Abstract
F-type phage tail-like bacteriocins (PTLBs) are high-molecular-weight protein complexes exhibiting bactericidal activity and share evolutionary similarities with the tails of non-contractile siphoviruses. In this study, we present the atomic structure of monocin, a genetically engineered F-type PTLB from Listeria monocytogenes. Our detailed atomic-level analysis, excluding two chaperone proteins, provides crucial insights into the molecular architecture of F-type PTLBs. The core structure of monocin resembles TP901-1-like phage tails, featuring three side fibers with receptor-binding domains that connect to the baseplate for host adhesion. Based on these findings, we propose a potential mechanism by which F-type PTLBs induce cell death, offering a foundation for developing targeted antibacterial therapies.
Similar content being viewed by others
Introduction
Bacteriocins are ribosomally synthesized peptides or proteins that bacteria secrete extracellularly to inhibit closely related bacterial strains, often at low concentrations1. These antimicrobial agents can be produced by both Gram-positive and Gram-negative bacteria, exhibiting a diverse range of morphological and biochemical characteristics2. The complexity of these molecules ranges from simple peptides to high-molecular-weight structures3. A particularly interesting subgroup of these high-molecular-weight bacteriocins is the phage tail-like bacteriocins (PTLBs), also known as tailocins, which structurally resemble bacteriophage tails4,5,6.
PTLBs, known for their ability to induce cell death upon interaction with bacterial cells, are a form of microbial altruism where host cells undergo autolysis to protect the population7,8. These bacteriocins are typically named after their bacterial producers, such as diffocin from Clostridium difficile9 and pyocin from Pseudomonas aeruginosa (formerly known as pyocyanea)10. PTLBs are grouped into two types: R-type (Rigid type), characterized by a non-flexible contractile nanotube similar to the contractile tails of myoviruses, and F-type (Flexible type), which consists of flexible, non-contractile rod-like structures akin to siphoviruses11,12.
R-type PTLBs function as molecular machines resembling contractile syringes and are evolutionarily related to myoviral tails13,14 and type VI secretion systems15. The atomic structures of R-type PTLBs from both Gram-negative16 and Gram-positive17 bacteria have been resolved in pre-contraction and post-contraction states, offering insights into their bactericidal mechanisms. These structures comprise a central core tube surrounded by a contractile sheath, with a baseplate structure at one end featuring receptor-binding proteins (RBPs). The bactericidal mechanism involves the RBPs binding to specific cell surface receptors, triggering sheath contraction, which drives the core tube into the bacterial cell membrane. The core tube is tipped with a needle-like spike that penetrates the membrane, creating a channel through which protons and small ions can flow, leading to membrane disruption. This mechanism mirrors the DNA injection process seen in myoviruses.
F-type PTLBs, in contrast, possess a simpler structure consisting of a flexible, non-contractile tube connected to a baseplate where RBPs are attached. Unlike R-type PTLBs, F-type PTLBs do not have a core tube that pierces the cell membrane. However, studies suggest that the tape measure protein in siphoviruses may embed in the membrane to facilitate DNA translocation18,19,20,21. It is plausible that similar mechanisms exist in F-type PTLBs, where tape measure proteins could play a role in forming pores, though not necessarily in DNA transport. The F-type pyocin produced by Pseudomonas aeruginosa is one of the most studied F-type PTLBs and shares structural similarities with λ-like phage tails12.
Monocins, identified in Listeria during the 1960s, have been historically used for bacterial typing and occasionally in conjunction with phage typing22,23. The monocin gene cluster, referred to as ftb (F-type bacteriocin), consists of 18 genes, including those encoding structural components, regulatory regions, and a lysis cassette known as holin-lysin (Fig. 1a)24,25. Sequence homology and gene arrangement analyses suggest that monocins share a close evolutionary relationship with the tail structures of TP901-1-like siphovirus, with the ftpP gene encoding the receptor-binding protein26.
