Abstract
Post-translational cycles of α-tubulin detyrosination and tyrosination generate microtubule diversity, the cellular functions of which remain largely unknown. Here we show that α-tubulin detyrosination regulates kinetochore-microtubule attachments to ensure normal chromosome oscillations and timely anaphase onset during mitosis. Remarkably, detyrosinated α-tubulin levels near kinetochore microtubule plus-ends depend on the direction of chromosome motion during metaphase. Proteomic analyses unveil that the KNL-1/MIS12/NDC80 (KMN) network that forms the core microtubule-binding site at kinetochores and the microtubule-rescue protein CLASP2 are enriched on tyrosinated and detyrosinated microtubules during mitosis, respectively. α-tubulin detyrosination enhances CLASP2 binding and NDC80 complex diffusion along the microtubule lattice in vitro. Rescue experiments overexpressing NDC80, including variants with slower microtubule diffusion, suggest a functional interplay with α-tubulin detyrosination for the establishment of a labile kinetochore-microtubule interface. These results offer a mechanistic explanation for how different detyrosinated α-tubulin levels near kinetochore microtubule plus-ends fine-tune load-bearing attachments to both growing and shrinking microtubules.
Similar content being viewed by others
Introduction
Microtubule building blocks, α- and β-tubulin, are expressed by several different genes that give rise to distinct tubulin isotypes1,2. Additionally, α- and β-tubulin are subject to a wide range of post-translational modifications (PTMs)1,2. The combination of tubulin isotypes with specific PTMs generates microtubule diversity and functions as a ‘tubulin code’ that regulates intrinsic microtubule properties and behavior, as well as the interactions with several microtubule-associated proteins (MAPs)3. Most α-tubulin isotypes are genetically encoded with a tyrosine residue at their C-terminal tails. However, after incorporation into the microtubule polymer, this last tyrosine can be catalytically removed by the action of the recently identified tubulin carboxypeptidases Vasohibin-1 (VASH1), Vasohibin-2 (VASH2) and MATCAP4,5,6. As microtubules depolymerize, soluble α-tubulin is re-tyrosinated by a tubulin tyrosine ligase (TTL)7. This α-tubulin detyrosination/tyrosination cycle has been implicated in the regulation of critical physiological processes, including mitosis and meiosis, cilia/flagella properties, neuronal function and cardiac muscle contraction1. Moreover, alterations in α-tubulin detyrosination/tyrosination have been implicated in cancer, including therapy response, as well as fertility, neurological and muscle disorders1,2,8,9.
While the quest to identify long-sought molecular ‘writers’ of the α-tubulin detyrosination/tyrosination code has recently been accomplished4,5,6, we still lack fundamental knowledge on the full breadth of code ‘readers’ and their respective cellular roles. During mitosis, the α-tubulin detyrosination/tyrosination code was proposed to mediate a navigation system that assists polar chromosome alignment by regulating opposing motor protein activities at kinetochores10,11, and it has been implicated in mitotic error correction and spindle microtubule length control by regulating microtubule depolymerizing enzymes12,13,14,15. Because mitotic spindles are highly complex cytoskeletal structures assembled from hundreds of different proteins that associate with microtubules, centrosomes, and/or kinetochores16, we hypothesized that the α-tubulin detyrosination/tyrosination code regulates additional mitotic processes that remain elusive and uncharacterized at the mechanistic level. Here, we show that different detyrosinated α-tubulin levels near kinetochore microtubule plus-ends fine-tune load-bearing attachments to both growing and shrinking microtubules, thereby ensuring normal chromosome oscillations and timely anaphase onset during mitosis.
Results
Genetic manipulation of α-tubulin detyrosination in human cells
As a first step towards decoding how the α-tubulin detyrosination/tyrosination code impacts mitosis, we set to overcome previous limitations related with the use of tubulin carboxypeptidase inhibitors with limited specificity or TTL overexpression that could inadvertently hamper tubulin polymerization17,18. To do so, we used CRISPR-Cas9 gene editing in human U2OS cells to generate non-clonal TTL or VASH1 + VASH2 knockout (KO) lines19 (Supplementary Fig. 1a–c). TTL or VASH1/2 KO lines constitutively expressed high or low detyrosinated α-tubulin levels, respectively (Fig. 1a, b). For subsequent phenotypic analysis, we optionally used lentiviral-mediated transduction to stably drive the expression of exogenous GFP-tagged detyrosinated α-tubulin in TTL KO cells or GFP-tagged tyrosinated α-tubulin in VASH1/2 KO cells to visualize the microtubules, as well as Histone H2B-mRFP to visualize the chromosomes (Fig. 1a, b). In parallel, RNAi-mediated depletion of the recently identified α-tubulin detyrosinase MATCAP6 resulted in ~35% reduction of detyrosinated α-tubulin in control cells, but this treatment had only a marginal effect in the amount of detyrosinated α-tubulin in VASH1/2 KO cells, where the baseline levels were already below 10% (Supplementary Fig. 1d).
a Establishment of U2OS cell lines knockout (KO) for TTL or VASH1/2 by CRISPR-Cas9 gene editing. Histone H2B-mRFP and detyrosinated or tyrosinated GFP-α-tubulin were optionally transduced for imaging purposes. b Expression levels (relative quantifications, reference = 1.00) of detyrosinated and tyrosinated tubulin in the KO lines analyzed by western blotting (values refer to relative levels obtained from the illustrated western blot, which was qualitatively validated by at least 2 independent experiments). GAPDH and endogenous α-tubulin were used as loading controls (c) Representative super-resolution CH-STED microscopy images of metaphase U2OS cells generated in (a), after immunofluorescence with anti-detyrosinated α-tubulin (green), total α-tubulin (magenta) and anti-centromere antibody (ACA; cyan). Scale bar is 5 μm. Source data are provided as a Source Data file.
Super-resolution CH-STED microscopy20 confirmed a profound redistribution of detyrosinated α-tubulin in TTL KO cells, decorating the entire mitotic spindle (including astral microtubules), whereas this form of tubulin was almost undetectable in VASH1/2 KO cells (Fig. 1c). Consequently, TTL KO cells showed longer metaphase spindles, whereas in VASH1/2 KO cells spindles were shorter (further exacerbated upon MATCAP RNAi) (Supplementary Fig. 2a, b), as expected from the inhibitory role of α-tubulin detyrosination on the activity of microtubule depolymerizing enzymes12,13,14,15. Thus, the generated cell lines represent a powerful tool to dissect the role of α-tubulin detyrosination during mitosis.
α-tubulin detyrosination has a broader impact on mitosis than previously anticipated
Previous RNAi-based studies that aimed at interfering with TTL function indicated that the α-tubulin detyrosination/tyrosination code played a critical role in error correction during mitosis by regulating the activity of microtubule depolymerizing enzymes at centromeres/kinetochores12. However, compensatory or cellular adaptation mechanisms to the chronic loss of TTL appear to exist, possibly involving the contribution of genetically-encoded tyrosinated α-tubulin12,21. Indeed, in contrast to TTL KO cells expressing tyrosinated GFP-α-tubulin, TTL KO cells constitutively expressing detyrosinated GFP-α-tubulin showed ∼2-fold increase in the frequency of chromosome segregation errors, consisting mostly in transient anaphase lagging chromosomes, a minority of which resulted in the formation of micronuclei (Supplementary Fig. 3a, b and Supplementary Movie 1). Thus, under the forced expression of detyrosinated α-tubulin, cells are able to compensate/adapt to the chronic loss of TTL.
Conversely, a significant fraction of VASH1/2 KO cells stably expressing tyrosinated (but not detyrosinated) GFP-α-tubulin showed persistent misaligned chromosomes in the vicinity of the spindle poles (Supplementary Fig. 4a, b and Supplementary Movie 1), consistent with a role of α-tubulin detyrosination in the regulation of CENP-E-mediated alignment of polar chromosomes10. Other VASH1/2 KO cells eventually completed chromosome alignment and entered anaphase, despite a significant delay (Fig. 2a–c and Supplementary Movie 1). Once in anaphase, no obvious increase in chromosome segregation errors was observed (Supplementary Fig. 3b). MATCAP depletion by RNAi also increased the time from NEB to metaphase and anaphase relative to controls, but the effect was not additive to that of VASH1/2 KO (Fig. 2a–c and Supplementary Movie 1). These data suggest that, besides guiding polar chromosomes to the spindle equator, α-tubulin detyrosination levels controlled either by VASH1/2 or MATCAP contributes to the establishment of functional kinetochore-microtubule attachments necessary for timely SAC satisfaction and anaphase onset. However, it remains unclear whether both functions rely on the regulatory effect of α-tubulin detyrosination over CENP-E activity.
a Live-cell imaging of chromosome and microtubule behavior showing delayed chromosome alignment and anaphase onset in the VASH1/2 knockout (KO) with/without MATCAP RNAi; time is in h:min from nuclear envelope breakdown (NEB); scale bar is 5 µm. b Quantification of NEB-metaphase duration during live cell imaging; Control Tyrosinated (Tyr) GFP-α-tubulin = 57 cells, pool of 16 independent experiments, TTL KO Detyrosinated (Detyr) GFP-α-tubulin = 67 cells, pool of 18 independent experiments, VASH1/2 KO Tyr GFP-α-tubulin = 72 cells, pool of 20 independent experiments, TTL KO Tyr GFP-α-tubulin = 59 cells, pool of 12 independent experiments, Control Tyr GFP-α-tubulin + MATCAP RNAi = 68 cells, pool of 16 independent experiments, VASH1/2 KO Tyr GFP-α-tubulin + MATCAP RNAi = 102 cells, pool of 25 independent experiments; each data point represents an individual cell; bars represent mean and standard deviation; exact p-values are displayed above each respective data set; significance p < 0.05, unpaired two-tailed t-test. c Quantification of NEB-Anaphase onset duration from the same data set as in (b); each data point represents an individual cell; bars represent mean and standard deviation. Source data are provided as a Source Data file.
