Abstract
Ring-shaped DNA sliding clamps are essential for DNA replication and genome maintenance. Clamps need to be opened and chaperoned onto DNA by clamp loader complexes (CLCs). Detailed understanding of the mechanisms by which CLCs open and place clamps around DNA remains incomplete. Here, we present a series of six structures of the Escherichia coli CLC bound to an open or closed clamp prior to and after binding to a primer-template DNA, representing the most significant intermediates in the clamp loading process. We show that the ATP-bound CLC first binds to a clamp, then constricts to hold onto it. The CLC then expands to open the clamp with a gap large enough for double-stranded DNA to enter. Upon binding to DNA, the CLC constricts slightly, allowing clamp closing around DNA. These structures provide critical high-resolution snapshots of clamp loading by the E. coli CLC, revealing how the molecular machine works.
Similar content being viewed by others
Introduction
Rapid and processive replication of chromosomal DNA requires DNA polymerases to be tethered to primer-template (p/t) DNA by a replicative sliding clamp that encircles and slides on double-stranded (ds)DNA in a process that does not require ATP binding or hydrolysis1,2,3,4. These replicative sliding clamps also serve as tethers for proteins involved in many other aspects of DNA metabolism5. They are structurally and functionally distinct from more specialised sliding clamps, for example the ATP-dependent MutS/MutL cascading clamp system that ultimately manages DNA mismatch repair6,7.
The ring-shaped replicative clamps8,9,10 need to be opened and chaperoned onto a p/t DNA junction in an ATP-dependent process by a multi-subunit clamp loader complex (CLC)2,11. Frequent and rapid loading of sliding clamps onto newly-primed sites by the CLC is critical for timely production of Okazaki fragments during lagging-strand DNA synthesis12.
Structures and functions of both replicative clamps and CLCs from different organisms are conserved3,4. All sliding clamps contain six structurally similar domains (Fig. 1a). The bacterial sliding clamps contain two β subunits arranged as a head-to-tail dimer8, while the clamp from bacteriophage T4 and eukaryotic and archaeal PCNA (proliferating cell nuclear antigen) are trimers3,4. Whereas the Escherichia coli and eukaryotic clamps are stable closed rings, the T4 clamp can open spontaneously13,14.
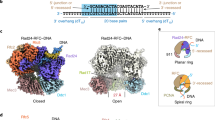
a Sliding clamps from E. coli45 (PDB 1MMI (E. coli β2 clamp)), phage T410 (1CZD (T4 gp45 clamp)) and human79 (1VYJ (human PCNA clamp)) are closed rings containing six structurally similar domains. The E. coli clamp is dimeric, while T4 and eukaryotic clamps are trimers. The N- and C-termini of the proteins are labelled as N and C, respectively. b Views of the clamp loader complex (CLC) from collar to AAA+ domain (top view, top panel) and front to back (side view, bottom panel). The E. coli CLC core subunits, δ, γ1, γ2, γ3 and δ’ (coloured salmon, medium pink, medium blue, deep sky blue and plum, respectively), are arranged anticlockwise in canonical positions A−E. ATP molecules (or analogues) are bound between Domains I and II of γ subunits at the γ1−γ2, γ2−γ3 and γ3−δ’ interfaces. c Domain organisation of E. coli CLC core subunits. All subunits contain AAA+ ATPase domains (I and II) and a Collar domain (III). The τ subunits contain an additional Domain IV that interacts with the DnaB helicase and Domain V that binds tightly to the Pol III α subunit. d Schematic of clamp loading in E. coli. To load a clamp, the ATP-bound CLC (state A) first binds to a β2 clamp (B)4,23,24,25 and opens it (C)25,26,27,28. Alternatively, the β2 clamp may open spontaneously and the CLC trap it in an open state (C). Then primer-template (p/t) DNA passes through the gap of the clamp and binds in the chamber of the CLC (D)30 either via its ssDNA or dsDNA portion depending on the size of the gap in β2. Upon binding and recognition of a p/t DNA junction, ATP hydrolysis occurs as the β2 clamp closes on DNA (E)31,32 and the CLC is ejected leaving the loaded clamp on p/t DNA for binding of the Pol III αεθ core to the clamp (F)3,4. Structures already reported for the E. coli CLC are in colours (states A and D without β2); structures only available for other organisms are in grey (B, yeast and human; C, yeast; D and E, phage T4 and yeast).
The CLCs are complexes comprised of a core of five structurally related AAA+ proteins (ATPases associated with various cellular activities)15 (Fig. 1b). The E. coli CLC has five core AAA+ subunits δτnγ3–nδ’ in positions A–E, respectively (Fig. 1b), and two smaller accessory subunits, ψ and χ12, whereas eukaryotic CLCs (RFC, replication factor C) contain five different proteins Rfc1−53,4, and the T4 CLC contains gp62 (in position A) and four gp44 proteins. The eukaryotic, E. coli and T4 clamp loader proteins have long been recognised as being homologous3,4. Multiple sequence alignment of the human RFCs 2–4, E. coli δ and γ, and T4 gp44 proteins16 showed that the E. coli γ protein shares ~23% sequence identity with human RFCs 2–4 and T4 gp44 in the N-terminal AAA+ domains (about 200 amino acids residues). These proteins utilise ATP to load their cognate clamps onto DNA, and they all possess consensus Walker A, Walker B, sensor 1 and Ser-Arg-Cys (SRC) motifs that are involved in nucleotide-binding and hydrolysis3,4.
The E. coli τ subunit is a longer version of γ with two additional C-terminal domains that interact with the DnaB helicase and the Pol III DNA polymerase α subunit, serving as an organisational hub in the Pol III holoenzyme12 (Fig. 1c). Each CLC protein consists of three domains. The N-terminal RecA-type ATPase domain (Domain I) and the middle lid domain (II) form a AAA+ module. The C-terminal domains (III) form a disc-like collar that caps and stabilises the AAA+ domain3,4 (Fig. 1b). Interactions of the CLC proteins with clamps are primarily mediated by a peptide with a consensus sequence called the clamp-binding motif (CBM) binding to surface crevices on the clamps17,18.
In the E. coli CLC, three ATP molecules bind at AAA+ subunit interfaces involving the three γ/τ subunits. In the absence of ATP, the AAA+ domains of the E. coli CLC are not well-organised19,20 and incapable of high-affinity binding to the clamp4,21. ATP induces conformational changes in the CLC that enable clamp binding22. To load a clamp (Fig. 1d), the ATP-bound CLC (state A) either binds to a clamp (B)4,23,24,25 and opens it22,25,26,27,28or traps a spontaneously opened clamp (C)29. The CLC-open clamp complex can directly bind to p/t DNA if the gap in the clamp is large enough for dsDNA to pass. Otherwise, the complex has to use a “filter and slide” mechanism in which only ssDNA can pass through the gap and p/t DNA has to slide up the chamber of the CLC (D)30,31. Upon binding and recognition of a p/t DNA junction, the clamp closes on DNA (E)31,32, followed by ejection of the CLC and binding of a DNA Pol III core to the clamp (F)3,4. The timing of hydrolysis of the three ATP molecules is uncertain. It seems likely that clamp closing and CLC ejection is a concerted process during which all three ATPs are hydrolysed rapidly and sequentially. Careful correlation of the pre-steady state kinetics of ATP hydrolysis with p/t DNA and clamp binding and release suggested the hydrolysis of two ATPs may occur during clamp closing and one during clamp ejection32,33,34,35, but the processes do not appear to be closely coupled. Moreover, which of the ATPs are hydrolysed during each of these steps is unknown.
High-resolution structures of CLC-clamp complexes are important to understand how CLCs load sliding clamps onto DNA. For the E. coli CLC, only structures of free CLC19,20 (A) and CLC bound to a p/t DNA (D) but without a β2 clamp30, are available (coloured structures in Fig. 1d). While related structures of CLC-clamp complexes from phage T431, yeast23,36,37,38,39 and human24 have provided snapshots of clamp loading intermediates for those systems, questions remain for the E. coli CLC, for which there has been no reported structure in complex with the clamp: Does the CLC actively open the clamp25,28 or does it trap a spontaneously-opened clamp29? If it actively opens the clamp, how does it achieve that? How does the clamp interact with CLC subunits other than δ? Is the gap on an open β2 clamp large enough to allow direct passage of dsDNA or only ssDNA? When is ATP hydrolysed and at which sites?
Here, we report a series of high-resolution cryogenic electron microscopy (cryo-EM) structures of E. coli CLC•β2 complexes both free in solution and bound to p/t DNA. These structures provide a comprehensive view of the clamp loading cycle (Supplementary Movie 1). Our studies reveal that the CLC first binds to β2 via the CBM of δ, then constricts to hold on to β2 through its simultaneous binding to subunits δ, γ1, γ3, and δ’; these interactions are revealed for the first time. The CLC then actively opens β2 to produce a gap large enough for direct passage of dsDNA. Surprisingly, the E. coli CLC opens β2 by moving the AAA+ modules of subunits A (δ) and B (γ1) outwards anticlockwise in hinge-like motions that are different from the yeast RFC that moves AAA+ modules C, D and E outwards clockwise.