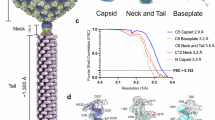
a The gene clusters responsible for monocin production exhibit similarities with those found in other phage tail-like bacteriocins. These clusters include a regulatory region, structural component genes, and a lysis cassette. The expression construct spans the ftbD to ftbP genes. Proteins encoded by the genes indicated by dashed lines within the structural component region were unresolved in the cryoEM reconstructions and are likely chaperones. b Side-view 2D class averages of the complete monocin reveal distinct regions, including the cap, tube, tip, and side fibers. The 22 hexameric rings of tail tube proteins are sequentially numbered from the distal to proximal ends. c Cross-sectional side view of the monocin surface, showing an inner lumen diameter of 2.4 nm. d Cartoon representation of monocin with each component color-coded as indicated in Fig. 1a. e Top-down (upper) and side (lower) views of the tip and side fiber complex. Three TMP-FtbJ and three TAP-FtbM are located within the DTP-FtbK hexamer. Three side fibers are connected to the tip via SFP-FtbN. f Tip components are color-coded in the side-view surface. g The three side fibers are color-coded in the side-view surface. Each side fiber consists of an SFP-FtbN trimer, an SFP-FtbO trimer, and three RBP-FtbP trimers. The attachment points of the side fibers to the tip complex are indicated by red circles. h Three-view zoom-in of one side fiber. The SFP-FtbN trimer (shown in slate blue) connects the fiber to the tip. SFP-FtbN features a “fishing rod” composed of 9.6 nm and 3.3 nm segments, with a “treble hook” outlined in a red dashed box. The treble hook connects to three RBP-FtbP trimers (green) and one SFP-FtbO trimer (saddle brown). The bottom of each RBP-FtbP trimer contains a trimeric RBD responsible for receptor binding.
In this study, we present the atomic structure of monocin, determined via cryogenic electron microscopy (cryo-EM), as a representative model of F-type bacteriocins. Through comparisons with other non-contractile nanomachines of the siphoviral family, we propose a penetration mechanism specific to F-type bacteriocins. This study aims to expand our understanding of the structural and functional diversity of PTLBs and their potential applications in antibacterial therapy.
Results
Overall structures of monocin
To explore the structural organization of monocin, we cloned 11 structural genes, ranging from ftbF to ftbP, along with the regulatory genes ftbD and ftbE, into Lactococcus lactis for expression24 (Fig. 1a). The monocin sample was purified for cryo-EM imaging via density gradient centrifugation. However, due to the intrinsic flexibility of monocin and the dynamic nature of siphoviral tails, only a 2D average representation of the entire monocin structure could be obtained (Fig. 1b).
To gain more detailed structural insights, we separately conducted cryo-EM analyses on three distinct regions: the cap, the tip, and the side fibers (Figs. S1, S2). These analyses yielded resolutions of 3.42 Å for the cap, 2.88 Å for the tip, and 3.23 Å for the side fibers (Supplementary Table S1). By mapping visible amino acid side chains in these cryo-EM structures, we identified FtbF as the cap protein, FtbG as the tube protein, FtbK and FtbL as components of the baseplate hub, and FtbN, FtbO, and FtbP as constituents of the side fibers (Figs. 1c, d, S3a). Within the tip lumen (Figs. 1c, S3b), the C-terminal segment of the tape measure protein (TMP), FtbJ (residues 573-622), and a protein of unknown function, FtbM (residues 70-99), were identified. The proximal end of the monocin structure is capped by a hexameric ring formed by the terminator protein (TrP) FtbF, while the flexible tube comprises 22 stacked hexameric rings of the tail tube protein (TTP) FtbG. Below the initial tube ring, the distal tail protein (DTP) FtbK forms another hexameric ring, with trimers of the baseplate hub protein (BHP) FtbL sealing the tube; their C-terminal spike domains extend from the baseplate. The overall structure of monocin measures approximately 103 nm in length, with a tube width of 9.1 nm and a lumen width of 2.4 nm.
Three side fibers are attached to the hexameric ring formed by FtbK at meta positions (Fig. 1e, f, g). The N-terminal region of the FtbN trimer resembles a fishing rod, while its C-terminal region adopts a treble hook shape (Fig. 1h). Three FtbP trimers, each featuring a receptor-binding domain (RBD) at the distal end, connect to this treble hook. Centrally, a trimer of FtbO forms a long rod bent at an angle of 138.6 degrees. Although the proteins FtbH and FtbI were not visible in the final structure, their gene location within the locus and comparisons with other PTLBs or phage tails suggest they may act as chaperones in tube assembly (Fig. S4).
Molecular organization of the monocin central core
The TTP fold is a conserved structural motif present in bacteriophage tail tubes and associated proteins27. Each TTP subunit is characterized by a β-sandwich structure flanked by an α-helix (Fig. S5a–e). The inner β-sheet comprises four β-strands, while the outer β-sheet consists of four shorter β-strands. Six TTP subunits assemble into a hexameric ring, forming a substantial β-barrel that lines the tube’s interior (Fig. S5f).
