Abstract
Microbial carbon use efficiency (CUE) is a key microbial trait affecting soil organic carbon (SOC) dynamics. However, we lack a unified and predictive understanding of the mechanisms underpinning the temperature response of microbial CUE, and, thus, its impacts on SOC storage in a warming world. Here, we leverage three independent soil datasets (n = 618 for microbial CUE; n = 591 and 660 for heterotrophic respiration) at broad spatial scales to investigate the microbial thermal response and its implications for SOC responses to warming. We show a nonlinear increase and decrease of CUE and heterotrophic respiration, respectively, in response to mean annual temperature (MAT), with a thermal threshold at ≈15 °C. These nonlinear relationships are mainly associated with changes in the fungal-to-bacterial biomass ratio. Our microbial-explicit SOC model predicts significant SOC losses at MAT above ≈15 °C due to increased CUE, total microbial biomass, and heterotrophic respiration, implying a potential abrupt transition to more vulnerable SOC under climate warming.
Similar content being viewed by others
Introduction
Microbial C use efficiency (CUE) – the ratio of organic C allocated to growth versus organic C uptake1 – has emerged as a critical community-level functional trait for predicting soil organic carbon (SOC) responses to climate warming in global biogeochemical models2,3,4. Yet, we lack a unified quantitative understanding of the mechanisms underpinning the temperature response of microbial CUE and its linkage with SOC responses to warming at broad spatial scales. So far, although some empirical studies show that biomass-specific heterotrophic respiration (i.e., respiration per unit of soil microbial biomass) is lower in soils from warmer climates, supporting the thermal adaptation theory5,6,7, other studies do not confirm this pattern8,9. From a conceptual perspective, the depletion of more labile compounds, thermal adaptation of microbial physiology (i.e., via enzyme and membrane structures), and changes in microbial community composition have been hypothesized as main drivers of reduced soil respiration in warmer climates5,6,10. If thermal adaptation reduces the biomass-specific microbial respiration rate, all else being equal, CUE would also be expected to adapt to warmer conditions and increase across spatial temperature gradients or in response to long-term warming. In fact, positive temperature effects were found when estimating CUE from a stoichiometric model11, but negative effects emerged from a meta-analysis and data assimilation3. Therefore, the CUE thermal response remains poorly understood, and the empirical evidence of increased CUE with temperature is still mixed. Moreover, while a previous global study has reported a threshold for temperature sensitivity of ecosystem respiration12, a key knowledge gap exists in whether the temperature-CUE relationship emerges in a similar nonlinear manner and how this would influence SOC responses to warming.
A deep understanding of the environmental controls and mechanisms governing the thermal response of CUE remains challenging due to two primary reasons. First, the methodological difficulty of estimating CUE in the field, which has relied heavily on laboratory studies using a range of different substrates, limits our understanding of the thermal response of CUE across broad spatiotemporal scales1,13. Second, interacting biotic and abiotic factors that covary with mean annual temperature (MAT) have led to inconsistent conclusions about the drivers of microbial CUE14,15. For instance, microbial community composition strongly influences community-level metabolism (CUE and heterotrophic respiration) and SOC dynamics16,17, as fungi have been hypothesized to have higher CUE than bacteria, owing to their more conservative resource use strategies18,19. However, empirical studies have shown neutral20 or negative21,22 relationships between microbial community CUE and fungal dominance. Furthermore, higher CUE has been found in conditions of abundant soil nutrients and moisture23. As such, to resolve the ongoing debate about the thermal response of microbial CUE, it is essential to account for the impacts of substrates alongside biotic and abiotic factors.
A unified and predictive understanding of the mechanisms underlying CUE thermal response could be important to improve predictions of SOC response in a warming world with biogeochemical models. An increase in CUE implies greater microbial growth for a given resource uptake rate, leading to higher microbial necromass that can contribute to SOC stability because of its long residence time3,24,25. Alternatively, higher CUE has the potential to increase the microbial biomass pool for enzyme production, which could accelerate SOC decomposition, decrease SOC stocks, and amplify the positive feedback to global warming2,26,27. Previous modeling studies have reported divergent responses, including positive or negative relationships between CUE and SOC under warming3,26, depending on model structure (specifically depending on whether microbial processes are implicit or explicit in the models) and spatial scales28. Moreover, previous studies have mainly focused on linear relationships among CUE, temperature and SOC25,26, but a strong nonlinear relationship could imply an abrupt change (i.e., decrease or collapse) of SOC in a warmer climate. Investigating whether nonlinear thermal response of CUE and respiration exists is urgently needed at global scales. Incorporating such nonlinear CUE-temperature relationships into biogeochemical models can increase our confidence on the prediction of temperature sensitivity of SOC turnover, which is a major source of uncertainty in projecting SOC feedbacks to the climate system16,29,30.
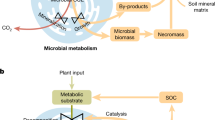
Here, to disentangle the potential nonlinear thermal response of CUE across a geographically-broad temperature gradient, we compiled a large-scale dataset (n = 618; see Supplemental Data 1 and Methods) of surface soil (0–30 cm depth) microbial CUE directly estimated in incubation conditions (Fig. 1a). Because CUE estimates depend on the type of substrates added in the incubations, we used two complementary approaches to account for substrate effects on CUE. In the first approach, we included substrate type as a categorical variable together with other complementary environmental conditions as covariates while examining the environmental controls on original CUE (hereafter original CUE approach). In the second approach, we standardized the varied substrate effects observed across sites/studies as a common substrate using the ‘detrend’ method in a partial least squares approach31, thus allowing us to link the detrended CUE with environmental conditions (hereafter detrended CUE approach; see Methods and Supplementary Fig. 1 for the comparison between original and detrended CUE). We then used two large-scale datasets of heterotrophic respiration from soil incubations under different temperatures (n = 660 for global Dacal dataset and n = 591 for regional Bradford dataset in North America)5,6 to examine the potential nonlinear thermal response of respiration (Fig. 1b). Finally, we included the nonlinear thermal response of CUE as estimated from our statistical models into a first order model32 and an explicit microbial model2,32,33 (Supplementary Fig. 2). These models were used to examine the changes of SOC under warming assuming that the CUE patterns we observed across climatic gradients still hold when temperature changes through time at a given location26,28. The model runs were spatially implicit without accounting for spatial heterogeneity by focusing on variations in CUE due to temperature, while other variables such as C inputs, climate and soil conditions were kept constant. As such, these model runs represent ‘all else being equal’ scenarios in which biotic and abiotic covariates needed to be controlled (i.e., by partial dependence analysis in machine learning)34 while deriving and incorporating the CUE-MAT equations into models (see Methods) – an approach motivated by coupling biogeochemical models with the data-driven machine learning35.
