Abstract
The morphological transformation of the pectoral/shoulder girdle is fundamental to the water-to-land transition in vertebrate evolution. Although previous studies have resolved the embryonic origins of tetrapod shoulder girdles, those of fish pectoral girdles remain uncharacterized, creating a gap in the understanding of girdle transformation mechanisms from fish to tetrapods. Here, we identify the embryonic origins of the zebrafish pectoral girdle, including the cleithrum as an ancestral girdle element lost in extant tetrapods. Our combinatorial approach of photoconversion and genetic lineage tracing demonstrates that cleithrum development combines four adjoining embryonic populations. A comparison of these pectoral girdle progenitors with extinct and extant vertebrates highlights that cleithrum loss, indispensable for neck evolution, is associated with the disappearance of its unique developmental environment at the head/trunk interface. Overall, our study establishes an embryological framework for pectoral/shoulder girdle formation and provides evolutionary trajectories from their origin in water to diversification on land.
Similar content being viewed by others
Introduction
The pectoral girdle anchors the pectoral appendage (pectoral fin or forelimb) to the body wall in jawed vertebrates1. The pectoral girdle comprises of dermal bones and cartilage or their replacement bones (endoskeleton), while cartilaginous fishes secondarily lost the dermal components2. In bony vertebrates with paired fins living in water (sarcopterygians and actinopterygians), the cleithrum3, one of the pectoral dermal bones, is both functionally and anatomically a major component. As vertebrates transitioned onto land, the relative size of the cleithrum in the pectoral girdle began to decrease and eventually disappeared independently in multiple lineages leading to extant tetrapods1,4,5. Consequently, except for some anuran amphibians6,7 and possibly turtles8, no extant tetrapods form the cleithrum, and enlarged endoskeletal elements are instead the dominant component of the pectoral girdle in tetrapods9. In contrast to these accumulating anatomical descriptions, the embryonic changes responsible for the evolutionary loss of the cleithrum and expansion of the endoskeleton remain elusive.
The major embryonic sources of the shoulder girdle in tetrapods have been identified by works in several model systems. Homotopic transplantations between chicken and quail embryos10 and genetic cell lineage tracing using Prx1:Cre in mice11 revealed that the clavicle, a dermal component of the shoulder girdle, derives mainly from the trunk lateral plate mesoderm (LPM). The same strategy of tissue transplantation and genetic cell lineage tracing using Mef2c-AHF:Cre mice (cardiopharyngeal marker) identified that the cardiopharyngeal mesoderm (CPM) also contributes to the medial extremity of the clavicle in chicken and mice10,12. The CPM or cardiopharyngeal field defines a deeply conserved progenitor field that forms at least in part within the anterior LPM13,14,15,16. Moreover, previous studies discovered that the scapula, an endoskeletal component of the girdle, in axolotls, chickens, and mice derives mainly from the trunk LPM and its dorsal portion from the somites, potentially due to the scapula’s developmental position across these mesodermal boundaries17,18,19,20. Consequently, the trunk LPM is an important contributor to both dermal and endoskeletal components in tetrapod shoulder girdles, while the CPM and somites contribute to their medial and dorsal margins to some extent.
Neural crest cells are migratory ectodermal cells with broad lineage potential that among its diverse descendant lineages contribute to canonical ectodermal tissues (neurons and gilia) and cranial mesenchymal tissues (skeletons and connective tissues)21,22. Previous work in axolotl tested the potential contribution of neural crest cells to the shoulder girdle, but did not detect any neural crest-derived cells in the shoulder girdle skeletons23. In contrast, tissue transplantations in chicken embryos24 and genetic lineage tracing using Sox10:Cre mice (with Sox10 being expressed in migratory neural crest cell4) identified that neural crest cells give rise to clavicles. More recent genetic tracing of Wnt1:Cre that marks pre-migratory neural crest cells in mice, however, found no neural crest cell-lineage in the shoulder girdle skeletons11,12. This inconsistency among lineage labeling studies may partially be due to the deployment of isolated cis-regulatory elements of different neural crest genes. The distribution of genetically labeled neural crest cell lineages in mice arguably varies depending on the cis-regulatory elements that drive Cre expression25. Consequently, the contribution of the neural crest cells to the shoulder skeletons must be resolved by additional cell lineage tracking such as region-specific photoconversions or tissue transplantations (see an example in ref. 26). Nonetheless, in sum, the contribution of neural crest cells to the tetrapod shoulder girdle has not been unambiguously determined and, if any, appears to be minor4,24.
Revealing the developmental processes forming the pectoral girdle in actinopterygians would provide mechanistic insights into how such intricate tetrapod shoulder development was established. However, in contrast to the accumulated, yet incomplete knowledge of the embryonic origins of tetrapod shoulder girdles, less is known about the developmental lineages contributing to the pectoral girdles of fishes with paired fins. Previous work assumed that the embryonic origin of the dermal pectoral components, including the fish cleithrum, is the neural crest, but this assumption has not yet been tested4. In contrast, Sox10:Cre-labeled lineages did not contribute to dermal components of the pectoral girdle in zebrafish27. However, this could be partially due to the deployment of Sox10 regulatory elements from mice in the used transgenics that might not fully recapitulate zebrafish sox10 expression or lineage. The endoskeletal scapulocoracoid, a cartilaginous precursor for the scapula and coracoid, seems to be formed within the fin bud mesenchyme in the larval zebrafish (reviewed in ref. 28), yet the embryonic origins of the endoskeletal component have not been fully identified either. Accordingly, the embryonic origins of the dermal and endoskeletal pectoral girdle components in bony fishes remain to be determined.
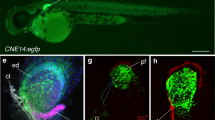
In zebrafish, the cleithrum lies between the head and trunk structures in the absence of a forming neck in fishes (Fig. 1a); the cleithrum posteriorly delineates the most caudal pharyngeal arch and the pericardium, the mesothelium-lined body cavity encapsulating the heart (Fig. 1b, c), and anteriorly delineates the trunk lateral body wall where the pectoral fin attaches (Fig. 1b, c). Moreover, the cleithrum develops closely associated with the pectoral musculature: cleithrohyoid, pectoral fin adductor/abductor, and posterior hypaxial muscles (Fig. 1b, c, see Supplementary Note 1 for nomenclature summary). Embryologically, these anatomical components associated with the cleithrum derive from distinct embryonic sources: the pharyngeal arch mesenchyme from cranial neural crest cells27,29, the pericardium from the LPM including tbx1-expressing CPM30,31, the lateral body wall (mesothelium and connective tissues of the posterior hypaxial muscle) and pectoral fin bud from the trunk LPM31,32,33, and the pectoral musculature from anterior somites34,35. Thus, given its anatomically unique position, cleithrum development in zebrafish potentially deploys any of these diverse embryonic cell populations (Fig. 1a–c).
a Left lateral view of the larval zebrafish. b, c Schematic view of horizontal sections obtained from two dorsoventrally different levels as indicated in (a), showing that the larval cleithrum is located at the interface made by multiple embryonic populations: anterior somites (pink), cranial neural crest cells (light blue), CPM (green), and trunk LPM (yellow). The embryonic origins of the pectoral girdle that develops at the boundary of fin-field LPM, cranial neural crest cells, and CPM remains to be identified. b, c Were drawn after open data sets available on Zebromes (https://neurodata.io/project/zebromes/) originally published in ref. 95. A anterior, ab abductor muscle, ad adductor muscle, cb 4–7 ceratobranchial 4–7, ccv common cardinal vein, cle cleithrum, clehy cleithrohyoid muscle, CPM cardiopharyngeal mesoderm, D dorsal, ed endoskeletal disc, ie inner ear, LPM lateral plate mesoderm, op operculum, pc pericardium, phy posterior hypaxial muscle, pn pronephros, sc scapulocoracoid.
Here, by combining region-specific photoconversions and genetic cell lineage analyses, we labeled the anterior somites, cranial neural crest cells, CPM, and trunk LPM to identify the embryonic lineage contributions to the pectoral girdle skeleton in zebrafish. Our results document that the cleithrum develops as a mosaic bone from multiple embryonic populations, while the scapulocoracoid exclusively derives from the fin field-associated LPM. A broad evolutionary comparison of the topographical position of the cleithrum implies that the multiple embryonic origins of the pectoral girdle skeletons are shared features among extinct and extant cleithrum-bearing species. Our data further propose that the evolutionary loss of the cleithrum in amniotes might have followed rearrangements of the ancestral developmental environment at the origins of the neck.