Results
The E. coli CLC binds, constricts and holds onto a closed β2 clamp before opening it
The β2 clamp is a very stable homodimer13,14,40,41 that needs to be opened for loading onto a p/t DNA by the CLC4. To understand how the CLC binds and opens the β2 clamp, we first used surface plasmon resonance (SPR) to test how strongly the CLC and β2 interact in the presence of ATP, ATPγS—an ATP analogue poorly hydrolysed by the CLC22,42, or ADP•AlFx43—a non-hydrolysable ATP transition-state mimic. The full seven-subunit δτ3δ’ψχ CLC was first bound to streptavidin on the SPR chip surface via the (biotinylated) χ subunit, then serially diluted β2 solutions were flowed over CLC to interact. The χ subunit forms a stable heterodimer with ψ44, which in turn binds tightly via its natively unstructured N-terminal region across the Collar domains of the three γ subunits of the CLC core30; χ is therefore remote from the site(s) of interaction of the CLC with the clamp.
We found that β2 bound to the CLC strongly with ATP and ATPγS (KD of 2.7 ± 0.3 and 13.3 ± 0.1 nM, respectively), while binding was weaker with ADP•AlFx (400 ± 15 nM) (Supplementary Fig. 1). Therefore, we first chose to use ATPγS as nucleotide and solved structures of δγ3δ’ψχ bound to β2 by single-particle cryo-EM using a sample pre-treated with the crosslinker bis(sulfosuccinimidyl)suberate (BS3)24,36 to further stabilise the complex.
The data reveal a mixture of CLC•β2 structures with open and closed β2 clamps (Supplementary Table 1 and Figs. 2,3), suggesting that ATPγS binding may not simply open the clamp, but produce an equilibrium mixture of open and closed states. Reconstructions of the CLC bound either to the closed (CLC•β2closed, refined to 3.7 Å resolution) (Fig. 2a and Supplementary Figs. 2b,3) or to the open β2 clamp (CLC•β2open, 2.7 Å) (Fig. 3a and Supplementary Figs. 2b,4) were made, and the structures of CLC•β2closed and CLC•β2open were then built using crystal structures of the E. coli CLC (PDB 3GLI (CLC•p/t DNA structure))30 and β2 (PDB 1MMI (E. coli β2 clamp))45. In both maps, the whole of the δ and δ’ subunits and Domains I−III of the γ subunits are resolved; flexible regions beyond Domain III of γ subunits (Fig. 1c) were not visible in the density maps. There is weak density of ψ beyond its well-resolved N-terminal γ-binding peptide (Fig. 3a) that binds across the collars of all three γ subunits as revealed previously (PDB 3GLI (CLC•p/t DNA structure))30, allowing placement of the ψχ complex using its reported crystal structure (PDB 1EM8 (ψχ structure))44. Density interpreted as ATPγS is present at all three ATP-binding sites on the CLC at the γ1−γ2, γ2−γ3 and γ3−δ’ interfaces in both structures (Supplementary Fig. 2d). There is also unassigned density ascribed to a short peptide nestling in a shallow groove on Domain I of δ’, next to its N-terminus and potentially stacking with the aromatic ring of Trp3 (Figs. 2a, 3a). This is likely a part of the flexible C-terminal region of one of the γ subunits; the role of this interaction is unknown.
a Cryo-EM map of E. coli CLC bound to a closed β2 clamp (CLC•β2closed) in the absence of p/t DNA. Cryo-EM densities corresponding to the δ, γ1, γ2, γ3 and δ’ subunits of the CLC are coloured salmon, medium purple, medium blue, deep sky blue and plum, respectively. The β-I subunit of β2 is coloured yellow and β-II orange. The colouring scheme is kept throughout unless stated otherwise. b The AAA+ modules (shown without the Collar domain) of γ1 and δ in CLC•β2closed encroach into the gap between δ and δ’. Structures are aligned on γ2, which is barely perturbed among aligned structures. The AAA+ domains of the free δγ3δ’ complex20 (PDB 1XXH (ATPγS bound E. coli clamp loader complex)) are disorganised, while those of CLC•β2closed and CLC•β2open are more regularly packed. c CLC−β interfaces on β2 (left) and CLC (right) of CLC•β2closed. CLC−β interactions are mediated by peptides of the CLC subunits that are located on an α-helix and the following loop in their Domains I, which are presented as white ribbons on the surfaces of β2. β2 and the CLC are presented side-by-side by separating and turning them 90° in opposite directions. Buried areas are coloured as for their interacting counterparts; those coloured in salmon (δ) and deep sky blue (γ3) are in the two symmetry-related canonical peptide binding sites of β2. d Dynamics of clamp binding revealed by 3D variability analysis (3DVA). The resolution at which eigenvectors of 3DVA are filtered was set to 6 Å. Volumes of the first (brick red) and last (blue) frames of principal component_001 of 3DVA that show the most significant and biologically relevant conformational changes are presented.
a Cryo-EM map of CLC bound to an open clamp (CLC•β2open) coloured as in Fig. 2, and with the highest confidence regions of ψ(χ) in orange red. Density next to Domain I of δ’, presumed to be from the unstructured C-terminal region of a γ/τ subunit is coloured white and indicated by a red asterisk. Top right, close-up view showing the unassigned EM density near the N-terminus of δ’. A moving window of 8-residue peptides selected from the C-terminal flexible region of γ were docked into δ’ and scored against the density. The best scoring peptide corresponded to the moderately-conserved γ residues Val403−Thr409. This assignment is yet to be verified experimentally. Bottom right, unsharpened EM density of the CLC-binding peptide of ψ (Thr2–Glu28) that bridges all three γ subunits30. b Interface areas between AAA+ modules increase significantly in the transition from CLC•β2closed to CLC•β2open. Source data are provided as a Source Data file. c Conformational changes of the AAA+ modules during transition from CLC•β2closed to CLC•β2open. While the AAA+ modules of γ2, γ3 and δ’ barely move, those of γ1 and δ swing outwards, resulting in expansion of the CLC and opening of β2. Structures were superimposed by aligning the γ2 subunit using MatchMaker in Chimera76. Arrows indicate movements of individual subunits. d Orthogonal views of conformational changes in β2 during transition from CLC•β2closed to CLC•β2open. The clamp remains mostly flat after opening. Most obvious motions are on the β-I subunit with Domain I rotating around Domain III of β-II and Domains II–III around Domain I. e Conformational changes in the final stage of clamp opening revealed by 3D variability analysis. The resolution at which eigenvectors of 3DVA are filtered was set to 5 Å. Volumes of the first and last frames of principal component_000 of 3DVA that illustrate the most significant and biologically relevant conformational changes are presented.
The E. coli CLC•β2closed structure represents the first stage of clamp loading (Fig. 1d, state B). Compared to the free “inactive” δγ3δ’ complexes, whose AAA+ domains are loosely packed and not arranged to allow simultaneous binding of multiple CLC subunits to β219,20, the AAA+ domains (Fig. 2b) of the CLC•β2closed complex are more regularly packed and roughly match the pseudo-6-fold symmetry of β2 (Fig. 2c). In this closed conformation, the CLC holds onto β2 by simultaneous binding of the δ, γ1, γ3 and δ’ subunits. The interactions are mediated mainly by δ and γ3 and to a lesser extent by γ1 and δ’. The δ subunit makes extensive contacts with β2 as seen in the crystal structure of δ−β (PDB 1JQJ (δβ structure))18. The interactions are mediated by the CBM motif 65-FSLCQAMSLF (canonical CBM underlined) binding in the canonical peptide-binding site of β2, burying an interface area of 717 Å2. The γ3 subunit occupies the canonical peptide-binding site in the other β subunit mostly via binding of peptide 109-DNVQYAPAR, burying 479 Å2. Although three distinct reconstructions of the yeast RFC•PCNAclosed complex were made36, they all contain a PCNA hanging on one side of RFC and none shows RFC held stably onto PCNA. Therefore, the CLC•β2closed structure represents the last step in clamp binding, visualised for the first time, in which the CLC has maximised contact with β2 and is about to open it.
However, the AAA+ domains of CLC•β2closed are not yet symmetrically packed. The AAA+ modules of δ and γ1 are displaced from ideal symmetrically-arranged positions, encroaching into the gap between δ and δ’, a space occupied by the sixth subunit in hexameric ring-shaped AAA+ proteins (Fig. 2b). As a result, only one of the arginine fingers from the γ2 (R169), γ3 (R169) and δ’ (R158) subunits that are vital to ATP hydrolysis is close enough to interact with the γ-(thio)phosphate of ATPγS (i.e., δ’R158 at the γ3–δ’ interface) and the β- and γ-phosphates are poorly resolved at this resolution (Supplementary Table 2 and Supplementary Fig. 2d, left). Therefore, the closed CLC complex is in an “auto-inhibited” state, similar to the E. coli δγ3δ’19,20 and eukaryotic RFC•PCNAclosed complexes24,36.
