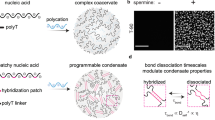
Abstract
Research on biomolecular liquid-state condensates (droplets) in cells has sparked burgeoning interest among synthetic biologists in programmable droplets assembled from synthetic nucleic acids––information-encoding biomolecules amenable to facile synthesis, versatile sequence design, and molecular decoration. Analogous to biological condensates, well-engineered nanostructures consisting of DNA or RNA strands, which are negatively charged, phase-separate into membrane-free droplets via weak multivalent specific interactions or via electrostatic attraction with positively charged peptides. The membraneless compartments of these droplets allow stimuli responsiveness to molecular cues (DNA/RNA, enzymes, etc.). Nucleic acid droplets thus offer a powerful platform for programming their various features, including hierarchical structuring, molecular recognition capabilities, droplet interactions, and physical properties. Specifically, we describe a DNA linker that serves as a programmable surfactant bridging immiscible DNA phases, which, upon molecular inputs, alters their separation level from mixed to divided states. Furthermore, a rational combination of these features can create intelligent liquid-state architectures capable of naturally unachievable functions and dynamics, such as Boolean operations and directional motion. To predict how molecular-level encoding leads to macroscopic characteristics, coarse-grained models, which treat nucleic acids as strings of interacting rigid beads, are widely utilized. This emerging field represents a cross-disciplinary integration of various fields, from biophysics to information science. This Focus Review highlights recent advances in synthetic nucleic-acid droplets and their far-reaching potential, concluding with perspectives on their future directions and challenges.
Similar content being viewed by others
Introduction
In recent years, the liquid–liquid phase separation (LLPS) of biomolecules has emerged as a pivotal concept in structural and synthetic biology. Since the discovery of liquid-like condensates, i.e., membraneless bodies of biomolecules within living cells [1], numerous studies have focused on unraveling their underlying mechanisms [2,3,4,5,6,7]. Their functions as membraneless cell organizers offer various properties, such as compartmentalization [8, 9], spatiotemporally isolated chemical reactions [10], DNA repair [11], and other potential applications [12,13,14,15]. In the cell nucleus, DNA condensates play a critical role in organization and gene expression. Their formation is based on multivalent interactions with partner molecules, such as transcription factors, DNA-repairing complexes or RNA [16,17,18,19]. These dynamic condensates exhibit liquid-like behavior (e.g., coalescence) and are often driven by π–π interactions among nucleic acids, specific RNA and peptide recognition sequences, and chromatin marks [20, 21]. Their functions range from simple DNA subcompartmentalization to reaction encapsulation and enhancement and even genome accessibility regulation [11].
Meanwhile, synthetic biomolecular condensates composed of DNA [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52] or RNA [53,54,55] have garnered particular attention because of their physicochemical similarity to biological condensates and their ability to form programmable structures. First reported in the 2010s [27, 56,57,58,59], these biocompatible programmable condensates show great promise across a range of studies, from fundamental, such as synthetic cells, microreactors, and molecular computations, to applied fields, including soft robotics and biomedical engineering, as highlighted in recent reviews [60,61,62,63].
Their formation involves a range of different mechanisms, from random multivalent electrostatic interactions of oppositely charged molecules, e.g., DNA and peptides (as in coacervates) [56, 58], to polymerization [58] with programmable multivalent Watson‒Crick (WC) base-pairing interactions, summarized in Fig. 1a–f. Specifically, DNA junctions, which are frequently used in DNA origami [64,65,66,67,68,69,70] as a step for the subunit (motif) formation, allow precise tuning of the condensate properties as encoded in the sequences. Typically, for ease of design, most previous works consider one or a few types of junctions (each assembled from up to a few strands) as modules well tailored to specific research purposes. This modularity contrasts with that of biological condensates as crucibles of diverse molecules.
Overview of DNA condensates: formation, functions, and programmability. DNA‒peptide multivalent interactions. a Liquid-crystal condensates formed through peptide‒DNA association mediated by temperature and ionic strength (salt concentration). Fusion events suggested that the condensates were in a liquid state, which maintained their anisotropic properties (spatially oriented). Reproduced with permission [56]. Copyright 2020, American Chemical Society. b, c Liquid-like condensates formed via electrostatic interactions of peptide‒DNA complexes. Dynamic and multidomain interactions lead to the formation of DNA‒peptide condensates, mediated by temperature and ionic strength, with ssDNA‒peptide complexes showing a greater tendency to form liquid‒shaped condensates. In contrast, (b) double-stranded DNA‒peptide (dsDNA‒peptide) complexes tend to form irregularly shaped solid-like structures (c). Reproduced with permission [57]. Copyright 2018, American Chemical Society. RNA-based. d Liquid‒crystal condensates. Oriented RNA strands formed through supramolecular nonenzymatic polymerization self-assemble into spatially oriented (anisotropic) condensates, which is mediated by temperature and crowding agents such as polyethylene glycol (PEG). Reproduced with permission [58]. Copyright 2018, American Chemical Society. DNA base-paired domains. e Liquid-like condensates formed by self-assembled multidomain DNA motifs, which are based on programmable complementary sequences, create short dsDNA and ssDNA domains along the motif. Each domain consists of complementary strands, orthogonal to neighboring domains, and free ssDNA arms that are able to rapidly bind/rebind with neighboring motifs, leading to condensate formation mediated by temperature and ionic strength. Reproduced with permission [59]. Copyright 2016, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. f Liquid-like condensates formed by self-assembled ssDNA into nanostar-shaped motifs sed on programmable complementary sequences, which, unlike in the previous example, form monomers composed of DNA junctions through the formation of structures named arms, which are orthogonal to each other and increase the specificity of DNA binding. Each nanostar is formed through simple WC base‒pairing interactions, and each arm is composed of complementary strands. Neighboring nanostars interact through overhang interactions at their ends, which are mediated by temperature and ionic strength. Reproduced with permission [27]. Copyright 2018, The Royal Society of Chemistry. Condensates. g Illustration of droplet-like condensates, highlighting their anisotropic and isotropic properties. Functions and programmability. h Designability of interactions. DNA droplets are highly programmable structures that can recognize and interact/fuse through sequence-specific interactions [22]. i Due to the specificity of DNA and RNA recognition enzymes and base-pairing interactions, droplet architecture, subcompartmentalization, and composition are designable through multistep reactions. Reproduced with permission [23]. Copyright 2024, American Chemical Society. j DNA computing is one of the possible functions within DNA droplets and is based on internal reactions such as strand displacement reactions (SDRs) [43], enzymatic degradation [51], and enzymatic and nonenzymatic DNA polymerization. k Molecular communication. Different DNA droplets might mutually exchange solutes, DNA/RNA strands, enzymes/proteins, and other molecules [149] on the basis of nonequilibrium diffusion patterns, designed WC base-pairing interactions, aptamer formation, etc. [34, 45]
Structurally, they are constructed hierarchically, similar to peptide organizational levels: primary, secondary, tertiary, and quaternary. (1) Primary structure: a DNA sequence designed according to each system’s features. (2) Secondary structure: the formation of motifs from single-stranded DNAs (ssDNAs) with a sticky end (SE), a single-stranded overhang, which is achieved via salt-dependent, thermally controlled self-assembly [22]. (3) Tertiary structure: network assembly from the motifs, driven by overhang recognition. The final structure possesses base-sequence programmability in the physical properties, such as fluidity (due to motif molecular reshuffling), a non-gating boundary (free exchange of solutes between inner and outer solutions), and internal molecular diffusion [25, 26, 44, 46, 71, 72]. These properties depend on parameters such as motif rigidity [26], solution ionic strength [71, 72], and motif multivalency [25, 44, 46], as highlighted in Fig. 1h–k. (4) Quaternary structure: compartmentalized condensates assembled from multiple immiscible DNA phases. Some researchers have shown that condensate associations [22, 24, 49, 73] and communication [39] are essential features of highly complex artificial biological systems.