Results
Anterior somites give rise to pectoral musculature and the dorsal cleithrum
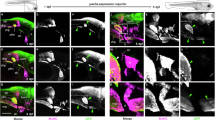
Previous work found that somites 1–4 are the major contributors to the pectoral musculature in zebrafish35. Of these, mesenchymal cells derived from somites 1-3 ventrally migrate over the prospective cleithrum area between the pharynx and the pectoral fin bud (Fig. 1a–c) and eventually develop into the cleithrohyoid muscle at the bottom of the pharynx33,35. We therefore tested whether these somitic mesenchymal cells also contribute to the cleithrum along their migratory route. To this end, we injected Kaede mRNA, encoding a photoconvertible protein, into one-cell stage embryos36 (see Methods for details) and labeled somites 1–3 by Kaede photoconversion from green to red fluorescence (Kaede-red) at the 10–12 somite stage (mid-segmentation period37) (Fig. 2a, b). We then examined the contribution of labeled cells to the pectoral region at 72 h post-fertilization (hpf) (protruding mouth stage37) when the bone matrix of the cleithrum, cartilage, and skeletal muscles become histologically and molecularly evident (see Supplementary Fig. 1). Due to laser illumination (405 nm) through the outside of the embryos to target somites, areas of the epidermis and neural tube dorsally and medially adjacent to the target somites, respectively, were also labeled (Fig. 2b). Given the lack of their contribution to skeletogenic mesenchyme38, these non-mesodermal cells were not expected to affect the analysis of somite-derived cell lineages (Fig. 2c). Labeled cells derived from the anterior three somites were found in the first to third myotomes (Fig. 2d, e) and the cleithrohyoid muscle (Fig. 2e, n = 8/8). The observed contributions of the anterior three somites to the myotomes and the cleithrohyoid muscle are consistent with previous reports34,35, supporting the efficacy of our photoconversion. Kaede-red signal was not found in the ventral cleithrum cells (n = 0/8; cleithrum positive/all labeled embryos), but in the cells delineating the dorsal cleithrum at the level of the myotome 2 (n = 4/8) (Fig. 2d–g; Supplementary Data 1). To comprehensively investigate the contribution of the anterior somites, we also photoconverted somites 2–4 at the 10–12 somite stage (Fig. 2h, i). At 72 hpf, labeled cells were found in the pectoral fin adductor/abductor muscles, posterior hypaxial muscles, myotomes 2–4, and cleithrohyoid muscle (Fig. 2j–l). These distributions also align with previous reports34,35. Again, cells labeled with Kaede-red were not found in the ventral cleithrum (n = 0/14) or scapulocoracoid (n = 0/14), but were distributed in the dorsal cleithrum (n = 4/14) (Fig. 2k–n; Supplementary Data 1).
a, b Photoconversion of the anterior three somites and adjacent neural tube at the 10–12 somite stage (ss), viewed from the left lateral side (a) and in horizontal confocal section (b) (n = 10). Kaede-red is pseudo-colored in magenta. c–e Confocal parasagittal sections of 72 hpf embryos from lateral (c) to medial (e) show that labeled cells are in the anterior three myotomes and cleithrohyoid muscle, but not in the ventral cleithrum (n = 8/8). f, g Transverse confocal sections obtained at the level of ventral (f) and dorsal (g) cleithrum found that labeled cells are in the dorsal cleithrum in a mosaic manner (arrowheads, n = 4/8), but not in ventral cleithrum. h, i Photoconversion of the somites 2–4 at the 10–12 ss (n = 14/14). j–l At 72 hpf, parasagittal confocal sections obtained from the medial (j) to lateral (l) levels show that labeled cells are in the adductor and abductor fin muscles (n = 10/14), posterior hypaxial muscles (n = 10/14), myotomes (n = 13/14), and cleithrohyoid muscle (n = 10/14), but not in the ventral cleithrum (n = 14/14). m, n Transverse confocal sections obtained at the level of ventral (m) and dorsal (n) cleithrum show that labeled cells are found in the dorsal cleithrum (arrowheads, n = 4/14), but not in ventral cleithrum.XII, hypoglossal nerve; A anterior, ab abductor muscle, ad adductor muscle, cle cleithrum, clehy cleithrohyoid muscle, ed endoskeletal disc, epi epidermis, gp posterior lateral line ganglion, gX vagus ganglia, i inner ear, op operculum, P posterior, pa pharyngeal arches, phy posterior hypaxial muscle, ppa primary pectoral artery, sc scapulocoracoid, m1–3 myotomes 1–3, nt neural tube, pn pronephros, s1–4 somites 1–4. Scale bars: (a and h) 200 µm, (b–g, i–n) 50 µm.
To genetically trace the somitic lineages in the pectoral region, we analyzed tbx6:cre; ubi:Switch embryos that switch the reporter expression from EGFP to mCherry upon Cre expression driven by the tbx6 paraxial mesoderm promoter (Supplementary Fig. 2a–g)38,39. At 72 hpf embryos, labeled cells were found in the anterior myotomes, adductor and abductor pectoral muscles, posterior hypaxial muscles, and cleithrohyoid muscles (Supplementary Fig. 2a–d). The tbx6:cre lineage cells were also sparsely distributed in the scapulocoracoid (Supplementary Fig. 2c) and ventral cleithrum (Supplementary Fig. 2e), and densely in the dorsal cleithrum (Supplementary Fig. 2f). The sparse labeled cells in the scapulocoracoid and ventral cleithrum could be a consequence of an additional labeling of the LPM lineage at the gastrulation stage, when tbx6 expression does not explicitly discern the LPM and paraxial mesoderm39. We confirmed that the labeled cells in the dorsal cleithrum were osteoblasts by colocalization of mCherry and sp7 expression (Supplementary Fig. 2g), supporting the somitic contributions to dorsal cleithrum osteoblasts. Since the labeled dorsal cleithrum cells were exclusively at the level of the paraxial mesoderm dorsoventrally, these cells seem to derive from the dermis of the epaxial region, not from migratory somitic cells in the hypaxial region. Furthermore, applying sp7 fluorescent in situ hybridization chain reaction (HCR) to various ontogenetic stages, we confirmed that the somitic dorsal cleithrum is formed appositionally, following the initiation of the cleithrum ossification at the pharyngeal arch level (see Supplementary Note 2 and Supplementary Fig. 2h–p). In summary, our photoconversion and genetic lineage tracing show that the first four anterior somites contribute to the pectoral musculature and dorsal extremity of the cleithrum.
Trunk LPM contributes to the fin mesenchyme, lateral body wall, scapulocoracoid, and cleithrum
Next, to query the contribution of the trunk LPM to the pectoral girdle, we photoconverted the LPM at the prospective pectoral fin region (hereafter “fin-field LPM” as a subpopulation of the trunk LPM) of Kaede-injected embryos at the 10–12 somite stage (Fig. 3a, b). To label the fin-field LPM, we photoconverted the LPM at the level from the first to the third somite32 (Fig. 3a, b). At 72 hpf, the photoconverted cells did not appear in skeletal muscles that arose from the anterior somites (Fig. 2), indicating that our photoconversion avoided ectopic labeling of adjacent somites (Fig. 3c–e). The labeled cells in the epidermis of the head and pectoral fin likely reflect ectopically labeled epidermal ectoderm (Fig. 3c, Supplementary Note 3). Labeled cells were also distributed in the endoskeletal disc, the scapulocoracoid (n = 7/7) (Fig. 3c; Supplementary Data 1), and additionally in the posterior margin of the cleithrum (n = 5/7) (Fig. 3f; Supplementary Data 1). These data indicate that LPM at the level of the prospective pectoral fin region contributes to the cleithrum.
a, b Photoconversion of the LPM adjacent to somites 1–3 at the 10–12 somite stage (ss), viewed from the left lateral side (a) and in horizontal confocal section (b) (n = 9). Kaede-red is pseudo-colored in magenta. Asterisk in (a) shows the ectopically labeled optic vesicle. c–e At 72 hpf, parasagittal confocal sections obtained from mediolaterally different levels [lateral (c) to medial (e)] show that labeled cells are in the endoskeletal disc (n = 7/7), scapulocoracoid (n = 7/7), and (f) in the posterior half of the cleithrum (n = 5/7). g Contributions of the genetically labeled drl:creERT2 lineage cells to the osteoblasts of the cleithrum. The obtained embryos were subjected to in situ HCR with sp7 probe and DAPI staining. EGFP signal is raw fluorescence. Arrowheads in (g) point drl:creERT2 lineage cells expressing sp7 (n = 3).A anterior, ab abductor muscle, ad adductor muscle, ccv common cardinal vein, cle cleithrum, clehy cleithrohyoid muscle, D dorsal, ed endoskeletal disc, epi epidermis, ie inner ear, LPM lateral plate mesoderm, M medial, m1–2 myotomes 1–2, op operculum, P posterior, pa pharyngeal arches, phy posterior hypaxial muscle, s1–4 somites 1–4, sc scapulocoracoid. Scale bars: (a) 200 µm, (b–e) 50 µm, (g) 20 µm.