α-tubulin detyrosination fine-tunes kinetochore-microtubule attachments
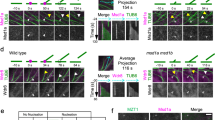
The important implication of the slow kinetics of α-tubulin detyrosination on polymerized microtubules is that this modification progressively accumulates along kinetochore microtubules, which are significantly more stable than astral microtubules that rapidly turn over. As tubulin heterodimers ‘age’ after their incorporation into the microtubule lattice at the kinetochore-bound end on metaphase chromosomes, the detyrosinated subunits “flux” toward the associated pole12. However, the newly incorporated tubulins at the kinetochore-associated ends are mostly tyrosinated, generating a gradient of detyrosinated/tyrosinated α-tubulin levels along the kinetochore fibers. Thus, kinetochores moving towards the associated pole at the end of depolymerizing microtubules are likely to encounter the ‘older’ lattice enriched in detyrosinated α-tubulin. Conversely, kinetochores moving away from the associated poles should encounter the ‘younger’ microtubule lattice with recently polymerized tyrosinated α-tubulin22. In agreement with this idea, we found a significant reduction in the levels of detyrosinated α-tubulin on growing, EB1-positive, kinetochore microtubules, when compared with shrinking, EB1-negative, microtubules on the corresponding kinetochore pair (Fig. 3a–d). These findings imply that oscillating kinetochores interact with microtubules with varying levels of detyrosinated α-tubulin, depending on the direction of their motion.
a Immunofluorescence analysis of a 3D projection from a U2OS Parental cell using anti-tyrosinated (tyr) α-tubulin (white), anti-centromere antibody (ACA; magenta), anti-detyrosinated (detyr) α-tubulin antibodies (blue) and anti-EB1 (green); Grayscale panels show single channels for tyrosinated α-tubulin and detyrosinated α-tubulin (max intensity projection vs. sum projection). Scale bars 5 µm. a’ Single section from the same cell in (a); individual signals for each channel are also displayed in insets. b Line scan profile of the fluorescence intensities (F.I.) from the highlighted region from the inset in (a’). c Representation of the model of the distribution of tyrosinated and detyrosinated α-tubulin near oscillating kinetochores (kMTs = kinetochore microtubules). AP = anti-poleward, P = Poleward. d Quantification of the ratio between detyrosinated α-tubulin fluorescence intensity and tyrosinated α-tubulin fluorescence intensity of the growing (EB1 + ) kMTs and shrinking (EB1-) kMTs near oscillating kinetochores in the parental cell line; error bars represent mean and standard deviation; each point represents a kinetochore; quantifications from a pool of 3 independent experiments; at least 1 kinetochore pair per cell was measured (total kinetochore pairs = 95) from a total of 50 cells; exact p-values are displayed above each respective data set; significance p < 0.05, unpaired two-tailed t-test. e Quantification of the ratio of the detyrosinated α-tubulin normalized fluorescence intensity between the growing (EB1 + ) and shrinking (EB1-) kMTs from the same kinetochore pair, in the same data set as in (d). Source data are provided as a Source Data file.
Based on these observations, we hypothesized that detyrosinated α-tubulin levels might serve as a regulatory cue that modulates kinetochore-microtubule interactions during chromosome oscillations. To test this hypothesis, we measured metaphase plate width in fixed TTL or VASH1/2 KO lines, and compared the respective chromosome dispersion relative to parental U2OS cells (Supplementary Fig. 5a). VASH1/2 KO cells had wider metaphase plates compared to parental controls, whereas TTL KO cells showed slightly more compact metaphase plates, consistent with our hypothesis (Supplementary Fig. 5a). To get quantitative insight into the dynamics of chromosomes at the spindle equator and rule out potential indirect effects due to incomplete chromosome congression upon experimental perturbation of α-tubulin detyrosination, we performed live-cell imaging, combined with 3D-tracking of sister kinetochore behavior over time during metaphase23 in parental, TTL or VASH1/2 KO cells stably expressing GFP-CENP-A. While no significant deviations from normal inter-kinetochore distances were found (Supplementary Fig. 5b), VASH1/2 KO cells showed more dispersed kinetochores leading to wider metaphase plates (Fig. 4a–c), in line with our fixed-cell analysis (Supplementary Fig. 5a). To quantify the regularity of sister-kinetochore oscillations, we calculated the autocorrelation function of sister-kinetochore movements along the spindle axis. Sister-kinetochores in parental cells behave as semi-regular oscillators (Fig. 4a−d), resulting in an autocorrelation curve in which the first negative side-lobe indicates the mean half-period, the second positive lobe indicates the mean full period of the oscillator, and the magnitude of the side-lobes reflects its regularity. In contrast, VASH1/2 KO cells were characterized by highly irregular and slower sister kinetochore oscillations, with several chromosomes coming off the plate and eventually recovering, suggesting transient kinetochore-microtubule detachment and re-attachment (Fig. 4a–f and Supplementary Movie 2). Indeed, a significantly higher percentage of VASH1/2 KO cells in metaphase showed one or more kinetochores that accumulated detectable MAD1 (Fig. 4g, h), a SAC protein that decorates unattached or partially attached kinetochores24.
a Live-cell imaging of MG132-treated (<1 h) GFP-CENP-A-expressing Parental and VASH1/2 knockout (KO) cell lines. Arrowheads indicate movement of kinetochore (KT) pairs off the metaphase plate. Time is in min:sec; scale bar is 5 µm. b Chromokymographs obtained from live-cell imaging of MG132-arrested GFP-CENP-A-expressing Parental and VASH1/2 KO cell lines. Arrowheads indicate movement of KT pairs off the metaphase and recovery events. c Frame-to-frame displacement of sister-KTs and their distance from the metaphase plate center for the control condition (Parental; black), VASH1/2 KO (green) and TTL KO (magenta); quantifications from: Parental = 59 cells, 7 independent experiments, TTL KO = 65 cells, 7 independent experiments; VASH1/2 KO = 47 cells, 5 independent experiments. d Cumulative autocorrelation curves representing sister-KT oscillations in control (black), VASH1/2 KO (green) and TTL KO (magenta). The vertical axis represents the regularity of the oscillations and the horizontal axis represents the time (quantifications from the same data set of (c)). e Sister-KT velocity in control (gray), VASH 1/2 KO (green) and TTL KO (magenta). The velocity was quantified as the mean of all sister-KT pairs per cell, multiplied by the number of frames within 1 min (quantifications from the same data set of (c)). Exact p-values are displayed above each respective data set; significance p < 0.05, Dunn’s multiple comparison test. f Quantification of the percentage of cells with KTs out of the metaphase plate from the same data set of (c). g Immunofluorescence of U2OS Parental, TTL KO and VASH1/2 KO cell lines using anti-MAD1 (green) and anti-centromere antibodies (ACA; magenta) antibodies; scale bar is 5 µm. h Quantification of the percentage of cells with MAD1 positive KTs per cell from a pool of 3 independent experiments: Parental = 95 cells, TTL KO = 111 cells, VASH1/2 KO = 92 cells. Source data are provided as a Source Data file.
The previous 3D-tracking of kinetochores on already aligned chromosomes rules out indirect effects on kinetochore-microtubule attachments caused by spatial constrains, such as those imposed by problems in chromosome congression. We also do not favor the idea that reduced α-tubulin detyrosination levels negatively impact chromosome positioning and kinetochore-microtubule attachments during metaphase either by inhibiting the activity of the kinetochore motor CENP-E10 or by enhancing the activity of the microtubule depolymerizing enzyme MCAK at centromeres/kinetochores12,14,15. This hypothesis seems unlikely because experimental modulation of α-tubulin detyrosination levels does not significantly affect microtubule dynamics in vitro25 or kinetochore microtubule dynamics upon chromosome bi-orientation12, whereas the inhibition of CENP-E motor activity with GSK-92329526 or enhancement of MCAK activity either with UMK57 or MCAK overexpression12 had no significant impact on the width of the metaphase plate or on the fraction of MAD1-positive kinetochores (note that only aligned chromosomes were considered in the case of CENP-E-inhibited cells due to well-established roles for this kinetochore motor in the alignment of peripheral polar chromosomes27) (Supplementary Fig. 6a–c; see also ref. 23 showing that CENP-E-depletion by RNAi had no effect on any of the kinetochore tracking parameters on aligned chromosomes). Taken together, these findings indicate that α-tubulin detyrosination promotes the formation of a functional kinetochore-microtubule interface necessary for normal metaphase chromosome dynamics. Because these phenotypes are not caused by the improper regulation of CENP-E and MCAK activity, additional ‘readers’ of α-tubulin detyrosination levels on kinetochore microtubules are likely to be involved.
Molecular readers of detyrosinated/tyrosinated α-tubulin during mitosis
To seek additional molecular ‘readers’ of the α-tubulin detyrosination/tyrosination code that might be involved in regulating kinetochore-microtubule attachments we isolated taxol-stabilized microtubules from mitotic cell extracts derived either from TTL KO or VASH1/2 KO cells, in the presence of ATP to enrich for non-motor microtubule-associated proteins (MAPs)19,28 (Fig. 5a, b). Our choice for TTL KO or VASH1/2 KO mitotic cell extracts was based on the rationale that they constitute two extreme conditions where microtubules can be experimentally enriched for detyrosinated and tyrosinated α-tubulin, respectively, and thus more likely to reveal meaningful differences on enriched MAPs. Staining with SiR-tubulin confirmed the successful isolation of mitotic microtubules using this approach (Fig. 5c). Moreover, their respective enrichment in detyrosinated or tyrosinated α-tubulin was confirmed by western blotting (Fig. 5d). Upon salt extraction, MAPs associated with microtubules enriched for either detyrosinated or tyrosinated α-tubulin were identified by mass spectrometry analysis (Fig. 5b). The results from three independent experiments identified 3954 proteins that were subsequently ranked by their mean relative abundance between VASH1/2 KO and TTL KO, and respective confidence level. This list was then filtered for known association with the cytoskeleton and/or cell cycle roles, resulting in a shortlist of 290 proteins (see Methods). An arbitrary threshold of 1.5-fold enrichment either on detyrosinated or tyrosinated microtubules was applied, leading to a final list of 31 and 77 proteins that were differentially enriched on detyrosinated and tyrosinated mitotic microtubules, respectively (Supplementary Data 1 and Supplementary Data 2).
a Western blot analysis of the expression levels (relative quantification, reference = 1.00) of detyrosinated (detyr) and tyrosinated (tyr) α-tubulin in the knockout (KO) U2OS cell lines used in the mass-spec analysis (values refer to relative levels obtained from the illustrated western blot, which was qualitatively validated by at least 2 independent experiments). α-tubulin and GAPDH were used as loading controls. b Schematic representation of the protocol for isolation of microtubules (MTs) and microtubule-associated proteins (MAPs) from mitotic extracts derived from U2OS TTL KO and VASH1/2 KO cell lines. STLC = S-trityl-L-cysteine; S = Supernatant; P = Pellet. c Wide-field fluorescence microcopy images of polymerized microtubules (stained with SiR-tubulin) obtained from the P3 fraction (see scheme in (b)) from the TTL KO and VASH1/2 KO cell lines; scale bar is 5 µm. d Western blot analysis of detyrosinated and tyrosinated α-tubulin in the P3 fractions from 3 independent experiments per condition. Total α- and β-tubulin were used as reference controls. e Summary of the most relevant mitotic hits obtained from mass-spec analysis of the isolated S3 fractions from the same 3 independent experiments as in (d), comparing abundance (Ab) ratios between VASH1/2 KO and TTL KO cell lines.