In this Focus Review, we briefly describe how the properties and structures of these condensates can be controlled and modulated, how to achieve dynamic regulation, and a nascent but growing body of applications in this field, concluding with some future perspectives. Specifically, this review highlights the synthetic nucleic acid linker system proposed by our group to drive the dynamic behavior of hierarchically structured DNA droplets in a sequence-specific manner.
Programmable nucleic-acid droplets
Design of DNA/RNA droplets
The basic engineering procedures largely begin with designing nanoscale building subunits (termed motifs or nanostars), which self-assemble into microscale condensates, similar to existing DNA/RNA nanoengineering methods. The subunits consist of multiple strands, which self-assemble into multibranched structures, each terminating in an SE. In the design steps, the component strands are carefully screened using numerical software (commonly, NUPACK [74]) and well-established thermodynamic models (e.g., nearest neighbor models [75]) to ensure high specificity in the construction and interaction of the subunits and suitable thermostability in SE hybridization. The design of DNA/RNA droplet behaviors primarily leverages the commonly accepted DNA/RNA nanoengineering methods. To predict how the component design leads to the phase behavior of the condensate level, coarse-grained models have been proposed because of their ability to simulate large-scale dynamics, as described later.
Programmability of physical properties on the basis of sequence information
The rheological properties of phase-separated droplets are essential for controlling droplet behavior. For example, the viscosity and surface tension of droplets influence their growth through coalescence [76] and the exchange rates of molecules with the surrounding environment [77]. A decrease in the internal dynamics of droplets, representing a state change from liquid to solid, has been suggested to be involved in neurodegenerative diseases [9, 78], such as amyotrophic lateral sclerosis (ALS) [79] and Alzheimer’s disease [80].
A significant advantage of DNA-based LLPS systems is that their physical properties, including rheological characteristics, can be controlled not only by external factors (temperature [81, 82], salt concentration [27], and pH [83]) but also by appropriate design of the DNA sequence. DNA condensates assembled from multidomain DNA strands exhibit programmable rheological properties and melting temperatures, which can be engineered by modifying the length and number of domains [59]. DNA nanostructures that undergo LLPS through SE hybridization serve as programmable templates for 1) valency, 2) flexibility, and 3) strength of SE interactions, which are involved in the networked structure of condensates. 1) Solutions of DNA motifs with multivalency phase-separate into DNA-poor and DNA-rich phases that behave as fluids (Fig. 2a-i). Notably, when the valency was increased, the phase-separated state was more accessible (Fig. 2a-ii, iii) [84]. As the valency increased, the mobility of the motifs decreased, resulting in a hydrogel-like state [22] and increased stiffness [25]. 2) The flexibility of DNA motifs was augmented by adding unpaired bases as spacers at joints and overhanging sections, which increased the conformational entropy of the DNA motifs (Fig. 2b-i) [33, 40]. Flexible SE enabled the formation of phase-separated droplets with fluid-like behavior (Fig. 2b-ii) [40]. 3) The strength of SE interactions is determined by the sequence-dependent \(-\varDelta {H}\) (\(\varDelta H\): binding–unbinding enthalpy difference). At a constant temperature, large \(-\varDelta H\), i.e., stable SE hybridization, promotes droplet formation, whereas a further increase in \(-\varDelta H\) leads to a state change from droplet to gel (Fig. 2c-iii) [22]. Notably, the thermally induced phase behavior and physical properties (e.g., viscosity and surface tension) [30] are determined by the SE sequence (Fig. 2c-iii). The driving force behind the relaxation process of coalescing droplets is surface tension, where viscosity slows down this process. The surface tension was estimated from both the SE sequence-dependent relaxation process (Fig. 2d-i) and viscosity. In a liquid state, the surface tension depends more on the SE sequence than does the viscosity (Fig. 2d-ii) [30].
Sequence-dependent physical properties of DNA droplets. a Phase behavior of DNA liquid condensates using multivalent DNA motifs. (i) DNA motifs characterized by valency \(f=3\) or \(f=4\) and phase behaviors of the \(f=4\) motif contained within a capillary tube. The DNA motif equilibrates in the dispersed phase at high temperatures (left) and in the liquid state at low temperatures (right). The green and pink frame colors correspond to the dots in (ii). (ii) Phase diagram plotting the concentration of each state. Phase separation occurs when the temperature decreases to the range within the curve on the plots. Adapted with permission [84]. Copyright 2013, National Academy of Sciences. b DNA droplets generated via the self-assembly of flexible DNA motifs. (i) A DNA motif featuring unpaired bases (cyan and pink letters) as spacers at joints or overhanging sections for flexibility. (ii) Phase separation using DNA motifs with unpaired bases (cyan and pink circles). The sodium chloride concentration is shown at the bottom of the figure. (iii) Confocal microscopy images of fully stiff (left) and fully flexible (right) DNA motif samples. Reproduced with permission [40] from the Royal Society of Chemistry. c SE sequence control of the temperature-dependent phase behavior of the Y-shaped DNA motif (Y-motif). (i) Schematic of the Y-motif. (ii) Temperature-dependent phase behavior of the Y-motifs with a specific length of the SE sequence. At high temperatures, Y-motifs are dispersed in the solution. As the temperature decreases, they change to a liquid or gel state. (iii) Each state diagram of Y-motifs with different lengths of SEs. As the length of the SE sequence increases, the binding affinity, as indicated by \(-\varDelta H\) (\(\varDelta H\): the enthalpy difference between binding and unbinding), also increases. Reproduced [22] under the terms of the Creative Commons CC BY-NC 4.0 license. Copyright 2020, American Association for the Advancement of Science. d Coalescence behavior of DNA droplets. (i) (Top) Representative sequential images of the relaxation process for DNA droplets composed of the Y motif with an SE length of 8 nt. Scale bar: 10 μm. (Bottom) Time-dependent representative aspect ratios of DNA droplets with various SE designs at different droplet formation temperatures. (ii) The surface tension was estimated from the relaxation process and viscosity. Reproduced [30] with permission from the Royal Society of Chemistry
DNA droplets as regulatory hosts for chemical reactions
Intracellular biomolecular condensates, as phase-separated droplets, selectively take up or exclude molecules depending on their charge or size, acting as scaffolds for downstream biochemical reactions [85]. This selectivity has been reconstructed in synthetic LLPS droplets primarily by modulating the physical parameters of the droplets, including charges, mesh sizes, and the binding of clients with molecules. DNA droplets can control molecule uptake or exclusion by utilizing sequence-dependent selectivity of molecules. This selective partitioning capability allows programmable control of chemical reactions by spatial control of reactants by selectively localizing client molecules, which is realized, e.g., accelerated reactions due to selective enrichment of client molecules [47] and spatially coupled enzymatic cascade reactions [48, 86].
Design of dynamic behavior
Membraneless synthetic LLPS droplets can easily exhibit dynamic behaviors in response to changes in external conditions or signals, serving as models of dynamic intracellular condensates to elucidate their functions and dynamic behaviors. Previously, synthetic complex coacervates, formed by the electrostatic assembly of polyanions and polycations, have demonstrated various dynamic behaviors, including formation and dissolution, growth, and division [87,88,89,90]. However, their electrostatic interactions are regulated by enzymatic reactions and environmental conditions, such as pH, temperature, and salt concentration. In contrast, DNA droplets (DNA coacervates) can dynamically behave as encoded by their designed sequences. By harnessing their physical programmability, DNA droplets exhibit various dynamic behaviors initiated by the external signals of nucleic acids or enzymes.