All proteins constituting the central core of monocin adopt this TTP fold (Fig. 2a). The terminator protein TrP-FtbF exhibits the TTP fold but lacks the outer β-sheets (Fig. S5a). In TrP-FtbF, the β-strand β6.1 connects with β6.2, forming part of the inner β-sheet, and an Ω-shaped loop between β6.1 and β6.2 protrudes into the hollow interior, narrowing the diameter of the monocin cap to 3 nm (Fig. S5f).
a The components of the central core of monocin, including TrP-FtbF, TTP-FtbG, DTP-FtbK, and BHP-FtbL, are depicted alongside their domain architectures. b Cartoon and surface representations of one asymmetric unit of the monocin tip in a side view. DTP-FtbK contains the TTP, rod holder, and galectin domains. DTP-FtbK, with or without (indicated by “*”) the active rod holder, is colored in wheat and salmon, respectively. The TTP-FtbG units in each layer are staggered by 35 degrees relative to one another. c Side-view surface representation of the monocin tip, showing DTP-FtbK positioned above HDI and DTP-FtbK* situated above HDIV. The hub, baseplate, and spike domains are illustrated with hydrophobic surfaces. d Structural alignment between the K (wheat) and K* (salmon) configurations of DTP-FtbK. The primary structural difference, highlighted in the red dashed box, is in the “rod holder” domain, which is critical for side fiber attachment. e Assembly mode of TMP-FtbJ and TAP-FtbM, with labeled C-terminal residues. f Channel radii along the potential transport path, calculated using HOLE software47. The white line represents the channel radii calculated without the spike domain.
TTP-FtbG also adopts the TTP fold (Fig. S5b) with an elongated N-terminal loop as observed in the helical reconstruction (PDB ID: 8QHS.). The hexameric ring of TTP-FtbG is 4 nm thick, and these rings stack helically with each upper layer rotating 35° clockwise relative to the layer below (Fig. 2b). Despite the classic interaction between the β3–β4 loop and a portion of the N-terminus observed in other siphoviral tails, TTP-FtbG also features a disulfide bond between N-arm Cys9 and Cys104 in the β5 strand of the adjacent upper layer. Although the Cys mutant strain is still released, it shows defective activity28, indicating that the disulfide bond plays a critical role in monocin function (Fig. S5g). Twenty-two hexameric units of TTP-FtbG stack from the tip of monocin up to TrP-FtbF, resulting in a negatively charged lumen (Fig. S5h). The N-terminal loop of the final TTP-FtbG unit remains unpaired due to the absence of a cysteine partner on TrP-FtbF.
The distal tail protein (DTP) FtbK consists of two domains: an N-terminal domain (NTD) with a TTP fold and a C-terminal domain (CTD) with a galectin-like β-sandwich structure (Fig. 2a). The loop connecting these domains serves as the attachment site for side fibers. Six units of DTP-FtbK form a hexameric ring through their NTDs (Fig. 2b, c), extending the tube structure. The six CTDs radiate outward from this ring. Within this hexameric ring, two distinct unit types, K and K*, alternate positions for side fiber attachment (Fig. 2d). The primary structural difference between K and K* lies in the “rod holder” domain, which is critical for side fiber attachment (Fig. 2d). Above the BHP-FtbL protein, the CTD galectin domain of K stabilizes the rod holder domain alongside the NTD TTP domain, facilitating side fiber attachment.
The baseplate hub protein (BHP) FtbL forms a trimer positioned beneath the DTP-FtbK ring (Fig. 2b, c). This trimer contains a structural scaffold known as the ‘Hub Domain’ (HD), characteristic of many sipho- and myoviral BHPs (Fig. 2b, c). The HDI (Fig. S5d) and HDIV (Fig. S4e) regions both exhibit a TTP fold, contributing to a pseudo-C6 symmetry within the baseplate hub (Fig. 2b, c). HDII and HDIII form the distal end of the BHP, with a spike insertion at the C-terminus effectively sealing the tube (Fig. 2c). The spike subunits consist of an α-helical coiled coil, forming a rod-like structure extending from the baseplate hub. Additionally, the galectin domains from all six DTP subunits create a “crown” over the baseplate trimer (Fig. 2c), and the interaction between the DTP and the baseplate is primarily hydrophobic. The galectin domain of the K DTP-FtbK is located above HDI, while the galectin domain of the K* subunit is positioned above HDIV.
The C-terminal α-helices of the three TMP-FtbJ subunits (residues 593–622) form a tripod structure intertwined with the N-terminal α-helices of the spike trimer (Fig. 2e). The distance between the C-terminal ends of TMP-FtbJ measures approximately 3.25 nm, which is larger than the channel diameter formed by the HDIII-HDII complex (Fig. 2f). This suggests that while the spike domain of the HD may extend beyond the baseplate to penetrate the membrane, the channel itself may not be wide enough to allow the full exit of the TMP-FtbJ C-termini. Therefore, it is likely that the HDIII-HDII complex must undergo expansion to accommodate this process, similar to the mechanism observed in bacteriophage tail infections. Furthermore, the C-terminal α-helices of three TAP-FtbM subunits (residues 70-99) form helix bundles with the α-helices of TMP-FtbJs (residues 573-588), resulting in a pseudo-C3 symmetric structure (Fig. 2e). Additional density corresponding to TMP-FtbJ and TAP-FtbM was observed within the tube lumen, though it was not interpretable at atomic resolution (Fig. S2b). In summary, TMP-FtbJs assemble into a trimer with a coiled-coil structure, with their C-termini anchored to the spike, while the coiled-coil TMP-FtbJs above the baseplate are decorated by the TAP-FtbM helix bundles.