It drew our attention that the KMN network components, including KNL1, the MIS12 complex proteins DSN1, NSL1 and MIS12, and the NDC80 complex proteins NUF2, SPC25 and HEC1/NDC80, that form the core microtubule-binding site at kinetochores29, were all enriched on tyrosinated microtubules during mitosis (Fig. 5e and Supplementary Data 1). A similar enrichment was found for the NDC80 and MIS12 complexes-interacting proteins ZWINT30 and SUGT131, several Augmin complex subunits (HAUS3, HAUS5 and HAUS8) and the γ-TuRC-targeting protein NEDD1 involved in branched microtubule nucleation32, the small GTPase RAN required for chromatin-mediated microtubule nucleation and organization33, as well as the microtubule destabilizing protein Stathmin, consistent with its capacity to prevent microtubule polymerization by binding preferentially to (tyrosinated) tubulin dimers34,35. Importantly, the CAP-Gly domain protein CLIP1/CLIP-170, a well-established ‘reader’ of tyrosinated α-tubulin36, was also enriched, validating our approach. Interestingly, the microtubule-associated proteins EML1 and EML4, related with EML2 that preferentially associates with tyrosinated microtubules37, were also found enriched under our experimental conditions.
In contrast, the kinetochore and spindle-associated protein CLASP2 involved in the regulation of kinetochore microtubule dynamics38 stood out as one of the top hits enriched on detyrosinated microtubules during mitosis (Fig. 5e and Supplementary Data 2). Other proteins with well-established roles in spindle architecture and kinetochore microtubule organization/dynamics that showed a preference for detyrosinated microtubules included CKAP5/ch-TOG, TACC3 and CENP-F39,40,41, as well as the centriolar protein Centrin-242. Some established ‘readers’ of α-tubulin detyrosination/tyrosination, such as CENP-E, dynein/dynactin and MCAK10,14,15,43, were largely undetected in our preparations, as expected from the employed conditions that disfavored the association of motor proteins with microtubules, and hinting that this list is not fully exhaustive. Lastly, we note that the relative abundance of several signaling proteins appears to be modulated either by α-tubulin detyrosination or tyrosination levels during mitosis, but the significance of these findings remains to be explored. Overall, our results identify a large cohort of potential direct and indirect readers of the α-tubulin detyrosination/tyrosination code in mitosis, suggesting that this code regulates a broad range of mitotic processes, including spindle microtubule organization and dynamics, centriole assembly and the formation of a functional kinetochore-microtubule interface.
α-tubulin detyrosination enhances CLASP2 binding and NDC80 complex diffusion along the microtubule lattice
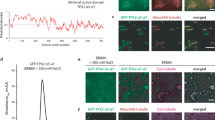
The association of HEC1/NDC80 and CLASPs with microtubules has been previously shown to depend on tubulin C-terminal tails44,45, but the functional significance of these interactions and whether they were regulated by the α-tubulin detyrosination/tyrosination code remained unclear. Given the well-established role of the NDC80 complex and CLASPs in the formation and regulation of kinetochore-microtubule attachments29,38,45,46,47,48,49,50, we focused on directly assessing how the α-tubulin detyrosination/tyrosination code impacts their association with microtubules. To do so, we reconstituted the binding of purified human NDC80 “Broccoli”-GFP51 (hereafter referred as NDC80-GFP) and human GFP-CLASP252 to taxol-stabilized porcine brain microtubules that have been fully detyrosinated or tyrosinated in vitro using either recombinant CPA10 or TTL53, respectively (Fig. 6a, b). To avoid interfering with microtubule binding affinity, we used a BRB80-based buffer without NaCl, despite some evident aggregation of GFP-CLASP2 (Fig. 6a). These experiments revealed a clear preference for GFP-CLASP2 to bind detyrosinated microtubules, with an estimated KD value of 0.027 μM vs. 0.64 μM for tyrosinated microtubules (Fig. 6a). In contrast, purified NDC80-GFP showed only a slight (25%) preference for binding to tyrosinated microtubules in vitro, with an estimated KD value of 0.90 μM vs. 1.2 μM for detyrosinated microtubules (Fig. 6b).
a–c Microtubule (MT) co-pelleting assay using purified GFP-CLASP2 (a) or NDC80 “broccoli”-GFP (c) and tyrosinated (tyr) or detyrosinated (detyr) microtubules. P = pellet; S = supernatant. The ratios of GFP-CLASP2 and NDC80 “broccoli”-GFP between the pellet and supernatant fractions were quantified on the BlueSafe gel. Expression levels of detyrosinated and tyrosinated α-tubulin were confirmed by western blot (WB) analysis. MW Molecular weight. b–d Quantifications of GFP-CLASP2 (b) or NDC80 “broccoli”-GFP (d) in the pellet fractions. Bound fraction (%) to microtubules were plotted after baseline subtraction (no microtubules). Estimated KD (equilibrium dissociation constant) was obtained after fitting the data to a non-linear regression (2 independent experiments with 2 replicates each). Error bars indicate standard deviation of the mean. Source data are provided as a Source Data file.
To shed light into how the α-tubulin detyrosination/tyrosination code may impact additional NDC80 complex properties, we reconstituted its association with coverslip-immobilized microtubules enriched either for detyrosinated or tyrosinated α-tubulin purified from HeLa cells and stabilized with GMPCPP10 (Fig. 7a, b). Consistent with our microtubule pelleting assays, we could not detect significant differences in the residence time of single NDC80-GFP molecules on both microtubule lattices (Fig. 7c, d). However, we found that NDC80-GFP molecules diffused slower on tyrosinated microtubules (Fig. 7e, f), suggesting that NDC80 may form a different kinetochore-microtubule interface depending on α-tubulin detyrosination levels. Altogether, these in vitro results establish CLASP2 and the NDC80 complex as direct readers of the α-tubulin detyrosination/tyrosination code, which modifies the strength of their association with and mobility along the microtubule lattice, respectively.
a Schematic of the single molecule experimental setup to visualize diffusion of NDC80 “broccoli”-GFP along coverslip-immobilized microtubules (MTs). b Representative kymographs for diffusing NDC80 “broccoli”-GFP molecules on detyrosinated (detyr) and tyrosinated (tyr) MTs. Scale bars are 0.5 sec and 2 μm. c Cumulative distribution of the residence time for NDC80 “broccoli”-GFP from experiments using either detyrosinated (magenta, n = 1138 molecules) or tyrosinated microtubules (green, n = 693). Lines are exponential fits. d Column graph shows characteristic residence times (indicated by the bar height, which corresponds also to the center of the error bars) derived from exponential fit in panel (c) with error bars showing standard error of the mean (SEM), and p value determined from two-tailed t-test, see Materials and Methods section “Analysis of NDC80 diffusion”. e Mean square displacement (MSD) with standard deviations for NDC80 “broccoli”-GFP molecules diffusing on detyrosinated (n = 2278 molecules) and tyrosinated (n = 1442) microtubules. f Column graph shows diffusion coefficients (indicated by the bar height, which corresponds also to the center of the error bars) derived from the MSD graph in panel (e) with error bars showing SEM for the molecules with longest diffusion time (n = 481) for detyrosinated and n = 281 for tyrosinated microtubules, and p value determined from two-tailed t-test. p-values are displayed above each respective data set; significance p < 0.05. Source data are provided as a Source Data file.
CLASP2 and NDC80 association with spindle microtubules depends on detyrosinated/tyrosinated α-tubulin levels
To translate the biological significance of these in vitro results, we investigated CLASP2 and HEC1/NDC80 association with detyrosinated or tyrosinated microtubules in mitotic cells by quantifying the spindle accumulation of these proteins in parental, TTL KO and VASH1/2 KO cell lines by immunofluorescence. In line with our biochemical purifications and in vitro reconstitution studies, CLASP2 levels on metaphase spindle microtubules were significantly increased in TTL KO cells with high levels of detyrosinated α-tubulin (Fig. 8a, b). Conversely, and in addition to kinetochores, HEC1/NDC80 was found slightly enriched on metaphase spindle microtubules from VASH1/2 KO cells composed essentially of tyrosinated α-tubulin, (Fig. 8c, d). Importantly, the spindle microtubule enrichment of both proteins was not caused by marked changes in microtubule density under the different conditions, as revealed by quantifying total β-tubulin levels in the corresponding spindle regions (Supplementary Fig. 7). This quantification rules out a potential impact of molecular readers involved in microtubule amplification (e.g. Augmin). Therefore, different levels of CLASP2 and HEC1/NDC80 on mitotic spindles with altered α-tubulin detyrosination/tyrosination reflect their preference for the corresponding microtubule lattices in unperturbed cells.
a Immunofluorescence analysis of the U2OS Parental, TTL KO and VASH1/2 KO cell lines using anti-NDC80 (green) and anti-β-tubulin (magenta) antibodies; NDC80 signal is presented in grayscale for better visualization (max = max intensity projection vs. sum = sum projection); scale bar is 5 µm (b). Quantification of the NDC80 fluorescence intensity normalized for the β-tubulin fluorescence intensity for each cell line (NDC80/β-tubulin); error bars represent mean and standard deviation; quantifications from a pool of 3 independent experiments: Parental = 30 cells, TTL KO = 30 cells, VASH1/2 KO = 30 cells; Exact p-values are displayed above each respective data set; significance p < 0.05, unpaired two-tailed t-test. c Immunofluorescence analysis of the U2OS Parental, TTL KO and VASH1/2 KO cell lines using anti-CLASP2 (green) and anti-α-tubulin (magenta) antibodies; CLASP2 signal is presented in grayscale for better visualization (max = max intensity projection vs. sum = sum projection); scale bar is 5 µm. (d) Quantification of the CLASP2 fluorescence intensity normalized for the α-tubulin fluorescence intensity for each cell line (CLASP2/α-tubulin); error bars represent mean and standard deviation; quantifications from a pool of 3 independent experiments: Parental = 51 cells, TTL KO = 47 cells, VASH1/2 KO = 38 cells; Exact p-values are displayed above each respective data set; significance p < 0.05, unpaired two-tailed t-test. Arrowheads in (a) and (c) draw attention to the spindle pool. Source data are provided as a Source Data file.
