA editing.
a Schematic diagram for searching CTSB RBPs by ChIRP-MS method in endothelial cells with Ang II treatment. b Bioinformatic analysis of CTSB RBPs detected by ChIRP-MS in a. c Western blot analysis of RBPs obtained from ChIRP-MS in endothelial cells with Ang II treatment. d RIP and real-time PCR analyses of CTSB mRNA enrichment by RBP antibodies in endothelial cells with Ang II treatment. e Western blot analysis of RBPs obtained from ChIRP-MS in endothelial cells with Ang II treatment. f, g Western blot analysis of MATR3 (f) or SFPQ (g) levels in endothelial cells with MATR3 or SFPQ knockdown. h RNA editing rate analysis of CTSB mRNA in endothelial cells with SFPQ or MATR3 knockdown followed by Ang II treatment. i Bioinformatic analysis of MATR3-binding elements in CTSB mRNA (n = 5). j RIP and real-time PCR analyses of CTSB 3’UTR enrichment in endothelial cells with stressor treatment. The CTSB mRNA was pulled down by anti-MATR3 antibody. k Co-IP analysis of the interaction between ADAR1 and MATR3 in endothelial cells with stressor treatment. For h, data were shown as violin plot of indicated independent experiments. For other panels, data represent means ± SEM of three independent experiments. Statistical analyses were performed using Student’s t-test for d, f, g, one-way ANOVA for the rest. *P < 0.05, **P < 0.01, ***P < 0.001.
S-nitrosylation of CTSB promotes nuclear translocation of alpha-adducin to facilitate the recruitment of MATR3/ADAR1 to CTSB mRNA
Our data clearly linked S-nitrosylation of CTSB with an increase in MATR3 binding to CTSB mRNA. However, S-nitrosylated CTSB itself does not enter the nucleus (Supplementary information, Fig. S5a). To understand this further, we next sought to determine whether this downstream signaling required the WT CTSB protein or whether a smaller fragment was sufficient. We thus constructed plasmids containing an HA-tagged WT or mutant (C319A) CTSB R domain (Tyr227–Asp333, CTSBR) linked with a nuclear export signal (NES) sequence (LPPLERLTL) to prevent aberrant nuclear localization of the CTSB truncated proteins CTSBRW and CTSBRM (Fig. 5a). The CTSBRW or CTSBRM plasmids were transfected into CTSB-MUT endothelial cells. To confirm the cellular localization of the CTSBR proteins, cell fractionation and western blot assay were performed, and we found that CTSBRW and CTSBRM were detected only in the cytoplasm regardless of Ang II treatment (Fig. 5b). Additionally, S-nitrosylation of the CTSBRW fragment was detected, but not the CTSBRM fragment, in cells after Ang II treatment (Fig. 5c). We next tested whether the S-nitrosylation of CTSBRW protein was sufficient to induce endogenous CTSB A-to-I RNA editing. Measurement of RNA editing rate demonstrated that Ang II treatment increased the RNA editing rate of CTSB mRNA in CTSBRW-expressing cells, but not in CTSBRM-expressing cells (Fig. 5d). Comparison of these results with WT CTSB transfection revealed that the levels of CTSB S-nitrosylation and CTSB mRNA editing in CTSBRW-expressing cells was similar with those of WT CTSB-transfected cells (Fig. 5e, f). Moreover, the RIP assay showed that CTSBRW S-nitrosylation increased the binding of CTSB mRNA with MATR3, ADAR1 and HuR (Fig. 5g). These data suggest that the R domain of CTSB, when S-nitrosylated, is sufficient to induce the downstream phenotype.
a Sequence and 3D structure of CTSB R domain. b Distribution of exogenous CTSB (CTSBR) between cytoplasm and nucleus in CTSB-MUT cells with CTSBRW or CTSBRM transfection followed by Ang II treatment. c Biotin switch and immunoblot analysis of exogenous CTSB (CTSBR) S-nitrosylation (SNO) in CTSB-MUT cells with CTSBRW or CTSBRM transfection followed by Ang II treatment. d RNA editing rate analysis of CTSB mRNA in CTSB-MUT cells with CTSBRW or CTSBRM transfection followed by Ang II treatment (n = 6). e Biotin switch and immunoblot analysis of exogenous CTSB S-nitrosylation in CTSB-MUT cells with CTSBRW or CTSB (WT) transfection followed by Ang II treatment. f RNA editing rate analysis of CTSB mRNA in CTSB-MUT cells with CTSBRW or CTSB (WT) transfection followed by Ang II treatment (n = 6). g RIP and real-time PCR analyses of CTSB 3’UTR enrichment in CTSB-MUT cells with CTSBRW and CTSBRM transfection followed by Ang II treatment. The CTSB mRNA was pulled down by anti-MATR3, anti-ADAR1, or anti-HuR antibody. h IP/MS analysis of the interaction proteins between CTSBR (HA) and MATR3 in CTSB-MUT cells with CTSBRW or CTSBRM transfection followed by Ang II treatment. i, j Co-IP analysis of the interaction between ADD1 and exogenous CTSB (CTSBR) (i) or MATR3 (j) in CTSB-MUT cells with CTSBRW or CTSBRM transfection followed by Ang II treatment. k Co-IP analysis of the interaction between ADD1 with MATR3 and CTSB in endothelial cells with Ang II treatment. l Western blot analysis of ADD1 level in endothelial cells with ADD1 knockdown. m RNA editing rate analysis of CTSB mRNA in endothelial cells with ADD1 knockdown followed by Ang II treatment (n = 5). n Real-time PCR analysis of CTSB mRNA in CTSB-WT and CTSB-MUT cells with ADD1 knockdown after Ang II treatment. o Western blot analysis of CTSB in CTSB-WT and CTSB-MUT cells with ADD1 knockdown followed by Ang II treatment. p RIP and real-time PCR analyses of CTSB 3’UTR enrichment in CTSB-WT and CTSB-MUT cells with ADD1 knockdown followed by Ang II treatment. The CTSB mRNA was pulled down by anti-MATR3 antibody. q Distribution of ADD1 between cytoplasm and nucleus in CTSB-WT and CTSB-MUT cells with Ang II treatment. r Immunofluorescence analysis of ADD1 distribution between cytoplasm and nucleus in CTSB-WT and CTSB-MUT cells with Ang II treatment. Scale bars, 5 μm. s S716 phosphorylation of ADD1 in CTSB-WT and CTSB-MUT endothelial cells with Ang II treatment. t Immunofluorescence analysis of ADD1 S716 phosphorylation in CTSB-WT and CTSB-MUT cells with Ang II treatment. Scale bars, 5 μm. For d, f, m, data were shown as violin plot of indicated independent experiments. For other panels, data represent means ± SEM of three independent experiments. Statistical analyses were performed using Student’s t-test for f, l, m, one-way ANOVA for the rest. **P < 0.01, ***P < 0.001.