Assembly and interaction of side fibers
The six N-terminal domains (NTDs) of the distal tail protein (DTP) FtbK form a central core, while the six C-terminal domains (CTDs) radiate outward. Among these proteins, three (designated as K) adopt conformations suitable for side fiber attachment (Fig. 3a). These side fiber trimers encircle the baseplate core, forming the primary visible components of the peripheral structures (Fig. S1). The side fiber proteins, SFP-FtbN, assemble into a trimer composed of two domains: an N-terminal α-helical coiled-coil “rod” domain that bends 90° at its central hinge and a “treble hook” domain that serves as the anchoring platform for receptor-binding proteins (RBPs) (Fig. 3b, c, d). The “rod holder” is situated between the TTP and galectin domains of DTP, projecting a loop into the gap at the top of the rod domains to secure them in place. This interface exhibits remarkable shape complementarity and electrostatic potential (Fig. S6a). Additionally, several aliphatic and aromatic residues (F144, M151, and S156) from the rod holder fit into the central cup of the N-terminal rod handle. A comparable loop mechanism is also identified in bacteriophage 80α for attaching RBPs to DTP29 (Fig. S6b). The rod helices are positioned at approximately a 60° angle relative to the plane of the DTP hexameric ring, terminating in a kink (Fig. 1h). The residues of SFP-FtbN following the rod domain form a threefold symmetric trimer resembling a “treble hook,” stabilized by a three-helix bundle (shank). Each of the three peripheral hooks on the SFP-FtbN platform (Fig. 3c) connects to individual RBP-FtbP tripods (Fig. 1e, f).
a The lower diagram illustrates how three side fibers form a triangular structure surrounding the central tip. Each side fiber consists of one SFP-FtbN trimer, one SFP-FtbO trimer, and three RBP-FtbP trimers. b Side and top views of the SFP-FtbN trimer (violet) with attached DTP-FtbK (wheat). The three SFP-FtbN subunits are colored distinctly. c Zoom-in view of the SFP-FtbN platform. Each of the three lateral hooks of SFP-FtbN can independently connect to an RBP-FtbP tripod, while the SFP-FtbO trimer attaches to the central junction of the treble hook. d Zoom-in top view revealing the interactions between the rod holder of DTP-FtbK (wheat) and the coiled-coil stem region of SFP-FtbN (violet), with side chains shown in ball-and-stick form. e Structural organization of RBP-FtbP. The three chains of RBP-FtbP are shown in different shades of green. Each RBP-FtbP trimer consists of a P-latch domain, a P-stalk domain, and a RBD domain. f Side and top views illustrating the interaction between the RBP-FtbP-latch (green) and the SFP-FtbN-hook (violet).
The receptor-binding proteins (RBPs), RBP-FtbP, form a trimeric structure comprising three domains (Fig. 3e): (i) an N-terminal P-latch domain composed of three β-hairpins; (ii) a central triple P-stalk domain formed by bundles of three helices; and (iii) a C-terminal receptor-binding domain (RBD) composed of three β-barrels that establish three receptor-binding sites at their interface (Fig. S7). Each of the three RBPs in the tripod associates with the hooks of the treble hook domain by complementing the β-sheets (Fig. 3f). The RBD of each FtbP monomer adopts a β-sandwich structure with the topology shown in Fig. S7b, featuring two sheets of four β-strands each, with β3 and β4 on the outside and β6 and β7 on the inside of the trimer. A DALI search30 correlates the RBD to the C-terminal domain of bacteriophage K gp144 (Z-score: 7.3, PDB ID: 5M9F), as well as to the C-terminal domain of bacteriophage T7 fiber gp17 (Z-score: 5.3, PDB ID: 4A0U)31.
SFP-FtbO consists of three domains: (i) an N-terminal O-latch domain composed of three β-hairpins resembling the P-latch domain (Figs. 4a, S8a); (ii) an O-stalk 1 domain; and (iii) an O-stalk 2 domain. The O-stalk 1 and O-stalk 2 segments both coordinate ferric ions at their tips through pairs of histidine residues (His67, His69, and His122, His124) sourced from the β-hairpins (Fig. 4a). The SFP-FtbO trimer attaches to the center junction of the treble hook (Fig. 4b), with the O-latch connecting to the treble hook via the shank tip. The three RBPs are designated P1, P2, and P3 (Fig. 4c, d). The distances between each pair of RBPs measure at least 7 Å. The SFP-FtbO trimer interacts with the RBDs of both P1 and P2, providing greater stability compared to P3 (Fig. 4d, e).