On the other hand, the ATPγS binding sites in the CLC•β2open structure (especially at the γ1−γ2 and γ2−γ3 interfaces) are much better organised as would be expected since ATP binding promotes clamp opening; all three arginine fingers are close enough to the γ-thiophosphate of ATPγS to participate in ATP binding (Supplementary Fig. 2d, right). Note however that the CLC•β2 complex has only weak ATPase activity in the absence of p/t DNA42, so none of the ATP-binding sites is yet poised for efficient ATP hydrolysis.
We considered that the consensus reconstruction of CLC•β2closed may contain a mixture of similar conformations, which may explain why the refinement stalled at ∼3.7 Å resolution despite there being more particles than for our other higher-resolution reconstructions (Supplementary Fig. 2). To probe these conformations, we used 3D variability analysis (3DVA)46 of the refined particles. The results show that the consensus reconstruction indeed contains an ensemble of continuously changing and significantly different conformations (Fig. 2d and Supplementary Movie 2). The process of clamp binding is thus highly dynamic with a wide range of different motions of the CLC. The most significant and biologically relevant insights gained from 3DVA are: (i) δ must be the first subunit to engage in β2 binding. (ii) The CLC then gradually constricts to allow γ1, γ3 and δ’ to interact simultaneously with β2. (iii) Meanwhile, β2 and the CLC also move gradually closer as their interactions become stronger until the CLC holds stably onto β2.
E. coli CLC opens β2 by crab claw-like motions
In the CLC•β2open structure, β2 is open with a gap of ∼20 Å, large enough for dsDNA to enter (Fig. 3a), representing the stage where β2 is fully opened and ready to be transferred to p/t DNA (Fig. 1d, state C). The structure is similar to the yeast RFC•PCNAopen structures36 and the crystal structure of E. coli δγ3δ’ (without β2) bound to p/t DNA (CLC•DNA; PDB 3GLI (CLC•p/t DNA structure))30. The CLC in the two E. coli structures, with and without bound β2, superimpose with an RMSD of 1.09 Å over 1417 Cα atoms across all five subunits. Compared to CLC•β2closed, the AAA+ modules of CLC•β2open are more tightly packed (Figs. 2b,3b). The total interface area between the AAA+ modules increases by 52% from 2781 Å2 in CLC•β2closed to 4225 Å2. The most dramatic change occurs at the γ1−γ2 interface with the buried area increasing by more than 900 Å2 (Fig. 3b), transforming the most loosely packed interface to the most tightly packed. There is also a significant increase at the γ2−γ3 interface, whereas the δ−γ1 interface barely changes and these two modules move almost as a rigid body. In the transition from CLC•β2closed to CLC•β2open, the chamber of the CLC expands mainly by outward rotation of the AAA+ modules of δ and γ1 pivoting around that of γ2 (Fig. 3c and Supplementary Movie 3).
The β2 ring opens mostly by in-plane outward rotation of individual β domains and remains mostly flat after opening. Significant movements occur between Domains I and II of the β-I subunit that move outwards by 14˚, and at the unbroken β2 interface that swings out by 11˚, together providing greatest leverage for clamp opening at the opposite interface. A smaller change occurs between Domains I and II of β-II (Fig. 3d and Supplementary Movie 3). Domains II and III of both β subunits move mostly as rigid bodies. Their connection is either rigid or held together by δ and γ3 binding in the canonical CBM peptide-binding sites.
During opening of β2, the AAA+ modules of γ2, γ3 and δ’ hold onto the β-II subunit, resembling the fixed finger of a crab claw, while those of δ and γ1 bound tightly to the β-I subunit rotate outwards, resembling the moving finger, causing rotation of β-I to pull it away from β-II (Fig. 3c, d and Supplementary Movie 3). Upon breaking of the dimer interface, Domain I of β-II also extends as if to release spring tension of the closed clamp14,18,19, leading to wider opening. During the opening, all CLC subunits interact with β2 more and more extensively until they tightly hold the clamp open. 3DVA of all open particles for the last 3D classification also captured similar movements at the final stage of clamp opening from partly to fully open (Fig. 3e and Supplementary Movie 4). Therefore, the E. coli CLC opens β2 by a crab-claw mechanism1,29.
All CLC core subunits bind to the clamp in a similar fashion
Although CLC−β2 interactions are important for clamp loading and each of δ, γ and δ’ can interact with β47, only the δ−β interaction has been visualised structurally18. Our CLC•β2open structure shows that all five subunits interact extensively with β2 (Fig. 4a,b), burying interface areas of 757, 709, 382, 989 and 847 Å2 between β2 and δ, γ1, γ2, γ3 and δ’, respectively (Supplementary Table 3 and Supplementary Fig. 5). Like δ, all CLC subunits bind to surface crevices between pairs of structurally similar β domains through peptides located at the C-terminal ends of clamp-interacting helices and the following loops. The δ subunit binds to the canonical peptide-binding site on β-I via peptide 65-FSLCQAMSLF similar to that seen in CLC•β2closed and the structure of δ−β18. The γ1 subunit binds to a site between Domains I and II of β-I and γ2 binds mainly to Domain I of β-I near the dimer interface, while γ3 binds to the second CBM-binding site on β-II. Binding of the γ subunits to β2 is primarily mediated by different parts of peptides 109-DNVQYAPAR. In each case, residue Tyr113 of γ serves as an anchor for binding by nestling in a cavity in β2. Peptide 86-RFVLIEI of γ3 provides additional nearby contacts with β, and δ’ binds between Domains I and II of β-II through peptide 66-QLMQAGTHP at a site involving the outside rim surface of β2 and peptide 101-NEHARLG at a nearby site. In comparison, the canonical CBM peptide of δ provides an interface with β-I that is more hydrophobic, contributing more binding energy than the other, more polar interfaces (Supplementary Table 3).
a CLC−β interfaces on β2 and the CLC of CLC•β2open. CLC−β interactions are mediated by peptides of the CLC subunits that are presented on the surfaces of β2 with their secondary structures depicted (white). Interface areas are coloured as their interacting counterparts. Peptide-binding surfaces of the δ, γ3 and δ’ subunits on β2 are more extensive than those of γ1 and γ2. b Close-up views of clamp-binding peptides (ribbons with side chains as sticks) in binding pockets of β2 (surface representation). Sequences of peptides involving in clamp binding are shown above. Important residues, Leu73 and Phe74 of δ and Tyr113 of γ subunits that anchor the peptides in the binding sites, are labelled. The surface of β2 is coloured by hydrophobicity with hydrophobic areas in orange, neutral areas in white and hydrophilic areas in blue.
Closing of β2 around a p/t DNA junction
To delineate the events after p/t DNA binding, we determine structures of CLC•β2 assembled on p/t DNAs. First, we tested how different nucleotides affected the stability of the CLC•p/t DNA complex by SPR. We found that the CLC did not bind stably in the presence of ATP to a p/t DNA containing 23 bp dsDNA and a 5’-dT50 overhang (to which single-stranded DNA-binding protein SSB can also bind) (Supplementary Fig. 6a), as expected since p/t DNA-stimulated ATP hydrolysis by the CLC helps to quickly eject it from the DNA22,32,33,34,35,42,48. While the CLC with either ATPγS or ADP•AlFx bound to p/t DNA, the ADP•AlFx-bound CLC•p/t DNA complex was very much more stable than that with ATPγS (Supplementary Fig. 6b−f). This suggests that ADP•AlFx may act truly as an analogue of the transition state in ATP hydrolysis43, where it stabilises the ATP-binding sites and freezes the whole CLC•p/t DNA complex. Moreover, both the β2 clamp and SSB were able to bind sequentially to the ADP•AlFx-bound CLC•p/t DNA complex, forming very stable complexes with both the γ3 and τ3 CLCs (Supplementary Fig. 6g–i).
Therefore, samples containing the CLC (δτ3δ’ψχ instead of the γ3 CLC used previously), β2, ADP•AlFx and two different p/t DNAs were used for cryo-EM. The p/t DNAs contained a 23 bp dsDNA and either a 5’-dT10 overhang or a 5’-dT45 overhang bound to SSB; SSB binds more stably to dT45 than to dT10. In addition, a sample of the δτ3δ’ψχ CLC alone with ADP•AlFx and a p/t DNA containing 15 bp of dsDNA and 5’-dT10 overhang was used. These complexes were stable and did not require crosslinking for data collection. Note that although the τ3-CLC was used for all following complexes with p/t DNA, in no case did we observe any additional cryo-EM density beyond that for the common Domains I–III of γ/τ seen in the γ3-CLC structures above. For consistency, these regions are referred to hereafter as γ rather than τ.