a A total of 618 samples of surface soil microbial original CUE were collected from published references with information provided on substrates added and incubation temperature. These data were strictly screened from a larger database of CUE (n = 1145), whereby the soil microbial CUE without information on substrates added and incubation temperature, with soil depths out of surface (i.e., with sampling depths > 30 cm), and measured with 18O-H2O method were not included. b A total of 1,251 soil heterotrophic respiration measurements (μg C-CO2 g dry-weight soil−1 h−1) were used from published references by Bradford et al. (n = 591) and Dacal et al. (n = 660)5,6. The Bradford dataset was located in North America and the Dacal dataset was distributed globally. The datasets have been previously used to examine the linear thermal adaptation of heterotrophic respiration, and here we focused on examining the nonlinear thermal response of heterotrophic respiration.
Results and Discussion
Nonlinear thermal response of CUE and heterotrophic respiration along climatic gradients
The random forest model analysis indicated that MAT was one of the most important factors influencing CUE, following substrate for original CUE (Fig. 2a) and followed by F:B ratio for detrended CUE (Supplementary Fig. 3a). Instead, the simple ordinary least squares linear regression analysis showed that both original and detrended CUE were independent of MAT (Fig. 2b; Supplementary Fig. 3b). The partial dependence analysis confirmed a strong nonlinear relationship between both original and detrended CUE and MAT and identified two apparent threshold temperatures (≈0 and 15 °C) that shape the slope of CUE-MAT relationships (Fig. 2c; Supplementary Fig. 3c). The further analyses using segmented regressions quantitively showed the two MAT threshold values as 1.2 (standard error - SE = 0.3) and 16.3 °C (SE = 0.4) for original CUE (Supplementary Fig. 4a) versus 1.3 (SE = 0.7) and 16.5 °C (SE = 0.2) for detrended CUE (Supplementary Fig. 4b), respectively.
a Mean decrease in accuracy (%IncMSE, mean and SD) estimated from 100 simulations of random forests. This was used to evaluate the importance of top environmental drivers on the original CUE. All variables were significant as evaluated using rfPermute package. b The linear relationships between original CUE and incubation temperature, mean annual temperature (MAT), aridity index, soil C:N and fungal:bacterial (F:B) ratio fitted by simple linear ordinary Least Squares regressions. c Partial feature contributions of MAT to original CUE by controlling effects of F:B ratio, incubation temperature, aridity index, SOC, and soil C:N.
Our CUE datasets were largely limited to temperate regions in the Northern Hemisphere (Fig. 1a), so other MAT threshold values may occur in warmer tropical regions. We attempted to increase the spatial coverage of our CUE datasets by including CUE measures from 18O-H2O studies (n > 200) (Supplementary Fig. 5), while we caution that the 18O-H2O incorporation method tends to yield lower CUE estimates in comparison to the traditional C isotopic method26. Our results after including the 18O-H2O CUE data showed a robust pattern of CUE with a MAT threshold of ≈15 °C, while the breakpoint of ≈0 °C disappeared in detrended CUE presumably because of lower CUE sample sizes at high latitude, low temperature zones12 (Supplementary Fig. 6). Because soil microbial communities experience soil rather than air temperature (MAT)36, we further examined the original and detrended CUE responses to soil temperature. The results by using soil temperature demonstrated the robustness of the MAT threshold of ≈15 °C for both original and detrended CUE (Supplementary Fig. 7), but the temperature threshold at ≈0 °C was not apparent, presumably because of the less broad soil temperature gradient than the MAT gradient36 (Supplementary Fig. 8). The uncertainty about the pattern at low temperatures highlights the need for more empirical evidence of CUE in both low and high latitude regions to better constrain microbial CUE response to temperature. To assess whether the uneven sample distribution has any consequence on the MAT thresholds, we further used bootstrapping (200 iterations) to randomly sample 90% CUE values for estimating the probability distribution of the mean MAT threshold value, showing robustness of the MAT threshold ≈15 °C (Supplementary Fig. 9).
Collectively, our results using both original and detrended CUE with and without inclusion of CUE measures from 18O-H2O method, and using either air or soil temperature showed a nonlinear increase of microbial CUE when MAT increased above a threshold value of ≈15 °C. This strongly nonlinear increase of CUE with MAT contradicts previous studies at a broad spatial scale, which predicted a nonlinear but weak increase of CUE11 (estimated with a stoichiometric model) or even a decrease of CUE3 (estimated from data assimilation) with MAT. Our results of non-linear increase of CUE with MAT also differ from short-term warming experiments in which CUE was found to decrease at higher incubation temperatures37,38 (Fig. 2b). Our study thus provides the empirical evidence of a strong nonlinear CUE thermal response and also highlights the divergent response of CUE to short-term versus long-term warming.