a Structural organization of SFP-FtbO. Each SFP-FtbO trimer consists of an O-latch domain, an O-stalk1 domain, and an O-stalk2 domain. The O-stalk1 and O-stalk2 segments each coordinate ferric ions at their tips through pairs of histidine residues (His67, His69, and His122, His124) derived from the β-hairpins. b Side and top views illustrating the interaction between the SFP-FtbO-latch (brown) and the tip of the SFP-FtbN shank (violet). c Schematic showing the spatial arrangement of the side fibers. Each side fiber attaches to the tip (gray) via three SFP-FtbN trimers (slate blue). Each SFP-FtbN trimer has three hooks that connect to three distinct RBP-FtbP trimers. An SFP-FtbO trimer is positioned at the center of each SFP-FtbN trimer within the side fiber. The three RBP-FtbP trimers in each side fiber are asymmetric (P1, P2, P3). d Side view of a side fiber, with each RBP-FtbP trimer colored differently. The bent O-stalk2 domain of SFP-FtbO (brown) bridges the RBDs of the P1 and P2 RBP-FtbP trimers. e Top-down view showing the interaction between the RBD of RBP-FtbP and the hub domain of the baseplate. The P1 RBD exhibits a strong interaction with the FtbL-HDII domain.
However, the alignment of O-stalk 1 within SFP-FtbO is not vertical relative to the equilateral triangle formed by the three RBDs. Instead, the C-terminal region of O-stalk 1 tilts toward the P1 RBD, subsequently establishing contact with the P1-O monomer of the P1 RBD (Fig. 4d, e). The P1-L monomer from the P1 RBD interacts with the HDII domain of the baseplate central core, specifically through loop L56 (Fig. 4e). This dual interaction helps stabilize P1’s positioning, which exhibits the highest density quality in the cryo-EM map. O-stalk 2 then sharply turns to engage with the P2 RBD via its P2-O monomer (Fig. 4d, e), with P2 showing the next highest density quality. In contrast, P3 does not establish any contacts and appears highly mobile, as indicated by weaker density in the cryo-EM reconstruction.
A series of truncations were performed on SFP-FtbO, and negative staining analyses revealed that none of these truncations affected particle assembly (Figs. S10a, b). However, the resulting particles exhibited no antibacterial activity, underscoring the critical role of SFP-FtbO in the functional integrity of the monocin tail (Fig. S10c).
Discussion
This study presents the comprehensive structural analysis of an F-type phage tail-like bacteriocin (PTLB), focusing on monocin. F-type PTLBs, recognized for their structural simplicity compared to R-type PTLBs, have emerged as promising tools for targeting pathogenic bacteria32.
Despite significant divergence in the primary sequences of structural tail proteins between siphoviruses and F-type PTLBs, both groups exhibit a remarkably conserved gene locus organization, particularly regarding the positioning of genes encoding tail structural proteins. This conservation implies a shared evolutionary origin33,34, further supported by the accurate correlation between predicted tail assembly chaperones in F-type PTLBs and their respective gene locations.
The core structure of monocin is composed of multiple proteins that share a common tail tube protein fold35. Key components include TrP-FtbF, functioning as the tail terminator protein; TTP-FtbG, serving as the tail tube protein; the N-terminal domain of DTP-FtbK as the distal tail protein; and the HDI and HDIV domains of BHP-FtbL, which act as the baseplate hub protein. The TTP subunits form a hexameric or pseudo-hexameric ring, creating a substantial β-barrel lined with an inner β-sheet along the tube’s interior. Similar tail tube protein folds are present in other bacteriophage tail proteins35. However, monocin’s tail exhibits increased rigidity, attributed to a disulfide bond between Cys9 in TTP-FtbG and Cys104 in the upper layer outer β-sheet, distinguishing it from other bacteriophage tails, such as that of λ bacteriophage21,36.
The attachment of side fibers to the central core involves interactions with the DTP protein or related tube proteins. In Gram-positive siphoviruses, a galectin domain at the C-terminus of the TTP domain stabilizes a loop that anchors side fibers (Fig. S4). This mechanism is observed in monocin and the receptor-binding proteins (RBPs) of bacteriophage 80α (Fig. S6b). Some side fiber proteins feature an Ig-like domain at their N-terminus, contributing to a dodecameric ring around the DTP or tube protein hexameric core, anchoring side fibers in bacteriophages such as FibL (Fig. S9a), FibU, (Fig. S9b) in bacteriophage 80α and TP901-1 (Fig. S9c). In contrast, Gram-negative infecting phages like bacteriophage lambda lack the galectin domain, favoring dodecamerization through Ig-like domains, as seen in bacteriophage T5 (Fig. S9d)18.