A functional interplay between NDC80 and α-tubulin detyrosination promotes the establishment of a labile kinetochore-microtubule interface
Whereas the function of CLASP proteins in the mitotic spindle extends well beyond their roles at the kinetochore-microtubule interface48,54, the NDC80 complex is a strong candidate to fine-tune load-bearing attachments in response to the α-tubulin detyrosination state. This is supported by our finding that the rate of NDC80 complex diffusion along the microtubule lattice is sensitive to α-tubulin detyrosination, while prior reports have linked NDC80 diffusion to chromosome alignment and oscillations in metaphase55,56,57. To test whether there is a functional interplay between NDC80 microtubule-wall diffusion and α-tubulin detyrosination, we examined kinetochore-microtubule attachments in cells overexpressing wild-type HEC1/NDC80, or its mutant version (HEC1/NDC80-9A) with a slower diffusion rate57. Whereas the overexpression of wild-type HEC1/NDC80 in both parental and VASH1/2 KO cells was well tolerated and did not result in any obvious aggravation of chromosome alignment defects, overexpression of HEC1/NDC80-9A compromised chromosome alignment in ∼90% of the cases in both parental and VASH1/2 KO cells, in agreement with previous reports55. However, in those cells that managed to complete chromosome alignment, overexpression of both NDC80 variants suppressed chromosome dispersal, resulting in tighter metaphase plates in both parental and VASH1/2 KO cells (Fig. 9a, b). These results are consistent with previous observations of induction of effective end-on kinetochore-microtubule attachments after HEC1/NDC80-9A overexpression, but at the price of dampening metaphase chromosome oscillations55,56. Indeed, when we probed the state of kinetochore microtubule attachments in these cells using antibodies against MAD1, overexpression of HEC1/NDC80-9A was found to reduce the frequency of unattached (MAD1-positive) kinetochores in VASH1/2 KO cells with lower detyrosinated α-tubulin levels, but not in normal cells (Fig. 9a–c). These data support the importance of a proper balance between NDC80 activity and α-tubulin detyrosination for fine-tuning kinetochore-microtubule attachments.
a Immunofluorescence of the U2OS Parental cell line and VASH1/2 KO cell line overexpressing GFP-HEC1 wild-type (WT) and phosphorylation mutants (9 A), using anti-MAD1 (green) and anti-centromere antibodies (ACA; magenta) antibodies; GFP-HEC1 is in white; scale bar is 5 μm. b Quantification of the metaphase plate width (ACA signal) from parental and VASH1/2 KO cell lines, with/without overexpression of GFP-HEC1 WT and 9 A mutant in both cell lines; each data point represents an individual cell; bars represent mean and standard deviation; 3 independent experiments, Parental = 95 cells; Parental+Hec1 WT = 52 cells; Parental+Hec1 9 A = 52 cells; VASH1/2 KO = 92 cells; VASH1/2 KO+Hec1 WT = 53 cells; VASH1/2 KO+Hec1 9 A = 60 cells; exact p-values are displayed above each respective data set; significance p < 0.05, unpaired two-tailed t-test. c Comparative analysis of the percentage of cells with one or more MAD1 positive kinetochores (KTs) per cell from the same pool of (b); exact p-values are displayed above each respective data set; significance p < 0.05, non-parametric Mann-Whitney two-tailed test. Source data are provided as a Source Data file.
Discussion
The tubulin code has emerged as a powerful cellular strategy to regulate a broad range of critical cellular functions through the generation of microtubule diversity. Progress in recent years has unveiled the key enzymatic machinery – the so-called code ‘writers’ – underlying specific tubulin PTMs, opening up the exciting possibility of genetically manipulating the tubulin code to understand its functional implications, while exploring possible therapeutic opportunities. Here we used genetic manipulations to accurately control the enzymatic machinery underlying α-tubulin detyrosination/tyrosination in human cells and inquire about its roles during mitosis. Our approach was validated by confirming a role for the α-tubulin detyrosination/tyrosination code in error correction, polar chromosome alignment and the regulation of spindle microtubule length10,12,13. In addition, by combining quantitative immunofluorescence analyses with 3D kinetochore tracking at high spatial and temporal resolution in dividing human cells, we uncovered a previously overlooked role for the α-tubulin detyrosination/tyrosination code in the regulation of the kinetochore-microtubule interface. This role was manifested in two ways: (1) kinetochores on metaphase chromosomes interact with microtubules with varying levels of detyrosinated α-tubulin near their plus-ends, depending on the direction of their motion; and (2) experimental attenuation of α-tubulin detyrosination delayed chromosome alignment and anaphase onset, while causing highly irregular and slower sister kinetochore oscillations on already aligned chromosomes, with several chromosomes coming off the plate after completing alignment. Importantly, this role was independent from previously established roles of α-tubulin detyrosination on polar chromosome congression and the regulation of CENP-E and MCAK activity10,58, suggesting the involvement of additional ‘readers’ of α-tubulin detyrosination/tyrosination. To seek possible candidates, we took advantage of our genetically modified cells to identify MAPs that were enriched on either detyrosinated or tyrosinated mitotic microtubules. Some of these MAPs, such as CENP-F and CLIP1/CLIP-170, showed an enrichment on detyrosinated and tyrosinated microtubules, respectively, in line with previous studies using asynchronous HeLa cell extracts37. However, other MAPs, such as EML-1 and EML-4, which did not show any specific enrichment on the different microtubule sub-types in these previous studies, were now found enriched on tyrosinated mitotic microtubules, whereas Centrin-2 showed opposite preferential enrichment between the two studies. While the differences found for EML-1 and EML-4 might reflect additional factors (e.g. Cdk1-mediated phosphorylation) that differentiate their binding to microtubules during interphase and mitosis, the opposite preference found for the centriolar protein Centrin-2 is more difficult to explain, given the well-established enrichment of centrioles in detyrosinated microtubules10,59. Most striking, several KMN network components, including NDC80, which establishes critical load-bearing contacts with the ends of dynamic kinetochore microtubules60, were consistently found enriched on tyrosinated microtubules isolated from mitotic cells and in situ on mitotic spindles from VASH1/2 KO cells. Interestingly, subsequent in vitro reconstitution experiments with purified NDC80 complex revealed no significant preference for its association with tyrosinated microtubules, suggesting that enrichment of the NDC80 complex on tyrosinated spindle microtubules might be indirect and/or involve additional factors. For example, we note that the ZWINT/KNL1 sub-complex or the CAP-Gly proteins CLIP1/CLIP170 and Dynactin are also implicated in kinetochore-microtubule attachments30,61,62, and they may cooperate with the NDC80 complex to favor its interaction with tyrosinated microtubules in mitotic cells36,63. Another non-mutually exclusive possibility is that the preferential association of the NDC80 complex with tyrosinated microtubules is modulated by other tubulin PTMs, such as polyglutamylation64. Of note, NDC80 is also known to associate with other factors, such as CKAP5/ch-TOG65, which we found enriched on detyrosinated microtubules, and deciphering how these readers are affected by the tubulin code may be challenging. Indeed, CLASP2 was found enriched on detyrosinated microtubules, but its well-established interacting protein CLIP1/CLIP170, appears to prefer tyrosinated microtubules36. Such contrasting associations suggest a fine regulation that likely reflects specific mechanisms underlying the association of these proteins with different microtubule sub-structures (e.g. lattice vs. plus-ends). Overall, these data support the idea that the combined action of multiple factors with opposite preferences for tyrosinated or detyrosinated microtubules, together with the relative levels of tyrosinated or detyrosinated α-tubulin, ultimately determine how kinetochores interact with microtubules.
Regardless of the detailed mechanism, we speculate that the enrichment of the KMN network on ‘younger’ tyrosinated microtubules reflects its key role in the establishment of the initial contacts during microtubule capture and subsequent stabilization of the nascent end-on attached microtubules early in mitosis. At metaphase, the situation is more complex, as the oscillating kinetochores on bi-oriented chromosomes maintain end-on attachments, while translocating along microtubule lattices with different detyrosinated α-tubulin levels, depending on the direction of motion (Fig. 10). In this respect, we were fascinated to discover that NDC80 complex diffusion is modulated by α-tubulin detyrosination levels. While the effect appears subtle for single NDC80 molecules, the mobility of multiple NDC80 molecules, such as found at the kinetochores, may be affected more significantly. At the trailing kinetochore, which moves away from its pole at the newly growing microtubule plus-ends enriched in tyrosinated α-tubulin, the slower diffusion of the NDC80 complex along this lattice may help to sustain end-on attachments and prevent this kinetochore from slipping under the force of the leading kinetochore (Fig. 10), in a way that mimics the increased friction observed on the trailing kinetochore upon HEC1/NDC80 tail dephosphorylation66. Since these microtubule plus-ends are polymerizing, the activity of the microtubule rescue factor CLASP2 might be dispensable, consistent with its lower affinity to this type of microtubule lattice. The leading kinetochore, on the other hand, moves poleward under the microtubule depolymerizing force on a lattice enriched for detyrosinated α-tubulin (Fig. 10). The faster diffusion of NDC80 on detyrosinated α-tubulin may facilitate and smooth kinetochore poleward motion during metaphase chromosome oscillations, a function that likely involves additional players, such as the Astrin-SKAP complex67. The enhanced affinity of CLASP2 to microtubules enriched in detyrosinated α-tubulin may prevent uncontrolled microtubule depolymerization and eventual kinetochore detachment68, which occurs with higher probability during microtubule disassembly than assembly60. In agreement, chromosome oscillations are more irregular and microtubule detachments are increased during metaphase in VASH1/2 KO cells. Under these conditions, microtubule friction at kinetochores might be too low to sustain regular chromosome oscillations, while maintaining load-bearing end-on association. The preferential association of CLASP2 with detyrosinated spindle microtubules might additionally contribute to distinguish older from newly incorporated tubulin subunits at kinetochores and/or depolymerizing from polymerizing kinetochore-attached microtubules that were recently shown to have distinct nucleotide-binding states68,69. These findings might have implications for our understanding of non-Mendelian segregation of selfish centromeres that are known to detach at higher frequency from cortical facing spindle microtubules enriched in tyrosinated α-tubulin during female meiosis70. Overall, the present work unveils a level of complexity and regulation of the kinetochore-microtubule interface that was not previously anticipated, while providing evidence that an intricate gradient of detyrosinated/tyrosinated α-tubulin along kinetochore microtubules promotes efficient alignment of chromosomes and their normal oscillations during metaphase.
The NDC80 complex has a preference for ‘younger’ tyrosinated α-tubulin. Black arrows show direction of chromosome motion. As the initial end-on microtubule (MT) contacts become stabilized, gradual α-tubulin detyrosination would lead to a different makeup of the kinetochore (KT)-bound MT ends, depending on the direction of motion. NDC80 complex diffusion along the ‘older’ MT lattice towards the pole is facilitated, whereas stronger binding to “younger” polymerizing end help preventing MT detachment from the trailing KT. CLASP2 might prevent MT detachments under higher friction with detyrosinated microtubules at KTs.