We interpret these large-scale patterns in soil microbial CUE as results of ecological and evolutionary processes leading to changes in microbial growth or respiration that determine CUE. As higher respiration for given C uptake implies lower CUE, we expect a sharp decrease in respiration at the MAT threshold where CUE increases (assuming the CUE thermal response is due to variations in respiration). Confirming this expectation, a nonlinear relationship was also found between soil heterotrophic respiration and MAT using two large-scale independent (global Dacal and regional Bradford) datasets5,6, with apparently similar threshold values (≈0 or 15 °C) of thermal adaptation as found with CUE (Fig. 3). The segmented regression analysis on the global Dacal dataset estimated these thresholds of MAT to be 4.9 (95% confidence interval = 0.8) and 13.3 °C (95% confidence interval = 2.8) (Fig. 3a). These results agree with those of a recent study using 210 eddy covariance sites and showing the temperature threshold at 15 °C for ecosystem respiration12. By comparison, the segmented regression analysis on the regional Bradford dataset in North America identified a lower MAT threshold value of 11 °C (95% confidence interval = 2.8) (Fig. 3b) presumably because of their regional distribution in North America.
Partial feature contributions of MAT to heterotrophic respiration rate at a standard incubation temperature (natural log-transformed, μg C-CO2 g dry-weight soil−1 h−1) in global Dacal et al. (2019)5 (a) and regional Bradford et al. (2019)6 (b) datasets after controlling for effects of microbial biomass, substrates, incubation temperature, and soil properties. The percent of variance explained increased from 59% and 67% by multivariate linear regression in the original studies5,6 to 75% and 91%, respectively, when using the Random Forest machine learning algorithm in this study. Microbial biomass is included in the analysis to represent the heterotrophic respiration rates at a controlled biomass.
Besides the MAT threshold for respiration, microbial growth could also influence CUE13,22,39 and contribute to this nonlinear behavior. Indeed, the rate of increase of CUE was steeper than the rate of decrease of respiration over ≈15 °C (Fig. 2c; Fig. 3), implying that microbial growth rates could have further contributed to the nonlinear increase of CUE at higher temperatures. So far, however, the lack of full report on the matched/paired measures of microbial growth rate, respiration, and CUE across the meta-data compiled in this study (see Methods) hinders our ability to robustly examine the role of microbial growth, highlighting the need of future research endeavors on this topic. Despite the limitations, our results based on the three independent datasets consistently confirm that microbial CUE thermal response was nonlinear along a temperature gradient. Yet, it remains largely understudied whether and how CUE thermal response covaries with other biotic and abiotic factors.
Soil microbial composition associated to nonlinear thermal response of CUE
Multiple interacting and causal variables such as soil microbial community composition and abiotic conditions covarying with MAT could have contributed to the emergent nonlinear thermal response of CUE. Soil communities with a greater relative dominance of fungi tended to exhibit a lower original or detrended CUE, as shown by analysis on original CUE in incubation conditions (Supplementary Fig. 11), using nonlinear random forest models (Fig. 2a; Supplementary Fig. 3a; Supplementary Fig. 12a, b) on original and detrended CUE by controlling other natural environmental conditions. These results suggest that the thresholds observed in the thermal response of CUE and respiration could be related to shifts in microbial community composition5,6,40. Evolutionary trade-offs in enzyme and membrane structure and function might manifest at the community scale through shifts in microbial taxa, suggesting that the F:B ratio – CUE relationship could arise from turnover of species which are physiologically adapted to warmer conditions41. We suspect that the lower CUE observed in fungal-dominated communities may be partly explained by fungi dominating in soils with more recalcitrant substrates or limited nutrient availability42,43, as evidenced by the negative relationship between soil C:N and CUE (Supplementary Fig. 12c, d). As such, fungi spend more energy to decompose recalcitrant substrates22,44 or locally adapt to nutrient-poor soils by respiring more C per unit of C acquired45. Moreover, greater investment in enzyme production to compensate lower enzymatic activity at cooler temperatures, which needs to be tested in future field studies, could also lead to lower CUE at high latitudes46. Future studies should also examine whether competitive interactions among different fungal taxa47 or trophic interactions48 exert a stronger influence on CUE than temperature alone. As such, multiple mechanisms could contribute to the shift towards bacterial dominance at warmer locations and thus to the CUE changes across the MAT gradient (Fig. 2c; Supplementary Fig. 3c).
Besides the biotic factors, aridity index and soil C:N were identified as important abiotic factors in influencing original (Fig. 2b) and detrended CUE (Supplementary Fig. 3b). Our findings thus supported the resource acquisition and stress tolerance hypothesis23, which state that the energy cost (and thus respiration) is high and thus CUE is lower in drier climates (i.e., lower aridity index; Fig. 2b for original CUE, Supplementary Fig. 3b for detrended CUE) or under lower soil nutrient availability (i.e., high soil C:N; Fig. 2b and Supplementary Fig. 12c for original CUE; Supplementary Figs. 3b and 12d for detrended CUE).
The detrended CUE approach standardizes substrate effects as a common substrate, thus enabling further investigation in the role of underlying environmental factors by accounting for spatial autocorrelation of sample distributions (i.e., using spatial error models). Spatial error model analysis on the whole dataset or the dataset with MAT < 15 °C showed the robustness of lacking linear relationships between MAT and detrended CUE, while F:B ratio was a strong predictor of detrended CUE (Supplementary Figs. 13a, b). By comparison, spatial error model analysis on the dataset with MAT > 15 °C showed the positive and linear relationship between MAT and detrended CUE (Supplementary Fig. 13c), thus highlighting the dependence of spatial scale in deriving the MAT-CUE relationship.
Given the observed substantial role of biotic and abiotic factors in affecting the CUE-temperature relationship, we propose that future studies adopt a systemic scheme to test the adaptation of CUE to changing temperatures by subjecting soil samples from different natural climates to a consistent incubation temperature gradient while controlling for covarying factors. Future advances in this research area should further disentangle the mechanisms underlying microbial adaptation to global warming, such as individual physiological changes in enzyme and membrane structures at evolutionary timescales versus species composition shifts at ecological timescales. This emergent knowledge would be highly valuable to improve the accuracy of biogeochemical models used to estimate the magnitude of climate-C cycle feedback, and subsequently, a crucial step to establish more appropriate greenhouse gas emission targets in a future warmer climate.