While the precise nature of this receptor remains unclear, previous studies involving functional experiments with domain swaps in the receptor-binding domain (RBD) of Listeria phage A118 suggest that RBP-FtbPs are key to this interaction24. Identifying the specific receptor and elucidating how it is recognized by RBP-FtbPs will be crucial for understanding monocin targeting, and future studies should aim to characterize this receptor through functional assays or binding studies. Another open question is how the receptor engagement induces structural changes of the monocin tip to form a functional pore across the bacterial membrane once the monocin penetrates the cell wall. High-resolution structure of monocin in its open state, combined with functional assays that identify the membrane receptor, will be essential to validate this model in the future.
In summary, our study of monocin, an F-type PTLB, provides valuable insights into its structure, offering a promising foundation for future applications. These findings not only advance our understanding of PTLBs but also open avenues for the development of antimicrobial agents aimed at combating pathogenic bacteria, potentially reducing the incidence of related diseases. However, several critical questions remain. Answering these questions will provide a deeper understanding of the bactericidal mechanism of monocin and may have broader implications for phage tail-like bacteriocins in general.
Methods
Plasmid construction, expression and purification
The monocin gene cluster, encompassing 13 essential genes from ftbD to ftbP24 whose CDSs were identical to Listeria monocytogenes ATCC 35152, was amplified from L. monocytogenes (provided by the Jiang Lab) using colony PCR. The amplified gene cluster was then inserted into the PCR-amplified pNZ8148 vector. The resulting plasmid was introduced into competent Lactococcus lactis NZ9000 cells via electroporation.
Lactococcus lactis was cultured in M17 medium supplemented with 0.05% glucose and 10 µg/mL chloramphenicol at 30 °C with shaking at 100 rpm until an optical density of 0.5 was achieved. Monocin expression was induced by adding 200 ng/mL nisin and incubating at 18 °C for over 20 h. The induced culture was harvested by centrifugation at 4600 × g for 20 min, and the pellet was resuspended in 8 mL TN50 buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl) per liter of the original culture. This reconstituted pellet was subjected to digestion with 2 mg/mL lysozyme at 37 °C for 30 min, followed by sonication for 40 s per liter of the original culture. After centrifugation at 22,000 × g for 30 min, 10% (v/v) 5 M NaCl and 10% (w/v) PEG8000 were dissolved in the supernatant, which was then incubated on ice for 2.5 h. PEG8000 precipitation was achieved by centrifugation at 20,000 × g for 15 min, and the pellet was gently resuspended in TN50 buffer (1.5 mL per liter of the original culture). The suspension was loaded onto a 10%–40% (v/v) glycerol density gradient and centrifuged at 109,585 × g for 16 h. The gradient was divided into four equal fractions. The second upper fraction was subjected to further centrifugation at 145,161 × g for 3.5 h, and the resulting pellet was diluted in 25 mM Tris-HCl (pH 7.5). Negative ion-exchange chromatography was performed twice to remove PEG8000, and monocin-containing fractions were verified by negative staining.
Preparation of cryo-EM samples and data acquisition
A 4 μL aliquot of the concentrated monocin solution was applied to glow-discharged grids (Quantifoil Au R1.2/1.3, 300 mesh). The grids were then gently blotted for 3.5 s at 8 °C and 100% humidity before being rapidly frozen in liquid nitrogen-cooled ethane using a Vitrobot Mark IV (Thermo Fisher). The quality of the prepared grids was assessed with a Tecnai Arctica electron microscope operating at 200 kV, equipped with a K2 camera (Gatan). Approved grids were then transferred to a Titan Krios electron microscope operating at 300 kV, fitted with a Gatan Gif Quantum energy filter. A total of 2699 movies were recorded using a K3 Summit counting camera (Gatan) controlled by AutoEMation37. During data acquisition, motion correction was performed automatically with MotionCorr238, utilizing a calibrated pixel size of 0.5371 Å and applying a 2-fold binning process. This resulted in a final pixel size of 1.0742 Å for the captured images.
Image processing and 3D reconstruction
Image processing and 3D reconstruction were performed using cryoSPARC v4.1.2 software39. The contrast transfer function (CTF) was recalculated using the Patch-CTF estimation method. Micrographs of insufficient quality or containing a limited number of monocin particles were manually excluded, resulting in a total of 2351 micrographs being retained.
332,840 particles were initially picked automatically using a blob picker tool, with further manual filtering to remove featureless particles. Given the distinctive morphological features of the monocin tip, subsequent 2D classification identified 44,283 particles with clear characteristics of the monocin tip. For the reconstruction of the monocin tip, particles were extracted with a box size of 512 pixels. An initial model was generated through ab-initio reconstruction. Multiple rounds of refinement were conducted to tentatively determine the particle orientations. To improve resolution, particle orientations were expanded around the C3 symmetry axis, followed by additional local refinement to produce the final map of the monocin tip.