DNA linkers as programmable surfactants
Droplets of immiscible DNA phases favor separation into multiple compartments. Their spatial coupling is necessary for their immediate interactions that enable their distinct functions and behaviors to be associated. In biological systems, RNA functions as a surfactant in the formation of the nucleolus, which appears as a tripartite layered liquid condensate concentrically composed of an immiscible fibrillar center (FC), dense fibrillar center (DFC), and granular component (GC) (Fig. 3a) [91, 92]. For dynamic control of the cohesivity between immiscible DNA phases, our group proposed a DNA linker system, a multibranched DNA nanostructure, which binds to multiple orthogonal DNAs [22, 31, 43, 53].
DNA linker as programmable surfactant. a RNA transcripts that act as surfactants in the nucleolus [91, 92]. The nested nucleolus comprises immiscible liquid-state condensates: (from the core) a fibrillar center (FC), a dense fibrillar component (DFC), and a granular component (GC). Ribosomal RNA (rRNA) is transcribed at the FC–DFC interface. As nascent rRNA transcripts undergo processing and ribosomal subunit assembly, they are preferentially expelled outside the nucleus. When the transcription machinery is switched off, the tripartite layered organization is disfavored and replaced with a pancake-like conformation. Adapted [92] with permission. Copyright 2021, Nature Publishing Group. b Sequence-specific interaction of DNA droplets [22]. (i) Different liquid condensates of orthogonal DNA Y-motifs (YA and YB) favor segregation while fusing with those of the same sequence. (ii) Time-sequential snapshots of selective droplet fusion events. c Linker DNA, LAB, which binds to both YA and YB. (i) Linker SEs hybridize with motif SEs labeled in the corresponding color. (ii) In the presence of LAB, the orthogonal DNA motifs favor the formation of mixed-state droplets, which can (iii) fuse with each other. d Cleavage of the linker leads to dynamic phase separation. (i) Chimeric linker, which includes two RNA sequences at the center junctions. Upon the input of ribonuclease A (RNase A), the linker favors cleavage. (ii) In the presence of RNase A, mixed-state droplets favor phase separation into immiscible phases. Adapted [22] under the terms of the Creative Commons CC BY-NC 4.0 license. Copyright 2020, American Association for the Advancement of Science. Scale bars: (b, c) 10 µm; (d) 20 µm. e Molecular input of DNAs (RNAs) functioning as fuel thermodynamically drives linker cleavage. The linker is designed to undergo SDRs upon inputs of targeted sequences. In the free-energy landscape, the fuel lowers the energy barrier (dashed curve in magenta). \(\varDelta G\) is the free-energy gap between the local minima
In the six-branched linker, LAB, three consecutive SEs and the others bind exclusively to the orthogonal motifs YA and YB, respectively (Fig. 3c-i). In the absence of LAB, the immiscible DNA liquid-state condensates prefer to coalesce with the same-sequence droplets and remain segregated with the other type (Fig. 3b). In the presence of LAB, YA and YB favor the formation of single-phase droplets (Fig. 3c-ii), which can fuse with each other (Fig. 3c-iii). Furthermore, the concentration of LAB determines the compartmentalized state of droplets: at a lower concentration, the two immiscible DNA phases favor adherence to the distinctly compartmentalized structure; at increased concentrations, the YA–YB interface increases; and at a sufficiently high concentration, YA and YB equilibrate in an entirely mixed state [22, 24, 31].
Conversely, well-mixed DNA droplets of multiple orthogonal DNA motifs phase-separate into a demixed state upon molecular input. This phase separation behavior can be programmed by encoding an input-responsive cleavage in the linker, which is triggered by, e.g., enzymes and DNA/RNA strands. Our group designed a chimeric linker, which includes RNA sequences in the junctions bridging the YA- and YB-binding domains (Fig. 3d-i). Upon the input of ribonuclease A (RNase A), an RNA-degrading enzyme, the chimeric linker undergoes cleavage. As a result, the mixed-state droplets favor irreversible division into multiple droplets of distinct phases (Fig. 3d-ii). The active control over biomolecular condensates reported thus far has focused on the switching formation (‘ON’) or dissolution (‘OFF’) of condensate bodies [93]. In contrast, our linker system can fine-tune the wetting property between the immiscible DNA phases.
Thermodynamically, the linker breakup can be understood as a fuel reshaping the energy landscape (Fig. 3e). Now, we take as an example another design of a DNA linker, L’AB, in which two strand displacement reactions (SDRs) are encoded in the sequence design, which is a widely used technique for dynamic nucleic acid molecular restructuring [94]. In an SDR, one strand of double-stranded DNA (dsDNA) is replaced by another ssDNA [94]. The incoming ssDNA, called the input DNA, hybridizes to a dangling single-stranded portion of the dsDNA, called a toehold, and displaces the incumbent strand via branch migration, releasing the single-stranded displaced DNA, called the output DNA. Upon inputs of two targeted sequences (DNA or RNA), the resulting SDRs lead to a breakup of the L’AB into two substructures. In the intermediate steps of one SDR, the branch migration assisted by toehold–input hybridization proceeds like a random walk in the middle of a local minimum in the energy landscape (Fig. 3e), finally reaching the lowest minimum. Without the inputs, the energy barrier thermodynamically prohibits the initial-state L’AB from reaching the final equilibrium. By lowering the energy barrier, the inputs enable L’AB to transition to the lower minimum. Furthermore, since the inputs drive the phase dynamics by releasing the binding energy in L’AB–YA and L’AB–YB binding, the inputs can be considered to be fuel [61]. By encoding complementary sequences for specific target sequences in the stem regions, the linker can be provided with sequence-recognition capabilities, serving molecular computing purposes, which we describe later.
Division of DNA droplets
To elucidate the roles of coacervate-based protocells in origin-of-life research, researchers have reconstructed cell-like growth and division, which are essential for cellular sustainability and evolution [95], using synthetic LLPS droplets as protocell models [96, 97]. The cell-like division of DNA droplets has been achieved by inserting enzymatically cleavable (Fig. 3d-ii) [22, 42] or photocleavable [42] linkers within multibranched DNA surfactants.
Recently, our group demonstrated temporally controlled DNA droplet division by regulating the cleavage rate of linkers [43]. Input DNAs, which can cleave linkers by SDRs, initially exist as RNA/DNA hybrids that bind with protector RNAs. These input DNAs are gradually released through RNA decomposition catalyzed by ribonuclease H (RNase H), which selectively degrades RNA in RNA/DNA hybrids. By adjusting the release rate via changes in the concentration of protector RNAs or RNase H, the cleavage rate of the linkers was regulated, enabling control over the timing of DNA droplet division. Furthermore, controlling the cleavage order of multiple linkers enabled multistep droplet division, effectively achieving pathway control of droplet division in a reaction landscape [43] (Fig. 4a, b). Mixed DNA droplets, formed by the assembly of three orthogonal motifs (A-, B-, and C-motifs) and two linkers (AB- and AC-linkers), were divided into orthogonal droplets (A-, B-, and C-droplets) through one of two pathways, depending on the cleavage order of the two linkers. The C-droplet divided first when the AC-linker was cleaved first (Fig. 4a), whereas the B-droplet divided first when the AB-linker was cleaved first (Fig. 4b). Regulating the cleavage rate of these linkers by adjusting protector RNAs or RNase H enabled control over their cleavage order and thus the selection of division pathways.