Implication of nonlinear thermal response of soil microbial physiology in SOC responses to warming
We also examined how the identified nonlinear microbial thermal CUE response could improve our understanding of SOC responses to warming. To this end, we incorporated this thermal CUE response to MAT into a first-order SOC model, where soil microbes are implicitly represented49, and a four-pool microbial explicit SOC model2,32,33. The microbial-explicit model accounts for soil microbial physiology, community composition or biomass and its interactions or feedbacks to SOC decomposition2,26,27. Specifically, the segmented linear regressions of original or detrended CUE as a function of MAT (R2 = 0.96 for original CUE; R2 = 0.92 for detrended CUE; see legend in Supplementary Fig. 4) were incorporated into both SOC models. We run these models in transient conditions without accounting for spatial heterogeneity (see Methods) to assess how MAT affects SOC mass loss, as quantified by the decay constant (proportional to total heterotrophic respiration) emerging from model simulations.
Both the first-order SOC and soil microbial models predicted a nonlinear decrease in mass-specific heterotrophic respiration rate and nonlinear increase in microbial biomass C with increasing MAT (Supplementary Figs. 14a–d), as expected from our statistical analysis (Fig. 3). However, the decay constant (Fig. 4a) or the total heterotrophic respiration rate (mass-specific respiration times microbial biomass) (Supplementary Figs. 14e–f) nonlinearly decreased with MAT only in the first-order model. Therefore, all else being equal, higher MAT slowed down decomposition and SOC loss in the first-order model, but only above the ≈0 or 15 °C thresholds (Fig. 4b for original CUE; Supplementary Fig. 15b for detrended CUE). The microbial-explicit model behaved in the opposite way, predicting higher decay constant and SOC losses above a MAT of ≈0 or 15 °C (Fig. 4a, b for original CUE; Supplementary Figs. 15a, b for detrended CUE). This divergence in predictions is caused by different roles of CUE in the SOC dynamics of the two models. In the microbial model (see Supplementary Fig. 2 for model structure), the higher CUE in warmer climates promoted microbial growth and biomass (Supplementary Fig. 14c, d), which in turn accelerated decomposition with increased effective K and thus decrease SOC (Fig. 4; Supplementary Fig. 16). Similarly, a recent empirical study showed the negative relationship between CUE and SOC at regional scales50.
The decay constant (effective K, 1/yr) (a) used to quantify SOC mass loss (b) in response to air mean annual temperature (MAT) emerging from model simulations by microbial-explicit versus first-order models at transient conditions (i.e., 1 year) by including the segmented equations of original CUE as a function of air MAT fitted using segmented regressions (see Supplementary Fig. 4 legend for the segmented equations) (see Methods for modeling details). The relative changes (%) of both variables (effective K and SOC) as compared with equilibrium values at 20 °C were presented to facilitate comparisons between models.
Focusing on results from the microbial model—which accounts for known substrate-microbe interactions—our analysis highlights the risk for the potential abrupt SOC losses in boreal and temperate ecosystems under a warmer climate. However, we clarify that our modeling results were spatially implicit without accounting for spatial heterogeneity. Thus, controlling for covarying biotic and abiotic factors (i.e., through partial dependence analysis34) is necessary when deriving and incorporating the emerging nonlinear MAT-CUE equation into the microbial model (see Methods). Further analysis on examining the sensitivity of this partial dependence approach to covarying variables suggested that the emerging nonlinear MAT-CUE relationships were generally robust in scenarios of 1) excluding each of these covarying variables (incubation temperature, soil C:N, SOC, aridity index, and F:B ratio in Fig. 2b) (Supplementary Figs. 17a–e), and 2) including only F:B ratio and substrate (Supplementary Fig. 17f). Our approach thus advocates for the integration of data-driven machine learning approach35,51,52 (e.g., identifying and incorporating the emerging nonlinear equations as shown in this study) with biogeochemical models. While applying our approach across a heterogeneous landscape, ‘scale transition theory’ could provide the insight or tool to scale-up process functions in heterogeneous systems by adopting the mean-field equation and also accounting for different types of heterogeneity53. With lacking of previous theoretical and empirical applications, however, we would like to also highlight the caveats about formulating the partial dependence plot (PDP) as equations incorporated into biogeochemical models. As such, we suggest that our partial dependence plot approach and segmented linear regression derived equations—despite its high accuracy in fittings in our study (R2 > 0.9)—require additional research to refine their future applications, whereby spatial scales would be critical. Our results also suggested the uncertainty of the ≈ 0 °C MAT or SOC thresholds in boreal regions and potential uncertainty in applying the space-time substitution approach54. Moreover, as demonstrated by this study (Supplementary Fig. 12), CUE is a crucial microbial trait likely shaped by shifts in microbial species composition and soil conditions (e.g., soil C:N). Therefore, future studies should explicitly link microbial species composition (e.g., fungi vs. bacteria), MAT, and CUE with SOC in biogeochemical models16.
In conclusion, our study leveraged three independent empirical datasets at broad spatial scales that collectively highlight the presence of nonlinear trends in the thermal response of soil microbial physiology. Above the threshold value of ≈15 °C, there is a potential decoupling of heterotrophic respiration and growth, whereby warming-induced increases in microbial CUE correspond with decreased heterotrophic respiration rates and potentially increased microbial growth. The results underscore the heterogeneity of SOC responses to warming, with high temperature sensitivity in temperate zones that could drive positive SOC feedbacks to climate warming. These findings also emphasize the necessity of incorporating nonlinear warming responses into biogeochemical models to accurately evaluate the sensitivity of SOC dynamics and its feedbacks to climate warming. The ≈15 °C threshold of CUE could be linked to the thermal adaptation of heterotrophic respiration, which is likely driven by shifts in microbial community composition. By including the thermal thresholds of CUE into a microbial model, our findings suggest the likely potential for abrupt SOC losses under a warming climate, especially in temperate regions.