The side fiber region of the tip map exhibited poor quality due to inherent flexibility and heterogeneity, exacerbated by physical damage during purification, particularly affecting distal receptor-binding proteins (RBPs) and FtbO. To address this, numerous refinement cycles were performed using a mask focused on one of the three side fiber regions. For enhanced quality of the particularly flexible FtbO fiber, 132,849 symmetry-expanded particles were repositioned at the center of the FtbO fiber and re-extracted with a reduced box size of 256 pixels. Multiple rounds of 3D classification, constrained to the FtbO fiber and adjacent RBP, yielded 42,169 particles with relatively consistent fiber conformation, which were then subjected to local refinement.
To reconstruct the cap region of the monocin, the tip particles were iteratively re-centered upwards, gradually approaching the cap region. After several rounds of 2D classification, 13,473 particles centered on the cap were obtained. Final refinement iterations were based on an initial model reconstructed using orientation parameters derived from the tip.
Protein model building and structure refinement
Protein models were built de novo using EMBuilder40. Subsequent manual adjustments to the models were performed using UCSF Chimera41 or COOT42. The refinement of the protein structures was conducted using either PHENIX43 or Refmac544. Statistical details regarding the 3D reconstruction and model refinement processes are provided in Supplementary Table S1. Visual representations of the structures were generated using PyMol45 or UCSF ChimeraX46.
Mutation and Bacteriocin killing assays
The mutants were constructed based on the pNZ8148-monocinWT plasmid. Fragments taking mutations which resulted in FtbO truncation or deletion were PCR-amplified from the pNZ8148-monocinWT plasmid, ligated through homologous recombination and then transformed into NZ9000 competent cells. Mutations were verified by colony PCR and Sanger sequencing.
Wild type and mutated monocins’ activity was tested with dish diffusion bioassay. Monocin expression was described above and the cell lysis supernatant filtration (by 0.22 μm filter) before PEG8000 precipitation was used for the killing assays. The lysis was further concentrated by PEG8000 precipitation and density gradient centrifugation and then checked by negative-stain EM to confirm monocin expression and assembly. 4b serotype Listeria monocytogenes strain CGMCC 1.10753 (from China General Microbiological Culture Collection Center) was cultured in BHI medium for 24 h at 37 °C, 220 rpm. The saturated culture was then diluted with 1:30 ratio in warm BHI medium with 0.7% arga (w/v) and then overlaid on 10 cm dish. Wells were made on the solid bacteria-containing medium and filled with 80 uL monocin solutions. The dish was then incubated static at 30 °C for 12 h and examined for growth inhibition zones.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-61073 (Cap); EMD-61074 (Tip); and EMD-61075 (Side fiber). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes 9J1J (Cap); 9J1K (Tip); and 9J1L (Side fiber).
References
Sugrue, I., Ross, R. P. & Hill, C. Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-024-01045-x (2024).
Ongpipattanakul, C. et al. Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides. Chem Rev. https://doi.org/10.1021/acs.chemrev.2c00210 (2022).
Cotter, P. D., Ross, R. P. & Hill, C. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol. 11, 95 (2013).
Scholl, D. Phage Tail-Like Bacteriocins. Annu Rev. Virol. 4, 453–467 (2017).
Patz, S. et al. Phage tail-like particles are versatile bacterial nanomachines - A mini-review. J. Adv. Res 19, 75–84 (2019).
Ghequire, M. G. K. & De Mot, R. The Tailocin Tale: Peeling off Phage Tails. Trends Microbiol. 23, 587–590 (2015).
Uratani, Y. & Hoshino, T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J. Bacteriol. 157, 632–636 (1984).
Bhattacharjee, R. et al. Phage-tail-like bacteriocins as a biomedical platform to counter anti-microbial resistant pathogens. Biomed. Pharmacother. 155, 113720 (2022).
Gebhart, D. et al. Novel high-molecular-weight, R-type bacteriocins of Clostridium difficile. J. Bacteriol. 194, 6240–6247 (2012).
Michel-Briand, Y. & Baysse, C. The pyocins of Pseudomonas aeruginosa. Biochimie 84, 499–510 (2002).
Nakayama, K. et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol 38, 213–231 (2000).
Saha, S. et al. F-Type Pyocins Are Diverse Noncontractile Phage Tail-Like Weapons for Killing Pseudomonas aeruginosa. J. Bacteriol. 205, e0002923 (2023).
Kanamaru, S. et al. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 (2002).
Leiman, P. G. & Shneider, M. M. Contractile tail machines of bacteriophages. Adv. Exp. Med Biol. 726, 93–114 (2012).