Methods
Cell line maintenance
All cell lines used in this work were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Corning) with 10% Fetal Bovine Serum (FBS; Gibco), supplemented with 10 µg/µl of antibiotic-antimycotic mixture (Gibco). Cell lines were maintained at 37 °C in a 5% CO2, humidified atmosphere.
CRISPR-Cas9 generation of KO cells
The TTL and VASH1/2 knockout cells were generated by CRISPR/Cas9-mediated gene editing, using a lentiviral backbone containing both the Streptococcus pyogenes Cas9 (spCas9) nuclease and the single guide RNA scaffold (lentiCRISPRv2). For TTL gene knockout, two 20-bp single guide RNA: 5′-CAC CGA ACA GCA GCG TCT ACG CCG-3′ and 5′-AAA CCG GCG TAG ACG CTG CTG TTC-3′ were designed to target the TTL gene as previously described (Ref. 19); for VASH1 knockout the oligonucleotides used were 5’-CACCGACGGCTTCCAGGCATTTGAT-3’ and 5’-AAACATCAAATGCCTGGAAGCCGTC-3’; for VASH2 knockout 5′‐ CACCGAATGGCCGCTATGGCTCATT‐3′ and 5′‐ AAACAATGAGCCATAGCGGCCATTC‐3′. [Sigma-Aldrich; (Ref. 4)]. Previous rescue experiments in human Hap1 cells validate specificity of the VASH1/2 sgRNAs (Ref. 4), which were cloned into the lentiviral vector pLenti-CRISPR-v2 (no. 52961, Addgene). Non-clonal knockout cells were selected by their resistance to puromycin (2 µg/mL, Calbiochem) and Blasticidin S (10 μg/mL, Invitrogen), and protein depletion inferred by western blot analysis. To validate CRISPR/Cas9 editing of VASH2 gene due to lack of a specific antibody, a PCR of the targeted genomic region was optimized in order to detect any alteration and concomitant loss of primer homology to the equivalent wild type version of the genomic loci affected in the edited region. The forward primer OHM302 5´-ATAATCACACAGGGACCCAGTTCTT-3´recognizes the exon 3 of VASH2 genomic locus. The reverse primer OHM298 5´-GGATGACAGCTTCAAGGCATTTGAT-3´ recognizes the targeted region of the VASH2 CRISPR guide described previously (Ref. 4). The optimized genomic PCR amplifies a band of 614 bp from any unmodified genomic DNA in the gene VASH2. The PCR was performed using the same starting amount of genomic DNA supplemented with Betaine (0.2 M, Sigma-Aldrich) to enhance the reaction. The same result was consistently obtained at different melting temperatures, genomic DNA quantities (between 30–250 ng per 25 cycles) and different DNA extraction protocols.
Lentiviral transduction
All lentiviral particles were produced by co-transfection of HEK293T cells with the lentiviral vectors, psPAX2 (Gag, Pol, Rev, and Tat expressing packaging vector), pMD2.G (VSV-G expressing envelope vector) and with plasmids bearing the gene of interest: pIRESneo3-EGFP-Tubalpha-GEEY, pIRESneo3-EGFP-Tubalpha-GEE* (obtained by direct mutagenesis of the previous), LV-H2B-RFP #26001 (Addgene) and pRRL-2xGFP-CENP-A generated by Gibson assembly (Ref. 19). After 72 h supernatants containing lentiviral particles were collected. For lentiviral transduction, lentiviral particles were added to U2OS cell lines with DMEM 10% FB with 1:2000 Polybrene (Sigma-Aldrich) for 24 h.
Plasmid transfection
Plasmids (~1 µg) containing EGFP-MCAK full-length (kind gift from Linda Wordeman, University of Washington, Seattle, WA, USA) and pCDNA5-pEGFP-HEC1, both WT and 9 A (lacking the Aurora B phosphorylation sites in the tail; kind gift from Geert Kops, Oncode Institute, Hubrecht Institute, Utrecht, the Netherlands) were dissolved in 1 mL of Opti-MEM (Gibco) and then mixed with 1 mL with other solution of Opti-MEM containing 5 µL of Lipofectamine 2000 (Invitrogen). The solution mix was incubated for 30 min at room temperature and then added to ~0.2 × 106 cells seeded in 6 well plate. After a 6-h incubation at 37 °C, the medium was removed and replaced by 2 mL of DMEM with 10% FBS. The cells were fixed for immunofluorescence after 36-48 h.
RNAi
RNAi experiments were performed in 0.2 × 106 cells cultured in 1.5 mL DMEM with 5% FBS. A solution containing 2 µL of Lipofectamine RNAiMax (Invitrogen) diluted in 250 µL Opti-MEM (Gibco) was mixed with another containing 100 pmol of siRNA (Integrated DNA Technologies-IDT) diluted in 250 µL Opti-MEM. The solution mix was incubated for 30 min at room temperature and then added dropwise to the cells. The siRNA oligonucleotides used were the pre-designed siRNAs hs.Ri.KIAA0895L.13.1 5’-CAGUUGGAAAGUCCUUAUUCAUUTA-3’ and hs.Ri.KIAA0895L.13.2 5’-AUAUGGUCGAUUGUGCAAAUACA-3’ from Integrated DNA Technologies-IDT. After a 6-h incubation at 37 °C, the medium was removed and replaced by 2 mL of DMEM with 10% FBS.
Western blotting
Protein extracts were obtained from cells by addition of lysis buffer (20 mM Hepes/KOH, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 2 mM DTT at −20 °C, and protease inhibitor 4 C + PMSF 0.1 mM at −20 °C [1:100], pH 7.9) and frozen with liquid N2. The suspension was centrifuged for 5 min at 20817 g at 4 °C, collecting the supernatant. Total protein levels were quantified using Bradford reagent (Thermo Scientific) and BSA (Thermo Scientific) solutions as standards. 25-50 µg of total protein per sample were mixed with sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue) and denatured at 95 °C for 5 min. Samples were loaded in a 10% acrylamide gel mounted in a Mini-PROTEAN vertical electrophoresis apparatus (Bio-Rad) using an NZYColour Protein Marker II (NZYTech). Blotting was performed with a Transfer-Blot Turbo transfer system (Bio-Rad). Membranes were blocked with 5% powder milk in PBS Tween 0.1% for 45 min. For primary antibodies, it was used rabbit anti-detyrosinated tubulin 1:5000 (kind gift from Marin Barisic, Danish Cancer Institute, Copenhagen, Denmark), rat anti-tyrosinated tubulin 1:5000 (BioRad), mouse anti-TTL 1:5000 (Proteintech), mouse anti-VASH1 1:1000 (C-6, sc-365541, Santa Cruz), mouse anti-FLAG 1:10000 (M-2, Sigma-Aldrich), mouse anti-α-tubulin B-512 1:10,000 (Sigma-Aldrich), mouse anti-GAPDH 1:40000 (Proteintech) and rabbit anti-β-tubulin 1:2000 (Abcam) diluted in 5% powder milk in PBS Tween 0.1%, and incubated overnight (4 °C) with agitation. Anti-rat HRP, anti-rabbit HRP and anti-mouse HRP secondary antibodies (Jackson ImmunoResearch) were used at 1:5000 dilution in 5% powder milk in PBS Tween 0.1% and incubated for 1 h. Signal was developed with Clarity Western ECL Blotting Substrate (Bio-Rad) and detected/quantified in a Bio-Rad Chemidoc XRS system, with Image Lab software. Minor inter-experimental differences after MATCAP RNAi might be due to the detection limit of detyrosinated tubulin by western blotting that changes as a function of the total protein loaded in the gel, or due to different depletion efficiency between experiments.
Fixed cell analysis and immunofluorescence
Cells were fixed using paraformaldehyde (PFA; Electron Microscopy Sciences) at 4% in cytoskeleton buffer (CB; 137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 0.4 mM KH2PO4, 2 mM EGTA, 2 mM MgCl2, 5 mM PIPES, and 5 mM glucose, pH = 6.1) for 10 min or in alternative in methanol (Merck) at −20 oC. In the cases of inducing metaphase arrest, cells were treated with 5 µM MG132 (Merck) for 1 h prior fixation. To inhibit CENP-E, 20 nM GSK-923295 (MedChemexpress) were added to the cells for 1 h prior fixation. To enhance MCAK activity 250 µM of UMK57 (Ref. 71) was added to the cells for 12–16 h before fixation. Extraction was accomplished using CB–Triton X-100 0.5% for 10 min. Primary antibodies used in this work were mouse anti-α-tubulin clone B-512 at 1:1500 (Sigma-Aldrich), rat monoclonal anti-CLASP2 at 1:2 (Ref. 72), mouse monoclonal anti-Hec1/NDC80 at 1:1000 (Abcam), mouse anti-MAD1 clone BB3-8 at 1:500 (Millipore), mouse monoclonal anti-EB1 at 1:500 (BD Biosciences, clone 5/EB1 cat 610535), human anti-centromere antiserum 1:1000 (ACA; Fitzgerald), rabbit anti-detyrosinated tubulin (kind gift from Marin Barisic, Danish Cancer Institute, Copenhagen, Denmark) 1:1500, rat anti-tyrosinated tubulin 1:1000 (BioRad),and rabbit anti-β-tubulin at 1:1000 (Abcam) diluted in PBS–Triton X-100 0.1% with 10% FBS. Secondary antibodies Alexa Fluor anti-mouse 568, anti-rat 568, anti-rat 488, anti-rabbit 488, anti-human 647 and anti-rat 647 (Invitrogen) were diluted to 1:1000 in 0.1% PBS–Triton X-100 with 10% FBS. DNA staining was achieved by addition of 1 µg/ml 4’,6-Diamidino-2-Phenylindole (DAPI; SigmaAldrich). Coverslips were mounted using mounting medium (20 mM Tris, pH 8, 0.5 mM N-propyl gallate, and 90% glycerol). Images were acquired with an Axio imager Z1 (Carl Zeiss) equipped with an ORCA-R2 precooled charge-coupled device (CCD; Hamamatsu), using an immersion oil 63× 1.46 NA planapochromatic objective lens, controlled by Zen software. 3D deconvolution was performed using AutoQuant X Image Deconvolution software (Media Cybernetics). All images in figures are maximum intensity projections, except when indicated otherwise. Quantifications, histogram adjustments and panel construction were performed with Fiji-ImageJ software, Adobe Photoshop CS6 and Adobe Illustrator CS5 (Adobe Systems). Quantifications of fluorescence intensity of NDC80 and CLASP2 spindle levels were made using sum projections of the raw data in Fiji-ImageJ. A ROI was generated including the entire mitotic spindle -small area- and another bigger ROI around the small one defined as big area. Both integrated fluorescence intensities were measured. The Background signal was calculated using the equation = integrated fluorescence intensity of “big area” —integrated fluorescence intensity of “small area”)/(“big area” – “small area”). The corrected fluorescence intensity was calculated using the equation = integrated fluorescence intensity of “small area”—(background signal × “small area”).