While samples with the 5’-dT10 overhang and no SSB contained only CLC bound to a closed β2 clamp (CLC•DNA•β2closed; solved at 2.6 Å resolution; Supplementary Fig. 7), samples with 5’-dT45 and SSB contained CLCs bound either to a partially open (CLC•DNA•β2open) or a closed (CLC•DNA•β2closed) clamp (Fig. 5a,b, Supplementary Table 1 and Fig. 8). The CLC•DNA•β2open dataset was further separated into two similar complexes by 3D classification with β2 opened by ∼8 Å (CLC•DNA•β2open1; 2.9 Å resolution) and ∼6 Å (CLC•DNA•β2open2; 3.0 Å). These two structures are similar, representing equilibrated intermediates in the transition-state structure that occur prior to complete closure of β2 around DNA. Only CLC•DNA•β2open2 will be discussed.
a Cryo-EM map of the clamp loader complex (CLC) bound to a partially open clamp and p/t DNA (CLC•DNA•β2open). Densities corresponding to individual subunits are coloured as in Fig. 2. Density of the template DNA strand is coloured gold and the primer stand pink. Grey density at the top right corner belongs to SSB. b Cryo-EM map of the CLC bound to a closed clamp and p/t DNA (CLC•DNA•β2closed). c Cryo-EM map of the CLC bound to p/t DNA (CLC•DNA). d The CLC constricts slightly on binding of p/t DNA. The AAA+ modules of CLC•β2open (coloured grey) and CLC•DNA•β2open are superimposed to show small inward movements of the CLC upon p/t DNA binding. Arrows point to the tips of the δ subunits. e Orthogonal views showing β2 partially closes on p/t DNA and also becomes spiral. β2 of CLC•β2open in the left pane is coloured grey and CLC•DNA•β2open yellow and orange. Most movement occurs in the β-I subunit. β2 of CLC•DNA•β2open becomes lock washer-like (right). f,g CLC−β interfaces on β2 and CLC of CLC•DNA•β2open and CLC•DNA•β2closed, respectively, showing that the latter has much reduced contacts. h Mg•ADP•AlF4− molecules are present in all three ATP-binding sites of CLC•DNA•β2closed and the arginine finger residues: R169 of γ2 and γ3 and R158 of δ’, coordinate the AlF4− groups that mimic the γ-phosphates of ATP. Mg2+ (spheres) and AlF4– ions are coloured in shades of green and EM densities of Mg•ADP•AlF4− are shown as mesh. Hydrogen bonds between ADP•AlF4− and residues of the CLC are shown as red lines.
The structure of the CLC on p/t DNA alone (CLC•DNA; 2.6 Å) (Fig. 5c and Supplementary Fig. 9) is similar to CLC•DNA•β2open, except density for Domain I of δ is more poorly resolved in the absence of the β2 interaction. In all DNA-bound structures, the entire dsDNA and six dTs of the 5’ overhang, longer than the four nucleotides seen in the crystal structure of the δγ3δ’ CLC•DNA30, are resolved. The aromatic ring of Trp279 of δ that is conserved and important for p/t DNA binding49 sandwiches between the last two visible nucleotides of the 5’ ssDNA overhang that protrudes from the dsDNA-binding chamber of the CLC, i.e., the fifth and sixth unpaired nucleotides from the p/t junction (dT5th and dT6th). The CLC mainly interacts with the template DNA strand as described for the earlier δγ3δ’ CLC•DNA structure30 and β2 has only limited contacts with DNA. The AlF4− moiety of the ATP analogue ADP•AlF4− is clearly visible in all three ATP binding sites in all structures (Fig. 5h). That none of the sites contain only ADP suggests that the stabilised transition state conformations we observe are indicative of the concerted (rapidly sequential or simultaneous) hydrolysis of ATP at all three sites.
The CLCs in CLC•DNA, CLC•DNA•β2open and CLC•DNA•β2closed are very similar to one another and to the crystal structure of E. coli CLC•DNA (PDB 3GLI (CLC•p/t DNA structure))30; the latter superimposes with CLC•DNA•β2open with an RMSD of 1.13 Å over 1618 Cα atoms. Domain I of δ and the tips of β2 subunits in the CLC•DNA•β2open structures are more poorly ordered, suggesting that either these parts are more flexible or the reconstructions contain a mixture of similar conformations. 3DVA of all refined particles with dT45 and SSB revealed that these reconstructions are indeed intermediates in an ensemble of continuously changing conformations (Supplementary Movie 5). Compared to CLC•β2open, the CLC on DNA is slightly constricted (Fig. 5d). Consequently, while transitioning from CLC•β2open to CLC•DNA•β2open, the β-I subunit moves inwards and slightly downwards, so that β2 becomes spiral (Fig. 5e). For β2 to close around DNA, Domain I of δ moves farther inwards, allowing β-I to approach β-II, while β-II detaches from δ’, drops lower and re-closes the ring. The closed β2 is slightly distorted compared to the free clamp, with β-I more extended, as seen also in the structure of β−δ18. While the CLC of CLC•DNA•β2open maintains extensive interactions with β2 like in CLC•β2open, the interactions are much reduced in CLC•DNA•β2closed (Fig. 5f and Supplementary Fig. 7e,f). The total interface area between the CLC and β2 decreases from 3663 Å2 in CLC•DNA•β2open to 2496 Å2, which is compensated by the reformation of a β dimer interface (667 Å2) and more contacts with DNA.
Tethered SSB is distant from CLC and does not affect clamp closure on DNA
SSB is expected to bind rapidly and with very high affinity to the 5’ ssDNA overhang of a p/t DNA50. It also interacts with χ and many other proteins via a conserved protein-interacting motif within its C-terminal 9 residues: 169-MDFDDDIPF12,50,51 (referred to as the C-tail, SSB-Ct). On p/t DNA with a 5’ overhang long enough to stably bind SSB (e.g., 35 nt or longer), SSB modestly stabilises the CLC on p/t DNA through interactions with χ, and binding of the CLC to a preformed p/t DNA-SSB complex leads to rapid remodelling of SSB to move it farther from the 3’ end of the primer, presumably to accommodate interaction of the δ subunit with the immediately-downstream ssDNA52. In all our structures with SSB, the presence of SSB is evident (Fig. 5a) but appeared to be mobile, diffusing within a volume around the CLC at similar distances, as if being held remote from the CLC, perhaps by electrostatic repulsion. Surprisingly, there is no direct contact seen although SSB must be tethered to the CLC via interactions with χ52,53,54 and the ssDNA overhang.
We tried to uncover why β2 showed dynamic open and closed states after loading only in the presence of SSB and the dT45 overhang. To test if it is the interactions of SSB with χ or other CLC proteins, SSB binding to the 5’ ssDNA overhang, or the physical connection between the template DNA strand and ψχ mediated by SSB that enables β2 dynamics, several χ and SSB variants that are impaired in protein-protein interaction were used for cryo-EM studies. These proteins include χ∼SSB-Ct with the C-tail of SSB fused to χ by a flexible 9-residue linker that renders χ unable to interact with SSB55, SSB-T* containing only the ssDNA-binding structured OB-fold of SSB56, and SSBCysApp with a Cys residue appended to the C-terminus of SSB that prevents protein interaction through SSB-Ct57. Five combinations of the CLC•DNA•β2 complex with SSB variants were tested: (i) ψχ∼SSB-Ct without SSB; (ii) ψχ∼SSB-Ct plus SSB; (iii) ψχ∼SSB-Ct plus SSB-T*; (iv) ψχ plus SSB-T*; and (v) ψχ plus SSBCysApp. All five samples contained CLC complexes with both partially open and closed β2 in similar ratios of particles. Therefore, interaction of χ with SSB does not appear to affect β2 dynamics after DNA binding. Finally, we found that the dT45 ssDNA overhang alone was sufficient to keep a fraction of the clamp open. It seems likely therefore that the energy barrier between CLC•DNA•β2open and CLC•DNA•β2closed is so small that just a longer 5’ ssDNA overhang is enough to affect this equilibrium.
Discussion
Our structures of E. coli CLC•β2 complexes prior to and after binding to a p/t DNA provide critical information to understand the clamp loading process in E. coli. Now every step of clamp loading from clamp binding, opening and loading, has a representative structure. These structures have provided molecular and mechanistic details and insights into the clamp loading process, revealing the entire cycle of clamp loading in E. coli (Fig. 6 and Supplementary Movie 1).
Schematic of the clamp loading pathway in E. coli. To load a clamp, the ATP-bound clamp loader complex (CLC) (state A) first binds to it via the δ subunit. The CLC then constricts, holds onto the clamp by simultaneous binding of the δ, γ1, γ3 and δ’ subunits (B). The CLC next expands and opens the clamp (C). The CLC−open clamp complex then directly binds to a p/t DNA. The CLC slightly constricts, and the clamp first partly closes (D) before complete closure around the p/t DNA junction (E). ATP hydrolysis, which may occur at any stage after p/t junction recognition, is presumed to accelerate clamp closing and trigger ejection of the CLC to allow a DNA polymerase to bind to the loaded clamp for DNA synthesis.