To genetically corroborate the contribution of the LPM to the pectoral girdle, we labeled the LPM lineage using drl:creERT2 transgenic zebrafish, in which the draculin (drl) regulatory region drives (Z)−4-hydroxytamoxifen (4-OHT)-inducible CreERT2 recombinase expression in the LPM40 (see Methods, and Supplementary Table 1 for detailed strain information). Embryos obtained from crossing drl:creERT2 and hsp70l:Switch reporter (in which EGFP is expressed upon Cre-mediated loxP recombination) were treated by 4-OHT from the shield stage to 24 hpf and subjected to heat shock at 72 hpf as previously described30,41 (Supplementary Fig. 3). At 72 hpf, EGFP-labeled cells distributed in the cleithrohyoid muscle and the epithelium of the pharyngeal clefts (Supplementary Fig. 3a, c, d) are attributable to early-stage drl enhancer activity in the somitic lineage and endodermal epithelium of the pharynx, respectively33,40. Consistent with previous studies, EGFP-positive cells were observed in the pectoral fin and aortic arches40,41 (Supplementary Fig. 3a, b, d). EGFP-labeled cells were enriched in the cartilage of the endoskeletal disc, scapulocoracoid, and cleithrum (Supplementary Fig. 3b, c). Additionally, in situ HCR confirmed that EGFP-positive cells in the cleithrum are osteoblasts expressing an osteoblast marker sp7 (Fig. 3g, Supplementary Fig. 3c). Taken together, these results indicate that the region-specific photoconversion and genetic lineage labeling consistently identified the contribution of the LPM to the endoskeletal disc, scapulocoracoid, and cleithrum.
The LPM containing CPM-associated cells contributes to the cleithrum
To further confirm our finding using drl:creERT2-based LPM lineage labeling, we sought to refine the spatial photoconversion and genetic labeling into anterior LPM including prospective CPM-derived cells30,31,40. We photoconverted the ventrolateral head region at the post-otic level (hereafter “ventrolateral head region”), which contains the CPM, at the 10–12 somite stage (Fig. 4a). Immediately after photoconversion, we confirmed that the labeled ventrolateral head region contained the prospective CPM, epidermal ectoderm, and ventrolateral half of the post-otic placode (Fig. 4a, b; see also Supplementary Note 3). At 72 hpf, we found labeled cells in the epidermis in the head and pectoral fin, presumably due to ectopic photoconversion of the epidermal ectoderm (Fig. 4c–e). The entire vagus ganglia (derivatives of the epibranchial placode: n = 8/8) and the ventral portion of the posterior lateral line ganglion (a derivative of the lateral line placode: n = 7/8) were also labeled (Fig. 4d, e; Supplementary Data 1), which probably derived from the ectopically labeled ventrolateral half of the post-otic placode at the 10–12 somite stage (Fig. 4b). This result is consistent with the previously reported developmental fate of the post-otic placode42,43. Labeled cells were also found to delineate the bone matrix of the cleithrum without specific spatial distribution (n = 7/8) (Fig. 4f; Supplementary Data 1). In horizontal confocal sections, we found that the labeled cells surrounding the bone matrix of the cleithrum (Fig. 4g, h; white arrowheads) are contiguous with the mesenchymal cells at the posterior edge of the pericardial wall (Fig. 4g, h; black arrows) that itself originates from the LPM including tbx1-expressing presumptive CPM31,41. Overall, these results indicate that anterior LPM including CPM-associated cells (Fig. 4b) generates osteoblasts contributing to the cleithrum.
a, b Photoconversion of the ventrolateral head region at the 10–12 somite stage (ss), viewed from lateral (a) and transverse confocal section (b) (n = 10). The photoconverted area (pseudo-colored in magenta) in (b) contains CPM. Dotted line in (b) depicts the interface between ectodermal epithelium and its underlying tissues. Asterisk in (a) shows the ectopically labeled optic vesicle. c–e At 72 hpf, parasagittal confocal sections at mediolaterally different levels [lateral (c) to medial (e)] show that labeled cells are in the ventral part of the posterior lateral line ganglion (n = 7/8), vagus nerve ganglia (n = 8/8), and (f) in the cleithrum (n = 7/8). g, h Horizontal confocal sections at different dorsoventral levels also show labeled cells in the cleithrum (white arrowheads) laterally adjacent to the common cardinal vein (outlined by a dotted line) (n = 7/8). Labeled cells are also observed in the pericardial wall (black arrows). i Contribution of the genetically labeled tbx1:creERT2 lineage cells to osteoblasts of the cleithrum. The obtained embryos were subjected to in situ HCR with sp7 probe and DAPI staining. EGFP signal is raw fluorescence. Arrowheads in (i) point tbx1:creERT2 lineage cells expressing sp7 (n = 3). A anterior, ab abductor muscle, ad adductor muscle, bncc branchial neural crest cells, ccv common cardinal vein, cle cleithrum, clehy cleithrohyoid muscle, CPM cardiopharyngeal mesoderm, D dorsal, ed endoskeletal disc, epi epidermis, gp posterior lateral line ganglion, gX vagus nerve ganglia, ie inner ear, L lateral, nt neural tube, op operculum, otv otic vesicle, P posterior, pa pharyngeal arches, phy posterior hypaxial muscle, pl placode, m1–2 myotomes 1–2, s1 first somite. Scale bars: (a) 200 µm, (b–e) 50 µm, (i) 20 µm.
To corroborate the result of the anterior LPM and CPM photoconversion, we subsequently performed a genetic cell lineage tracing of tbx1:creERT2;hsp70l:Switch embryos (see Methods and Supplementary Table 1). The T-box transcription factor gene Tbx1 shows evolutionarily conserved expression patterns in the pharyngeal mesoderm across chordates44,45. As such, the zebrafish tbx1:creERT2 transgene indelibly labels cardiopharyngeal lineages, endoderm in the pharynx, and possibly trigeminal neural crest lineages in zebrafish embryos30,41. We treated embryos obtained from crossing tbx1:creERT2 and hsp70l:Switch zebrafish with 4-OHT from the shield stage to 24 hpf and observed the distribution of EGFP-positive cells at 72 hpf as previously described41 (Supplementary Fig. 4). Consistent with previous studies30,41, EGFP-positive cells were observed among mesenchymal cells surrounding the inner ear and in pharyngeal musculature (Supplementary Fig. 4a–c). Additionally, labeled cells sparsely delineated the cleithrum bone matrix. In situ HCR confirmed that these cells are sp7-positive osteoblasts (Fig. 4i, Supplementary Fig. 4c), demonstrating tbx1-positive lineage contribution to the cleithrum. Labeled cells were also found in the epithelium of the pharyngeal clefts (Supplementary Fig. 4d), reflecting early tbx1:creERT2 activity in the endodermal lineage, akin to Tbx1 in mouse30,46. Taken together, the results of both photoconversion and genetic lineage tracing of the tbx1:creERT2-labeled lineage further support a contribution of the anterior LPM and presumptive CPM-associated cells to the zebrafish cleithrum (Fig. 4, Supplementary Fig. 4).