We next sought to find a mechanistic link between MATR3 and CTSB S-nitrosylation. Towards this end, we immunoprecipitated the S-nitrosylated CTSBR fragment and identified co-precipitating proteins by mass spectrometry (IP/MS). These results were compared to a similar IP/MS assay targeting MATR3 (Fig. 5h; Supplementary information, Table S5). The Venn diagram of these results suggested that alpha-adducin (ADD1) interacted with both proteins. The known function of ADD1 is as an actin-binding protein that regulates the structure of the actin cytoskeleton.38 To further confirm the interaction of S-nitrosylated CTSB and MATR3 with ADD1, IP analysis was performed in CTSBRW and CTSBRM cells with Ang II treatment. We showed that only S-nitrosylated CTSBRW could bind ADD1 and promote the recruitment of MATR3 to ADD1 (Fig. 5i, j). Additionally, endogenous ADD1 interacted with CTSB and MATR3 under the condition of CTSB S-nitrosylation with or without allopurinol treatment (Fig. 5k; Supplementary information, Fig. S5b). Furthermore, ADD1 knockdown blocked CTSB A-to-I RNA editing in Ang II-treated cells (Fig. 5l, m). ADD1 knockdown had little effect on the baseline expression of CTSB but abolished the increase of CTSB expression induced by Ang II (Fig. 5n, o). Finally, the RIP assay showed that ADD1 knockdown abolished the binding of CTSB mRNA with MATR3 (Fig. 5p). These data strongly support a model in which S-nitrosylated CTSB interacts with ADD1, and ADD1 is required for stimuli-enhanced binding of MATR3 to CTSB mRNA, eventually leading to increasing A-to-I RNA editing and stabilization of the CTSB mRNA.
To further understand the connection between these interactions, we noted that ADD1 has been shown to contain a nuclear localization signal (NLS) and a NES sequence, and is detectable both in the nucleus and cytoplasm.39 Therefore, we speculated that CTSB S-nitrosylation may promote ADD1 nuclear translocation and its subsequent interaction with MATR3, which localizes in the nucleus. We examined the localization of ADD1 pre- and post-Ang II treatment and found an increase of ADD1 nuclear translocation under Ang II treatment in WT cells, but not in the CTSB-MUT cell line (Fig. 5q, r; Supplementary information, Fig. S5c). Furthermore, we examined the level and distribution of phosphor-Ser716 of ADD1, which antagonizes the NLS function of ADD1.40 We found that the S716 phosphorylation of ADD1 from WT cells was much lower than that from CTSB-MUT cells upon Ang II treatment (Fig. 5s). As expected, the phosphor-Ser716 form of ADD1 was predominantly localized to cytoplasm (Fig. 5t). These results suggest that the binding of S-nitrosylated CTSB with ADD1 induces Ser716 dephosphorylation of ADD1, thus causing ADD1 nuclear translocation.
Mutation of the phosphorylation site (S716A) leads to ADD1 nuclear translocation (Supplementary information, Fig. S5d). To further explore the functional role of ADD1 nuclear translocation, WT and CTSB-MUT cells were transfected with WT ADD1 or mutant ADD1 plasmids (S716A), then CTSB mRNA editing, mRNA stability, and protein expression were examined. Interestingly, despite the efficient binding of MATR3 with mutant ADD1 (S716A), the levels of CTSB mRNA enrichment in anti-MATR3 or anti-ADAR1 pull-downs, CTSB mRNA editing, mRNA stability, and protein expression were not changed by exogenous ADD1 (S716A) expression in WT cells, CTSB-MUT cells, and Ang II-treated CTSB-MUT cells (Supplementary information, Fig. S5e–p). Therefore, while ADD1 is necessary, ADD1 nuclear translocation alone is not sufficient to trigger CTSB A-to-I RNA editing (see Discussion later). Taken together, S-nitrosylation of CTSB promotes nuclear translocation of dephosphorylated ADD1, which is required for the enhanced recruitment of MATR3 to CTSB mRNA.
Additionally, the results of mass spectrum analysis suggested that PP1C (PPP1CC, protein phosphatase PP1-gamma catalytic subunit), PP2A-B55α (PPP2R2A, protein phosphatase 2A 55 kDa regulatory subunit B alpha), or PP6 (PPP6C, protein phosphatase 6 catalytic subunit) might interact with S-nitrosylated CTSBRW protein and dephosphorylate ADD1 (Supplementary information, Table S5). Indeed, only PP6 knockdown blocked the dephosphorylation of ADD1 in Ang II-treated cells (Supplementary information, Fig. S6a–c). PP6, a Ser/Thr protein phosphatase, is involved in inflammatory signaling, DNA damage repair, cell cycle regulation, lymphocyte development, and tumor progression.41 Moreover, we showed that PP6 could interact with S-nitrosylated CTSBRW and ADD1 (Supplementary information, Fig. S6d). Additionally, PP6 knockdown blocked CTSB A-to-I RNA editing in Ang II-treated cells (Supplementary information, Fig. S6e, f). Finally, PP6 knockdown had little effect on the baseline expression of CTSB but abolished the increase of CTSB expression induced by Ang II (Supplementary information, Fig. S6g, h). These data suggest that S-nitrosylated CTSB promotes dephosphorylation of ADD1 via PP6.
Finally, we examined these molecular events at various time points in Ang II-treated endothelial cells. The results suggested that the S-nitrosylation of CTSB and dephosphorylation of ADD1 occurred 12 h post Ang II treatment (Supplementary information, Fig. S7a–c). MATR3 and ADAR1 bound to CTSB mRNA 18 h after treatment (Supplementary information, Fig. S7d, e), and the CTSB mRNA editing rate increased at around the same time (Supplementary information, Fig. S7f). Lastly, the interaction of HuR with CTSB mRNA and the increasing of CTSB mRNA and protein levels were observed 18–24 h after stimulation (Supplementary information, Fig. S7g–j).