Methods
Soil microbial CUE and respiration datasets
We compiled a dataset of soil microbial CUE at cross-biome scale. Part of dataset was derived from Qiao et al., 2019, which included 780 observations in incubation experiments from 98 sites across the globe. It reported the metadata of coordinates (latitude and longitude) of soil samples, incubation temperature, incubation time, and the substrate added, but some information of sampling depth was lacked. As such, we added the information of sampling depth from original references37. We further added and updated this dataset of soil microbial CUE from peer-published references before 1st August 2022 using the Web of Science, Google Scholar, and the China National Knowledge Infrastructure. We used the following combinations of keywords for searches: (“soil microbial” OR “soil microbe”) AND (“carbon use efficiency” OR “CUE” OR “microbial carbon accumulation” OR “cumulative heterotrophic respiration” OR “carbon substrate consumption”). Soil microbial CUE was directly reported from references or any of microbial C accumulation, cumulative heterotrophic respiration, C substrate consumption in lab conditions were clearly described so that CUE could be estimated based on its definition37. When needed, the microbial CUE data were extracted directly from figures or using the GetData Graph Digitizer (version 2.0). This ultimately led to an expanded CUE with 1145 observations).
We used the following criteria to select the studies or CUE values that were used in our study: (1) no disturbance and contamination occurred during soil sampling (thus also excluding measurements on microbial isolates); (2) if the experiments included other treatments, such as warming, elevated CO2, nitrogen addition, or other experimental treatments, only data from the control were used; (3) soil microbial incubation temperature, the type of substrate added, and soil sampling depth had to be provided; and (4) for standardization purposes, we limited the data of soil microbial CUE experiments to surface samples (≈0-30 cm; no samples from the litter layer) and further excluded the observations of CUE with missing information of incubation temperature and/or the substrate added. This ultimately led to a total of 618 (with C-based substrate) (Fig. 1a) and 826 (Supplementary Fig. 5) (with C-based substrate and water-based substrate - 18O-H2O method26) samples of surface soil microbial CUE. Overall, the studies in the dataset were located between 71.9°S to 78.9°N, with MAT ranging from –5.6 °C to 27.4 °C and MAP from 32.1 mm to 3147 mm (Fig. 1a and Supplementary Data 1). Collectively, the meta-data of CUE were compiled from several studies (n = 68) in which various types of substrates were added. Following previous studies37,55, we classified them into 16 types (see Supplementary Data 1 for details).
We used two large-scale published datasets of soil heterotrophic respiration rates and both of these two datasets have been used to investigate the thermal adaptation of respiration. The first set of data was from Dacal et al.5, which sampled surface soils across 110 global dryland ecosystems located in 19 countries across all continents except Antarctica between June 2006 and December 2013. In each dryland site, five replicates of soil were randomly sampled in vegetated microsite and open microsite, respectively. Soil samples were then bulked and homogenized to derive two composite samples per site, thus leading to 220 soil samples in total. These 220 soil samples were incubated in three profiling assay temperatures (10, 20, 30 °C) to measure soil heterotrophic respiration rates with substrate in excess (glucose). This ultimately led to sample size of 660 used in both the study by Dacal et al.5 and in our study (Fig. 1b). The second set of data was from Bradford et al.6, which sampled surface soils in two vegetation types (forest vs herb) across 3 years (2010, 2011, 2012) from 11 locations (including 10 US Long Term Ecological Research [LTER] stations) ranging from Hawaii to Northern Alaska in North America. The soil samples were incubated in profiling assays to measure soil heterotrophic respiration rates with controlled temperature (12, 20, 28 °C) and substrates in excess (glucose, oxalic acid, glycine). This ultimately led to sample size of 591 (after excluding three outlier values of respiration) for the study led by Bradford et al.5 and in our study (Fig. 1b). More details of soil samplings and measures of soil heterotrophic respiration in incubation conditions were presented in Dacal et al5 and Bradford et al.6. Different experimental designs, methods of measuring soil heterotrophic respiration and microbial biomass (which needed to be included in statistical analysis to estimate mass specific respiration)6,7 precluded the capability of merging these two independent datasets into a harmonized global dataset of soil heterotrophic respiration. Moreover, the lack of full reports on matched/paired measures of soil microbial growth, soil heterotrophic respiration and CUE also precluded the possibility of merging the three independent datasets of CUE and soil heterotrophic respiration.
Drivers of CUE
Incubation temperatures, substrates, and other environmental conditions such as mean annual temperature (MAT), aridity index, soil organic carbon (SOC) and soil C:N ratio as well as microbial composition (F:B ratio) were expected to influence CUE. As clarified above, incubation temperatures and substrate types were compiled from the literature. Since the majority of sites were lacking field measurements of environmental variables, we extracted the other environmental variables from global gridded rasters17 based on georeferenced CUE data. This approach provided standardized environmental variables across geographical areas, and avoided the inconsistence of data sources (field vs. rasters) with different spatial scales. As such, F:B ratio was derived from the recently published global maps17 based on the georeferenced coordinates of CUE. Climate variables – MAT and aridity index and soil properties – SOC and soil C:N ratio were derived from world climate and SoilGrids56, respectively, based on the georeferenced coordinates of CUE. Mean annual soil temperature (MAT_soil) was derived from the recently published global map of soil temperature based on the georeferenced coordinates of CUE36.