Branchial neural crest cells give rise to the pharyngeal mesenchyme and the anterior half of the cleithrum
Several elements of the tetrapod shoulder girdle integrate cells of different lineage origins10,11,12,17,18,19,20. In our LPM-focused experiments above, we noted highly mosaic lineage labeling in the cleithrum, indicating that the cleithrum might also integrate cells from additional lineage origins. The neural crest contributes to numerous features in the craniofacial skeleton as major evolutionary innovation in vertebrates47,48. To date, a possible contribution of the neural crest to the cleithrum remains unclear. We therefore used photoconversion to revisit a potential cleithrum contribution of branchial neural crest cells, an evolutionarily conserved, posterior-most cranial neural crest stream in jawed vertebrates49. We first photoconverted the dorsomedial region of the embryonic head at the post-otic level (hereafter referred to as “dorsomedial head region” corresponding to a previously identified cardiac neural crest population50) in embryos ubiquitously expressing Kaede-green at the 10–12 somite stage (Fig. 5a). In situ HCR discerned a cell population positive for foxd3, a migrating neural crest cell marker51, between the otic vesicle and first somite, confirming that the photoconverted dorsomedial head region includes the branchial neural crest cells (compare Fig. 5b with 5c). The adjacent otic vesicle, neural tube, epidermal ectoderm, and dorsomedial portion of the post-otic placode were also ectopically labeled (Fig. 5a, b, Supplementary Note 3). The lateral migrating frontier of the branchial neural crest stream at this stage does not reach beyond the placodal region (Fig. 5c) and does not enter into the ventrolateral head region (compare Fig. 4b with Fig. 5c), indicating that our photoconversions of the branchial neural crest cells (in the dorsomedial head region) and prospective CPM (in the ventrolateral region) were mutually exclusive.
a, b Photoconversion of the dorsomedial head region at the 10–12 somite stage (ss) labels the branchial neural crest cells and adjacent neural tube, epidermis, and placode (n = 15). Asterisk in (a) points the faintly labeled optic vesicle. c foxd3 in situ HCR image obtained from approximately at the same transverse level in (b). The lateral migratory frontier of branchial neural crest cells does not laterally exceed the level of the placode at this stage (n = 3). d–f At 72 hpf, parasagittal confocal sections at mediolaterally different levels [lateral (d) to medial (f)] show that labeled cells are in the dorsal part of the posterior lateral line ganglion (n = 11/12), mesenchymal cells in the pharyngeal arches (n = 12/12), and the anterior half of the cleithrum (g) (n = 10/12). h, i Horizontal confocal sections at different dorsoventral levels also show labeled cells in the cleithrum (white arrowheads). Labeled cells are continuously distributed from the pharyngeal arch mesenchyme to the cleithrum (black arrows) (n = 10/12). j Contribution of the genetically labeled sox10:creERT2 lineage to osteoblasts of the cleithrum (n = 3). The obtained embryos were subjected to in situ HCR with sp7 probe and DAPI staining, and then stained by DsRed immunofluorescence (see Methods). Arrowheads in (j) point sox10:creERT2 lineage cells expressing sp7. A anterior, bncc branchial neural crest cells, cle cleithrum, clehy cleithrohyoid muscle, CPM cardiopharyngeal mesoderm, D dorsal, ed endoskeletal disc, epi epidermis, gp posterior lateral line ganglion, gX vagus nerve ganglia, ie inner ear, L lateral, nt neural tube, op operculum, otv otic vesicle, P posterior, pa pharyngeal arches, phy posterior hypaxial muscle, pl placode, m1–2 myotomes 1–2, s1 first somite, sc scapulocoracoid. Scale bars: (a) 200 µm, (b–f) 50 µm, (j) 20 µm.
At 72 hpf, Kaede-red labeled cells were found in the posterior wall of the inner ear, epidermis, and posterior lateral line ganglion (Fig. 5d–f), consistent with ectopic photoconversion of the otic vesicle, epidermal ectoderm, and dorsolateral half of the post-otic placode, respectively, using our laser-based approach (Fig. 5a, b). Nonetheless, the labeled mesenchymal cells around the vagus ganglia in the posterior pharyngeal arches match previously observed descendants of cranial neural crest cells52, supporting our photoconversion efficacy (Fig. 5d–f). We also found that labeled cells delineate the anterior margin of the cleithrum (n = 10/12) (Fig. 5e, white arrowheads in Fig. 5g–i, Supplementary Data 1) and disseminate toward the labeled pharyngeal arch mesenchyme (black arrows in Fig. 5h). We also corroborated this result by conducting neural crest cell-specific photoconversion with sox10:Kaede embryos (see Supplementary Table 1). We altered the color of Kaede from green to red in the branchial neural crest region by UV illumination at the 12 somite stage (Supplementary Fig. 5a) and identified the photoconverted cells in the anterior margin of the cleithrum at 72 hpf (Supplementary Fig. 5b, c; white arrowheads. For our interpretations on other labeled tissues, see Supplementary Note 4.). All these observations persistently indicate a branchial neural crest cell contribution to the cleithrum in addition to our previously established somitic and LPM contribution.
To complementarily determine the neural crest distribution with genetic labeling, we first used crestin:creERT2 with ubi:Switch as loxP reporter (GFP to mCherry change upon Cre activity; see Methods, and Supplementary Table 1). The crestin regulatory element deployed in this transgenic fish is selectively active in the premigratory and migratory neural crest cells after the somitogenesis stage53,54. We treated embryos with 4-OHT at the shield stage, imaged at 96 hpf, and observed mCherry-labeled cells in the opercle (n = 4/6), a pharyngeal arch derivative, and the cleithrum (n = 2/6; Supplementary Fig. 6a). Second, we obtained embryos from crossing sox10:creERT2 and actb2:Switch reporter zebrafish (BFP to DsRed change upon Cre activity; see Methods, and Supplementary Table 1). The deployed zebrafish sox10 promoter is active in premigratory/early migratory neural crest cells and the otic epithelium at 11–16 hpf and chondrocytes after 48 hpf55,56,57. To predominantly label migrating neural crest cells, we treated the embryos with 4-OHT from 11 – 24 hpf (Supplementary Fig. 6). At 72 hpf, DsRed-positive cells were found in mesenchymal cells in the operculum, pharyngeal arch mesenchyme, epithelium of the inner ear, and cleithrum (Supplementary Fig. 6b–e). In situ HCR confirmed that these DsRed-positive cells express sp7 and reside in only the anterior half of the cleithrum (Fig. 5j, Supplementary Fig. 6d), consistent with the Kaede-based photoconversion result (Fig. 5g). Overall, the comprehensive and meticulous photoconversions and genetic labeling consistently demonstrated the contribution of cranial neural crest cells to the anterior half of the cleithrum at the posterior to the pharyngeal arch mesenchyme (Fig. 5g–j, Supplementary Figs. 5, 6).
Discussion
Our photoconversion and genetic-cell lineage tracing experiments here provide evidence that the zebrafish cleithrum develops as a composite bone of somitic cells, LPM-derived cells (the fin-field LPM and CPM-associated cells), and cranial neural crest cells, while the endoskeletal scapulocoracoid derives solely from the fin-field LPM (Fig. 6). The conflicting data of neural crest contribution from previous work27 may be attributed to an activity difference between the mouse Sox10 enhancer and the here deployed zebrafish sox10 promoter57, observations with which we further support using crestin-based genetic lineage labeling and targeted Kaede photoconversion (Fig. 5, Supplementary Figs. 5 and 6). Intriguingly, photoconverted fin-field LPM cells were found at the posterior margin of the cleithrum (Figs. 3 and 6) while branchial neural crest cells were found only at the anterior margin (Figs. 5 and 6). In contrast, tbx1 lineage-labeled and generally CPM-associated LPM cells distributed throughout the cleithrum (Figs. 4 and 6). The spatially confined aggregation of the fin-field LPM and branchial neural crest cells in the cleithrum might result from the position of the cleithrum primordium which lies anteriorly to the pectoral fin bud and posteriorly to the pharynx58. Later, the cleithrum dorsally extends into the epaxial region and incorporates somitic cells, which become topographically available (Fig. 6b, Supplementary Fig. 2p). Intriguingly, the tetrapod endoskeletal scapula also arises from two distinct cell populations: the LPM and somitic/paraxial mesoderm17,18,20. Although the somitic contribution into the endoskeletal scapula could be a derived character in tetrapods due to the dorsal expansion of the scapula2,20, similar position-dependent incorporation of the somitic cells may also be the case in the ontogenetic dorsal expansion of the zebrafish cleithrum (Fig. 6b). Formulating available evidence and our results, we postulate that the zebrafish larval cleithrum develops at the head/trunk interface by assembling topographically nearby cell populations at each ontogenetic stage, such as the head (CPM and cranial neural crest cells), trunk (fin-field LPM) mesenchyme, and later somitic cells (Fig. 6, Supplementary Fig. 7a). As previous research found indispensable genes for normal cleithrum formation58,59,60, future genetic and molecular studies are warranted to functionally test these genes or identify the previously unknown molecular machinery that converges on the assembly of distinct embryonic cell populations into the cleithrum.