S-nitrosylation of CTSB and CTSB mRNA editing may be critically involved in vascular diseases
To explore the clinical significance of our findings, we examined S-nitrosylation of CTSB in vascular tissues from patients with vascular diseases (coronary artery disease, aortic dissection and diabetes mellitus) and control subjects. Our results showed that the S-nitrosylated CTSB levels significantly increased in patients with vascular diseases (Fig. 6a, b; Supplementary information, Fig. S8a). Additionally, the mRNA levels of CTSB, ADAR1, and MATR3 significantly increased in patients with vascular diseases compared with the control subjects (Fig. 6c–e). Moreover, MATR3 mRNA levels were positively associated with CTSB mRNA expression (Fig. 6f). We next investigated the A-to-I RNA editing rate of CTSB mRNA, and found that it significantly increased in patients with vascular diseases (Fig. 6g). Notably, the distribution and pattern of RNA-editing sites were similar across all three vascular diseases (Fig. 6g, h). Furthermore, we also tested the expression levels of ADAR1 and CTSB proteins, and NLRP3 inflammasome activation in vascular tissues of patients with vascular diseases and control subjects. Our results showed that the levels of ADAR1, CTSB, caspase-1 p20, and IL-1β proteins significantly increased in patients with vascular diseases (Fig. 6i–k; Supplementary information, Fig. S8b). Notably, the concentration of serum CTSB also increased in patients with vascular diseases, as compared to that in control subjects (Fig. 6l). Additionally, the A-to-I RNA editing rate of the CTSB mRNA significantly increased in peripheral blood mononuclear cells (PBMCs) from patients with vascular diseases (Fig. 6m). Collectively, the results suggest that S-nitrosylation of CTSB and CTSB A-to-I RNA editing may be critically involved in vascular diseases.
a, b Western blot analysis of CTSB S-nitrosylation (SNO) in vascular tissues from patients with vascular diseases and from control subjects (n = 3 in a, b; n = 2 in Supplementary information, Fig. S8a). CAD coronary artery disease, AD aortic dissection, DM diabetes mellitus. c–e Real-time PCR analysis of the expression of CTSB, ADAR1, and MATR3 mRNA in vascular tissues from patients with vascular diseases and from control subjects (Ctrl, n = 7; CAD, n = 6; DM, n = 6; AD, n = 10). f Linear regression of MATR3 with CTSB mRNA expression in vascular tissues from patients with vascular diseases. g, h RNA editing rate and distribution analysis of CTSB 3’UTR in vascular tissues from patients with vascular diseases and from control subjects (n = 10). i–k Western blot analysis of ADAR1, CTSB, caspase-1 p20, and IL-1β in vascular tissues from patients with vascular diseases and from control subjects (AD, n = 5; Ctrl, CAD, DM, n = 3 in i, j; n = 2 in Supplementary information, Fig. S8b). l ELISA analysis of the concentrations of serum CTSB in patients with vascular diseases and control subjects (Ctrl, n = 15; CAD, n = 83; DM, n = 21; AD, n = 27). m RNA editing rate analysis of CTSB 3’UTR in PBMCs from patients with vascular diseases and from control subjects (n = 5). For g, m, data were shown as violin plot of indicated independent experiments. For other panels, n means the number of samples, and data represent means ± SEM of indicated independent experiments. Statistical analyses were performed using correlation analysis for f, Student’s t-test for the rest. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In this study, we revealed that the S-nitrosylation of CTSB protein invokes ADD1 dephosphorylation and translocation into the nucleus, followed by the recruitment of MATR3 and ADAR1 to CTSB mRNA. Through A-to-I RNA editing, ADAR1 enhances the stability of CTSB mRNA by facilitating HuR recruitment. The activation of this pathway leads to increased steady-state levels of CTSB protein, which contributes to the enhanced inflammation in patients with vascular diseases (Fig. 7). Our findings uncover a feedforward mechanism: S-nitrosylation of CTSB exclusively controls its own mRNA editing and mRNA stability via an unexpected flow of gene expression information from protein to RNA.
Pathological stimuli induce CTSB cytoplasm translocation and S-nitrosylation at Cys319 site. S-nitrosylated CTSB promotes dephosphorylation and nuclear translocation of ADD1. Then, ADD1 recruits MATR3 and ADAR1 to CTSB mRNA, inducing A-to-I RNA editing of CTSB mRNA. Finally, edited CTSB mRNA is recognized by HuR, which promotes the stability of CTSB mRNA, enhancing the steady-state levels of CTSB. The continuous accumulation of CTSB aggravates disease progression.
Surprisingly, our current study illustrates that the post-translational modification of a protein accurately induces the editing of its own RNA without affecting the editing of others. We employed RNA-seq and GIREMI assays to comprehensively identify all the edited genes and found that only the A-to-I editing rate of CTSB mRNA was dependent on S-nitrosylation of CTSB. However, ADARs regulate multiple RNA targets.42,43,44 The interesting question is: by what mechanism does CTSB S-nitrosylation exclusively regulate its own mRNA editing? Our data demonstrates that the CTSB A-to-I editing triggered by S-nitrosylation of CTSB is dependent on the ADD1/MATR3/ADAR1 molecular complex.
ADD1 is known as an actin-binding protein that regulates the structure of actin cytoskeleton.38,45 It has been reported to play a critical role in regulating diverse physiological and pathological processes including mitotic spindle assembly, synaptic activity, neurodegeneration and essential hypertension.46,47,48,49 Our work is the first to implicate ADD1 in RNA editing. Our results revealed that ADD1 interacts with S-nitrosylated CTSB, which promotes the Ser716 dephosphorylation of ADD1 and facilitates its nuclear translocation, where ADD1 enhances the recruitment of MATR3 to the loci of RNA editing. Taken together, S-nitrosylated CTSB promotes nuclear translocation of ADD1, where it helps recruit MATR3/ADAR1 to the CTSB mRNA. However, S-nitrosylated CTSB itself does not enter the nucleus, and the inability of the phospho-null ADD1-S716A on its own to trigger CTSB mRNA editing suggests that an additional factor or factors may also be involved (Fig. 7). Regardless of the exact process of CTSB mRNA target specification, the resulting complex formation and RNA editing enhances the binding of HuR and improves CTSB mRNA stability, consequently causing CTSB overexpression and vascular inflammation (Fig. 7).
Changes in structure or function of a protein could modulate nucleic acid homeostasis. Transcription factors are proteins that interact with DNA in a sequence-specific manner to regulate information flow from DNA to RNA.50 RBPs and ribonucleoproteins coordinate RNA processing and post-transcriptional gene regulation.51 However, the correlation between protein modification and its own RNA editing has rarely been reported. Very recently, the Ou’s group has shown that the ADAR2-mediated RNA editing of DYF-5CA restricts its own protein kinase hyperactivity via its antisense pair. However, in that case ADAR2 causes RNA editing of multiple targets.24 The ability of a post-translational modification of a protein to induce RNA editing of its own mRNA as reported in the current study is unique. Our data showed that S-nitrosylated CTSB modulates its own mRNA stability by ADAR1-dependent RNA editing of Alu regions in its 3′UTR. It is known that post-transcriptional RNA editing could be regulated by ADARs.52 Moreover, ADAR1 is responsible for a majority of RNA-editing activity in noncoding parts of the transcriptome.53,54 As a general RNA-editing enzyme, ADAR1 is unable to specially identify CTSB mRNA. In addition, ADAR1-mediated A-to-I RNA editing events are dynamically regulated, which require cofactors. Recent studies have identified a number of RBPs as key regulators of ADAR1-mediated editing.36 For example, Zinc finger RBP Zn72D interacts with ADAR and ADAR target mRNAs, thus facilitating RNA editing in neurons.55 In order to identify cofactors of ADAR1, ChIRP-MS assay was employed. Strikingly, our study discovered that MATR3 binds to CTSB mRNA and recruits ADAR1, thus promoting CTSB A-to-I editing. MATR3 harbors zinc finger domains and tandem RNA-recognition motifs, which possess DNA- and RNA-binding capacity.56 As a nuclear-localized protein, MATR3 appears to be involved in multiple processes, including chromatin architecture control, gene expression regulation, DNA repair, and RNA splicing.57 Due to its RNA-binding property, MATR3 recognizes a consensus AUCUU sequence in substrate RNAs.58 Based on our analysis, CTSB mRNA contains the elements necessary for MATR3 recognition. Herein, it is reasonable to suggest that MATR3 binds to CTSB mRNA and recruits ADAR1 for CTSB mRNA editing. Our work reveals a previously unrecognized role of MATR3 in ADAR1-mediated A-to-I RNA editing and responses to pathological stimulations.