Statistical analysis
CUE in incubation conditions
We first investigated the relationship between fungal:bacterial ratio and incubation temperature and CUE under laboratory conditions4,43. Generalized linear mixed models using Markov Chain Monte Carlo (MCMCglmm) were used to examine the dependence of soil microbial CUE on fungal: bacterial ratio, by controlling for the effects of incubation temperature and the substrate. We thus have
where \(F:B\) is fungal:bacterial ratio, \({T}_{{incubation}}\) is the incubation temperature, \({\beta }\) is the standardized fixed effect associated with an individual model parameter, \(b\) represents the random effect of the substrate, and \({\varepsilon }\) is random error which is assumed to follow a normal distribution with mean zero and standard deviation σ.
Effects of substrate and environmental conditions on CUE using original CUE
We assess whether CUE variations could be linked with natural environmental conditions after controlling for the effects of substrates and incubation temperature under lab conditions. A similar approach was previously used to examine the thermal adaptation of soil heterotrophic respiration when using a common or single substrate in lab incubation conditions6. To this end, while examining the relationships between original CUE and natural environmental conditions, we first used simple ordinary least squares linear regressions. Moreover, a random forest machine learning algorithm using randomForest package was applied to determine the importance of different variables including substrate, incubation temperature (\({T}_{{incubation}}\)), MAT, F:B ratio, aridity index, SOC and soil C:N57 on original CUE. Substrate types and incubation temperature were included as categorical and continuous variables, respectively, to control their effects on original CUE. As such, we have
where CUE refers to ‘original CUE’ as opposed to the ‘detrended CUE’ presented below. Mean decrease in accuracy (%IncMSE) was calculated to identify the more important variables as those with greater values of %IncMSE. The significance of variables was evaluated using rfPermute package. Moreover, partial functions of the most important variables (MAT and F:B ratio) were plotted using pdp package to examine their influences on original CUE by controlling other variables in Eq. 2. Based on the partial relations between original CUE and MAT, segmented linear regressions (also used in previous studies58) were used to identify the threshold values of MAT. The obtained segmented linear regressions were incorporated into microbial-explicit and first-order models (see Section ‘SOC models’ for details) to examine their implications on SOC dynamics in a warmer climate. This approach could be feasible in spatially implicit model runs without accounting for spatial heterogeneity (see the following section of SOC models for details), thus requiring to control the biotic and abiotic factors (see Eq. 2) covarying with temperature in influencing CUE in the partial dependence analysis34. The sensitivity of nonlinear CUE and MAT relations and MAT threshold values were examined by using more datasets of compiled CUE which include 18O-H2O method26 or using MAT_soil.
Effects of substrate and environmental conditions on CUE using detrended CUE
Because CUE was substantially influenced by the type of substrates added (Fig. 2a) and that varied across meta-data studies59, here we used a ‘detrend’ approach to standardize the effects of substrates. The principle of ‘detrend’ was to remove the influence of individual substrate effects on the overall relationship using a partial least squares approach31. As such, CUE measured in incubation conditions with the 16 types of substrates added across studies could be regarded as the systematic study of using the common type of substrates. We then evaluated the thermal response of detrended CUE across the climate gradient—an approach previously used to examine the thermal adaptation of soil heterotrophic respiration from laboratory incubations with a common or single substrate5,6. The detrended CUE with values lower than 0 or higher than 1 (n = 11 out of 826 samples) were not used. The relationship between the detrended CUE and original CUE was examined using the simple ordinary least squares regression. The detrended CUE led to overestimated or underestimated values as compared to the original low or high CUE, respectively, thus generally decreasing the range of CUE values (Supplementary Fig. 1).
As in the analysis of original CUE, simple ordinary least squares linear regressions and random forest models (%IncMSE and a feature partial dependence analysis) were used to examine the dependence of detrended CUE on \({T}_{{incubation}}\) and natural environmental conditions (MAT, F:B ratio, aridity index, SOC and soil C:N). In the random forest models, we have
Similarly to the approach used for the original CUE, the segmented regressions were used to identify the threshold values of MAT in detrended CUE-MAT relations. The segmented linear regressions were incorporated into microbial-explicit and first-order models to examine their implications of the nonlinear detrended CUE thermal response on SOC dynamics under climate warming. The full CUE datasets including 18O-H2O method26 or MAT_soil were used to examine the sensitivity of nonlinear detrended CUE and MAT relations and MAT threshold values.
The detrended CUE approach standardized the CUE values by controlling the effects of different substrates, thus allowing to further use spatial error models accounting for spatial autocorrelation of sample distribution to examine environmental controls on detrended CUE. The spatial error models used a spatial weights matrix with neighborhoods defined as cells within a certain distance of the focal cell to remove the impacts of the spatial autocorrelation in the residuals (Moran’s I test; all P > 0.05).
Nonlinear thermal response of soil heterotrophic respiration
We used the soil heterotrophic respiration datasets from Dacal et al. and Bradford et al.5,6 to inspect the potential nonlinear thermal response. To this end, we used the random forest analysis and feature partial dependence analysis to investigate the nonlinear thermal response of respiration by controlling the covariates presented in the linear analysis by Dacal et al. and Bradford et al.5,6. Similarly, the segmented regressions58 were performed to identify the threshold values of MAT in correlating with soil microbial mass-specific heterotrophic respiration.