a Four embryonic populations reside at the prospective pectoral girdle region at the 10–12 somite stage. They establish the embryonic head/trunk interface in the zebrafish embryo by the pharyngula stage. b Schematic drawing of the left pectoral region of a zebrafish larva. Based on its embryonic origin, each element follows the same color code as in (a): somites-derived, pink; fin-field LPM-derived, yellow; CPM-derived, green; branchial neural crest cell-derived, light blue. The anterior somites contribute to pectoral muscles and dorsal cleithrum. The branchial neural crest cells contribute to ceratobranchial cartilages and anterior ridge of the cleithrum. The CPM gives rise to mesenchymal cells surrounding the cleithrohyoid muscle and is mosaically distributed throughout the cleithrum. The fin-field LPM forms the posterior half of the cleithrum. The scapulocoracoid and endochondral disc originate exclusively from the fin-field LPM. The oval cells surrounding the bone matrix of the cleithrum represent osteoblasts. The distal portion of the pectoral fin elements and the posterior portion of the posterior hypaxial muscle is partially removed (cut surfaces are shaded). ab abductor muscle, ad adductor muscle, bncc branchial neural crest cells, cb ceratobranchial, cle cleithrum, CPM cardiopharyngeal mesoderm, ed endoskeletal disc, gp posterior lateral line ganglion, gX vagus ganglia, ie inner ear, m myotomes, opv optic vesicle, otv otic vesicle, phy posterior hypaxial muscle, sc scapulocoracoid.
The head/trunk interface generates multiple structures, including the neck, that grants independent mobility to the skull from the pectoral girdle61. Neck musculoskeletal components have been reported to originate from a mixture of multiple embryonic cell populations in a variety of jawed vertebrates. In mice, neck musculatures, such as the trapezius, sternocleidomastoid, and infrahyoid muscles, contain connective tissues derived from a combination of cranial neural crest cells and LPM11,12. Moreover, the gill arch skeletons in cartilaginous fishes and amphibians and the laryngeal skeleton in amniotes develop as composites of cranial neural crest cells and CPM62,63,64,65. Likewise, the cleithrum develops between the pharyngeal arches and pectoral fin, deploying distinct cellular sources and, possibly, developmental genetic programs. Although the functional contributions or properties of these mixed embryonic origins in neck and shoulder girdle evolution remain limited, functional diversity of osteoblasts derived from several progenitor cell populations may be critical for development and evolution of the cleithrum, which was lost in amniotes during the posterior shift of the pectoral girdle due to the neck elongation (see below).
The detailed analysis of the embryonic origins of the cleithrum and scapulocoracoid illuminates the intricate developmental mechanisms of the pectoral girdle in bony fishes. Several genetic mutant zebrafish with a complete loss of the pectoral fin and scapulocoracoid develop a completely or almost normal cleithrum (reviewed in ref. 28), suggestive of a certain degree of ontogenetic independency between the cleithrum and scapulocoracoid, at least during early pectoral girdle development. For example, ikarus-mutant zebrafish, which possess a stop codon mutation in fgf24, lack the scapulocoracoid with morphologically normal cleithrum66. Fgf24 is expressed in the LPM and regulates mesenchyme migration and gene expressions indispensable for fin development. Consistent with our cell fate tracing demonstrating that the endoskeletal disc and scapulocoracoid arise exclusively from the LPM, fgf24 mutation seems to chiefly affect the scapulocoracoid formation but not the adjacent cleithrum. The fin-field LPM contributes only to the posterior portion of the cleithrum and could be compensated by the CPM that also forms the posterior cleithrum (Fig. 6b). Concomitantly, the LPM could be responsible for the integration of cleithrum and scapulocoracoid developmental processes. A recent study identified that LPM cells differentiate to either osteoblasts in the cleithrum or chondrocytes in the scapulocoracoid depending on the intensity of hedgehog signaling67. This bipotency of LPM cells constructs the trade-off of development and evolution of the cleithrum and scapulocoracoid. Overall, the investigation of fish pectoral girdle origins explains a wide range of phenotypes that have been difficult to interpret by genetic knockout or genomics alone.
Multiple embryonic origins of the cleithrum provides novel insights into the origin of the pectoral girdle in fin-bearing gnathostomes. The head/trunk interface where the zebrafish cleithrum develops (Fig. 6) is embryologically defined as the distribution boundary between the head and trunk mesenchyme68. Importantly, anatomical structures around the interface (e.g., circumpharyngeal ridge, common cardinal vein, hypoglossal nerve, and hypoglossal cord) are highly preserved across all jawed vertebrates68,69; thus, the relative position of cleithrum with these anatomical structures serves as a means to infer the embryonic environment of the cleithrum in extinct and extant species. For instance, in Australian lungfish (Neoceratodus), an extant fin-bearing sarcopterygian, the topographical relationship among the circumpharyngeal ridge, common cardinal vein, pronephros, and cleithrum is comparable to that of zebrafish (see refs. 70,71. and compare with Fig. 1b). This conserved topographical relationship was likely established and fixed in the course of the stem-group gnathostomes45,69,72. Moreover, their tight topographical association even dates back to extinct osteostracans, jawless gnathostomes that possess early paired appendages and dermal pectoral girdle73 (discussed in ref. 74). Thus, despite the anatomically derived characteristics of the teleost pectoral girdle among actinopterygians6, the embryonic environment of the cleithrum identified in this study (Fig. 6) seems to be conserved across fishes bearing dermal pectoral girdles (also see Supplementary Discussion). We propose that, since its emergence, the pectoral girdle and appendage position has been confined to the posterior edge of the head and heart72,75. This ancestral developmental environment of the dermal pectoral girdle at the posterior edge of the head aligns with a recently proposed posterior pharyngeal origin of the pectoral girdle in placoderms76. Our findings also supports the dual embryonic origins of the pectoral complex (i.e., pectoral girdle and fin) as pharyngeal and trunk domains, bridging two mutually exclusive hypotheses on the evolutionary origin of pectoral appendages: the archipterygium hypothesis77 and the fin-fold hypothesis78,79 (see also discussion in ref. 10).
Early Synapsida and Sauropsida, such as Edaphosaurus (stem Synapsida) and Procolophonoidea (stem Sauropsida), retained the cleithrum even in their entirely terrestrial habitats80,81. However, along with increases in the cervical vertebral number and extension of the neck82,83, their crown-groups, including all extant amniotes, completely lost the cleithrum6,9. In extant amniotes, the long neck integrates cells from a specialized trunk LPM region, namely the neck LPM84, which is located between the head/trunk interface and forelimb bud (Supplementary Fig. 7b, c). The expansion of the neck LPM during development is critical for the posterior relocation of the heart and providing the mesenchymal environment for the diaphragm, which is a mammalian-specific structure85,86,87 (Supplementary Fig. 7c). Given these developmental shifts, we hypothesize that the loss of the cleithrum in all extant amniotes may be attributed to the distant location of the pectoral girdle from the branchial neural crest cell source (i.e., the head/trunk interface). Alternatively, the dispensability of the gill respiratory system supported by the cleithrum in terrestrial habitats might cause a loss of the genetic program for cleithrum formation. As a consequence, the cleithrum and associated dermal plates might have disappeared, and the functional neck originated in terrestrial vertebrates.
Recent studies have demonstrated that embryonic sources and morphological homology can be evolutionarily decoupled20,21,62,88,89. If this is the case in pectoral/shoulder girdle evolution, the embryonic sources for the cleithrum could vary in different fish species. Moreover, contrary to the above discussion, the cleithrum may not simply be a lost component in extant tetrapods; its developmental program may be maintained in the shoulder girdle even with its distinct embryonic environment4. Further studies of the cellular trajectories and molecular mechanisms underlying these cell differentiations in various cleithrum-bearing fishes are warranted to determine the possibility of decoupling of ontogenetic origins and homological traits over the course of pectoral/shoulder girdle evolution.