Pathway control of multistep DNA droplet division by controlling the cleavage order of linkers. a Pathway 1 was selected when the AC-linker was cleaved first. b Pathway 2 was selected when the AB-linker was cleaved first. Scale bar: 20 µm. Adapted with permission [43]. Copyright 2024, Nature Publishing Group
Dynamic structural changes in DNA droplets
Dynamic structural changes, such as dynamic formation/dissolution, hierarchical structure formation, or the patterning of synthetic liquid condensates akin to intracellular condensates, facilitate the sequestration of molecules and the spatial regulation of chemical reactions [98, 99]. Dynamic control over the formation and dissolution of DNA droplets has been achieved by changing the binding ability of DNA motifs via protection control of the SE as a binding site. The dissolution [44, 47, 50] and reassembly of DNA droplets are facilitated by SDR [44, 47], light irradiation [46], and enzymes [50]. Additionally, time-fluctuating protection through RNA binding and unprotection via ribonuclease reactions enables the transient formation and dissolution of DNA droplets [50]. Enzymatic reactions with motifs also permit control over formation and dissolution. Hierarchical structures of DNA droplets have been engineered by combining multiple motifs, e.g., core–shell structures [49], concentric multiphase structures [35], and the transient formation of reaction‒diffusion patterns [23].
Directional motion of DNA droplets
Membraneless LLPS droplets allow the free diffusion of molecular components between the interior and exterior of the droplet. This stimuli-responsive property allows the dynamic motion of phase-separated droplets. Among LLPS droplets, those utilizing aqueous two-phase systems have been extensively studied, with several reports focusing on combinations of polyethylene glycol (PEG) and dextran (DEX). For example, PEG/DEX droplets have demonstrated directional motion by forming concentration gradients in their surroundings, enabling surface tension differences that drive their motion through the Marangoni effect [100, 101]. However, these LLPS droplets possess low molecular programmability in their driving mechanisms.
In contrast, nucleic acid-based droplets leverage the base-sequence programmability of nucleic acids, enabling programmable motion that is unattainable in conventional LLPS droplets. By encoding the recognition site of the restriction enzyme in the motif sequences, DNA droplets were transiently yet directionally expelled by the resulting vacuole formation [51].
Computational nucleic acid droplets
Boolean operation on DNA droplets
Since the fusion of DNA engineering and computer science by Adleman [102], DNA has been recognized as a promising material for molecular computing owing to its high specificity and ease of massive parallelization. DNA computing and DNA storage have been studied as emerging information processing technologies exploiting DNA base sequence complementarity. The field involves solving computational problems [102,103,104], constructing logic elements [105,106,107], building neural networks [108,109,110] and storing information [111,112,113]. Nucleic acid sequences are treated as inputs in SDRs [105, 106, 114,115,116], enzyme reactions [107, 117,118,119,120], or DNAzymes [104], which are accepted by various DNA structures, including the i-motif for changes in pH [121, 122], the G-quadruplex that detects ion concentrations [123], and the aptamer that captures specific particles [124].
Recently, our group exploited the DNA linker system to realize DNA-based computational architectures. We defined the demixed state of droplets as the Boolean variable [1] and the mixed state as [0]. This bit representation of the phase state of DNA droplets was used to construct a DNA droplet-based AND gate (Fig. 5a-i) [31]. A mixed droplet was prepared by mixing two orthogonal DNAs and a linker DNA. Upon inputs of two targeted DNA or RNA sequences (input [1, 1]), this single-phase droplet phase-separated into two distinct phases of orthogonal DNAs as a result of SDRs at the linker binding sites (Fig. 5a-ii). This means that the DNA droplet generated an output of [1] upon inputs of [1, 1], corresponding to the AND operation (Fig. 5a-iii). A further complex combination of AND and NOT gates was demonstrated using triple DNA droplets for the detection of early-stage breast cancer. The linker-based molecular computing using DNA droplets has great potential for broader implementation in more complex logic operations, such as a neural network classifier and random access memory.
DNA droplets computing an AND gate. a Scheme of a DNA droplet AND gate. The mixed droplets are defined as [0] state, whereas the demixed droplets are the [1] state. The state change from a mixed state to a demixed state occurs only upon the input of two specific nucleic acid sequences. b Cleavage process of a six-branched linker. Toehold sequences for the SDR were applied to the receptor mechanism (Receptor 1, Receptor 2). Input DNAs (Input 1, Input 2) start SDR from the receptors, deactivating the cross-bridging capability. c Microscopy images of a DNA droplet AND gate for each input pattern. Scale bar: 10 μm. Reproduced [31] with permission. Copyright 2022, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim
Boolean operation on RNA droplets
Increasing evidence underscores the RNA-centric view that intrinsic RNA self-assembly is the major contributor to the formation and function of intracellular condensates [125,126,127,128,129,130]. In protein-free environments, RNA homopolymers (e.g., poly(rU)) [130] and RNA polymers with trinucleotide repeats (e.g., CAG repeats linked to neurodegenerative diseases) [130,131,132] can phase separate into droplets in the presence of divalent Mg2+ ions [133].
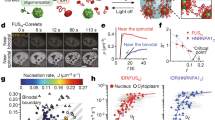
Recently, RNA base-sequence programmability has been used to engender functional RNA droplets assembled from designed RNA nanostructures. Our group has demonstrated computational RNA droplets capable of the ‘AND’ operation (Fig. 6a, b) [53]. The multibranched nanostructure building block (‘computational motif’) comprised six single-stranded RNAs of well-engineered sequences base-paired in the stem regions. Each branch terminating in a stem loop was designed to enable significantly stable coaxial base stacking between two stem loops [134], known as the kissing loop (KL) interaction [135, 136]. The designed RNA motifs optimized for KL interactions underwent phase separation into liquid-state RNA droplets as a result of network-like structuring. Notably, the computational motif possessed two extensions terminating in a single-stranded toehold to perform the ‘AND’ logic operation (Fig. 6c); only upon the input of two target microRNAs (miRNAs) (inputs [1, 1], Fig. 6e) could the quadrivalent motif break up into two divalent substructures through SDRs. The consequent remodeling of the network-like structure into a linear chain-like structure (Fig. 6d) led to a liquid-to-dispersed phase-state change in the condensates (output [1]). Conversely, the other combinations of inputs ([1, 0], [0, 1], and [0, 0]) did not allow a reduction in the motif valency, conserving the phase state as a liquid state (Fig. 6f, g). Furthermore, by leveraging macroscopic phase-state alterations, naked-eye-visible detection of the target miRNAs was demonstrated without the use of costly imaging setups (Fig. 6h).
Computational RNA droplets capable of ‘AND’ logic operation [53]. a Truth table of ‘AND’ logic operation. The input of a target miRNA corresponds to ‘1’. b Illustration of the computational RNA droplets that sequence-specifically favor a liquid-to-dispersed phase-state change upon inputs of two target miRNAs via c strand displacement reactions at the toehold regions. d A reduction in the motif valency from \(f=4\) to \(f=2\) enables a structural rearrangement of liquid condensates from network-like to chain-like structures. e Sequences of miRNAs linked to early-stage breast cancer [150] (m1, 2: targets; m3, 4: nontargets). f Schematics of motif structures before and after various inputs. g Snapshots of the RNA droplets captured before and after ( + 3.5 h) the inputs. The dye used for staining was SYBR Gold. Scale bars: 50 µm. h Naked-eye detection of the ‘AND’ operation. A mixture of RNA droplets and inputs was centrifuged in PCR tubes to yield pellets at the bottom. Methyl-green pyronin, as a staining solution, can detect the phase state of the condensates. [0, 0] refers to an input of a buffer. Scale bars: (zoomed-out; Rows 1, 3) 5 mm, (zoomed-in; Rows 2, 4) 2 mm
The multistranded RNA motif adopted above is suitable for SDR-mediated motif breakup. Other groups have reported the cotranscriptional formation of RNA condensates, where the processes of transcription from a specific DNA template and the self-folding of the single-stranded transcript into a prescribed motif cooccur [54, 55]. However, the single-stranded self-folded structure has limited flexibility in incorporating SDR sites, presumably restricting the applicability of the resulting condensates to computational use.