SOC models
We used two soil organic carbon (SOC) models: i) a conventional first-order decomposition model49 and ii) a four-pool microbial-explicit model2 to examine how the CUE-MAT relationships identified in this study (the segmented linear regressions in relating MAT with original or detrended CUE; Supplementary Fig. 4) influence the response of SOC to warming. To do this, we used a space-for-time approach to simulate the effects of CUE under future warming on SOC dynamics3,26. The first-order model includes three C pools, (i.e., SOC, dissolved organic C (DOC), and microbial biomass C (MBC), with each pool’s decomposition rate represented as a first-order decay function. The first-order model lacks an explicit CUE, but the transfer coefficients that indicate flux partitioning into CO2 vs. soil C pools are conceptually comparable2,32,33. The four-pool microbial model adds an enzyme pool (ENZ) with the enzymatic decomposition of SOC and subsequent assimilation (i.e., uptake) of DOC by microbes2 (see Supplementary Fig. 2 for model structure). As such, the four-pool microbial-explicit model allows the microbial pool to feed back to SOC decomposition. To examine the influence of the CUE-MAT relation on respiration and SOC dynamics, only CUE was assumed to vary with MAT in the model parameterization and setup with other variables such as C inputs, climate, and soil conditions being setup as constants. The detailed equations and parameters in the two models can be found in Ye et al. (2019)33.
It is important to emphasize that the model runs did not account for spatial heterogeneity across landscapes where C inputs, climate and soil conditions could vary substantially. To this end, any spatially varying predictor of CUE in Eq. (2) and (3) needed to be considered in partial dependence plots to derive the relationships between original or detrended CUE and MAT while incorporating nonlinear MAT-CUE equations into the biogeochemical models. As such, it leveraged the data-driven machine learning approach to be integrated with biogeochemical models35 while examining the SOC dynamics in a warming climate. To test the sensitivity of this partial dependence approach to covarying abiotic and biotic variables, we considered two scenarios: (1) running several models in each of which we excluded one of different covariates shown in Fig. 2b (i.e., incubation temperature, soil C:N, SOC, aridity index, F:B ratio; Supplementary Figs. 17a–e), and (2) building the model including only F:B ratio and substrate as covariates (Supplementary Fig. 17f).
To assess the thermal sensitivity of decomposition, we ran the SOC models without litter input in the transient scenario by following a unit of C as it is decomposed at different MAT. We defined the effective decomposition rate (effective ke) as an indicator of thermal sensitivity of decomposition at different MAT:
where t is the time that models run, \({{SOC}}_{0}\) is the initial SOC, and \({{SOC}}_{t}\) is the remaining SOC after decomposition within time t. The pattern shown in Fig. 4 refers to the long-term ke values after they have approximately stabilized (i.e., 1 year).
Data availability
Data of original and detrended CUE, the associated environmental variables and the original references as well as data of respiration in Bradford et al and Dacal et al. and associated environmental variables are deposited in https://github.com/KailiangYu/Microbial-carbon-use-efficiency.
Code availability
Codes used to derive detrended CUE and generate Figs. 1–4 in main text are deposited in https://github.com/KailiangYu/Microbial-carbon-use-efficiency.
References
Bradford, M. A. & Crowther, T. W. Carbon use efficiency and storage in terrestrial ecosystems. New Phytol. 199, 7–9 (2013).
Allison, S. D., Wallenstein, M. D. & Bradford, M. A. Soil-carbon response to warming dependent on microbial physiology. Nat. Geosci. 3, 336–340 (2010).
Tao, F. et al. Microbial carbon use efficiency promotes global soil carbon storage. Nature 618, 981–985 (2023).
Wieder, W. R., Grandy, A. S., Kallenbach, C. M. & Bonan, G. B. Integrating microbial physiology and physio-chemical principles in soils with the MIcrobial-MIneral Carbon Stabilization (MIMICS) model. Biogeosciences 11, 3899–3917 (2014).
Dacal, M., Bradford, M. A., Plaza, C., Maestre, F. T. & García-Palacios, P. Soil microbial respiration adapts to ambient temperature in global drylands. Nat. Ecol. Evol. 3, 232–238 (2019).
Bradford, M. A. et al. Cross-biome patterns in soil microbial respiration predictable from evolutionary theory on thermal adaptation. Nat. Ecol. Evol. 3, 223–231 (2019).
Luo, Y., Wan, S., Hui, D. & Wallace, L. L. Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413, 622–625 (2001).
Hartley, I. P., Heinemeyer, A. & Ineson, P. Effects of three years of soil warming and shading on the rate of soil respiration: substrate availability and not thermal acclimation mediates observed response. Glob. Chang. Biol. 13, 1761–1770 (2007).
Karhu, K. et al. Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 513, 81–84 (2014).
Moinet, G. Y. et al. Soil microbial sensitivity to temperature remains unchanged despite community compositional shifts along geothermal gradients. Glob. Chang. Biol. 27, 6217–6231 (2021).
Sinsabaugh, R. L. et al. Stoichiometry of microbial carbon use efficiency in soils. Ecol. Monogr. 86, 172–189 (2016).
Johnston, A. S. et al. Temperature thresholds of ecosystem respiration at a global scale. Nat. Ecol. Evol. 5, 487–494 (2021).
Manzoni, S. et al. Optimal metabolic regulation along resource stoichiometry gradients. Ecol. Lett. 20, 1182–1191 (2017).
Domeignoz-Horta, L. A. et al. Microbial diversity drives carbon use efficiency in a model soil. Nat. Commun. 11, 3684 (2020).
Malik, A. A. et al. Land use driven change in soil pH affects microbial carbon cycling processes. Nat. Commun. 9, 3591 (2018).
Crowther, T. W. et al. The global soil community and its influence on biogeochemistry. Science 365, eaav0550 (2019).
Yu, K. et al. The biogeography of relative abundance of soil fungi and bacteria in top surface soil. Earth Syst. Sci. Data Discuss. 14, 4339–4350 (2022).
Kreft, J.-U. & Bonhoeffer, S. The evolution of groups of cooperating bacteria and the growth rate versus yield trade-off. Microbiology 151, 637–641 (2005).
Pfeiffer, T., Schuster, S. & Bonhoeffer, S. Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507 (2001).
Thiet, R. K., Frey, S. D. & Six, J. Do growth yield efficiencies differ between soil microbial communities differing in fungal: bacterial ratios? Reality check and methodological issues. Soil Biol. Biochem. 38, 837–844 (2006).