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J. & Mekalanos, J. J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012).
Ge, P. et al. Action of a minimal contractile bactericidal nanomachine. Nature 580, 658–662 (2020).
Cai, X. et al. Atomic structures of a bacteriocin targeting Gram-positive bacteria. Nat. Commun. 15, 7057 (2024).
Linares, R. et al. Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation. Sci. Adv. 9, eade9674 (2023).
Ge, X. & Wang, J. Structural mechanism of bacteriophage lambda tail’s interaction with the bacterial receptor. Nat. Commun. 15, 4185 (2024).
Degroux, S., Effantin, G., Linares, R., Schoehn, G. & Breyton, C. Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger. J. Virol. e0158422. https://doi.org/10.1128/jvi.01584-22. (2023).
Wang, C. et al. Architecture of the bacteriophage lambda tail. Structure 32, 35–46.e33 (2024).
Lee, S. Bacteriocins of Listeria monocytogenes and Their Potential as a Virulence Factor. Toxins (Basel) 12. https://doi.org/10.3390/toxins12020103 (2020).
Bannerman, E., Boerlin, P. & Bille, J. Typing of Listeria monocytogenes by monocin and phage receptors. Int J. Food Microbiol 31, 245–262 (1996).
Lee, G. et al. F-Type Bacteriocins of Listeria monocytogenes: a New Class of Phage Tail-Like Structures Reveals Broad Parallel Coevolution between Tailed Bacteriophages and High-Molecular-Weight Bacteriocins. J. Bacteriol. 198, 2784–2793 (2016).
Zink, R., Loessner, M. J. & Scherer, S. Characterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiol. (Read.) 141, 2577–2584 (1995).
Veesler, D. et al. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc. Natl Acad. Sci. USA 109, 8954–8958 (2012).
Veesler, D. & Cambillau, C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol. Biol. Rev. 75, 423–433 (2011).
Sigal, N. et al. Specialized Listeria monocytogenes produce tailocins to provide a population-level competitive growth advantage. Nat. Microbiol 9, 2727–2737 (2024).
Kizziah, J. L., Manning, K. A., Dearborn, A. D. & Dokland, T. Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. PLoS Pathog. 16, e1008314 (2020).
Holm, L. DALI and the persistence of protein shape. Protein Sci. 29, 128–140 (2020).
Garcia-Doval, C. & van Raaij, M. J. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl Acad. Sci. USA 109, 9390–9395 (2012).
Woudstra, C., Sørensen, A. N., Sørensen, M. C. H. & Brøndsted, L. Strategies for developing phages into novel antimicrobial tailocins. Trends Microbiol. https://doi.org/10.1016/j.tim.2024.03.003 (2024).
Linares, R., Arnaud, C. A., Degroux, S., Schoehn, G. & Breyton, C. Structure, function and assembly of the long, flexible tail of siphophages. Curr. Opin. Virol. 45, 34–42 (2020).
Davidson, A. R., Cardarelli, L., Pell, L. G., Radford, D. R. & Maxwell, K. L. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med Biol. 726, 115–142 (2012).
Zinke, M., Schröder, G. F. & Lange, A. Major tail proteins of bacteriophages of the order Caudovirales. J. Biol. Chem. 298, 101472 (2022).
Xiao, H. et al. Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly. PLoS Biol. 21, e3002441 (2023).
Lei, J. & Frank, J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 69–80 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zhou, N., Wang, H. & Wang, J. EMBuilder: A Template Matching-based Automatic Model-building Program for High-resolution Cryo-Electron Microscopy Maps. Sci. Rep. 7, 2664 (2017).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol. 74, 531–544 (2018).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D. Biol. Crystallogr. 71, 136–153 (2015).
DeLano, W. L. The PyMOL Molecular Graphics System. on World Wide Web http://www.pymol.org (2002).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph 14, 354–360 (1996).
Acknowledgements
We extend our gratitude to Professor Peng Jiang from Tsinghua University for providing the Listeria monocytogenes strain. We are also thankful to the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for their generous assistance with cryo-EM facility support and computational resources on the Bio-Computing Platform cluster. Additionally, we appreciate the valuable technical support from J. Lei, X. Li, and F. Yang. This research was funded by grants from the National Natural Science Foundation of China, specifically under grant numbers 32371254 and 32171190.
Author information
Authors and Affiliations
Contributions
Z.G. and J.W. conceived the project. Z.G. optimized the preparation of cryo-grids, recorded the cryo-EM data, and processed this data. J.W. built the atomic models. X.G. and J.W. wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, Z., Ge, X. & Wang, J. Structure of an F-type phage tail-like bacteriocin from Listeria monocytogenes. Nat Commun 16, 1695 (2025). https://doi.org/10.1038/s41467-025-57075-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57075-3