Application in biomedical engineering
Carefully engineered bioapplications integrate cargo transport, durability, selective targeting, and stimulus responsiveness, making them ideal for drug delivery and imaging applications. For widespread implementation, these biobased applications rely on precise molecular selectivity, which can be effectively realized by DNA droplets [60]. Previous work has demonstrated bacterial immobilization [34, 122] via the dynamic aggregation of core–shell DNA particles, enabled by input-triggered disruption of the outer shell, and the recruitment and release of guest molecules using DNA–RNA-aptamer hybrid condensates [45].
Additionally, the most recent work from our group demonstrated that computational DNA droplets coupled with a pathway-controlled division mechanism could be used to compare miRNA concentrations [43] (Fig. 7). As discussed earlier, the concentration of the protector RNA, which delays linker cleavage, determined the cleavage order of two linkers (AB- and AC-linkers) in ternary-mixed DNA droplets, dictating the division pathway (Fig. 4a, b). For example, when the concentration of protector RNA for the AB-linker was greater than that for the AC-linker, the AC-linker was cleaved first, leading to Pathway 1 division. Thus, the observed division pathway indicated the relative levels of protector RNA concentrations for the AB-linker and AC-linker (Fig. 7), serving as a miRNA concentration comparator. Thus, the programmable phase separation of DNA droplets has great potential as a tailor-made diagnostic tool.
miRNA concentration comparator using the pathway-controlled division mechanism of DNA droplets. The observed division pathway indicated the relative levels of miRNAs (miRNA AB and miRNA AC) used as protector RNAs. Adapted with permission [43]. Copyright 2024, Nature Publishing Group
Simulation modeling
The various behaviors of DNA droplets involve bottom-up contributions from motif-level reactions. Owing to the large number of motifs of interest, all-atom molecular dynamics models are limited by current computational capabilities. Instead, coarse-grained models are commonly applied to simulate higher-order DNA structures. oxDNA [137] is the best-known coarse-grained model and effectively reduces computational complexity by coarse-graining nucleotides as rigid bodies with multiple interaction sites connected via strings. Owing to its accessibility as a standalone package or as a module of LAMMPS [138] and its track record in the field of DNA nanotechnology, oxDNA has been widely applied to simulate DNA motif condensates (Fig. 8a) [33, 71, 72, 139, 140]. Using LAMMPS, a more advanced model coarse-grained a Y-shaped nanostar as a module consisting of seven rigid particles and three specific patches at the arm tips, and was used to evaluate the rheological properties of hydrogels (Fig. 8b) [141,142,143,144,145]. These coarse-grained models are still limited by computational power when modeling the microscale droplet dynamics.
Numerical simulation using the coarse-grained model and continuum model. a (i) NDA nanostars with a valence of 2–4 modeled by oxDNA. (ii) Simulation snapshot of a binary mixture composed of 50 trimers (violet) and 100 dimers (green). Reproduced [72] under the terms of the Creative Commons CC BY 4.0 license. b (i) Y-shaped nanostars modeled with 10 beads: one central bead, six structural beads arranged in three arms (blue), and three patch beads (red and yellow). The interactions of neighboring beads and angle constraints are described by harmonic potentials, and the attraction between complementary patches representing SEs is described by Lennard–Jones potentials. (ii) Simulation snapshots of the fully associated system in the unsheared (left) and sheared states (right). Reproduced with permission [145]. Copyright 2019, American Chemical Society. c (i) Coarse-grained motifs with X-shaped particles can diffuse in four directions on a two-dimensional plane. The system free energy is reduced only for arm-to-arm configurations and is not affected by any other contact. (ii) DNA droplet segregation dynamics under confinement in simulation (top) and wet (bottom) experiments. The time of the simulation experiment refers to the number of simulation steps. Scale bars (wet experiments, bottom): 10 µm. Reproduced [42] under the terms of the Creative Commons CC BY 4.0 license. d Snapshot of simulation of condensate formation by DNA subunits of different sizes. The subunit size on the right is five times larger than that on the left. Reproduced with permission [41]. Copyright 2022 American Chemical Society. e (i) Droplets are randomly displaced with Stokes drag and coalesce probability when they overlap. The amount of displacement and the probability of coalescence depend on the size of the droplet, and the volume and center of mass are conserved when they merge. (ii) Snapshots of droplet growth simulation. Reproduced with permission [146]. f Snapshots of a simulation combining the Cahn–Hilliard equation and a chemical reaction. An inhibitor deactivates the phase-separating material, causing droplets to shrink over time (left); droplets regrow after the addition of an activator (right). Reproduced [44] under the terms of the Creative Commons CC BY 4.0 license
A complex of two orthogonal motifs and a splittable linker [22] was coarse-grained as a two-dimensional X-shaped particle (splitting into two L-shaped particles) to model the segregation of the droplet by the splitting of the linker [42] (Fig. 8c). The effect of motif size on the aggregation process was investigated on 10,000 particles, each represented by a three-branched motif of various sizes (Fig. 8d) [41].
In addition to the coarse-grained model, coalescence-driven droplet growth was modeled, starting with 40,000,000 particles 10 nm in radius, confirming both nucleation and spinodal dynamics (Fig. 8e) [146]. Furthermore, the phase separation behavior, involving monomers and their activators/inhibitors, was continuously modeled using the Cahn‒Hilliard equation, which describes spinodal dynamics (Fig. 8f) [44]. Nevertheless, there is a need for cross-scale models that connect microscale phenomena with the structural and thermodynamic properties of diverse motifs. Such models will accelerate droplet dynamics research.
Conclusions
This Focus Review highlights the key features of DNA liquid condensates, such as their dynamic regulation, programmability, designability of chemical modifications, and ability to host diverse reactions. These properties and their programmability through information encoding via base-sequence design enable a broad range of applications across various fields.
Their dynamics are time-controllable, with kinetic mechanisms tailored to specific inputs, such as SDRs and enzymatic reactions. Our group has introduced the linker system as an efficient means of programming the phase dynamics of multiple DNA phases, with a thermodynamic description of the fueling roles of input molecules. These tunable features make DNA condensates versatile tools for applications ranging from nanotechnology to biomedicine. SDRs enable different applications, such as DNA logic operations (e.g., AND gates), nonenzymatic cascade reactions, and biomedical uses, including HIV detection and antimicrobial activity. However, interactions between living cells and DNA condensates are still a new topic, awaiting deeper investigation of in vivo applications and further functionalization.
With respect to motif associations, condensates with heterogeneous subcompartments are still limited to concentric multilayered architectures [23], patchwork subcompartmentalization via cross-linked motifs [22, 24], and core–shell structures [34, 50]. To advance synthetic cell development, more sophisticated architectures are essential to create organelle-like molecular carriers for different biomolecules, storage, reaction encapsulation, and biomimetic autophagy [147, 148]. These architectures would then enable complex metabolism, fulfilling critical functions that increase synthetic cell applicability.
References
Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–32.
Linsenmeier M, Hondele M, Grigolato F, Secchi E, Weis K, Arosio P. Dynamic arrest and aging of biomolecular condensates are modulated by low-complexity domains, RNA and biochemical activity. Nat Commun. 2022;13:3030.
Weis K. Dead or alive: DEAD-box ATPases as regulators of ribonucleoprotein complex condensation. Biol Chem. 2021;402:653–61.
do Amaral MJ, Mohapatra S, Passos AR, Lopes da Silva TS, Carvalho RS, da Silva Almeida M, et al. Copper drives prion protein phase separation and modulates aggregation. Sci Adv. 2023;9:eadi7347.
Fuxreiter M, Vendruscolo M. Generic nature of the condensed states of proteins. Nat Cell Biol. 2021;23:587–94.
Mittag T, Pappu RV. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol Cell. 2022;82:2201–14.