CH-STED
The U2OS cell lines expressing the different detyrosinated/tyrosinated GFP-tubulin and H2B-mRFP were fixed using PFA (Electron Microscopy Sciences) at 4% and Glutaraldehyde (Electron Microscopy Sciences) at 0.2% in CB for 10 min. For autofluorescence quenching it was used 0.1% sodium borohydride solution (Sigma-Aldrich) diluted in PBS, for 10 min. Extraction was performed with CB-0.5%Triton (Sigma-Aldrich), for 10 min. Primary antibodies used: rabbit anti-detyrosinated tubulin (kind gift from Marin Barisic, Danish Cancer Institute, Copenhagen, Denmark) 1:150, mouse anti-α-tubulin clone B-512 at 1:150 (Sigma-Aldrich), human anti-centromere antiserum 1:1000 (ACA; Fitzgerald) diluted in PBS 0.1% Tween with 10% FBS. Secondary antibodies used were anti-mouse STAR 580 (Abberior Instruments); anti-rabbit STAR 635 (Abberior Instruments) at 1:100 and Alexa Fluor anti-human 405 (Invitrogen) at 1:1000, diluted in CB-0.1% Tween with 10% FBS. Coverslips were mounted using mounting medium. For Coherent-Hybrid STED (CH-STED) imaging (Ref. 20), an Abberior ‘Expert Line’ gated-STED microscope was used, equipped with a Nikon Lambda Plan-Apo 1.4 NA 60x objective lens. The depletion beam was generated by a bivortex phase mask (radii ratio = 0.88) to improve suppression of background fluorescence in comparison to conventional 2D-STED. This performance stems from the capacity of a bivortex to generate a depletion dip in 3D that, unlike z-STED or 2D+z-STED combinations, preserves resilience to spherical aberration. All acquisition channels (confocal and STED) were performed using a 0.8 Airy unit pinhole. A time-gate threshold of 500 ps was applied to the STED channel. Fixed-cell images were acquired using excitation wavelengths at 561 nm and 640 nm. Excited volumes were doughnut-depleted with a single laser at 775 nm. All images in the figures represent maximum intensity projections. Quantifications, histogram adjustments and panel construction were performed with Fiji-ImageJ software, Adobe Photoshop CS6 and Adobe Illustrator CS5 (Adobe Systems)
Live-cell imaging
The U2OS cell lines expressing the different detyrosinated/tyrosinated GFP-tubulin and H2B-RFP were cultured in glass coverslips using DMEM without phenol red and supplemented with 25 mM Hepes (Gibco) and 10% FBS. Time-lapse imaging was performed in a heated chamber (37 °C) using a 60× oil immersion 1.40 NA plan-apochromatic objective mounted on an inverted microscope (Eclipse TE2000U; Nikon) equipped with a CSU-X1 spinning-disk confocal head (Yokogawa Corporation of America) controlled by NIS-Elements software and with two laser lines (488 nm, 561 nm). Images were detected with an iXonEM+ EM-CCD camera (Andor Technology). 11 z-planes separated by 1 µm were collected every 2 min. All images in the figures represent maximum intensity projections. Histogram adjustment, quantifications and panel construction were performed using Fiji-ImageJ software, NIS Viewer (Nikon), Adobe Photoshop CS6 and Adobe Illustrator CS5 (Adobe Systems).
Isolation of mitotic microtubules and MAPs
U2OS TTL KO and VASH1/2 KO cell lines were grown in T175 flasks until they reach ~70% confluence and then incubated with 5 μM of STLC for 14 h. The mitotic cells were collected by shake-off and pelleted at 259 g for 5 min. It was gathered ~1 g of cell pellet for each condition. The cell pellet was resuspended in 1.5 mL of K-PIPES buffer (100 mM PIPES pH 6.9, 1 mM EGTA, 1 mM MgSO4,1 mM DTT, 0.1 mM GTP) supplemented with 1 mM Mg-ATP and protease inhibitors at 4 °C, and incubated on ice for 15 min. The resulted mixture was gentle sonicated and then centrifuged at 100000 g for 1 h at 4 °C. At the resulted supernatant was added 40 μM taxol and incubated for 30 min at 37 °C. The assembled MTs were pelleted over a 10% sucrose cushion at 45000 g for 30 min at 25 °C (supernatant S1 + pellet P1). The pellet was resuspended in K-PIPES buffer supplemented with 20 μM taxol. The resuspended MTs were pelleted again over a 10% sucrose cushion at 45000 g for 30 min at 25 °C (supernatant S2) + pellet P2. To extract MAPs from the assembled MTs, the pellet was resuspended in 100 μL of K-PIPES buffer supplemented with 20 μM taxol and 350 mM NaCl and then centrifuged over a 10% sucrose cushion at 45,000 g for 30 min at 25 °C (Ref. 19). The resulted supernatant S3 was analyzed by Mass Spec; the pellet P3 was isolated for subsequent analysis by western blot and wide-field microscopy.
Detection of microtubules extracted from P3
The P3 pellet resulting from the microtubule isolation protocol, representing the polymerized microtubules stripped from MAPs, was diluted in K-PIPES buffer with 10 µM taxol and incubated with 20 nM of SiR-Tubulin (Spirochrome) for 30 min. A small sample of this solution was mounted in a glass slide and observed in Axio imager Z1 (Carl Zeiss) in the 647 nm channel.
Mass–spec analysis
Each sample was processed for proteomic analysis following the solid-phase-enhanced sample-preparation (SP3) protocol and enzymatically digested with Trypsin/LysC as previously described73. Protein identification and quantitation were performed by nanoLC-MS/MS. This equipment is composed of an Ultimate 3000 liquid chromatography system coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). Five hundred nanograms of peptides of each sample were loaded onto a trapping cartridge (Acclaim PepMap C18 100 Å, 5 mm × 300 μm i.d., 160454, Thermo Scientific, Bremen, Germany) in a mobile phase of 2% ACN, 0.1% FA at 10 μL/min. After 3 min loading, the trap column was switched in-line to a 50 cm × 75 μm inner diameter EASY- Spray column (ES903, PepMap RSLC, C18, 2 μm, Thermo Scientific, Bremen, Germany) at 250 nL/min. Separation was achieved by mixing A: 0.1% FA and B: 80% ACN, 0.1% FA with the following gradient: 5 min (2.5% B to 10% B), 120 min (10% B to 30% B), 20 min (30% B to 50% B), 5 min (50% B to 99% B), and 10 min (hold 99% B). Subsequently, the column was equilibrated with 2.5% B for 17 min. Data acquisition was controlled by Xcalibur 4.0 and Tune 2.11 software (Thermo Scientific, Bremen, Germany). The mass spectrometer was operated in the data-dependent (dd) positive acquisition mode alternating between a full scan (m/z 380-1580) and subsequent HCD MS/MS of the 10 most intense peaks from a full scan (normalized collision energy of 27%). The ESI spray voltage was 1.9 kV. The global settings were as follows: use lock masses best (m/z 445.12003), lock mass injection Full MS and chrom. peak width (FWHM) of 15 s. The full scan settings were as follows: 70 k resolution (m/z 200), AGC target 3 × 106, maximum injection time 120 ms; dd settings: minimum AGC target 8 × 103, intensity threshold 7.3 × 104, charge exclusion: unassigned, 1, 8, >8, peptide match preferred, exclude isotopes on and 45 s of dynamic exclusion. The MS2 settings were as follows: microscans 1, resolution 35 k (m/z 200), AGC target 2× 105, maximum injection time 110 ms, isolation window 2.0 m/z, isolation offset 0.0 m/z, dynamic first mass, and spectrum data type profile. The raw data was processed using the Proteome Discoverer 2.5.0.400 software (Thermo Scientific) and searched against the UniProt database for the Homo sapiens reviewed Proteome (2021_03 with 20371 entries) together with a spectral library database (NIST Human Orbitrap HCD 20160923). A common protein contaminant list from MaxQuant was also considered in the analysis. The MSPepSearch and Sequest HT search engines were used to identify tryptic peptides. The ion mass tolerance was 10 ppm for precursor ions and 0.02 Da for fragment ions. Maximum allowed missing cleavage sites was set to two. Cysteine carbamidomethylation was defined as constant modification. Methionine oxidation, deamidation of glutamine and asparagine, peptide terminus glutamine to pyroglutamate, and protein N-terminus acetylation, Met-loss, and Met-loss+acetyl were defined as variable modifications. Peptide confidence was set to high. The processing node Percolator was enabled with the following settings: maximum delta Cn 0.05; decoy database search target false discovery rate 1%, validation based on q-value. Protein label-free quantitation was performed with the Minora feature detector node at the processing step. Precursor ions quantification was performed at the consensus step with the following parameters: unique plus razor peptides were considered, precursor abundance based on intensity, and normalization based on total peptide amount. For hypothesis testing, protein ratio calculation was pairwise ratio-based and an t-test (background based) hypothesis test was performed. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE74 partner repository with the dataset identifier PXD047133.
Quantification of α-detyrosinated tubulin levels at the sister kinetochore vicinity of metaphase cells
Parental U2OS cells were fixed and immune-stained (as previous described). Single-plane, in focus, sisters KT pairs were chosen with a k-fiber growing in one side (with a positive EB1 signal) and the other k-fiber shrinking in the opposite side (with a negative EB1 signal), for fluorescence intensity quantifications. A ROI with the same area of 1 µm x µm was drawn in the vicinity of the KT-MT attachment in both sister KTs and detyrosinated α-tubulin fluorescence intensity levels were measured and background corrected. Tyrosinated α-tubulin levels were also measured and background corrected in the same ROIs, in order to calculate the ratio between detyrosinated and tyrosinated α-tubulin. All the quantifications were made in Fiji-ImageJ.
Quantification of metaphase spindle length
Metaphase spindle length was measured in the cell lines in fixed material (as previous described). Pole to pole distance was measured in the xy plane; z-stack measurement was also acquired, considering the spindle tilt. The overall length was calculated using the Pythagorean theorem: length = √[(xy)2 + z2]. All the quantifications were made in Fiji-ImageJ.