E. coli β2 is a very stable dimeric ring that rarely opens in solution13,14,40,41. Although an early fluorescence study showed that the E. coli CLC only underwent limited conformational changes during clamp opening and may passively template an open β229, it is now certain that ATP binding to the CLC actively opens bound β2 clamps25,28. Here, we provide direct evidence that the CLC indeed opens the clamp in a process that does not require ATP hydrolysis to produce a gap wide enough to allow direct passage of dsDNA using a crab-claw mechanism. 3D variability analysis (3DVA) of our reported structures suggests that δ is the first subunit of the CLC to bind to β2. Upon binding via δ, the CLC gradually constricts until it can hold onto β2 by additional interactions with the γ1, γ3 and δ’ subunits (Fig. 2d). In contrast to yeast RFC that is constricted even prior to PCNA binding, the conformation of the free E. coli CLC appears more flexible and likely can spontaneously expand and constrict. Although δ binding alone is not sufficient to open β2, it must help to destabilise the clamp. Similar to the crystal structure of β−δ18, the β-I subunit in CLC•DNA•β2closed is more extended than in the free clamp, and this may indicate some spring tension in the closed β2 ring that distorts it and weakens the subunit interface. The CLC then undergoes further conformational changes in which its AAA+ domains become tightly packed, especially at the γ1−γ2 interface. As a result, the AAA+ modules of δ and γ1 rotate outwards as a rigid body while γ3 holds the β-II subunit stationary, causing the β-I subunit to be pulled away in a hinge-like motion. Such a mechanism is consistent with the proposed role of δ as a wrench19 that is actively involved in clamp opening, but it is γ3 rather than δ’ that has the important role as a stator19 in holding the β-II subunit.
Like the E. coli CLC, yeast RFC also opens PCNA using a crab-claw mechanism36. However, RFC opens PCNA with larger scale conformational changes in a direction opposite to that of the E. coli CLC. The AAA+ domains of E. coli CLC•β2closed and yeast RFC•PCNAclosed are overtwisted and the gaps in otherwise symmetrical CLCs are invaded. However, it is the AAA+ modules of subunits D and E in yeast RFC that are most displaced from ideal positions rather than those of A (δ) and B (γ1) in the E. coli CLC. Consequently, yeast RFC opens a PCNA clamp by moving AAA+ modules C, D and E outwards in a clockwise direction as viewed from the collar, while E. coli CLC opens β2 by moving A (δ) and B (γ1) anticlockwise. Thus, despite conserved structures and functions, the mechanisms of clamp opening by CLCs of E. coli and Saccharomyces cerevisiae have diverged.
Like the yeast complexes, the conformational changes from CLC•β2closed to CLC•β2open are accompanied by substantial increases in protein-protein interactions both among CLC subunits and between the CLC and β2; hence clamp opening is primarily driven by the free energy of protein interactions. The crab-claw mechanism used by E. coli CLC is not contradicted by the results of a fluorescence assay that suggest the CLC only undergoes limited conformational changes during clamp opening29. First, the conformational changes of the CLC are indeed smaller than yeast RFC. Secondly, the distances between the FRET pairs introduced in the CLC fall between those on CLC•β2closed and CLC•β2open. To open β2, CLC first constricts to bind, then expands to open. The net distance changes between the fluorophores from free CLC to CLC•β2closed then CLC•β2open are too small to be accurately detected.
Our studies also suggest that E. coli CLC can directly load clamps onto p/t DNA junctions. Previously, a “filter and slide” mechanism of clamp loading onto p/t DNA was proposed based on the crystal structure of the T4 CLC•DNA•clampopen complex4,31, in which only ssDNA can pass through the gap in the clamp and p/t DNA has to screw up the chamber of CLC for binding. However, it has since been shown that the E. coli CLC and yeast RFC both open their cognate clamps laterally with a gap large enough for dsDNA to enter. Moreover, this is also the case for the Rad24-RFC CLC that loads the 9-1-1 checkpoint clamp onto ssDNA or dsDNA with a 5’ recessed end58. This may therefore be a common characteristic in clamp opening, including in phage T4. Like the E. coli CLC•DNA•β2open structure, the T4 CLC•DNA•clampopen structure4,31 is likely a partially closed intermediate. As (i) the conformation of the E. coli CLC only changes slightly before (CLC•β2open) and after DNA binding (CLC•DNA•β2open) and (ii) the critical residue Tyr316 of δ that directly stacks on the 3’ base of the primer strand to recognise the p/t junction is well poised to interact, CLC•β2open must be able to bind directly to a p/t junction. Compared to the filter and slide mechanism, direct clamp loading is more efficient and feasible since the E. coli CLC is tethered to the DnaB helicase through Domain IV of τ12 and hence is very close to priming sites at replication forks, perhaps not requiring an extensive search for a p/t terminus on ssDNA. The τ subunit can thus chaperone the CLC•β2open complexes onto a newly primed site, then the Pol III core to the loaded clamp. Compared to the direct recognition of a p/t DNA junction by the E. coli CLC, yeast RFC has to partly melt the p/t junction, by which the 3’ base of the primer strand is flipped out to a pore where it stacks with Phe582 of Rfc1, a residue that is conserved in eukaryotes36. The p/t DNAs are also likely to bind to exterior sites on Rfc1 before being transferred into the central chamber of RFC, as do nicked and gapped DNAs37,38,39.
It is well established that hydrolysis of the three bound molecules of ATP by the E. coli CLC occurs at some stage during clamp closure and subsequent ejection of the CLC31,32,33,34,35. In one of the crystal structures of T4 CLC•DNA•clampclosed (PDB 3U61 (T4 CLC•DNA•clampclosed))31, the ATP-binding site at the A–B subunit interface is occupied by ADP. It was proposed that ATP hydrolysis at this site triggers conformational changes in the T4 CLC and clamp closure. In contrast, ATPγS was present in the four main ATP-binding sites in the yeast RFC•DNA•PCNAclosed complexes and all ATP-binding sites in an alternate T4 CLC•DNA•clampclosed structure (PDB 3U5Z (T4 CLC•DNA•clampclosed))31 are occupied by ADP•BeF4.
We expected that one or more of the ATP-binding sites in our CLC•DNA•β2closed structure would be occupied by ADP while the other(s) are bound to the ATP analogue ADP•AlF4−, reflecting the order of ATP hydrolysis at the three sites during clamp closing. This was not the case. Indeed, in all of our structures of the CLC bound to p/t DNA (with or without β2), the ATP binding sites are fully occupied by ADP•AlF4−, consistent with this nucleotide acting as an analogue of the transition state on the pathway of ATP hydrolysis at all three sites (Fig. 5h; Supplementary Table 2). This observation suggests that ATP hydrolysis at all three sites occurs simultaneously (or nearly so) during clamp closing and/or CLC ejection. Moreover, rather than the trapping of the ATPase sites in transition state structures leading to a single intermediate state of the CLC and bound clamp, it leads to a delicately poised dynamic equilibrium between open, closed and intermediate state(s) of the CLC and the clamp itself.
The energy differences among these states must be close to zero, so close that the equilibrium is affected even by the presence (remote from the sites of action) of a (dT45) single-stranded extension of the template DNA strand (whether or not SSB is bound to it or interacts with the χ subunit of the CLC). This implies that the clamp may be completely closed before the first ATP is hydrolysed, and that hydrolysis of the three ATPs probably occurs rapidly and sequentially to drive ejection of the CLC immediately from the closed clamp (Fig. 6). This interpretation is not inconsistent with single-turnover kinetic measurements of clamp closing and hydrolysis of the first ATP32,35, which show that these two processes are not closely coupled.
We note that both the full CLC complexes and even the δ subunit acting alone59 also promote clamp unloading, and recycling of clamps during lagging-strand DNA synthesis from the ends of completed to subsequent Okazaki fragments has been demonstrated directly by single-molecule studies60. Clamp unloading by the CLC, but not δ acting alone, is also ATP dependent59. Since the same ATP-bound CLC•DNA•β2closed to CLC•DNA•β2open structures must be on the pathway of clamp unloading from a p/t DNA and this reverse reaction presumably requires ATP binding rather than ATP synthesis, the principle of microscopic reversibility dictates that clamp closing must not have an obligate requirement for ATP hydrolysis.
Methods
Proteins
All proteins had native E. coli sequences, without purification tags. They were overproduced individually in appropriate E. coli strains using plasmids that directed their expression from phage T7 ϕ10 (induced with IPTG) or phage λ tandem PRPL (induced by temperature shift from 30 to 42 °C) promoters. Unless specified otherwise, cells were lysed in optimised buffers (usually containing spermidine to compact nucleic acids) using a French press (two passages at 12,000 psi). Cell lysates were fractionated by ammonium sulphate precipitation, and proteins were isolated by chromatography using appropriate resins with optimised buffers. All steps were carried out at 4–6 °C. Protein concentrations were estimated spectrophotometrically from measured values of A280 using calculated values of ε280. Proteins were stored at –80 °C until use.