Pappu RV, Cohen SR, Dar F, Farag M, Kar M. Phase transitions of associative biomacromolecules. Chem Rev. 2023;123:8945–87.
Antifeeva IA, Fonin AV, Fefilova AS, Stepanenko OV, Povarova OI, Silonov SA, et al. Liquid-liquid phase separation as an organizing principle of intracellular space: overview of the evolution of the cell compartmentalization concept. Cell Mol Life Sci. 2022;79:251.
Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science 2017:357:eaaf4382.
Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39–58.
Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–98.
Hen Y, Posavec L, Bolisetty S, Hilty FM, Nyströ MG, Kohlbrecher J, et al. Amyloid fibril systems reduce, stabilize and deliver bioavailable nanosized iron. Nat Nanotechnol. 2017;12:642–7.
Hudalla GA, Sun T, Gasiorowski JZ, Han H, Tian YF, Chong AS, et al. Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat Mater. 2014;13:829–36.
Dler-Abramovich L, Gazit E. The physical properties of supramolecular peptide assemblies: from building block association to technological applications. Chem Soc Rev. 2014;43:6881–93.
Zhou X-M, Entwistle A, Zhang H, Jackson AP, Mason TO, Shimanovich U, et al. Self-assembly of amyloid fibrils that display active enzymes. ChemCatChem. 2014;6:1961–8.
Strom AR, Emelyanov AV, Mir MR, Fyodorov DV, Darzacq XR, Karpen GH. Phase separation drives heterochromatin domain formation. Biophys J. 2018;114:445a.
Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–40.
Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833–40.
Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F, et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021;220:e202008030.
Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018;361:eaar3958.
Erdel F, Rippe K. Formation of chromatin subcompartments by phase separation. Biophys J. 2018;114:2262–70.
Sato Y, Sakamoto T, Takinoue M. Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets. Sci Adv. 2020;6:eaba3471.
Bucci J, Malouf L, Tanase DA, Farag N, Lamb JR, Rubio-Sánchez R, et al. Enzyme-responsive DNA condensates. J Am Chem Soc. 2024;146:31529–37.
Jeon B-J, Nguyen DT, Saleh OA. Sequence-controlled adhesion and microemulsification in a two-phase system of DNA liquid droplets. J Phys Chem B. 2020;124:8888–95.
Conrad N, Kennedy T, Fygenson DK, Saleh OA. Increasing valence pushes DNA nanostar networks to the isostatic point. Proc Natl Acad Sci Usa. 2019;116:7238–43.
Nguyen DT, Jeon B-J, Abraham GR, Saleh OA. Length-dependence and spatial structure of DNA partitioning into a DNA liquid. Langmuir. 2019;35:14849–54.
Jeon B-J, Nguyen DT, Abraham GR, Conrad N, Fygenson DK, Saleh OA. Salt-dependent properties of a coacervate-like, self-assembled DNA liquid. Soft Matter. 2018;14:7009–15.
Wilken S, Chaderjian A, Saleh OA. Spatial organization of phase-separated DNA droplets. Phys. Rev. X. 2023;13:031014.
Sato Y, Takinoue M. Capsule-like DNA hydrogels with patterns formed by lateral phase separation of DNA nanostructures. JACS Au. 2022;2:159–68.
Sato Y, Takinoue M. Sequence-dependent fusion dynamics and physical properties of DNA droplets. Nanoscale Adv. 2023;5:1919–25.
Gong J, Tsumura N, Sato Y, Takinoue M. Computational DNA droplets recognizing miRNA sequence inputs based on liquid–liquid phase separation. Adv. Funct. Mater. 2022;32:2202322.
Saleh OA, Jeon B-J, Liedl T. Enzymatic degradation of liquid droplets of DNA is modulated near the phase boundary. Proc Natl Acad Sci USA. 2020;117:16160–6.
Lee T, Do S, Lee JG, Kim D-N, Shin Y. The flexibility-based modulation of DNA nanostar phase separation. Nanoscale. 2021;13:17638–47.
Walczak M, Brady RA, Mancini L, Contini C, Rubio-Sánchez R, Kaufhold WT, et al. Responsive core-shell DNA particles trigger lipid-membrane disruption and bacteria entrapment. Nat Commun. 2021;12:4743.
Leathers A, Walczak M, Brady RA, Al Samad A, Kotar J, Booth MJ, et al. Reaction–diffusion patterning of DNA-based artificial cells. J. Am. Chem. Soc. 2022 https://doi.org/10.1021/jacs.2c06140.
Samanta A, Sabatino V, Ward TR, Walther A. Functional and morphological adaptation in DNA protocells via signal processing prompted by artificial metalloenzymes. Nat Nanotechnol. 2020;15:914–21.
Samanta A, Hörner M, Liu W, Weber W, Walther A. Signal-processing and adaptive prototissue formation in metabolic DNA protocells. Nat Commun. 2022;13:3968.
Liu W, Samanta A, Deng J, Akintayo CO, Walther A, Mechanistic insights into the phase separation behavior and pathway-directed information exchange in all-DNA droplets. Angew. Chem. Weinheim Bergstr. Ger 2022. https://doi.org/10.1002/ange.202208951.
Zhao Q-H, Cao F-H, Luo Z-H, Huck WTS, Deng N-N. Photoswitchable molecular communication between programmable DNA-based artificial membraneless organelles. Angew Chem Int Ed Engl. 2022;61:e202117500.
Nguyen DT, Saleh OA. Tuning phase and aging of DNA hydrogels through molecular design. Soft Matter. 2017;13:5421–7.
Agarwal S, Osmanovic D, Klocke MA, Franco E. The growth rate of DNA condensate droplets increases with the size of participating subunits. ACS Nano. 2022;16:11842–51.
Tran MP, Chatterjee R, Dreher Y, Fichtler J, Jahnke K, Hilbert L, et al. A DNA segregation module for synthetic cells. Small. 2023;19:e2202711.
Maruyama T, Gong J, Takinoue M. Temporally controlled multistep division of DNA droplets for dynamic artificial cells. Nat Commun. 2024;15:7397.
Agarwal S, Osmanovic D, Dizani M, Klocke MA, Franco E. Dynamic control of DNA condensation. Nat Commun. 2024;15:1915.
Dizani M, Sorrentino D, Agarwal S, Stewart JM, Franco E. Protein recruitment to dynamic DNA-RNA host condensates. J Am Chem Soc. 2024;146:29344–54.
Agarwal S, Dizani M, Osmanovic D, Franco E. Light-controlled growth of DNA organelles in synthetic cells. Interface Focus. 2023;13:20230017.
Do S, Lee C, Lee T, Kim D-N, Shin Y. Engineering DNA-based synthetic condensates with programmable material properties, compositions, and functionalities. Sci Adv. 2022;8:eabj1771.
Deng J, Walther A. Programmable and chemically fueled DNA coacervates by transient liquid-liquid phase separation. Chem. 2020;6:3329–43.
Malouf L, Tanase DA, Fabrini G, Brady RA, Paez-Perez M, Leathers A, et al. Sculpting DNA-based synthetic cells through phase separation and phase-targeted activity. Chem. 2023;9:3347–64.
Liu W, Deng J, Song S, Sethi S, Walther A. A facile DNA coacervate platform for engineering wetting, engulfment, fusion and transient behavior. Commun Chem. 2024;7:100.
Saleh OA, Wilken S, Squires TM, Liedl T. Vacuole dynamics and popping-based motility in liquid droplets of DNA. Nat Commun. 2023;14:3574.
Ohno H, Kijima J, Ochi Y, Shoji M, Taira J, Mabuchi T, et al. Oligolysine enhances and inhibits DNA condensate formation. ACS Omega. 2025 https://doi.org/10.1021/acsomega.5c01928.
Udono H, Fan M, Saito Y, Ohno H, Nomura S-IM, Shimizu Y, et al. Programmable computational RNA droplets assembled via kissing-loop interaction. ACS Nano. 2024;18:15477–86.