Quantification of MAD1 levels and metaphase plate width
Cells arrested in metaphase with MG-132 (as previous described) were selected to quantify MAD1 positive KTs per cell. In the same data set, metaphase width was measured taking in account the farthest KTs from the center of the metaphase plate from both sides. All the quantifications were made in Fiji-ImageJ.
Chromokymograph generation
Chromomokymographs from cells derived from the same data set used in the characterization of kinetochore oscillations patterns were generated as previously described75.
Proteins for in vitro single-molecule assays
Bovine tubulin was purified from cow brains by thermal cycling and chromatography as in ref. 76, and labeled with HiLyte647 (HiLyte647-SE, Anaspec) as in ref. 77. Post-translationally modified human tubulin was prepared as in ref. 10. Human Broccoli NDC80-GFP construct containing well-behaved truncated versions of NDC80 and NUF2 that include all microtubule-binding and adjacent regions and behave identically to full-length NDC80 complex was generated by cloning Nuf2 (residues 1-348) and Hec1 (residues 1-506-7xHis) into pET3aTr and then into pST39 for co-expression. To generate hNdc80 “Broccoli”, Nuf2 (residues 1-348) was cloned into a modified pET3aTr containing sequence encoding a C-terminal GFP-tag. ceNDC80 “Broccoli”-sfGFP was generated by cloning HIM-10 (residues 1-392) into a modified version of pET3aTr containing sequence encoding a C-terminal sfGFP-tag and NDC80 (residues 1-507-7xHis) into pET3aTr and then into pST39 for co-expression51. Human GFP-CLASP2 was expressed and purified using the MultiBac expression system. Sf21 cells were lysed by sonication in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 7 mM β-mercaptoethanol supplemented with protease inhibitors (Complete EDTA-free, Roche, Basel, Switzerland). Protein extracts clarified by centrifugation at 20,000 × g for 30 min were loaded onto a HisTrap HP column (GE Healthcare, Chicago, IL, USA) preequilibrated in 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole, 7 mM52. MTs were prepared from a mixture of unlabeled human tubulin (tyrosinated or detyrosinated) and Hilyte647-labeled bovine tubulin at ratio 10:1. Tubulin at the final concentration 72.5 µM and 1 mM GMPCPP (Jena Bioscience) was incubated at 37 °C for 30 min, microtubules were spun at 16100 g, resuspended in Mg-BRB80 (K-Pipes 80 mM, pH 6.9, 4 mM MgCl2, 1 mM EGTA) and stored at room temperature for a maximum of 3 days.
TIRF microscopy
Imaging was performed on a Nikon Eclipse Ti-E inverted microscope57. Coverslips (22 × 22 mm) were silanized and assembled in custom-made flow chambers. Anti-tubulin antibodies (Serotec, Raleigh, NC) diluted 1:50 in BRB80 were flowed into the chamber and incubated for 10 min at room temperature, then blocked with 1% Pluronic F-127 (Sigma-Aldrich, St. Louis, MO) for 15 min. GMPCPP-stabilized MTs in BRB80 were flowed into the chamber and incubated for 10–15 s to promote binding to the coverslip, then chamber was washed. NDC80-GFP (100 pM) in imaging buffer (Mg-BRB80 supplemented 4 mg/ml bovine serum albumin, 2 mM DTT, 0.1 mg/ml glucose oxidase, 68 μg/ml catalase, 20 mM glucose, 0.5% 2-mercaptoethanol) was perfused continuously at 15 μl/min during data acquisition57. Images of NDC80-GFP were acquired continuously for 120 s with 40 ms exposure.
Analysis of NDC80 diffusion
Kymographs of diffusing NDC80 molecules were made from original stacks (acquired with NIS elements, Nikon) using Metamorph. Individual tracks were selected and molecular displacements for each track were calculated to plot MSD as a function of time57. The diffusion coefficient was determined as one-half of the slope of linear fitting based on 7 bins. Because confidence interval for a fitting is very small, we used the experiment-based standard error represented by the diffusion coefficient of the motionless GFP-tagged molecules imaged under the same experimental conditions (0.0026 μm2 s−1); this rate of diffusion results from thermal motion of the experimental chamber and tracking errors. To determine NDC80 molecular residence time for each type of microtubules, the durations of individual diffusion tracks that lasted at least 120 ms were used to build a cumulative distribution. The characteristic time was determined from a fit with one-phase association function, and duration of a single imaging frame (40 ms) was used to represent the standard error.
Microtubule pelleting assays using purified proteins
Porcine brain tubulin (Cytoskeleton) was tyrosinated or detyrosinated by combining it with either recombinant TTL (1:0.25; kind gift from Michel Steinmetz, Paul Scherrer Institut, Villigen, Switzerland) or carboxypeptidase A (1:0.004 – Sigma-Aldrich), respectively. Tyrosination assay was performed for 1 h at 37 °C78 and detyrosination assay for 5 min at 30 °C79. Microtubule assembly was initiated by adding 1 mM of GTP and 1 mM of DTT to 1.5 mg/ml of either tyrosinated or detyrosinated tubulin in BRB80 buffer (80 mM PIPES, 1 mM EGTA and 1 mM MgCl2), pH 6.9, and incubated for 5 min on ice. The mixture was cleared by ultracentrifugation at 488400 g at 4 °C for 5 min. Taxol was added to the reaction at final concentrations of 0.015, 0.15, and 2 mM successively with 5 min intervals. After 15 min of final incubation, 40% glycerol in BRB80 was added to the mixture and microtubules were pelleted by ultracentrifugation at 385900 g at 35 °C for 10 min and resuspended in BRB80 supplemented with 15 mM taxol. Microtubule concentration was measured after taking a small volume of microtubule solution and resuspended with 10 mM CaCl2 in BRB80 and incubated on ice for 10 min to complete microtubule disassembly. Tubulin concentration was calculated considering the Abs280 measurements and the extinction coefficient of tubulin (115000 M-1cm-1). To perform protein binding to microtubules, NDC80-GFP and GFP-CLASP2 proteins were previously pre-cleared by ultracentrifugation at 280000 g for 10 min at 25 °C. Microtubule-binding assay was carried out by incubating 500 nM of NDC80 brocoli protein and 0.5, 1, 2, 4 and 6 µM of either tyrosinated or detyrosinated microtubules in BRB80 with 15 mM of taxol for 10 min. CLASP2 protein was incubated with 0.1, 0.25, 0.5, 1 and 2 µM of microtubules in the same buffer. Supernatant (unbound fraction) and pellet (bound fraction) were separated by ultracentrifugation at 280000 g at 25 °C for 10 min. For each reaction, fractions were run on SDS-PAGE and gels were stained with BlueSafe (NZYTech) overnight, destained with distilled water and scanned with GS-900 Calibrated Densitometer (BioRad). Densitometer analysis were performed using ImageLab software (BioRad). Microtubule post-translational state was confirmed by western blot. Data analysis was performed on GraphPad Prism 9.
Kinetochore tracking
Time-lapse imaging of Parental, TTL KO and VASH1/2 KO cell lines stably expressing GFP-CENP-A was performed in a heated chamber (37 °C) using a 60 × oil immersion 1.40 NA plan-apochromatic objective mounted on an inverted microscope (Eclipse TE2000U; Nikon) equipped with a CSU-X1 spinning-disk confocal head (Yokogawa Corporation of America) controlled by NIS-Elements software and with two laser lines (488 nm, 561 nm). Images were detected with an iXonEM+ EM-CCD camera (Andor Technology). 10 z-planes separated by 1 µm were collected every 7.5 s for 5 min. The movies were converted with Softworx (GE Healthcare) into DeltaVision microscope format. The images were cropped in Softworx and analyzed blindly using an automated Kinetochore tracking code written in MATLAB 2013b (The Math Works, Natic, USA). The code is available under https://github.com/cmcb-warwick. The analysis output is the frame-to-frame displacement of sister-KTs and their relative distance from the metaphase plate center. It uses an auto-correlation function to quantify the regularity of the sister-kinetochore oscillations along the spindle central axis80.
Statistical analysis
Statistical analysis was performed with Graphpad Prism, version 9. The statistical significance of spindle length, segregation errors, NEB to anaphase duration, NEB to metaphase duration, percentage of MAD1 positive cells, normalized corrected fluorescence intensities in all experimental conditions, NDC80-GFP residence times and diffusion coefficients, was determined using the parametric two-tailed Student’s t-test. Statistical analysis of the percentage of cells with one or more MAD1 positive KTs per cell was determined using Mann–Whitney test. Dunn’s multiple comparison test was used to compare KT oscillation velocities in the different experimental conditions.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analysed during this study are included in this published article (and its supplementary information files). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD047133. Source data are provided with this paper.
Code availability
The kinetochore tracking code used in this published article is available under https://github.com/cmcb-warwick.
References
Janke, C. & Magiera, M. M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. 21, 307–326 (2020).
McKenna, E. D., Sarbanes, S. L., Cummings, S. W. & Roll-Mecak, A. The tubulin code, from molecules to health and disease. Annu Rev. Cell Dev. Biol. 39, 331–361 (2023).
Verhey, K. J. & Gaertig, J. The tubulin code. Cell Cycle 6, 2152–2160 (2007).
Nieuwenhuis, J. et al. Vasohibins encode tubulin detyrosinating activity. Science 358, 1453–1456 (2017).
Aillaud, C. et al. Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science 358, 1448–1453 (2017).
Landskron, L. et al. Posttranslational modification of microtubules by the MATCAP detyrosinase. Science 376, eabn6020 (2022).
Ersfeld, K. et al. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120, 725–732 (1993).
Lopes, D. & Maiato, H. The tubulin code in mitosis and cancer. Cells 9–2356 (2020).
Lopes, D., Seabra, A. L., Orr, B. & Maiato, H. alpha-Tubulin detyrosination links the suppression of MCAK activity with taxol cytotoxicity. J. Cell Biol. 222, e202205092 (2023).
Barisic, M. et al. Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015).
Barisic, M. & Maiato, H. The tubulin code: a navigation system for chromosomes during mitosis. Trends Cell Biol. 26, 766–775 (2016).
Ferreira, L. T. et al. alpha-Tubulin detyrosination impairs mitotic error correction by suppressing MCAK centromeric activity. J. Cell Biol. 219, e201910064 (2020).
Liao, S. et al. Molecular basis of vasohibins-mediated detyrosination and its impact on spindle function and mitosis. Cell Res. 29, 533–547 (2019).
Sirajuddin, M., Rice, L. M. & Vale, R. D. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344 (2014).