Essential aspects of isolation procedures are given below; full details have been described elsewhere: the δτ3δ’ and δγ3δ’ minimal clamp loader complexes40,61; bioχψδτ3δ’ full clamp loader complex with N-terminally biotinylated χ62; the χψ and bioχψ clamp loader accessory complexes61,62; β2 sliding clamp45; single-stranded DNA-binding protein SSB63 and SSB-T*, its OB-domain56. Proteins χ∼SSB-Ct with the C-tail of SSB (MDFDDDIPF) fused to χ by a flexible 9-residue linker (TRESGSIGS)55, and SSBCysApp with a cysteine residue appended to the C-terminus of SSB57 were isolated similarly to the wild-type proteins62,63.
Core subunits of the clamp loader complexes (CLCs)
Overproduction of CLC subunits δ and δ’ used T7 promoter plasmids64 while overproduction of γ65 and τ61 used tandem phage λ PRPL promoter plasmids. After the ammonium sulphate fractionation step, all four proteins were purified similarly by two steps of chromatography through columns of Toyopearl DEAE-650M resin (Supelco, Bellefonte, PA); the first isocratic step at high ionic strength removed nucleic acids and the second fractionated proteins in a salt gradient. The final chromatographic step used heparin agarose to give highly purified samples.
Accessory CLC subunits χ, bioχ, and χ∼SSB-Ct
Production of the wild-type χ subunit and variants of it used expression of holC genes in pET-χ66 and similar T7-promoter plasmids. Wild-type χ61 and N-terminally biotinylated χ (bioχ)62 were purified by sequential chromatography through columns of Toyopearl DEAE-650M resin (at high and low salt) and heparin agarose. For χ∼SSB-Ct55, chromatography on a Mono Q column (GE Healthcare) was used instead of heparin agarose. Since the bioχ apoprotein was not biotinylated in vivo, its complete biotinylation was accomplished by reaction in vitro with ATP and biotin, promoted by E. coli biotin ligase62.
Accessory CLC complexes χψ, bioχψ, and ψχ∼SSB-Ct
The bioχψ complex was produced by refolding of urea-solubilised ψ in the presence of bioχ61,62. The T7 promoter plasmid pET-ψ66 was used for high-level overproduction of the ψ subunit; ψ is expressed in insoluble form. Cells were lysed and the pellet was homogenised using a French press in buffer containing 1 M NaCl. The pellet was again homogenised similarly in buffer with 1 M NaCl, and then without added salt. The washed pellet containing ψ was solubilised in 6 M urea. A portion (6 mL) of purified bioχ (4.8 mg) was diluted with 9 mL of refolding buffer (20 mM Tris-HCl pH 7.6, 100 mM NaCl, 2 mM dithiothreitol, 0.5 mM EDTA), then 1 mL (10 mg) of ψ in 6 M urea was added dropwise to the solution with stirring. The solution was stirred for 4 h, then dialysed overnight in 2 L of 25 mM Tris-HCl pH 7.6, 90 mM NaCl, 2 mM dithiothreitol, 0.5 mM EDTA, 10% (v/v) glycerol. The solution was clarified by centrifugation and the supernatant loaded onto a Toyopearl DEAE-650M column equilibrated in the same buffer. Fractions of bioχψ that did not bind to the resin were collected. A similar procedure was used to prepare highly purified samples of the wild-type χψ and ψχ∼SSB-Ct complexes.
Preparation of the δγ3δ’ and δτ3δ’ minimal clamp loader complexes (CLCs)
The γ3 and τ3 CLCs were assembled from individually purified subunits and isolated chromatographically40,61. First, τ or γ was mixed with equimolar amounts (i.e., 3-fold excess) of δ and δ’. The mixture was then dialysed in 30 mM Tris-HCl pH 7.6, 2 mM dithiothreitol, 1 mM EDTA, 50 mM NaCl and loaded onto a 5 mL column of Q-Sepharose (GE Healthcare) equilibrated with the same buffer. After column washing with 10 mL of the buffer, the δτ3δ’ and δγ3δ’ complexes were eluted with a linear gradient (190 mL) of 50–450 mM NaCl.
Preparation of the full bioχψδτ3δ’ clamp loader complex
Chromatographic separation of three different CLC complexes of composition bioχψδγnτ3–nδ’ (where n = 0, 1 or 2), following the method described by Tanner et al.61. yielded the bioχψδτ3δ’ (n = 0) clamp loader complex in a well resolved peak62. Purified τ, γ, δ, δ’ and bioχψ were separately dialysed into a reconstitution buffer, then mixed in selected molar ratios in a defined order. A 1.8:1.0 mixture of τ and γ was treated for 2 h at 17 °C, while a 1.0:1.0:1.2 mixture of δ, bioχψ and δ’ was treated for 2 h at 4 °C. The two mixtures were then combined and set aside overnight at 4 °C before they were diluted and loaded onto a 1 mL MonoS HR column (GE Healthcare). The three CLC complexes of different τ/γ stoichiometry were eluted with a linear gradient of NaCl in buffer.
Purification of β
The E. coli β sliding clamp was overproduced using a phage λ promoter plasmid45. It was purified after ammonium sulphate precipitation by two chromatography steps through columns of Toyopearl DEAE-650M resin, as above. A flow-through step using hydroxyapatite (BioRad) removed a minor protease contaminant45.
Purification of E. coli SSB, SSB-T*, and SSBCysApp
SSB variants, including wild-type SSB, SSBT* and SSBCysApp were overproduced using phage λ promoter plasmids56,57,63, and purified using similar procedures. Following French press lysis, SSB and variants were precipitated from the cell-free extract with an (optimised for each) low concentration of ammonium sulphate (0.075–0.14 g/mL). The precipitated proteins were dissolved in a high salt buffer (300 mM NaCl) and dialysed to low ionic strength, resulting in precipitation/coacervation of SSB proteins. The precipitates were dissolved in a minimal volume of buffer with 300 mM NaCl and 30% (v/v) glycerol and added drop wise to the surface of a phosphocellulose P11 (Whatman) column equilibrated at low ionic strength. After column wash, SSB was eluted with a gradient of NaCl. Fractions containing SSB were pooled and SSB proteins were precipitated again by dialysis into a low salt buffer. The pellet was dissolved in buffer containing 500 mM NaCl and 30% (v/v) glycerol, and gel filtered through a Hiload 26/60 Superdex 75 column (GE Healthcare) equilibrated in buffer containing 500 mM NaCl and 10% (v/v) glycerol.
Oligonucleotides
Primer-template (p/t) DNAs (all from Integrated DNA Technologies, HPLC purified) for cryo-EM were prepared by annealing a primer oligonucleotide 862 (5ʹ-GAGATAGTTACAACATACGATCG) to template strands 864 (5ʹ-T10-CGATCGTATGTTGTAACTATCTC, for experiments of CLC•DNA•β2 without SSB) or 863 (5ʹ-T45-CGATCGTATGTTGTAACTATCTC, for experiments of CLC•DNA•β2 with SSB); and 629 (5’-TAGTTACAACATACT) to 883 (5’-T10-AGTATGTTGTAACTA) for CLC•DNA. For SPR studies, the biotinylated template strand 865 (5ʹ-biotin-dT50-CGATCGTATGTTGTAACTATCTC) was annealed to primer strand 862.
Surface plasmon resonance
SPR experiments were carried out on a Biacore T200 instrument (Cytiva) operated at 20 °C using a streptavidin-coated (SA) chip (Cytiva). The SPR buffer contained 30 mM Tris-HCl pH 7.6, 400 μM ATP, 8 mM MgCl2, 30 mM NaCl, 0.5 mM dithiothreitol, 0.005% surfactant P20; or 30 mM Bis-Tris pH 6.4, 0.4 mM ADP, 3 mM NaF, 0.3 mM AlCl3, 8 mM MgCl2, 30 mM NaCl, 0.5 mM dithiothreitol, 0.005% surfactant P20. Bis-Tris at pH 6.4 was used to increase the solubility of AlFx.
For interaction of β2 with the immobilised CLC complex, bioχψδτ3δ’ or bioχψδγ3δ’ containing N-terminally biotinylated χ (20 nM) was loaded onto a flow-cell of the SA chip to achieve a response of ∼1400 RU. Serially diluted solutions of β2 in buffer containing desired nucleotides were made to flow over the surface at 10 μL/min for desired durations. The bound protein was allowed to dissociate until the signal nearly returned to baseline. To determine the KD value of bioχψδτ3δ’−β2 interaction by kinetics fitting (Supplementary Fig. 1a), β2 was injected for 40 s, then its dissociation from the immobilised CLC was monitored for 700 s. The entire set of zero-subtracted data were fit simultaneously (globally) using a 1:1 Langmuir binding model with mass transfer correction to derive association and dissociation rate constants, ka and kd. The dissociation constant KD was calculated from Eq. 1:
To determine KD values of the bioχψδγ3δ’−β2 interaction by fitting using a 1:1 steady-state affinity (SSA) model (Supplementary Fig. 1b–d), β2 was injected for durations required to reach equilibrium with ATP, 100 s with ATPγS (400 μM) or 40 s with ADP•AlFx. The equilibrium responses of β2 interacting with the immobilised bioχψδγ3δ’ were fit using Eq. 2:
where Rmax corresponds to the response when all immobilised ligand on the surface (bioχψδγ3δ’) is saturated with the analyte (β2), KD is the dissociation constant and [A] is the concentration of analyte in solution.