Fabrini G, Farag N, Nuccio SP, Li S, Stewart JM, Tang AA, et al. Co-transcriptional production of programmable RNA condensates and synthetic organelles. Nat. Nanotechnol. 2024 https://doi.org/10.1038/s41565-024-01726-x.
Stewart JM, Li S, Tang AA, Klocke MA, Gobry MV, Fabrini G, et al. Modular RNA motifs for orthogonal phase separated compartments. Nat Commun. 2024;15:6244.
Fraccia TP, Jia TZ. Liquid crystal coacervates composed of short double-stranded DNA and cationic peptides. ACS Nano. 2020;14:15071–82.
Vieregg JR, Lueckheide M, Marciel AB, Leon L, Bologna AJ, Rivera JR, et al. Oligonucleotide-peptide complexes: phase control by hybridization. J Am Chem Soc. 2018;140:1632–8.
Todisco M, Fraccia TP, Smith GP, Corno A, Bethge L, Klussmann S, et al. Nonenzymatic polymerization into long linear RNA templated by liquid crystal self-assembly. ACS Nano. 2018;12:9750–62.
Jiang H, Pan V, Vivek S, Weeks ER, Ke Y. Programmable DNA hydrogels assembled from multidomain DNA strands. Chembiochem. 2016;17:1156–62.
Udono H, Gong J, Sato Y, Takinoue M. DNA droplets: Intelligent, dynamic fluid. Adv Biol (Weinh). 2023;7:e2200180.
Takinoue M. DNA droplets for intelligent and dynamical artificial cells: from the viewpoint of computation and non-equilibrium systems. Interface Focus. 2023;13:20230021.
Sato Y, Takinoue M. Pioneering artificial cell-like structures with DNA nanotechnology-based liquid-liquid phase separation. Biophys Physicobiol. 2024;21:e210010.
Abraham GR, Chaderjian AS, Nguyen ABN, Wilken S, Saleh OA. Nucleic acid liquids. Rep Prog Phys 2024:87:066601.
Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–44.
Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B-Z transition of DNA. Nature. 1999;397:144–6.
Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–20.
Tomaru T, Suzuki Y, Kawamata I, Nomura S-IM, Murata S. Stepping operation of a rotary DNA origami device. Chem Commun (Camb). 2017;53:7716–9.
Iwabuchi S, Kawamata I, Murata S, Nomura S-IM. A large, square-shaped, DNA origami nanopore with sealing function on a giant vesicle membrane. Chem Commun (Camb). 2021;57:2990–3.
Sato Y, Hiratsuka Y, Kawamata I, Murata S, Nomura S-IM. Micrometer-sized molecular robot changes its shape in response to signal molecules. Sci Robot. 2017;2:eaal3735.
Kuzuya A, Ohya Y. Nanomechanical molecular devices made of DNA origami. Acc Chem Res. 2014;47:1742–9.
Locatelli E, Rovigatti L. An accurate estimate of the free energy and phase diagram of all-DNA bulk fluids. Polymers (Basel). 2018;10:447.
Locatelli E, Handle PH, Likos CN, Sciortino F, Rovigatti L. Condensation and demixing in solutions of DNA nanostars and their mixtures. ACS Nano. 2017;11:2094–102.
Fisher RS, Elbaum-Garfinkle S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat Commun. 2020;11:4628.
Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–3.
SantaLucia J Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–62.
Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci Usa. 2015;112:E5237–45.
Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, et al. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60:220–30.
Mathieu C, Pappu RV, Taylor JP. Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science. 2020;370:56–60.
Li Z, Liu X, Liu M. Stress granule homeostasis, aberrant phase transition, and amyotrophic lateral sclerosis. ACS Chem Neurosci. 2022;13:2356–70.
Boyko S, Qi X, Chen T-H, Surewicz K, Surewicz WK. Liquid-liquid phase separation of tau protein: The crucial role of electrostatic interactions. J Biol Chem. 2019;294:11054–9.
Fernandez-Castanon J, Bianchi S, Saglimbeni F, Di Leonardo R, Sciortino F. Microrheology of DNA hydrogel gelling and melting on cooling. Soft Matter. 2018;14:6431–8.
Xing Z, Caciagli A, Cao T, Stoev I, Zupkauskas M, O’Neill T, et al. Microrheology of DNA hydrogels. Proc Natl Acad Sci Usa. 2018;115:8137–42.
Guo W, Lu C-H, Orbach R, Wang F, Qi X-J, Cecconello A, et al. pH-stimulated DNA hydrogels exhibiting shape-memory properties. Adv Mater. 2015;27:73–8.
Biffi S, Cerbino R, Bomboi F, Paraboschi EM, Asselta R, Sciortino F, et al. Phase behavior and critical activated dynamics of limited-valence DNA nanostars. Proceedings Natl Acad Sci. 2013;110:15633–7.
Laflamme G, Mekhail K. Biomolecular condensates as arbiters of biochemical reactions inside the nucleus. Commun Biol. 2020;3:773.
Mameuda A, Takinoue M, Kamiya K. Control of reversible formation and dispersion of the three enzyme networks integrating DNA computing. Anal Chem. 2023;95:9548–54.
Koga S, Williams DS, Perriman AW, Mann S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat Chem. 2011;3:720–4.
Aumiller WM Jr, Keating CD. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat Chem. 2016;8:129–37.
Donau C, Späth F, Sosson M, Kriebisch BAK, Schnitter F, Tena-Solsona M, et al. Active coacervate droplets as a model for membraneless organelles and protocells. Nat Commun. 2020;11:5167.
Nakashima KK, van Haren MHI, André AAM, Robu I, Spruijt E. Active coacervate droplets are protocells that grow and resist Ostwald ripening. Nat Commun. 2021;12:3819.
Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581:209–14.
Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22:165–82.
Söding J, Zwicker D, Sohrabi-Jahromi S, Boehning M, Kirschbaum J. Mechanisms for active regulation of biomolecular condensates. Trends Cell Biol. 2020;30:4–14.
Simmel FC, Yurke B, Singh HR. Principles and applications of nucleic acid strand displacement reactions. Chem Rev. 2019;119:6326–69.
Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–90.
Lin Z, Beneyton T, Baret J-C, Martin N. Coacervate droplets for synthetic cells. Small Methods 2023;7:e2300496.
Slootbeek AD, van Haren MHI, Smokers IBA, Spruijt E. Growth, replication and division enable evolution of coacervate protocells. Chem Commun 2022. https://doi.org/10.1039/d2cc03541c.
Lu T, Spruijt E. Multiphase complex coacervate droplets. J Am Chem Soc. 2020;142:2905–14.
Yewdall NA, André AAM, Lu T, Spruijt E. Coacervates as models of membraneless organelles. Curr Opin Colloid Interface Sci. 2021;52:101416.
Furuki T, Sakuta H, Yanagisawa N, Tabuchi S, Kamo A, Shimamoto DS, et al. Marangoni droplets of dextran in PEG solution and its motile change due to coil-globule transition of coexisting DNA. ACS Appl Mater Interfaces. 2024;16:43016–25.
Zhang S, Contini C, Hindley JW, Bolognesi G, Elani Y, Ces O. Engineering motile aqueous phase-separated droplets via liposome stabilisation. Nat Commun. 2021;12:1–11.
Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–4.
Lipton RJ. DNA solution of hard computational problems. Science. 1995;268:542–5.
Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat Biotechnol. 2003;21:1069–74.
Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–8.
Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–201.
Song T, Eshra A, Shah S, Bui H, Fu D, Yang M, et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat Nanotechnol. 2019;14:1075–81.
Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–72.
Okumura S, Gines G, Lobato-Dauzier N, Baccouche A, Deteix R, Fujii T, et al. Nonlinear decision-making with enzymatic neural networks. Nature. 2022;610:496–501.