Methods
Zebrafish lines
Animal husbandry and experiments were carried out in accordance with the protocol approved by Institutional Animal Care and Use Committee (IACUC) of Rutgers University (protocol #: 201702646) and of the University of Colorado School of Medicine (protocol #: 00979). Adult zebrafish [Danio rerio (Hamilton, 1822)90] were kept at 28.5 °C on a 14 h light/10 h dark cycle. Wild-type zebrafish embryos were obtained from intercross of *AB (star-AB) line. Zebrafish transgenic lines used in this study include: Tg(−6.3drl:creERT2, cryaa:Venus), Tg(−3.2tbx1:creERT2, cryaa:Venus), Tg(−1.5hsp70l:loxP-STOP-loxP-EGFP, cryaa:Venus), Tg(−3.5ubi:loxP-GFP-loxP-mCherry), and Tg(−4.9sox10:creERT2;actb2:loxP-BFP-loxP-DsRed)30,40,56,91. Additional transgenic lines are listed in the Supplementary Table 1. sox10:creERT2 fish were identified by PCR (Forward primer: 5’-TGCTGTTTCACTGGTTATGCGG−3’ and reverse primer: 5’-TTGCCCCTGTTTCACTATCCAG−3’). Other transgenic fishes were screened by fluorescence markers—cryaa:Venus, cryaa:YFP, ubi:Switch, and actb2:Switch.
Staging
Since experimental conditions, such as photoconversion, overnight shipment, and heat shock may affect the developmental speed of zebrafish embryos, embryonic stages were determined based on external morphology not on absolute time, with reference to the staging table provided by ref. 37.
Photoconversion-based spatial lineage tracing
To prepare the DNA template for Kaede mRNA synthesis, the coding sequence of Kaede was subcloned from pME-Kaede vector into pCS2+ expression vector. Kaede mRNA was synthesized in vitro using mMESSAGE mMACHINE SP6 kit (AM1340, Invitrogen) following the manufacturer’s instructions. To obtain zebrafish embryos with ubiquitous Kaede expression, Kaede mRNA diluted at 100 ng/µl in RNase-free water containing 0.005% phenol red (P0290-100ML, Sigma) was injected into the cytoplasm of wild-type embryos at one-cell stage using an MPPI-3 microinjector (ASI) under a stereomicroscope (S9E, Leica). Injected embryos were incubated in the embryo medium (10% Hank’s with full strength calcium and magnesium: 13.7 mM NaCl, 0.54 mM KCl, 0.025 mM Na2HPO4, 0.044 mM KH2PO4, 1.3 mM CaCl2, 1.0 mM MgSO4, and 0.42 mM NaHCO3). To photoconvert target embryonic regions at the 10–12 somite stage or 48 hpf, we embedded embryos into 0.7% low-gelling temperature agarose (50100, FMC BioProducts) in the embryo medium on a glass bottom dish (150680, Thermo Fisher Scientific) filled by phenol red-free L-15 medium (21083-027, Gibco). We then conducted photoconversion by illuminating 405 nm laser to the target regions for ~60 s on LSM510 Meta inverted confocal microscope (Zeiss). Labeled embryos were removed from the gel and incubated in six-well plates (657160, Greiner bio-one) filled with the embryo medium containing 0.003% 1-phenyl 2-thiourea (PTU; P7629, Sigma-Aldrich) at 28.5 °C until 72 hpf. Labeled embryos were anesthetized with 0.17 mg/ml ethyl 3-aminobenzoate methanesulfonate MS-222 (E10521-50G, Sigma-Aldrich), embedded in 0.7% agarose gel, and imaged alive with the confocal microscope. All labeled results are listed in Supplementary Data 1. For the consistency of the data, individuals with weak Kaede fluorescence or an excess amount of ectopic labeling (shown with asterisks in Supplementary Data 1) were omitted from the analysis (individuals highlighted in gray in Supplementary Data 1).
Photoconversions in sox10:Kaede embryos
Embryos obtained from sox10:Kaede adult fishes were photoconverted at the 10–12 somite stage as described above with the following modifications. To visualize tissues other than the neural crest cells during the photoconversion, sox10:Kaede embryos were raised in the embryo medium containing 2.5 µM BODIPY FL C5-Ceramide (D3521, Invitrogen) from immediately after fertilization to 10–12 somite stage.
Genetic lineage tracing
For heat-shock induction of loxP reporter cassettes driven by the hsp70l promoter and activation of CreERT2 in drl:creERT2 and tbx1:creERT2 lineages, we followed the previously reported procedures41. For CreERT2 activation in the crestin:creERT2 and sox10:creERT2 lineage, we added 5 µM 4-OHT to the embryo medium containing the dechorionated embryos from the shield stage to 24 hpf and 11 – 24 hpf, respectively. After the 4-OHT treatment, embryos were rinsed, raised to desired stages (72 or 96 hpf) in the embryo medium containing 0.003% PTU at 28.5 °C, and fixed by 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) at 4 °C overnight.
In situ hybridization
Probe sets for HCR RNA fluorescence in situ hybridization were designed to be complementary to target transcripts and synthesized by Molecular Instruments (Supplementary Table 1). For the whole-mount in situ HCR, fixed embryos were washed with PBST (PBS containing 0.1% Tween 20 [P20370-1.0, RPI Research Products International]) at least three times. Subsequent procedures followed the manufacturer’s instructions (MI-Protocol-RNAFISH-Zebrafish, Rev.#9)92. After the HCR, nuclei and actin were stained in PBST containing 5 µg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; D9542, Sigma-Aldrich) and Alexa Fluor 488 phalloidin (diluted in 1/100; A12379, Life Technologies), respectively, without light exposure at room temperature for two overnights.
Immunofluorescence
A duplexed in situ HCR and immunofluorescence has been performed as previously described93 with minor modifications. Briefly, we omitted the methanol and acetone permeabilization steps before the whole mount HCR from the original protocol. Embryos after in situ HCR were washed with PBST and soaked in blocking solution [5% heat inactivated (56 °C, 30 min) sheep serum (S2263, Sigma-Aldrich) in PBST] at room temperature for 2 h. The blocking solution was replaced by a primary antibody solution [anti-mCherry rabbit polyclonal antibody (GTX128508, GeneTex) diluted in 1/200 in blocking solution]. Then, we incubated embryos with the primary antibody solution at 4 °C overnight. We washed the excess antibody solution by PBST and replaced the solution with a secondary antibody solution [anti-rabbit IgG Alexa Fluor 594 antibody (A11037, Invitrogen) diluted in 1/200]. Subsequently, we incubated embryos with a secondary antibody solution at 4 °C overnight. The excess secondary antibody solution was removed and embryos were washed with PBST and mounted in the 0.7% low-gelling temperature agarose gel in PBST. Finally, mounted samples were subjected to a graded series of glycerol/PBST and transferred into 75% glycerol in PBST and imaged.
Imaging
Bright-field and fluorescence whole-mount images were photographed an upright microscope MZ16 (Leica) equipped with a digital camera MC170HD (Leica). Confocal sections were acquired by LSM510 Meta, LSM 800, and LSM 880 inverted confocal laser microscope equipped with a Plan-Apochromat 20x/0.8 M27, LD LCI Plan-Apochromat 25x/0.8 Imm Korr DIC M27, or C-Apochromat 40x/1.20 W Korr UV-VIS-IR M27 objective (Zeiss). The whole-mount and confocal images were processed by ImageJ (NIH) and ZEN 3.5 software (Zeiss), respectively. Volumetric 3D reconstructions from serial confocal images were performed by ZEN 3.5 software. Segmentation and 3D mesh generation were performed by Drishti 3.194 (https://github.com/nci/drishti) and rendered by Blender 2.80 (https://www.blender.org). Exported images in TIFF or PNG format were then assembled into figures in Adobe Photoshop (Adobe Systems). Three or more replicates were obtained for each imaging, unless otherwise described.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data generated in photoconversion-based lineage tracing are available in the Supplementary Data 1. Abbreviations are listed within the file. Any imaging data generated in this study are available from the Figshare under the following (https://doi.org/10.6084/m9.figshare.26045854).
References
Goodrich, E. S. Studies on the Structure and Development of Vertebrates (Dover Publications Inc, 1930).
McGonnell, I. M. The evolution of the pectoral girdle. J. Anat. 199, 189–194 (2001).
Gegenbaur, C. Clavicula und Cleithrum. Morphol. Jahrb. 23, 1–20 (1895).
Matsuoka, T. et al. Neural crest origins of the neck and shoulder. Nature 436, 347–355 (2005).