For interaction of χψδγ3δ’ with immobilised p/t DNA (Supplementary Fig. 6), the biotinylated oligo 865 (3 nM) was loaded onto flow-cell 2 for 108 s, yielding a response of 85 RU. Oligo 862 at 1 μM was then injected to assemble p/t DNA on the chip surface in situ. This DNA template is comprised of a 23-bp dsDNA region positioned away from the surface and a (dT)50 5ʹ ssDNA overhang attached to the chip surface through the stable biotin-streptavidin interaction. CLC (δγ3δ’ψχ, 200 nM) in SPR buffer containing different nucleotides was injected for desired durations and the bound proteins were allowed to dissociate in buffer containing desired nucleotides. For assembly of the CLC complex on p/t DNA, 100 nM δγ3δ’ or δτ3δ’, 200 nM ψχ, 1.6 μM β2 and 20 nM SSB were injected sequentially.
Sample preparation for cryo-EM
To prepare samples for cryo-EM, 30 μL of 6 μM δγ3δ’ or δτ3δ’ was mixed with ψχ complex at a molar ratio of 1:1.2, β2 at 1:1.3, p/t DNA (annealed DNA oligonucleotides) at 1:1.3 if required, and dialysed twice at 4 °C against 250 mL of 30 mM Na.HEPES pH 7.5, 3 mM MgCl2, 2 mM dithiothreitol, 0.25 mM EDTA, 2% glycerol. Nucleotides (1 mM ATPγS or 2 mM ADP plus 5 mM NaF and 0.5 mM AlCl3) were then added to the dialysates. Samples without DNA were crosslinked by treatment with 2 or 5 mM bis(sulfosuccinimidyl)suberate (BS3) on the bench (∼21 °C) for 15 min. Reactions were quenched by adding 1 M Tris-HCl pH 7.6 to 25 mM. Samples (3 μL) were applied onto Quantifoil UltrAuFoil grids (R1.2/1.3) that were blotted for 4 s at 6 °C and zero force, then plunged into liquid ethane using a FEI Vitrobot Mark IV.
Data collection, image processing and model building
Micrographs were collected using a Thermo Fisher Titan Krios G3i microscope at 300 kV, equipped with a Gatan K2 camera in electron counting mode at a nominal defocus range from −0.4 to −1.3 μm, with a pixel size of 0.82 Å/pixel and a total dose of 50e− over 5 s fractionated across 50 frames, except that a pixel size of 0.66 Å/pixel and a total dose of 60e− over 6 s fractionated across 60 frames was used for the sample containing 23-bp dsDNA and a dT10 5ʹ-overhang for the reconstruction of CLC•DNA•β2closed. Images were collected without use of an energy filter.
For samples without DNA, movies were corrected for drift and aligned using MOTIONCOR267 with electron-dose weighting in RELION 3.168. Motion-corrected micrographs were then imported into cryoSPARC69. CTF parameters were estimated with CTFFIND4.170 and micrographs with estimated resolutions worse than 3.5 Å were excluded. Particles were first picked by blob-picker, extracted at 256-pixel box-size and subjected to 2D classification; classes showing high-resolution features were retained and used for Topaz71 training. The best Topaz training model was used to pick particles on all the micrographs. Particles were subjected to a round of 2D classification and good particles were retained for 3D classification by a combination of ab-initio reconstruction and heterogeneous refinement. First, ab-initio reconstruction generates de novo a user-specified number of maps (classes), then heterogeneous refinement classifies particles more accurately into each class. Particles of selected 3D classes were used for homogeneous 3D refinement. Refined particles were imported into RELION 3.1, extracted at 360-pixel box-size for 3D auto-refinement, CTF refinement, Bayesian polishing and final 3D auto-refinement and postprocess. For samples with DNA, data were processed in RELION 3.1. Particles were first picked from a subset of micrographs by Laplacian picking and subjected to 2D classification. Selected 2D classes were then used for template picking on all micrographs. Particles thus picked were classified by rounds of Class2D and Class3D. Particles from selected classes were used for 3D auto-refinement, followed by CTF refinement, Bayesian polishing, then final 3D auto-refinement and postprocess. Particles of the final 3D reconstructions were imported into cryoSPARC for 3D variance analysis (3DVA)46. Global resolution is reported according to the gold standard 0.143 FSC criterion72. Local resolutions of models were estimated using the RELION algorithm.
For model building, the structures of E. coli δγ3δ’•DNA complex (PDB 3GLI (CLC•p/t DNA structure))30 and β2 (PDB 1MMI (E. coli β2 clamp structure))45 were placed into density maps using Phenix.dock_in_map73. The resulting complexes were then refined in Phenix.real_space_refine73. After inspection and model building in COOT74, ATPγS or ADP•AlF4− was included in the models. The models were subjected to rebuilding in COOT and REFMAC75 refinement, followed by a final cycle of refinement in Phenix.real_space_refine.
Molecular graphics and analyses used Chimera76 and ChimeraX77. Protein interface areas were calculated by the PDBePisa server78.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Cryo-EM maps and atomic coordinates have been deposited with the Electron Microscopy Data Bank and Protein Data Bank under accession codes EMD-40079 and 8GIY for CLC•β2closed, EMD-40080 and 8GIZ for CLC•β2open, EMD-40081 and 8GJ0 for CLC•DNA•β2open1, EMD-40082 and 8GJ1 for CLC•DNA•β2open2, EMD-40083 and 8GJ2 for CLC•DNA•β2closed, and EMD-40084 and 8GJ3 for CLC•DNA. Raw movies of cryo-EM data have been deposited in the Electron Microscopy Public Image Archive under accession codes EMPIAR-12052 for CLC•β2closed and CLC•β2open, EMPIAR-12090 for CLC•DNA•β2open1 and CLC•DNA•β2open2, EMPIAR-12095 for CLC•DNA•β2closed, and EMPIAR-12096 for CLC•DNA. Source data are provided with this paper.
References
Stukenberg, P. T., Studwell-Vaughan, P. S. & O’Donnell, M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266, 11328–11334 (1991).
O’Donnell, M. & Kuriyan, J. Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 16, 35–41 (2006).
Hedglin, M., Kumar, R. & Benkovic, S. J. Replication clamps and clamp loaders. Cold Spring Harb. Perspect. Biol. 5, a010165 (2013).
Kelch, B. A. The lord of the rings: Structure and mechanism of the sliding clamp loader. Biopolymers 105, 532–546 (2016).
Vivona, J. B. & Kelman, Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 546, 167–172 (2003).
Liu, J. et al. Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair. Nature 539, 583–587 (2016).
Yang, X.-W. et al. MutS functions as a clamp loader by positioning MutL on the DNA during mismatch repair. Nat. Commun. 13, 5808 (2022).
Kong, X.-P., Onrust, R., O’Donnell, M. & Kuriyan, J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69, 425–437 (1992).
Krishna, T. S., Kong, X.-P., Gary, S., Burgers, P. M. & Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79, 1233–1243 (1994).
Moarefi, I., Jeruzalmi, D., Turner, J., O’Donnell, M. & Kuriyan, J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296, 1215–1223 (2000).
O’Donnell, M. E. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J. Biol. Chem. 262, 16558–16565 (1987).
Lewis, J. S., Jergic, S. & Dixon, N. E. The E. coli DNA replication fork. Enzymes 39, 31–88 (2016).
Yao, N. et al. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1, 101–113 (1996).
Oakley, A. J. Dynamics of open DNA sliding clamps. PLoS One 11, e0154899 (2016).
Erzberger, J. P. & Berger, J. M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006).
O’Donnell, M., Onrust, R., Dean, F. B., Chen, M. & Hurwitz, J. Homology in accessory proteins of replicative polymerases − E. coli to humans. Nucleic Acids Res. 21, 1–3 (1993).
Dalrymple, B. P., Kongsuwan, K., Wijffels, G., Dixon, N. E. & Jennings, P. A. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl Acad. Sci. USA 98, 11627–11632 (2001).
Jeruzalmi, D. et al. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106, 417–428 (2001).
Jeruzalmi, D., O’Donnell, M. & Kuriyan, J. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106, 429–441 (2001).
Kazmirski, S. L., Podobnik, M., Weitze, T. F., O’Donnell, M. & Kuriyan, J. Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex. Proc. Natl Acad. Sci. USA 101, 16750–16755 (2004).
Naktinis, V., Onrust, R., Fang, L. & O’Donnell, M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. II. Intermediate complex between the clamp loader and its clamp. J. Biol. Chem. 270, 13358–13365 (1995).
Hingorani, M. M. & O’Donnell, M. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 273, 24550–24563 (1998).