S-nitrosylation of cellular proteins is a known post-translational modification that regulates a plethora of pathophysiological states.5 Our data show that S-nitrosylation of CTSB is associated with changes in CTSB mRNA editing and increased steady-state levels of CTSB protein, leading to increased inflammasome activation in endothelial cells. Importantly, in patients with vascular diseases, including coronary artery disease, aortic dissection, and diabetes mellitus, we observed a similar increase of S-nitrosylation of CTSB, changes in CTSB mRNA editing and increased steady-state levels of CTSB protein. The widespread observation of S-nitrosylated CTSB in a variety of pathological conditions suggests an important role for CTSB and the newly identified pathway in endothelial dysfunction. Due to the multifaceted function of CTSB in maintaining cell homeostasis in normal and pathological states, CTSB itself may not be an ideal target for drug development.59 That being said, there are known selective and reversible inhibitors of CTSB, which are currently in clinical and preclinical studies.60 It remains to be seen whether they would have any impact in this case. The S-nitrosylation itself is also not an ideal target for drug development. Therefore, the development of small molecules that target CTSB mRNA editing pathways holds promise to improve strategies for disease treatment.
We usually think of genetic information flowing in one direction from DNA to RNA to protein according to the classic “Central Dogma”. However, feedback and feedforward mechanisms exist, which begin at the protein stage and can lead back to transcriptional or post-transcriptional regulations, such as alternative start sites for transcription, changes in RNA splicing, or in this case RNA editing. In this study, we show a novel flow of gene expression information from protein to RNA. Triggered by S-nitrosylation, the post-translational modification of CTSB enables the editing of its own transcripts in nucleus. Our study uncovers a novel pathway in which post-translational modification of a protein leads to the editing of its own mRNA. We propose to name this novel process “PEDORA”. As we all known, the “reverse” flow of genetic information from RNA back to DNA is considered an extraordinary process in molecular biology. The endogenous RNA can serve as a template to repair DNA double-strand breaks.61 In retrovirus, such as HIV, the reverse flow of genetic information from RNA to DNA is a hallmark of the retroviral replication cycle.62 The complexity of transcriptome-wide protein–RNA interaction networks has important biological significance.63 However, little is known about whether a protein could share or transfer the information back to its own mRNA. Here, our study demonstrates the reverse information flow, which is achieved via a novel ADD1/MATR3/ADAR1 signaling axis.
Although the current study identifies the mechanism of “PEDORA” downstream of CTSB S-nitrosylation, given the presence of various types of protein modifications in cells, it is tempting to speculate that “PEDORA” may operate in other settings where it can be triggered by diverse protein modifications. Indeed, “PEDORA” has the potential to be a wider mechanism underlying protein–mRNA feedforward regulation. For instance, in response to physio-pathological stimuli, “PEDORA” could induce RNA editing and consequently change protein expression. Together, our study adds a new layer of gene expression information flow to the classical model of information transfer from mRNA to protein.
Materials and methods
Cell culture and treatment
Endothelial cells immortalized with human telomerase reverse transcriptase (hTERT) HUVEC/TERT2 (CRL4053) were purchased from ATCC (Manassas, USA) and grown in Vascular Cell Basal Medium (PCS-100-030, ATCC) supplemented with Vascular Endothelial Cell Growth Kit-VEGF (PCS-100-041, ATCC). HUVECs were isolated from umbilical cords according to a previously described method and were grown in complete endothelial cell medium (ECM, ScienCell, Carlsbad, CA, USA) supplemented with endothelial growth factors, 10% (v/v) fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% penicillin/streptomycin. Confluent cells were treated with Ang II (10–7 M; Sigma-Aldrich), HG (25 mM; Sigma-Aldrich), OxLDL (50 μg/mL; Yiyuan Biotechnology, China), allopurinol (100 μM, 2 h pretreated; Selleck, Houston, TX, USA), DMSO (Sigma-Aldrich), acridine orange (5 μg/mL, 15 min; Invitrogen, Thermo Fisher, USA), l-NAME (100 μM, 24 h; Beyotime), MG132 (10 μM, 8 h; Selleck), CHX (50 μg/mL, 6 h; Selleck), chloroquine (100 μM, 2 h pretreated; GlpBio), leupeptin (100 μM, 2 h pretreated; Beyotime), ActD (1 μg/mL; Selleck).
Plasmid construction and dual-luciferase reporter assay
The CDS (860–1879 bp) and R domain (1542–1858 bp) sequences of human CTSB (NM_001384714.1) with the NES sequence of HIV-Rev (LPPLERLTL) and HA tag were cloned into pcDNA3.1 vector, and the constructed plasmids were purchased from GENEWIZ, Inc. (Suzhou, China). The WT ADD1 plasmids with HA tag were constructed by GENEWIZ. The single mutation at cysteine-108/211/319 in CTSB and serine-716 in ADD1 were generated by GENEWIZ. The promoter (–1285 to +109 bp) of human CTSB was cloned into pGL6 vector (Beyotime), and the plasmids were purchased from GENEWIZ, Inc. (Suzhou, China). The 3’UTR of human CTSB (NM_001384714.1) was cloned into pGL6-miR vector (Beyotime), and the plasmids were purchased from GENEWIZ, Inc. (Suzhou, China). Endothelial cells were transfected with the indicated plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s recommendations. Luciferase activity was measured by dual-luciferase assay system (Promega, WI, USA) according to the manufacturer’s instructions. Data were normalized by the activity of Renilla luciferase.
siRNA silencing
siRNAs were purchased from GenePharma (Shanghai, China) (Supplementary information, Table S6) or Santa Cruz Biotechnology (siRNAs of SFPQ, # sc-38304; MATR3, # sc-62604; PP6, # sc-76205; PP1γ, # sc-36297; PP2A-B55-α, # sc-39185). Endothelial cells were transfected with the indicated siRNAs using the Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s recommendations.