Gess, R. & Ahlberg, P. E. A tetrapod fauna from within the Devonian Antarctic circle. Science 360, 1120–1124 (2018).
Jollie, M. Chordate Morphology (Reinhold, 1962).
Romer, A. S. & Parsons, T. S. The Vertebrate Body 6th edn, Vol. 656 (Thomson Learning, 1986).
Lyson, T. R. et al. Homology of the enigmatic nuchal bone reveals novel reorganization of the shoulder girdle in the evolution of the turtle shell. Evol. Dev. 15, 317–325 (2013).
Romer, A. S. Osteology of the Reptiles Reprint edn (Krieger Publishing Company, 1997).
Nagashima, H. et al. Developmental origin of the clavicle, and its implications for the evolution of the neck and the paired appendages in vertebrates. J. Anat. 229, 536–548 (2016).
Heude, E. et al. Unique morphogenetic signatures define mammalian neck muscles and associated connective tissues. eLife 7, e40179 (2018).
Adachi, N., Bilio, M., Baldini, A. & Kelly, R. G. Cardiopharyngeal mesoderm origins of musculoskeletal and connective tissues in the mammalian pharynx. Development 147, dev185256 (2020).
Nomaru, H. et al. Single cell multi-omic analysis identifies a Tbx1-dependent multilineage primed population in murine cardiopharyngeal mesoderm. Nat. Commun. 12, 6645 (2021).
Prummel, K. D., Nieuwenhuize, S. & Mosimann, C. The lateral plate mesoderm. Development 147, dev175059 (2020).
Wang, W. et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat. Cell Biol. 21, 674–686 (2019).
Lescroart, F., Dumas, C. E., Adachi, N. & Kelly, R. G. Emergence of heart and branchiomeric muscles in cardiopharyngeal mesoderm. Exp. Cell Res. 410, 112931 (2022).
Shearman, R. M., Tulenko, F. J. & Burke, A. C. 3D reconstructions of quail-chick chimeras provide a new fate map of the avian scapula. Dev. Biol. 355, 1–11 (2011).
Valasek, P. et al. Somitic origin of the medial border of the mammalian scapula and its homology to the avian scapula blade. J. Anat. 216, 482–488 (2010).
Durland, J. L., Sferlazzo, M., Logan, M. & Burke, A. C. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat. 212, 590–602 (2008).
Piekarski, N. & Olsson, L. A somitic contribution to the pectoral girdle in the axolotl revealed by long-term fate mapping. Evol. Dev. 13, 47–57 (2011).
Fabian, P. & Crump, J. G. Reassessing the embryonic origin and potential of craniofacial ectomesenchyme. Semin. Cell Dev. Biol. 138, 45–53 (2023).
Etchevers, H. C., Dupin, E. & Le Douarin, N. M. The diverse neural crest: from embryology to human pathology. Development 146, dev169821 (2019).
Epperlein, H. H., Khattak, S., Knapp, D., Tanaka, E. M. & Malashichev, Y. B. Neural crest does not contribute to the neck and shoulder in the axolotl (Ambystoma mexicanum). PLoS One 7, e52244 (2012).
McGonnell, I. M., McKay, I. J. & Graham, A. A population of caudally migrating cranial neural crest cells: functional and evolutionary implications. Dev. Biol. 236, 354–363 (2001).
Aoto, K. et al. Mef2c-F10N enhancer driven β-galactosidase (LacZ) and Cre recombinase mice facilitate analyses of gene function and lineage fate in neural crest cells. Dev. Biol. 402, 3–16 (2015).
Masselink, W. et al. A somitic contribution to the apical ectodermal ridge is essential for fin formation. Nature 535, 542–546 (2016).
Kague, E. et al. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE 7, e47394 (2012).
Mercader, N. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev. Growth Differ. 49, 421–437 (2007).
Mongera, A. et al. Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development 140, 916–925 (2013).
Felker, A. et al. Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nat. Commun. 9, 2001 (2018).
Prummel, K. D. et al. Hand2 delineates mesothelium progenitors and is reactivated in mesothelioma. Nat. Commun. 13, 1677 (2022).
Mao, Q., Stinnett, H. K. & Ho, R. K. Asymmetric cell convergence-driven zebrafish fin bud initiation and pre-pattern requires Tbx5a control of a mesenchymal Fgf signal. Development 142, 4329–4339 (2015).
Sagarin, K. A., Redgrave, A. C., Mosimann, C., Burke, A. C. & Devoto, S. H. Anterior trunk muscle shows mix of axial and appendicular developmental patterns. Dev. Dyn. 248, 961–968 (2019).
Minchin, J. E. N. et al. Oesophageal and sternohyal muscle fibres are novel Pax3-dependent migratory somite derivatives essential for ingestion. Development 140, 2972–2984 (2013).
Talbot, J. C. et al. Muscle precursor cell movements in zebrafish are dynamic and require Six family genes. Development 146, dev171421 (2019).
Hatta, K., Tsujii, H. & Omura, T. Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 1, 960–967 (2006).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 (1995).
Lee, R. T. H., Knapik, E. W., Thiery, J. P. & Carney, T. J. An exclusively mesodermal origin of fin mesenchyme demonstrates that zebrafish trunk neural crest does not generate ectomesenchyme. Development 140, 2923–2932 (2013).
Wardle, F. C. & Papaioannou, V. E. Teasing out T-box targets in early mesoderm. Curr. Opin. Genet. Dev. 18, 418–425 (2008).
Mosimann, C. et al. Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 6, 8146 (2015).
Lalonde, R. L. et al. Heterogeneity and genomic loci of ubiquitous transgenic Cre reporter lines in zebrafish. Dev. Dyn. 251, 1754–1773 (2022).
Šestak, M. S., Božičević, V., Bakarić, R., Dunjko, V. & Domazet-Lošo, T. Phylostratigraphic profiles reveal a deep evolutionary history of the vertebrate head sensory systems. Front. Zool. 10, 18 (2013).
McCarroll, M. N. et al. Graded levels of Pax2a and Pax8 regulate cell differentiation during sensory placode formation. Development 139, 2740–2750 (2012).
Onimaru, K., Shoguchi, E., Kuratani, S. & Tanaka, M. Development and evolution of the lateral plate mesoderm: Comparative analysis of amphioxus and lamprey with implications for the acquisition of paired fins. Dev. Biol. 359, 124–136 (2011).
Adachi, N., Pascual-Anaya, J., Hirai, T., Higuchi, S. & Kuratani, S. Development of hypobranchial muscles with special reference to the evolution of the vertebrate neck. Zool. Lett. 4, 5 (2018).
Arnold, J. S. et al. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development 133, 977–987 (2006).
Kuratani, S. & Ahlberg, P. E. Evolution of the vertebrate neurocranium: problems of the premandibular domain and the origin of the trabecula. Zool. Lett. 4, 1 (2018).
Gans, C. & Northcutt, R. G. Neural crest and the origin of vertebrates: a new head. Science 220, 268–273 (1983).
Stundl, J. et al. Migratory patterns and evolutionary plasticity of cranial neural crest cells in ray-finned fishes. Dev. Biol. 467, 14–29 (2020).
Sato, M. & Yost, H. J. Cardiac neural crest contributes to cardiomyogenesis in zebrafish. Dev. Biol. 257, 127–139 (2003).
Montero-Balaguer, M. et al. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev. Dyn. 235, 3199–3212 (2006).
Abrial, M. et al. TGF-β signaling is necessary and sufficient for pharyngeal arch artery angioblast formation. Cell Rep. 20, 973–983 (2017).
Kaufman, C. K. et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197 (2016).
Luo, R., An, M., Arduini, B. L. & Henion, P. D. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev. Dyn. 220, 169–174 (2001).
Dutton, J. R. et al. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev. Biol. 8, 105 (2008).
Giovannone, D. et al. Programmed conversion of hypertrophic chondrocytes into osteoblasts and marrow adipocytes within zebrafish bones. eLife 8, e42736 (2019).
Carney, T. J. et al. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619–4630 (2006).
Heude, É., Shaikho, S. & Ekker, M. The dlx5a/dlx6a genes play essential roles in the early development of zebrafish median fin and pectoral structures. PLoS ONE 9, e98505 (2014).
Grandel, H. et al. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development 129, 2851–2865 (2002).