Bowman, G. D., O’Donnell, M. & Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 (2004).
Gaubitz, C. et al. Structure of the human clamp loader reveals an autoinhibited conformation of a substrate-bound AAA+ switch. Proc. Natl Acad. Sci. USA 117, 23571–23580 (2020).
Douma, L. G., Yu, K. K., England, J. K., Levitus, M. & Bloom, L. B. Mechanism of opening a sliding clamp. Nucleic Acids Res. 45, 10178–10189 (2017).
Turner, J., Hingorani, M. M., Kelman, Z. & O’Donnell, M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 18, 771–783 (1999).
Pietroni, P. & von Hippel, P. H. Multiple ATP binding is required to stabilize the “activated” (clamp open) clamp loader of the T4 DNA replication complex. J. Biol. Chem. 283, 28338–28353 (2008).
Paschall, C. O. et al. The Escherichia coli clamp loader can actively pry open the β-sliding clamp. J. Biol. Chem. 286, 42704–42714 (2011).
Goedken, E. R. et al. Fluorescence measurements on the E. coli DNA polymerase clamp loader: Implications for conformational changes during ATP and clamp binding. J. Mol. Biol. 336, 1047–1059 (2004).
Simonetta, K. R. et al. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137, 659–671 (2009).
Kelch, B. A., Makino, D. L., O’Donnell, M. & Kuriyan, J. How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680 (2011).
Hayner, J. N. & Bloom, L. B. The β sliding clamp closes around DNA prior to release by the Escherichia coli clamp loader γ complex. J. Biol. Chem. 288, 1162–1170 (2013).
Bertram, J. G. et al. Molecular mechanism and energetics of clamp assembly in Escherichia coli: The role of ATP hydrolysis when γ complex loads β onto DNA. J. Biol. Chem. 275, 28413–28420 (2000).
Williams, C. R., Snyder, A. K., Kuzmič, P., O’Donnell, M. & Bloom, L. B. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the γ complex. J. Biol. Chem. 279, 4376–4385 (2004).
Anderson, S. G., Thompson, J. A., Paschall, C. O., O’Donnell, M. & Bloom, L. B. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli γ complex. Biochemistry 48, 8516–8527 (2009).
Gaubitz, C. et al. Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader. eLife 11, e74175 (2022).
Liu, X., Gaubitz, C., Pajak, J. & Kelch, B. A. A second DNA binding site on RFC facilitates clamp loading at gapped or nicked DNA. eLife 11, e77483 (2022).
Zheng, F., Georgescu, R., Yao, N. Y., Li, H. & O’Donnell, M. E. Cryo-EM structures reveal that RFC recognizes both the 3’- and 5’-DNA ends to load PCNA onto gaps for DNA repair. eLife 11, e77469 (2022).
Schrecker, M. et al. Multistep loading of a DNA sliding clamp onto DNA by replication factor C. eLife 11, e78253 (2022).
Jergic, S. et al. A direct proofreader-clamp interaction stabilizes the Pol III replicase in the polymerization mode. EMBO J. 32, 1322–1333 (2013).
Binder, J. K. et al. Intrinsic stability and oligomerization dynamics of DNA processivity clamps. Nucleic Acids Res. 42, 6476–6486 (2014).
Onrust, R., Stukenberg, P. T. & O’Donnell, M. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J. Biol. Chem. 266, 21681–21686 (1991).
Lacabanne, D. et al. ATP analogues for structural investigations: case studies of a DnaB helicase and an ABC transporter. Molecules 25, 5268 (2020).
Gulbis, J. M. et al. Crystal structure of the chi:psi subassembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur. J. Biochem. 271, 439–449 (2004).
Oakley, A. J. et al. Flexibility revealed by the 1.85 Å crystal structure of the β sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr. D. 59, 1192–1199 (2003).
Punjani, A. & Fleet, D. J. 3D variability analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Leu, F. P. & O’Donnell, M. Interplay of clamp loader subunits in opening the β sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem. 276, 47185–47194 (2001).
Ason, B. et al. Mechanism of loading the Escherichia coli DNA polymerase III β sliding clamp on DNA. Bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J. Biol. Chem. 278, 10033–10040 (2003).
Chen, S., Coman, M. M., Sakato, M., O’Donnell, M. & Hingorani, M. M. Conserved residues in the δ subunit help the E. coli clamp loader, γ complex, target primer-template DNA for clamp assembly. Nucleic Acids Res. 36, 3274–3286 (2008).
Bonde, N. J., Kozlov, A. G., Cox, M. M., Lohman, T. M. & Keck, J. L. Molecular insights into the prototypical single-stranded DNA-binding protein from E. coli. Crit. Rev. Biochem. Mol. Biol. 59, 99–127 (2024).
Shereda, R. D., Kozlov, A. G., Lohman, T. M., Cox, M. M. & Keck, J. L. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318 (2008).
Newcomb, E. S. P., Douma, L. G., Morris, L. A. & Bloom, L. B. The Escherichia coli clamp loader rapidly remodels SSB on DNA to load clamps. Nucleic Acids Res. 50, 12872–12884 (2022).
Kelman, Z., Yuzhakov, A., Andjelkovic, J. & O’Donnell, M. Devoted to the lagging strand—the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 17, 2436–2349 (1998).
Marceau, A. H. et al. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 30, 4236–4724 (2011).
Lo, A. T. Y. Protein dynamics on the lagging strand during DNA synthesis. Ph.D. Thesis, University of Wollongong (2012).
Su, X.-C. et al. Bound or free: Interaction of the C-terminal domain of Escherichia coli single-stranded DNA-binding protein (SSB) with the tetrameric core of SSB. Biochemistry 53, 1925–1934 (2014).
Wang, Y. Single-stranded DNA-binding protein and its role in Okazaki fragment maturation. Ph.D. Thesis, University of Wollongong (2015).
Castaneda, J. C., Schrecker, M., Remus, D. & Hite, R. K. Mechanisms of loading and release of the 9-1-1 checkpoint clamp. Nat. Struct. Mol. Biol. 29, 369–375 (2022).
Leu, F. P., Hingorani, M. M., Turner, J. & O’Donnell, M. The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem. 275, 34609–34618 (2000).
Tanner, N. A. et al. E. coli DNA replication in the absence of free β clamps. EMBO J. 30, 1830–1840 (2011).
Tanner, N. A. et al. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 15, 170–176 (2008).
Monachino, E. et al. A primase-induced conformational switch controls the stability of the bacterial replisome. Mol. Cell 79, 140–154 (2020).
Mason, C. E. et al. Escherichia coli single-stranded DNA-binding protein: NanoESI-MS studies of salt-modulated subunit exchange and DNA binding transactions. J. Am. Soc. Mass Spectrom. 24, 274–285 (2013).
Wijffels, G. et al. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides derived from β2-binding proteins. Biochemistry 43, 5661–5671 (2004).
Ozawa, K. et al. Cell-free protein synthesis in an autoinduction system for NMR studies of protein–protein interactions. J. Biomol. NMR 32, 235–241 (2005).
Xiao, H., Crombie, R., Dong, Z., Onrust, R. & O'Donnell, M. DNA polymerase III accessory proteins. III. holC and holD encoding χ and ψ. J. Biol. Chem. 268, 11773–11778 (1993).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Scheres, S. H. W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Scheres, S. H. W. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. 60, 2126–2132 (2004).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. 67, 355–367 (2011).
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Kontopidis, G. et al. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl Acad. Sci. USA 102, 1871–1876 (2005).
Acknowledgements
We thank Yao Wang and Amy McGrath for gifts of proteins, Jacob Lewis and Sarah Henrikus for discussion on cryo-EM data processing, and Joshua Silver for IT support. We are especially grateful for mechanistic discussions with Linda Bloom. This work was supported by the Australian Research Council (DP180100805 to A. J. O. and N. E. D.; DP210100365 to H. G., P. J. L., G. T., A. J. O. and N. E. D.).
Author information
Authors and Affiliations
Contributions
Conceptualisation, Methodology, Z.Q.X., S.J., H.G., P.J.L., G.T., A.J.O., N.E.D.; Investigation: Z.Q.X. (EM), A.C.P., S.H.J.B., J.C.B. (EM support), A.J.O. (model building), Z.Q.X. (structure analysis), S.J., Z.Q.X., A.C.P. (SPR); Resources, S.J., Z.Q.X., A.T.Y.L; Writing—Original Draft, Visualisation, Z.Q.X., Writing—Review & Editing, Z.Q.X., S.J., A.J.O., N.E.D.; All authors have read and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Richard Fishel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, ZQ., Jergic, S., Lo, A.T.Y. et al. Structural characterisation of the complete cycle of sliding clamp loading in Escherichia coli. Nat Commun 15, 8372 (2024). https://doi.org/10.1038/s41467-024-52623-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52623-9
This article is cited by
-
Divide-and-conquer strategy for NMR studies of the E. coli γ-clamp loader complex
Journal of Biomolecular NMR (2025)