CRISPR/Cas9-based mutagenesis
Genetically edited cells were created using SaCas9 AAV vector (pX601-AAV-SaCas9), purchased from Genscript Inc. (Nanjing, China), which was composed of saCas9 nuclease and a guide RNA targeting CTSB gene (Supplementary information, Table S6) (Cys319 to Ser319). The shuttle RGDLRVS-AAV9-cap plasmid was a gift from O. J. Müller (Universität Heidelberg, Germany).64 Endothelial cell-enhanced AAV packaging was achieved according to a protocol from Prof. Yu Huang (Chinese University of Hong Kong, China).65 The single-stranded DNA (ssDNA) oligonucleotides with mutation (Supplementary information, Table S6) were purchased from Genscript Inc. (Nanjing, China) and transfected into AAV-SaCas9-HUVEC/TERT2 cells. Transfection was performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocols.
Biochemical assays
CTSB activity of whole cell extracts was measured in 100 mM sodium acetate and 5 mM CaCl2 (pH 5.5) containing 10 mM DTT. Substrates benzyloxycarbonyl-Arg-Arg-p-nitroaniline (Z-Arg-Arg-pNA) was purchased from Bachem (CA, USA). Cleavage of the substrate resulted in free pNA that was detected colorimetrically at 405 nm. XOR activity was measured by XOR Assay Kit (Jiancheng Biotech, Nanjing, China) according to the manufacturer’s recommendations. NO levels were measured by Total Nitric Oxide Assay Kit (Nitrate/Nitrite Assay Kit, Beyotime) according to the manufacturer’s recommendations. CTSB concentration in serum was measured by Human Cathepsin B ELISA kit (Cusabio, Wuhan, China) according to the manufacturer’s recommendations.
Measurement of RNA stability
The decay rate of CTSB mRNA was measured in a time-course assay after inhibiting transcription using ActD (Selleck). Briefly, endothelial cells were treated with 1 μg/mL ActD in culture medium. RNAs were extracted at 0 (without ActD addition), 6, 12, and 24 h after treatment of HUVECs with ActD. The level of CTSB mRNA was determined at each time point by real-time PCR.
Cross-linked RIP and real-time PCR
RIP was performed as described previously.66 HUVEC cell suspension was fixed in 3% formaldehyde (# 47608, Sigma-Aldrich, USA) in PBS for 10 min at room temperature. Formaldehyde was blocked with 0.125 M glycine for 5 min at room temperature. The cells were washed and cell pellets were resuspended with 1 mL IP lysis buffer (50 mM Hepes, pH 7.5, 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, 10% Glycerol) and 20 μL 0.1 M phenylmethylsulfonyl fluoride, 20 μL protease inhibitor cocktail (Thermo Fisher, USA), and 5 μL RNasin RNase inhibitor (40 U/μL, # DP418, Tiangen, Beijing). The cell suspension was lysed by sonication (10 pulses for 10 s) on ice. The lysates were centrifuged for 3 min at 14,000× g at room temperature, and 50 μL of the supernatant was kept as an input control (used to standardize the real-time PCR analysis). Then, 30 μL of protein G agarose (GE, USA) was blocked with 10 μg antibody (400 μL IP lysis buffer and 1% BSA) overnight at 4 °C. IP was performed overnight at 4 °C using antibodies-blocked protein G agarose (HuR, Santa cruz, # sc-5261; ADAR1, CST, # 81284; MATR3, Santa cruz, # sc-81318). After collection by centrifugation at 400× g and washing 4 times with IP lysis buffer, the complexes were reverse cross-linked with 100 μL of RIP buffer (50 mM Hepes, pH 7.5, 0.1 M NaCl, 5 mM EDTA, 10 mM DTT, 0.5% Triton X-100, 10% Glycerol, 1% SDS) and 1 μL RNasin RNase inhibitor for 1 h at 70 °C. After collection by centrifugation at 400× g, the supernatant was used for RNA extraction with TRIzol reagent (TAKARA, Japan) according to the manufacturer’s instructions. Isolated RNAs were treated with RNase-free DNase I (# RT411, Tiangen) to remove any genomic DNA contamination. Total RNAs from cells and tissues were extracted using TRIzol reagent (TAKARA). First-strand cDNAs were generated using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) followed by real-time PCR using AceQ qPCR SYBR Green Master Mix (Vazyme) and ABI QuantStudio 5 System (Applied Biosystems). The real-time PCR primers are listed in the Supplementary information, Table S7.
ChIRP-MS
ChIRP was performed as described previously.35 Cultured endothelial cells (20 15-cm dishes for each sample) were harvested and resuspended in 3% formaldehyde (diluted with PBS) for 30 min with mixing at room temperature. The crosslinking reaction was stopped by the addition of glycine to a final concentration of 0.125 M glycine for 5 min followed by wash and centrifugation. The cell pellets were resuspended by 5 mL cell lysis buffer (50 mM Tris-HCl, pH 7.0, 10 mM EDTA, 1% SDS, 1 mM PMSF (Sigma-Aldrich), protease inhibitor cocktail (Thermo Fisher), and RNasin RNase inhibitor (40 U/μL, # DP418, Tiangen). The lysates were sonicated using a Vibra Cell VCX130 (Sonics & Materials, Inc., Newtown, CT, USA) (amplitude 20%, 10 s ON, 20 s OFF, 20 cycles). The lysates were centrifugated at 16,000× g for 10 min and pre-cleared by adding 30 μL washed MagBeads (# MB1003, Nanoeast, Nanjing, China) per 1 mL lysate at 37 °C for 30 min on rotation. After preclearing, the lysates were mixed with 2× volume freshly prepared hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-Cl, pH 7.0, 1 mM EDTA, 15% formamide, 1 mM PMSF, protease inhibitor cocktail, and RNasin RNase inhibitor), and CTSB probe pool or control probes (Supplementary information, Table S8) (1 μL of 100 μM probes per 1 mL of lysate). U1 or U2 probes were used as positive control (Supplementary information, Table S8). Subsequently, the mixture was incubated at 37 °C for 12 h on rotation. The beads were then washed five times for 5 min at 37 °C with wash buffer (2× SSC, 0.5% SDS, and 1 mM PMSF). After washing, the beads were resuspended in 100 μL biotin elution buffer (12.5 mM d-biotin, 7.5 mM Hepes, pH 7.5, 75 mM NaCl, 1.5 mM EDTA, 0.15% SDS, 0.075% sarkosyl, and 0.02% Na-Deoxycholate) and incubated at room temperature with mixing for 20 min, and then at 65 °C for 10 min. The supernatant was transferred to a fresh tube and beads were eluted again in 100 μL biotin elution buffer. Subsequently, 50 μL trichloroacetic acid was mixed to the total eluent and the mixture was incubated at 4 °C overnight. The mixture was centrifugated at 16,000× g for 10 min, and the protein pellets were washed twice with ice-cold acetone. The protein pellets were solubilized in 150 μL of 1× Laemmli sample buffer and boiled at 95 °C for 30 min with shaking. For silver staining, the eluted proteins were separated by SDS-PAGE and stained using the Silver Staining Kit (Beyotime, Shanghai, China) according to the manufacturer’s manual. In-gel digestion of lanes were performed for LC-MS/MS analysis.