Evans CG, O’Brien J, Winfree E, Murugan A. Pattern recognition in the nucleation kinetics of non-equilibrium self-assembly. Nature. 2024;625:500–7.
Wang S, Mao X, Wang F, Zuo X, Fan C. Data storage using DNA. Adv Mater. 2024;36:e2307499.
Lin KN, Volkel K, Cao C, Hook PW, Polak RE, Clark AS, et al. A primordial DNA store and compute engine. Nat Nanotechnol. 2024;19:1654–64.
Zhang C, Wu R, Sun F, Lin Y, Liang Y, Teng J, et al. Parallel molecular data storage by printing epigenetic bits on DNA. Nature. 2024;634:824–32.
Yurke B, Turberfield A, Mills A, Simmel F, Neumann J. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–8.
Venkataraman S, Dirks RM, Rothemund PWK, Winfree E, Pierce NA. An autonomous polymerization motor powered by DNA hybridization. Nat Nanotechnol. 2007;2:490–4.
Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131:17303–14.
Takinoue M, Kiga D, Shohda K-I, Suyama A. RNA oscillator: Limit cycle oscillations based on artificial biomolecular reactions. New Gener Comput. 2009;27:107–27.
Baccouche A, Montagne K, Padirac A, Fujii T, Rondelez Y. Dynamic DNA-toolbox reaction circuits: a walkthrough. Methods. 2014;67:234–49.
Sharon JA, Dasrath C, Fujiwara A, Snyder A, Blank M, O’Brien S, et al. Trumpet is an operating system for simple and robust cell-free biocomputing. Nat Commun. 2023;14:2257.
Kawamata I, Nishiyama K, Matsumoto D, Ichiseki S, Keya JJ, Okuyama K, et al. Autonomous assembly and disassembly of gliding molecular robots regulated by a DNA-based molecular controller. Sci Adv. 2024;10:eadn4490.
Karna D, Stilgenbauer M, Jonchhe S, Ankai K, Kawamata I, Cui Y, et al. Chemo-mechanical modulation of cell motions using DNA nanosprings. Bioconjug Chem. 2021;32:311–7.
Walczak M, Mancini L, Xu J, Raguseo F, Kotar J, Cicuta P, et al. A synthetic signaling network imitating the action of immune cells in response to bacterial metabolism. Adv Mater. 2023;35:e2301562.
Wang D, Yang Y, Chen F, Lyu Y, Tan W. Network topology–directed design of molecular CPU for cell-like dynamic information processing. Science Adv. 2022;8:eabq0917.
Lai W, Xiong X, Wang F, Li Q, Li L, Fan C, et al. Nonlinear Regulation of Enzyme-Free DNA Circuitry with Ultrasensitive Switches. ACS Synth Biol. 2019;8:2106–12.
Tian S, Curnutte HA, Trcek T. RNA granules: A view from the RNA perspective. Molecules 2020;25:3130.
Bevilacqua PC, Williams AM, Chou H-L, Assmann SM. RNA multimerization as an organizing force for liquid–liquid phase separation. RNA. 2022;28:16–26.
Isiktas AU, Eshov A, Yang S, Guo JU. Systematic generation and imaging of tandem repeats reveal base-pairing properties that promote RNA aggregation. Cell Rep. Methods. 2022;2:100334.
Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci Usa. 2018;115:2734–9.
Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174:791–802.
Wadsworth GM, Zahurancik WJ, Zeng X, Pullara P, Lai LB, Sidharthan V, et al. RNAs undergo phase transitions with lower critical solution temperatures. Nat Chem. 2023;15:1693–704.
Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–7.
Ma Y, Li H, Gong Z, Yang S, Wang P, Tang C. Nucleobase clustering contributes to the formation and hollowing of repeat-expansion RNA condensate. J Am Chem Soc. 2022;144:4716–20.
Poudyal RR, Sieg JP, Portz B, Keating C, Bevilacqua P. RNA sequence and structure control assembly and function of RNA condensates. RNA. 2021;27:1589–601.
Li PTX, Bustamante C, Tinoco I Jr. Unusual mechanical stability of a minimal RNA kissing complex. Proc Natl Acad Sci Usa. 2006;103:15847–52.
Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl. 1999;38:2326–43.
Bose M, Rankovic B, Mahamid J, Ephrussi A. An architectural role of specific RNA-RNA interactions in oskar granules. Nat Cell Biol. 2024;26:1934–42.
Snodin BEK, Randisi F, Mosayebi M, Šulc P, Schreck JS, Romano F, et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J Chem Phys. 2015;142:234901.
Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J Comput Phys. 1995;117:1–19.
Rovigatti L, Smallenburg F, Romano F, Sciortino F. Gels of DNA nanostars never crystallize. ACS Nano. 2014;8:3567–74.
Brady RA, Kaufhold WT, Brooks NJ, Foderà V, Di Michele L. Flexibility defines structure in crystals of amphiphilic DNA nanostars. J Phys Condens Matter. 2019;31:074003.
Stoev ID, Cao T, Caciagli A, Yu J, Ness C, Liu R, et al. On the role of flexibility in linker-mediated DNA hydrogels. Soft Matter. 2020;16:990–1001.
Walia S, Morya V, Gangrade A, Naskar S, Guduru Teja A, Dalvi S, et al. Designer DNA hydrogels stimulate 3D cell invasion by enhanced receptor expression and membrane endocytosis. ACS Biomater Sci Eng. 2021;7:5933–42.
Naskar S, Bhatia D, Lin S-T, Maiti PK. Mechanistic insight into the structure, thermodynamics and dynamics of equilibrium gels of multi-armed DNA nanostars. Phys Chem Chem Phys. 2023;25:7847–58.
Gutiérrez Fosado YA. Nanostars planarity modulates the rheology of DNA hydrogels. Soft Matter. 2023;19:4820–8.
Xing Z, Ness C, Frenkel D, Eiser E. Structural and Linear Elastic Properties of DNA Hydrogels by Coarse-Grained Simulation. Macromolecules. 2019;52:504–12.
Wilken S, Gutierrez J, Saleh OA. Nucleation dynamics of a model biomolecular liquid. J Chem Phys. 2024;160:214903.
Gouveia B, Kim Y, Shaevitz JW, Petry S, Stone HA, Brangwynne CP. Capillary forces generated by biomolecular condensates. Nature. 2022;609:255–64.
Agudo-Canalejo J, Schultz SW, Chino H, Migliano SM, Saito C, Koyama-Honda I, et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021;591:142–6.
Jambon-Puillet E, Testa A, Lorenz C, Style RW, Rebane AA, Dufresne ER. Phase-separated droplets swim to their dissolution. Nat Commun. 2024;15:3919.
Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–34.
Acknowledgements
This work was supported by MEXT/JSPS KAKENHI (Nos. JP20H05701, JP20H00619, JP20H05935, and JP24H00070 to M.T., and No. JP22K14528 to H.U.), a Japanese Government (MEXT) Scholarship for foreign students to N.N.E., a JSPS Grant-in-Aid for JSPS fellows (No. JP22J00940 to H.U.), Research Fellowships of JSPS for Young Scientists (No. JP22KJ1346 to T.M. and No. 24KJ1076 to N.Y.), the Human Frontier Science Program (HFSP; RGP0016/2022-102 to M.T.), and JST Adopting Sustainable Partnerships for Innovative Research Ecosystem (ASPIRE) (No. JPMJAP24B4 to M.T.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Udono, H., Maruyama, T., Evangelista, N.N. et al. Synthetic nucleic-acid droplets: a bioprogramming platform for designer microliquids. Polym J 57, 845–862 (2025). https://doi.org/10.1038/s41428-025-01050-8
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41428-025-01050-8