Neumann, C. J., Grandel, H., Gaffield, W., Schulte-Merker, S. & Nüsslein-Volhard, C. Transient establishment of anteroposterior polarity in the zebrafish pectoral fin bud in the absence of sonic hedgehog activity. Development 126, 4817–4826 (1999).
Trinajstic, K. et al. Fossil musculature of the most primitive jawed vertebrates. Science 341, 160–164 (2013).
Sleight, V. A. & Gillis, J. A. Embryonic origin and serial homology of gill arches and paired fins in the skate, Leucoraja erinacea. eLife 9, e60635 (2020).
Evans, D. J. & Noden, D. M. Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev. Dyn. 235, 1310–1325 (2006).
Tabler, J. M. et al. Cilia-mediated Hedgehog signaling controls form and function in the mammalian larynx. eLife 6, e19153 (2017).
Sefton, E. M., Piekarski, N. & Hanken, J. Dual embryonic origin and patterning of the pharyngeal skeleton in the axolotl (Ambystoma mexicanum). Evol. Dev. 17, 175–184 (2015).
Fischer, S., Draper, B. W. & Neumann, C. J. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development 130, 3515–3524 (2003).
Wei, J. et al. Distinct ossification trade-offs illuminate the shoulder girdle reconfiguration at the water-to-land transition. bioRxiv https://doi.org/10.1101/2023.07.17.547998 (2023).
Kuratani, S. Spatial distribution of postotic crest cells defines the head/trunk interface of the vertebrate body: embryological interpretation of peripheral nerve morphology and evolution of the vertebrate head. Anat. Embryol. 195, 1–13 (1997).
Higashiyama, H. et al. On the vagal cardiac nerves, with special reference to the early evolution of the head-trunk interface. J. Morphol. 277, 1146–1158 (2016).
Hirasawa, T. et al. Development of the pectoral lobed fin in the Australian lungfish Neoceratodus forsteri. Front. Ecol. Evol. https://doi.org/10.3389/fevo.2021.679633 (2021).
Greil, A. Entwickelungsgeschichte des Kopfes und des Blutgefäßsystems von Ceratodus forsteri. II. Die epigenetischen Erwerbungen während der Stadien 39-48. Denkschriften der Medicinisch-Naturwissenschaftlichen Ges. zu Jena. 4, 935–1492 (1913).
Trinajstic, K. et al. Exceptional preservation of organs in Devonian placoderms from the Gogo lagerstätte. Science 377, 1311–1314 (2022).
Janvier, P., Percy, L. R. & Potter, I. C. The arrangement of the heart chambers and associated blood vessels in the Devonian osteostracan Norselaspis glacialis. A reinterpretation based on recent studies of the circulatory system in lampreys. J. Zool. 223, 567–576 (1991).
Adachi, N., Robinson, M., Goolsbee, A. & Shubin, N. H. Regulatory evolution of Tbx5 and the origin of paired appendages. Proc. Natl Acad. Sci. USA 113, 10115–10120 (2016).
Janvier, P. Early Vertebrates (Oxford University Press, 1996).
Brazeau, M. D. et al. Fossil evidence for a pharyngeal origin of the vertebrate pectoral girdle. Nature 623, 550–554 (2023).
Gegenbaur, C. Zur Morphologie der Gliedmaassen der Wirbeltiere. Morpho. Jahrb. 2, 396–420 (1876).
Thacher, J. K. Median and paired fins: a contribution to the history of vertebrate limbs. Trans. Conn. Acad. Arts Sci. 3, 281–308 (1877).
Balfour, F. M. On the development of the skeleton of the paired fins of Elasmobranchii, considered in relation to its bearings on the nature of the limbs of the Vertebrata. Proc. Zool. Soc. Lond. 49, 656–670 (1881).
Williston, S. W. The Osteology of the Reptiles, reprint edn (Krieger Publishing Company, 1925).
MacDougall, M. J. & Modesto, S. P. & Botha-Brink, J. The postcranial skeleton of the Early Triassic parareptile Sauropareion anoplus, with a discussion of possible life history. Acta Palaeontol. Polonica 58, 737–749 (2013).
Hirasawa, T. & Kuratani, S. A new scenario of the evolutionary derivation of the mammalian diaphragm from shoulder muscles. J. Anat. 222, 504–517 (2013).
Müller, J. et al. Homeotic effects, somitogenesis and the evolution of vertebral numbers in recent and fossil amniotes. Proc. Natl Acad. Sci. USA 107, 2118–2123 (2010).
Lours, C. & Dietrich, S. The dissociation of the Fgf-feedback loop controls the limbless state of the neck. Development 132, 5553–5564 (2005).
Hirasawa, T., Fujimoto, S. & Kuratani, S. Expansion of the neck reconstituted the shoulder-diaphragm in amniote evolution. Dev., Growth Differ. 58, 143–153 (2016).
Sefton, E. M., Gallardo, M. & Kardon, G. Developmental origin and morphogenesis of the diaphragm, an essential mammalian muscle. Dev. Biol. 440, 64–73 (2018).
Sefton, E. M. et al. Fibroblast-derived Hgf controls recruitment and expansion of muscle during morphogenesis of the mammalian diaphragm. eLife 11, e74592 (2022).
Schneider, R. A. Neural crest can form cartilages normally derived from mesoderm during development of the avian head skeleton. Dev. Biol. 208, 441–455 (1999).
Teng, C. S., Cavin, L., Maxson, R. E. J., Sánchez-Villagra, M. R. & Crump, J. G. Resolving homology in the face of shifting germ layer origins: Lessons from a major skull vault boundary. eLife 8, e52814 (2019).
Hamilton, F. An Account of the Fishes Found in the River Ganges and its Branches Vol. 415 (Isha Books, 1822).
Mosimann, C. et al. Ubiquitous transgene expression and cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 (2011).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018).
Ibarra-García-Padilla, R., Howard, A. G. A., Singleton, E. W. & Uribe, R. A. A protocol for whole-mount immuno-coupled hybridization chain reaction (WICHCR) in zebrafish embryos and larvae. STAR Protoc. 2, 100709 (2021).
Hu, Y., Limaye, A. & Lu, J. Three-dimensional segmentation of computed tomography data using drishti paint: new tools and developments. R. Soc. Open Sci. 7, 201033 (2020).
Hildebrand, D. G. C. et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349 (2017).
Acknowledgements
This research was supported by the National Science Foundation under Grant IOS 2210072 and the Institutional support provided by the Rutgers University School of Arts and Sciences and the Human Genetics Institute of New Jersey to T.N.; the Children’s Hospital Colorado Foundation and the Helen and Arthur E. Johnson Chair for the Cardiac Research Director, Additional Ventures SVRF2021-1048003, and the University of Colorado School of Medicine to C.M.; NIH/NHLBI 1K99HL168148-01 to R.L.L. We thank Gage Crump and Claire Arata for providing sox10:creERT2;actb2:Switch embryos; Eric C. Liao, Eileen L. Dalessandro, and Lisa Tsay for providing sox10:Kaede fish; Charles K. Kaufman for providing adult crestin:creERT2 and pioneering work on cleithrum labeling; Hannah Cohen, Kathleen Flaherty, and all other lab members in Nakamura lab for maintaining zebrafish colonies in the fish facilities at Rutgers University; Christine Archer, Ainsley Gilbard, and Olivia Gomez for excellent zebrafish husbandry support at CU Anschutz; Hiroyasu Kamei and his lab members for zebrafish husbandry support at Kanazawa University; Satoru Okuda for allowing us to access their laboratory equipment; and Kazutaka Hosoda and Oki Hayasaka for helpful discussion.
Author information
Authors and Affiliations
Contributions
S.K. and T.N. conceived the project and designed the experiments. S.K. performed molecular, lineage tracing, and histological experiments. T.N. performed lineage tracing experiments using the sox10:Kaede and tbx6:cre;ubi:Switch. R.L.L., T.A.M., and C.M. conducted genetic lineage tracing experiments with drl:creERT2, tbx1:creERT2, and crestin:creERT2 zebrafish. S.K., R.L.L., C.M., and T.N. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Joost Woltering and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kuroda, S., Lalonde, R.L., Mansour, T.A. et al. Multiple embryonic sources converge to form the pectoral girdle skeleton in zebrafish. Nat Commun 15, 6313 (2024). https://doi.org/10.1038/s41467-024-50734-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-50734-x
This article is cited by
-
Distinct ossification trade-offs illuminate the shoulder girdle reconfiguration at the water-to-land transition
Nature Communications (2025)