Irreversible biotinylation procedures and biotin switch assay
Irreversible biotinylation procedures were performed as described previously with modifications.28 In brief, lysates (500 μL) were incubated in Lysis buffer (50 mM NH4HCO3, 1 mM EDTA, 0.1 mM neocuproine) with SDS (2.5% final concentration), protease inhibitor cocktail (Thermo Fisher), and 50 mM N-ethylmaleimide (NEM, Sigma) at 50 °C for 30 min. Proteins were precipitated with cold acetone, washed three times, and resuspended in HEN buffer with SDS (2.5% final concentration), 0.4 mM biotin-maleimide (Sigma), and 10 mM ascorbate at room temperature for 2 h. All the above steps were done in dark. Proteins were precipitated with cold acetone, washed three times, and resuspended in 250 μL reducing buffer (2 M Thiourea, 7 M urea, 20 mM DTT) at 37 °C for 1 h. Then, we added the liquid to Amicon Ultra-0.5 Centrifugal Filter (Millipore) and centrifuged. Next, 2 M thiourea, 7 M Urea, 50 mM Iodoacetamide (IAM, Sigma) were added up to 400 μL into Amicon Ultra-0.5 Centrifugal Filter and centrifuged. Finally, 25 mM NH4HCO3 was added up to 400 μL into Amicon Ultra-0.5 Centrifugal Filter and centrifuged. The supernatants were digested by Trypsin protease and the S-nitrosylated peptides were purified by using NeutrAvidin Plus UltraLink Resin (Thermo Fisher). The supernatant was then removed; 100 μL 0.5% Formic Acid (Sigma) was added to the Resin, heated 5 min at 100 °C, and centrifuged. The supernatant was collected for LC-MS/MS analysis.
S-nitrosylated proteins were detected using the biotin switch assay with modifications.27 In brief, lysates were incubated in HEN buffer with 20 mM methylmethanethio-sulfonate at 50 °C for 20 min. Proteins were then resuspended in HEN buffer, and mixed with 0.2 mM biotin-HPDP (Pierce, Rockford, IL) and 2.5 mM ascorbate at room temperature for 1 h. All steps were carried out in the dark. Finally, biotinylated proteins were purified by using streptavidin-agarose beads (Sigma-Aldrich), separated by SDS-PAGE, and detected by immunoblotting.
RNA-seq and RNA editing analysis
Editing site identification was performed by GENEWIZ, Inc. (Suzhou, China) to detect A-to-I RNA-editing sites in endothelial cells using RNA-seq. Libraries with different indices were multiplexed and loaded on an Illumina HiSeq/Novaseq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was carried out using a 2 × 150 paired-end configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument; image analysis and base calling were conducted by the NovaSeq Control Software (NCS) + OLB + GAPipeline-1.6 (Illumina) on the NovaSeq instrument; image analysis and base calling were conducted by the Zebeacall on the MGI2000 instrument. In order to remove technical sequences, including adapters, polymerase chain reaction (PCR) primers, or fragments thereof, and bases with quality lower than 20, pass filter data of fastq format were processed by Cutadapt (v1.9.1) to generate high-quality clean data. The A-to-I RNA-editing sites from RNA-seq data were analyzed using GIREMI tool. To ensure adequate statistical power and avoid bias, only editing sites covered by at least ten reads in three duplicate samples were kept.
Sanger sequencing was used to validate the editing sites. We obtained cDNA from cells, and then with primers for Alu regions of the CTSB 3′ UTR (Supplementary information, Table S7). PCR amplicons were subcloned into the pMD19-T vector (TAKARA). Clones were randomly picked and analyzed by GENEWIZ, Inc. (Suzhou, China) using Sanger sequencing.
IP and western blot
Cell lysates were obtained by resuspending cell pellets in RIPA lysis buffer (Beyotime) with freshly added protease inhibitor cocktail (Thermo Fisher), RNase A (Beyotime) and PMSF. Nuclear and cytoplasmic proteins were extracted by Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime) according to the manufacture’s instructions. The cytosolic fractions (without lysosomes) were collected as described previously.67 For IP assay, cell lysates were incubated with Protein G Sepharose Beads at 4 °C for 30 min. Then, antibodies (anti-MATR3, # ad151714, Abcam; anti-HA, # 3724S, CST; anti-ADD1, # sc133079, Santa cruz) or preimmune IgGs (Beyotime) were added to the supernatant and the mixtures were incubated with gentle rocking overnight at 4 °C. The supernatant was incubated with Protein G Sepharose Beads (GE healthcare) at 4 °C for 2 h. After the beads were washed with lysis buffer for 5 times, bound proteins were eluted by boiling the beads with 1× SDS-PAGE Sample Loading Buffer (Beyotime) for 5 min. For silver staining, the eluted proteins were separated by SDS-PAGE gels and stained using the Silver Staining Kit (Beyotime, Shanghai, China) according to the manufacturer’s manual. In-gel digestion of lanes was performed for LC-MS/MS analysis. For western blot analysis, equal amounts of proteins were loaded and separated on SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were then blocked with 5% bovine serum albumin and incubated with the specific primary antibodies: anti-CTSB (# 31718S, CST), anti-β-Tubulin (# BS1482M, Bioworld), anti-caspase-1 (# sc56036, Santa cruz), anti-caspase-1 p20 (# AG-20B-0048, AdipoGen), anti-IL1β (# sc32294, Santa cruz), anti-β-actin (# 66009-1-Ig, Proteintech), anti-GAPDH (# 10494-1-AP, Proteintech), anti-LAMP-1 (# sc20011b, Santa cruz), anti-ADAR1 (# 14175S, CST), anti-ADAR2 (# sc73409, Santa cruz), anti-HuR (# sc5261, Santa cruz), anti-XRCC5/Ku86 (# sc5280, Santa cruz), anti-XRCC6/Ku70 (# sc17789, Santa cruz), anti-SFPQ/PSF (# sc374502, Santa cruz), anti-MATR3 (# sc81318, Santa cruz), anti-HA (# 3724S, CST), anti-ADD1 (# sc133079, Santa cruz), anti-ADD1 pS316 (Genscript), anti-PP6 (# sc393294, Santa cruz), at 4 °C overnight, followed by incubation with the specific secondary antibodies. Polyclonal anti-ADD1 pS716 antibody was generated using the synthetic peptide KSPpSKKKKKFRTC as the antigen by Genescript, Inc.