Peris, L. et al. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185, 1159–1166 (2009).
Sauer, G. et al. Proteome analysis of the human mitotic spindle. Mol. Cell. Proteom.: MCP 4, 35–43 (2005).
Hotta, T. et al. Parthenolide destabilizes microtubules by covalently modifying tubulin. Curr. Biol. 31, 900–907.e906 (2021).
Szyk, A., Deaconescu, A. M., Piszczek, G. & Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1258 (2011).
Ferreira, L. T., Figueiredo, A. C., Orr, B., Lopes, D. & Maiato, H. Dissecting the role of the tubulin code in mitosis. Methods Cell Biol. 144, 33–74 (2018).
Pereira, A. et al. Coherent-hybrid STED: high contrast sub-diffraction imaging using a bi-vortex depletion beam. Opt. Express 27, 8092–8111 (2019).
Erck, C. et al. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl Acad. Sci. USA 102, 7853–7858 (2005).
Tirnauer, J. S., Canman, J. C., Salmon, E. D. & Mitchison, T. J. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. cell 13, 4308–4316 (2002).
Jaqaman, K. et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. cell Biol. 188, 665–679 (2010).
Kuhn, J. & Dumont, S. Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J. cell Biol. 216, 1533–1542 (2017).
Chen, J. et al. alpha-tubulin tail modifications regulate microtubule stability through selective effector recruitment, not changes in intrinsic polymer dynamics. Dev. cell 56, 2016–2028.e2014 (2021).
Wood, K. W. et al. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl Acad. Sci. USA 107, 5839–5844 (2010).
Barisic, M., Aguiar, P., Geley, S. & Maiato, H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16, 1249–56 (2014).
Sloboda, R. D. Isolation of microtubules and microtubule-associated proteins using Paclitaxel. Cold Spring Harb Protoc. 2015, pdb.prot081190 (2015).
Cheeseman, I. M., Chappie, J. S., Wilson-Kubalek, E. M. & Desai, A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997 (2006).
Kops, G. J. et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169, 49–60 (2005).
Davies, A. E. & Kaplan, K. B. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. J. cell Biol. 189, 261–274 (2010).
Zhang, Y., Hong, X., Hua, S. & Jiang, K. Reconstitution and mechanistic dissection of the human microtubule branching machinery. J. Cell Biol. 221, e202109053 (2022).
Carazo-Salas, R. E. et al. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 (1999).
Belmont, L. D. & Mitchison, T. J. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 84, 623–631 (1996).
Holmfeldt, P. et al. The catastrophe-promoting activity of ectopic Op18/stathmin is required for disruption of mitotic spindles but not interphase microtubules. Mol. Biol. cell 12, 73–83 (2001).
Peris, L. et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174, 839–849 (2006).
Hotta, T. et al. EML2-S constitutes a new class of proteins that recognizes and regulates the dynamics of tyrosinated microtubules. Curr. Biol. 32, 3898–3910.e3814 (2022).
Girao, H. et al. CLASP2 binding to curved microtubule tips promotes flux and stabilizes kinetochore attachments. J. Cell Biol. 219, e201905080 (2020).
Booth, D. G., Hood, F. E., Prior, I. A. & Royle, S. J. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919 (2011).
Auckland, P., Roscioli, E., Coker, H. L. E. & McAinsh, A. D. CENP-F stabilizes kinetochore-microtubule attachments and limits dynein stripping of corona cargoes. J. Cell Biol. 219, e201905018 (2020).
Cheeseman, L. P., Harry, E. F., McAinsh, A. D., Prior, I. A. & Royle, S. J. Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. J. cell Sci. 126, 2102–2113 (2013).
Salisbury, J. L., Suino, K. M., Busby, R. & Springett, M. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287–1292 (2002).
McKenney, R. J., Huynh, W., Vale, R. D. & Sirajuddin, M. Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. EMBO J. 35, 1175–1185 (2016).
Patel, K., Nogales, E. & Heald, R. Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton (Hoboken) 69, 155–165 (2012).
Ciferri, C. et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427–439 (2008).
Maia, A. R. et al. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J. cell Biol. 199, 285–301 (2012).
Maiato, H. et al. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell 113, 891–904 (2003).
Pereira, A. L. et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. cell 17, 4526–4542 (2006).
DeLuca, J. G. et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982 (2006).
McCleland, M. L. et al. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14, 131–137 (2004).
Schmidt, J. C. et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. cell 23, 968–980 (2012).
Chakraborty, M. et al. Microtubule end conversion mediated by motors and diffusing proteins with no intrinsic microtubule end-binding activity. Nat. Commun. 10, 1673 (2019).
Vemu, A., Garnham, C. P., Lee, D. Y. & Roll-Mecak, A. Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol. 540, 149–166 (2014).
Logarinho, E. et al. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat. cell Biol. 14, 295–303 (2012).
DeLuca, K. F., Lens, S. M. & DeLuca, J. G. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. cell Sci. 124, 622–634 (2011).
Zaytsev, A. V., Sundin, L. J., DeLuca, K. F., Grishchuk, E. L. & DeLuca, J. G. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol. 206, 45–59 (2014).
Zaytsev, A. V. et al. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. cell 26, 1829–1844 (2015).
Ferreira, L. T. et al. α-Tubulin detyrosination impairs mitotic error correction by suppressing MCAK centromeric activity. J.Cell Biol. 219, e201910064 (2020).
Gundersen, G. G. & Bulinski, J. C. Microtubule arrays in differentiated cells contain elevated levels of a post-translationally modified form of tubulin. Eur. J. Cell Biol. 42, 288–294 (1986).
Powers, A. F. et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136, 865–875 (2009).
Tanenbaum, M. E., Galjart, N., van Vugt, M. A. & Medema, R. H. CLIP-170 facilitates the formation of kinetochore-microtubule attachments. EMBO J. 25, 45–57 (2006).
Amin, M. A., Kobayashi, K. & Tanaka, K. CLIP-170 tethers kinetochores to microtubule plus ends against poleward force by dynein for stable kinetochore-microtubule attachment. FEBS Lett. 589, 2739–2746 (2015).
Nirschl, J. J., Magiera, M. M., Lazarus, J. E., Janke, C. & Holzbaur, E. L. alpha-tubulin tyrosination and CLIP-170 phosphorylation regulate the initiation of dynein-driven transport in neurons. Cell Rep. 14, 2637–2652 (2016).
Ebberink, E. et al. Tubulin engineering by semi-synthesis reveals that polyglutamylation directs detyrosination. Nat. Chem. 15, 1179–1187 (2023).
Miller, M. P., Asbury, C. L. & Biggins, S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell 165, 1428–1439 (2016).
Long, A. F., Udy, D. B. & Dumont, S. Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr. Biol. 27, 1692–1699.e1693 (2017).
Rosas-Salvans, M., Sutanto, R., Suresh, P. & Dumont, S. The Astrin-SKAP complex reduces friction at the kinetochore-microtubule interface. Curr. Biol. 32, 2621–2631.e2623 (2022).
Luo, W. et al. CLASP2 recognizes tubulins exposed at the microtubule plus-end in a nucleotide state-sensitive manner. Sci. Adv. 9, eabq5404 (2023).
Castrogiovanni, C. et al. Evidence for a HURP/EB free mixed-nucleotide zone in kinetochore-microtubules. Nat. Commun. 13, 4704 (2022).
Akera, T. et al. Spindle asymmetry drives non-mendelian chromosome segregation. Science 358, 668–672 (2017).
Orr, B., Talje, L., Liu, Z., Kwok, B. H. & Compton, D. A. Adaptive resistance to an inhibitor of chromosomal instability in human cancer cells. Cell Rep. 17, 1755–1763 (2016).
Maffini, S. et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 19, 1566–1572 (2009).
Osório, H. et al. Proteomics analysis of gastric cancer patients with diabetes mellitus. J. Clin. Med. 10, 407 (2021).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50, D543–D552 (2022).
Pereira, A. J. & Maiato, H. Improved kymography tools and its applications to mitosis. Methods 51, 214–219 (2010).
Miller, H. P. & Wilson, L. Preparation of microtubule protein and purified tubulin from bovine brain by cycles of assembly and disassembly and phosphocellulose chromatography. Methods Cell Biol. 95, 3–15 (2010).
Hyman, A. et al. Preparation of modified tubulins. Methods Enzymol. 196, 478–485 (1991).
Szyk, A., Deaconescu, A. M., Piszczek, G. & Roll-Mecak, A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind modify tubulin. Nat. Struct. Mol. Biol. 18, 1250–1259 (2011).
Souphron, J. et al. Purification of tubulin with controlled post-translational modifications by polymerization-depolymerization cycles. Nat. Protoc. 14, 1634–1660 (2019).
Olziersky, A. M., Smith, C. A., Burroughs, N., McAinsh, A. D. & Meraldi, P. Mitotic live-cell imaging at different timescales. Methods Cell Biol. 145, 1–27 (2018).
Acknowledgements
We thank Maxim Godzi, Ekaterina Tarasovetc and Fedor Balabin for assistance in single molecule experiments, Aaron Chen for purifying NDC80 Broccoli, Geert Kops for the HEC1 phosphorylation mutants, Michel Steinmetz for providing the TTL expression vector, Sandra Macedo-Ribeiro and Marco Novais-Cruz for help with TTL expression and purification, and members of the Maiato and Grishchuk Labs for discussions and the critical reading of the manuscript. This project was funded by European Research Council (ERC) consolidator grant 681443, under the European Union’s Horizon 2020 research and innovation program (HM), La Caixa Health Research Grant LCF/PR/HR21/52410025 (HM), Fundação para a Ciência e a Tecnologia of Portugal grants PTDC/MED-ONC/3479/2020 (HM), SFRH/BD/141066/2018 (HG), 2021.07945.BD (JMM), SFRH/BD/52044/2012 (RSS) and National Institutes of Health grant R35-GM141747 (ELG).
Author information
Authors and Affiliations
Contributions
Conceptualization: HM. Methodology: AJ, HO, ACF, JMM, ELG. Investigation: HG, JMM, RSS, VD, ED. Visualization: HG, HM, JMM, RSS, ED, ELG. Funding acquisition: HM, ELG. Project administration: HM. Supervision: HM, ELG, PM. Writing—original draft: HG, HM. Writing—review and editing: HG, JMM, HO, PM, ELG, HM
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Minhajuddin Sirajuddin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Girão, H., Macário-Monteiro, J., Figueiredo, A.C. et al. α-tubulin detyrosination fine-tunes kinetochore-microtubule attachments. Nat Commun 15, 9720 (2024). https://doi.org/10.1038/s41467-024-54155-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54155-8