Immunofluorescence assay
The 4% paraformaldehyde-fixed endothelial cells were blocked with 5% BSA and incubated with primary antibodies: anti-ADD1 (# sc133079, Santa cruz) and anti-ADD1 pS316 (Genscript, Nanjing, China) overnight at 4 °C. After several washes with PBS, cells were incubated with secondary fluorescent antibody for 1 h at room temperature. DAPI (Beyotime) was incubated with cells for 5 min prior to observation. Immunofluorescence was visualized on a confocal microscope (LSM 710, Zeiss).
Tissue collection and ethics statement
The clinical phenotype diagnosis of coronary artery disease, aortic dissection and diabetes mellitus was confirmed by standard histopathology at Nanjing Drum Tower Hospital. The study was approved by the Ethics Committee of Nanjing Drum Tower Hospital (2019-218-01, 2019-244-01, 2020-078-01) and performed in compliance with the Declaration of Helsinki Principles. Informed consent was obtained from all involved patients.
Statistical analysis
Data are presented as means ± SEM from three independent experiments unless otherwise stated. Statistical analysis was performed using GraphPad Prism 8.2 (GraphPad Software, San Diego, CA, USA). The mRNA levels over time were evaluated by using linear regression, and the slopes of the best linear fit lines were statistically analyzed. Pearson’s correlation analysis was used to determine the correlation indexes and P values of CTSB mRNA expression with MATR3 mRNA levels. Data were compared using ANOVA with Dunnett post hoc multiple-comparisons test or Student’s t-test as appropriate. For all tests, P values smaller than 0.05 were considered statistically significant. The number of asterisks indicates the P levels (*P < 0.05, **P < 0.01, ***P < 0.001).
References
Stamler, J. S. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell 78, 931–936 (1994).
Stamler, J. S., Singel, D. J. & Loscalzo, J. Biochemistry of nitric oxide and its redox-activated forms. Science 258, 1898–1902 (1992).
Zhao, Y., Vanhoutte, P. M. & Leung, S. W. Vascular nitric oxide: beyond eNOS. J. Pharmacol. Sci. 129, 83–94 (2015).
Hess, D. T. et al. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 (2005).
Lima, B., Forrester, M. T., Hess, D. T. & Stamler, J. S. S-nitrosylation in cardiovascular signaling. Circ. Res. 106, 633–646 (2010).
Stomberski, C. T., Hess, D. T. & Stamler, J. S. Protein S-nitrosylation: determinants of specificity and enzymatic regulation of S-Nitrosothiol-based signaling. Antioxid. Redox Signal. 30, 1331–1351 (2019).
Foster, M. W., Hess, D. T. & Stamler, J. S. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 15, 391–404 (2009).
Shin, M. K. et al. Reducing acetylated tau is neuroprotective in brain injury. Cell 184, 2715–2732.e23 (2021).
Zhou, H. L. et al. Metabolic reprogramming by the S-nitroso-CoA reductase system protects against kidney injury. Nature 565, 96–100 (2019).
Mort, J. S. & Buttle, D. J. Cathepsin B. Int. J. Biochem. Cell Biol. 29, 715–720 (1997).
Stamler, J. S. et al. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. USA 89, 444–448 (1992).
Moon, H. Y. et al. Running-induced systemic cathepsin B secretion is associated with memory function. Cell Metab. 24, 332–340 (2016).
Vasiljeva, O. et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 66, 5242–5250 (2006).
Matsunaga, Y., Saibara, T., Kido, H. & Katunuma, N. Participation of cathepsin B in processing of antigen presentation to MHC class II. FEBS Lett. 324, 325–330 (1993).
Chapman, H. A., Riese, R. J. & Shi, G. P. Emerging roles for cysteine proteases in human biology. Annu. Rev. Physiol. 59, 63–88 (1997).
Man, S. M. & Kanneganti, T. D. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy 12, 2504–2505 (2016).
Hamalisto, S. et al. Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat. Commun. 11, 229 (2020).
Olson, O. C. & Joyce, J. A. Cysteine cathepsin proteases: regulators of cancer progression and therapeutic response. Nat. Rev. Cancer 15, 712–729 (2015).
Werneburg, N. W., Guicciardi, M. E., Bronk, S. F. & Gores, G. J. Tumor necrosis factor-alpha-associated lysosomal permeabilization is cathepsin B dependent. Am. J. Physiol. Gastrointest. Liver Physiol. 283, G947–G956 (2002).
Liu, C. L. et al. Cysteine protease cathepsins in cardiovascular disease: from basic research to clinical trials. Nat. Rev. Cardiol. 15, 351–370 (2018).
Bruchard, M. et al. Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 19, 57–64 (2013).
McComb, S. et al. Cathepsins limit macrophage necroptosis through cleavage of Rip1 kinase. J. Immunol. 192, 5671–5678 (2014).
Nagakannan, P., Islam, M. I., Conrad, M. & Eftekharpour, E. Cathepsin B is an executioner of ferroptosis. Biochim. Biophys. Acta Mol. Cell Res. 1868, 118928 (2021).
Li, D. et al. RNA editing restricts hyperactive ciliary kinases. Science 373, 984–991 (2021).
Hayashida, K. et al. Improvement in outcomes after cardiac arrest and resuscitation by inhibition of S-nitrosoglutathione reductase. Circulation 139, 815–827 (2019).
Mehta, P. K. & Griendling, K. K. Angiotensin II cell signaling: physiological and pathological effects in the cardiovascular system. Am. J. Physiol. Cell Physiol. 292, C82–C97 (2007).
Pan, L. et al. S-nitrosylation of Plastin-3 exacerbates thoracic aortic dissection formation via endothelial barrier dysfunction. Arterioscler. Thromb. Vasc. Biol. 40, 175–188 (2020).
Huang, B. & Chen, C. Detection of protein S-nitrosation using irreversible biotinylation procedures (IBP). Free Radic. Biol. Med. 49, 447–456 (2010).
Musil, D. et al. The refined 2.15 A X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J. 10, 2321–2330 (1991).
Qian, Q. et al. S-nitrosoglutathione reductase dysfunction contributes to obesity-associated hepatic insulin resistance via regulating autophagy. Diabetes 67, 193–207 (2018).
Zhang, Y. et al. Contribution of cathepsin B-dependent Nlrp3 inflammasome activation to nicotine-induced endothelial barrier dysfunction. Eur. J. Pharmacol. 865, 172795 (2019).
Farah, C., Michel, L. Y. M. & Balligand, J. L. Nitric oxide signalling in cardiovascular health and disease. Nat. Rev. Cardiol. 15, 292–316 (2018).
Ghosh, S. M. et al. Enhanced vasodilator activity of nitrite in hypertension: critical role for erythrocytic xanthine oxidoreductase and translational potential. Hypertension 61, 1091–1102 (2013).
Stellos, K. et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 22, 1140–1150 (2016).
Chu, C. et al. Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015).
Quinones-Valdez, G. et al. Regulation of RNA editing by RNA-binding proteins in human cells. Commun. Biol. 2, 19 (2019).
Ramesh, N. et al. RNA-recognition motif in Matrin-3 mediates neurodegeneration through interaction with hnRNPM. Acta Neuropathol. Commun. 8, 138 (2020).
Hadziselimovic, N. et al. Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell 156, 1153–1166 (2014).
Liu, C. M., Hsu, W. H., Lin, W. Y. & Chen, H. C. Adducin family proteins possess different nuclear export potentials. J. Biomed. Sci. 24, 30 (2017).
Chen, C. L., Lin, Y. P., Lai, Y. C. & Chen, H. C. alpha-Adducin translocates to the nucleus upon loss of cell-cell adhesions. Traffic 12, 1327–1340 (2011).
Ohama, T. The multiple functions of protein phosphatase 6. Biochim. Biophys. Acta Mol. Cell Res. 1866, 74–82 (2019).
Freund, E. C. et al. Unbiased identification of trans regulators of ADAR and A-to-I RNA editing. Cell Rep. 31, 107656 (2020).
Ma, C. P. et al. ADAR1 promotes robust hypoxia signaling via distinct regulation of multiple HIF-1alpha-inhibiting factors. EMBO Rep. 20, e47107 (2019).
Hao, X. et al. ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16(INK4a) levels. Nat. Cell Biol. 24, 1202–1210 (2022).
Xu, K., Zhong, G. & Zhuang, X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013).
Chan, P. C. et al. Adducin-1 is essential for mitotic spindle assembly through its interaction with myosin-X. J. Cell Biol. 204, 19–28 (2014).
Zheng, K. et al. miR-135a-5p mediates memory and synaptic impairments via the Rock2/Adducin1 signaling pathway in a mouse model of Alzheimer’s disease. Nat. Commun. 12, 1903 (2021).
Gallardo, G. et al. An alpha2-Na/K ATPase/alpha-adducin complex in astrocytes triggers non-cell autonomous neurodegeneration. Nat. Neurosci. 17, 1710–1719 (2014).
Bayoumy, N. M. K., El-Shabrawi, M. M., Leheta, O. F. & Omar, H. H. alpha-Adducin gene promoter DNA methylation and the risk of essential hypertension. Clin. Exp. Hypertens. 39, 764–768 (2017).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Gerstberger, S., Hafner, M. & Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 (2014).
Keegan, L., Khan, A., Vukic, D. & O’Connell, M. ADAR RNA editing below the backbone. RNA 23, 1317–1328 (2017).
Gallo, A. et al. ADAR RNA editing in human disease; more to it than meets the I. Hum. Genet. 136, 1265–1278 (2017).
Roth, S. H., Levanon, E. Y. & Eisenberg, E. Genome-wide quantification of ADAR adenosine-to-inosine RNA editing activity. Nat. Methods 16, 1131–1138 (2019).
Sapiro, A. L. et al. Zinc finger RNA-binding protein Zn72D regulates ADAR-mediated RNA editing in neurons. Cell Rep. 31, 107654 (2020).
Malik, A. M. & Barmada, S. J. Matrin 3 in neuromuscular disease: physiology and pathophysiology. JCI Insight 6, e143948 (2021).
Cha, H. J. et al. Inner nuclear protein Matrin-3 coordinates cell differentiation by stabilizing chromatin architecture. Nat. Commun. 12, 6241 (2021).
Ray, D. et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013).
Mijanovic, O. et al. Cathepsin B: a sellsword of cancer progression. Cancer Lett. 449, 207–214 (2019).
Li, Y. Y., Fang, J. & Ao, G. Z. Cathepsin B and L inhibitors: a patent review (2010 - present). Expert Opin. Ther. Pat. 27, 643–656 (2017).
Keskin, H., Meers, C. & Storici, F. Transcript RNA supports precise repair of its own DNA gene. RNA Biol. 13, 157–165 (2016).
Wood, C. & Harrington, W. Jr. AIDS and associated malignancies. Cell Res. 15, 947–952 (2005).
Sun, L. et al. Predicting dynamic cellular protein-RNA interactions by deep learning using in vivo RNA structures. Cell Res. 31, 495–516 (2021).
Varadi, K. et al. Novel random peptide libraries displayed on AAV serotype 9 for selection of endothelial cell-directed gene transfer vectors. Gene Ther. 19, 800–809 (2012).
Wang, L. et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 540, 579–582 (2016).
Knuckles, P. et al. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 15, 962–969 (2012).
Nakayama, M. et al. Multiple pathways of TWEAK-induced cell death. J. Immunol. 168, 734–743 (2002).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82121001, 82030013, 91639204, 82241211, 81820108002, 82270421, 81970428, 31771334, 91649125, 81900262, 82100414, 81800385, 82270484), the National Key R&D Program of China (2019YFA0802704), and Jiangsu Provincial Natural Science Foundation (BK20190656).
Author information
Authors and Affiliations
Contributions
Y.J. and H.C. designed the research. Z.L., S.Z., X.L., and Y.H. designed the experiments. Z.L., S.Z., X.L., Z.M., J.C., Y.C., Z.S., and J.Z. performed the experiments. Z.L., S.Z., Z.M., A.G., and F.C. analyzed the data. Z.L., S.Z., X.L., and Z.M. wrote the manuscript. D.W., S.C., L.W., T.Y., and K.S. provided the clinical samples. All authors discussed and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, Z., Zhao, S., Li, X. et al. Cathepsin B S-nitrosylation promotes ADAR1-mediated editing of its own mRNA transcript via an ADD1/MATR3 regulatory axis. Cell Res 33, 546–561 (2023). https://doi.org/10.1038/s41422-023-00812-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-023-00812-4
This article is cited by
-
DDX1 methylation mediated MATR3 splicing regulates intervertebral disc degeneration by initiating chromatin reprogramming
Nature Communications (2025)
-
Reduced expression of cathepsin F predicts poor prognosis in patients with clear cell renal cell carcinoma
Scientific Reports (2024)
-
Hepatocellular carcinoma-specific epigenetic checkpoints bidirectionally regulate the antitumor immunity of CD4 + T cells
Cellular & Molecular Immunology (2024)
-
RNA editing: new roles in feedback and feedforward control
Cell Research (2023)