Six, J., Frey, S. D., Thiet, R. K. & Batten, K. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci. Soc. Am. J. 70, 555–569 (2006).
Soares, M. & Rousk, J. Microbial growth and carbon use efficiency in soil: links to fungal-bacterial dominance, SOC-quality and stoichiometry. Soil Biol. Biochem. 131, 195–205 (2019).
Anthony, M. A., Crowther, T. W., Maynard, D. S., van den Hoogen, J. & Averill, C. Distinct assembly processes and microbial communities constrain soil organic carbon formation. One Earth 2, 349–360 (2020).
Kallenbach, C. M., Frey, S. D. & Grandy, A. S. Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 7, 13630 (2016).
Wang, C. et al. Large‐scale importance of microbial carbon use efficiency and necromass to soil organic carbon. Glob. Chang. Biol. 27, 2039–2048 (2021).
García-Palacios, P. et al. Evidence for large microbial-mediated losses of soil carbon under anthropogenic warming. Nat. Rev. Earth Environ. 2, 507–517 (2021).
Wieder, W. R., Bonan, G. B. & Allison, S. D. Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Chang. 3, 909–912 (2013).
Tao, F. et al. Reply to: Model uncertainty obscures major driver of soil carbon. Nature 627, E4–E6 (2024).
Amundson, R. The carbon budget in soils. Annu. Rev. Earth Pl. Sc. 29, 535–562 (2001).
Crowther, T. W. et al. Quantifying global soil carbon losses in response to warming. Nature 540, 104–108 (2016).
Nocita, M. et al. Prediction of soil organic carbon content by diffuse reflectance spectroscopy using a local partial least square regression approach. Soil Biol. Biochem. 68, 337–347 (2014).
Georgiou, K., Abramoff, R. Z., Harte, J., Riley, W. J. & Torn, M. S. Microbial community-level regulation explains soil carbon responses to long-term litter manipulations. Nat. Commun. 8, 1223 (2017).
Ye, J. S., Bradford, M. A., Dacal, M., Maestre, F. T. & García‐Palacios, P. Increasing microbial carbon use efficiency with warming predicts soil heterotrophic respiration globally. Glob. Chang. Biol. 25, 3354–3364 (2019).
Georgiou, K. et al. Global stocks and capacity of mineral-associated soil organic carbon. Nat. Commun. 13, 3797 (2022).
Reichstein, M. et al. Deep learning and process understanding for data-driven Earth system science. Nature 566, 195–204 (2019).
Lembrechts, J. J. et al. Global maps of soil temperature. Glob. Chang. Biol. 28, 3110–3144 (2022).
Qiao, Y. et al. Global variation of soil microbial carbon-use efficiency in relation to growth temperature and substrate supply. Sci. Rep. 9, 5621 (2019).
Li, J. et al. Reduced carbon use efficiency and increased microbial turnover with soil warming. Glob. Chang. Biol. 25, 900–910 (2019).
Sinsabaugh, R. L., Manzoni, S., Moorhead, D. L. & Richter, A. Carbon use efficiency of microbial communities: stoichiometry, methodology and modelling. Ecol. Lett. 16, 930–939 (2013).
Hochachka, P. W. & Somero, G. N. Biochemical adaptation: mechanism and process in physiological evolution. (Oxford university press, 2002).
Tian, W. et al. Thermal adaptation occurs in the respiration and growth of widely distributed bacteria. Glob. Chang. Biol. 28, 2820–2829 (2022).
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A. & Cleveland, C. C. Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249 (2009).
Strickland, M. S. & Rousk, J. Considering fungal: bacterial dominance in soils–methods, controls, and ecosystem implications. Soil Biol. Biochem. 42, 1385–1395 (2010).
Roller, B. R. & Schmidt, T. M. The physiology and ecological implications of efficient growth. ISME J. 9, 1481–1487 (2015).
Schimel, J. P. & Weintraub, M. N. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: a theoretical model. Soil Biol. Biochem. 35, 549–563 (2003).
Melillo, J. M. et al. Long-term pattern and magnitude of soil carbon feedback to the climate system in a warming world. Science 358, 101–105 (2017).
Maynard, D. S., Crowther, T. W. & Bradford, M. A. Fungal interactions reduce carbon use efficiency. Ecol. Lett. 20, 1034–1042 (2017).
Kane, J. L. et al. Fungivorous nematodes drive microbial diversity and carbon cycling in soil. Ecology 104, e3844 (2023).
Parton, W. J., Stewart, J. W. & Cole, C. V. Dynamics of C, N, P and S in grassland soils: a model. Biogeochemistry 5, 109–131 (1988).
Shi, J. et al. Soil organic carbon increases with decreasing microbial carbon use efficiency during vegetation restoration. Glob. Chang. Biol. 30, e17616 (2024).
Liu, L. et al. Knowledge-guided machine learning can improve carbon cycle quantification in agroecosystems. Nat. Commun. 15, 357 (2024).
He, X. et al. Emerging multiscale insights on microbial carbon use efficiency in the land carbon cycle. Nat. Commun. 15, 8010 (2024).
Englund, G. & Leonardsson, K. Scaling up the functional response for spatially heterogeneous systems. Ecol. Lett. 11, 440–449 (2008).
Yu, K. et al. Pervasive decreases in living vegetation carbon turnover time across forest climate zones. Proc. Natl Acad. Sci. USA 116, 24662–24667 (2019).
Roels, J. Application of macroscopic principles to microbial metabolism. Biotechnol. Bioeng. 22, 2457–2514 (1980).
Hengl, T. et al. SoilGrids1km—global soil information based on automated mapping. PloSOne 9, e105992 (2014).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Berdugo, M. et al. Global ecosystem thresholds driven by aridity. Science 367, 787–790 (2020).
Frey, S. D., Lee, J., Melillo, J. M. & Six, J. The temperature response of soil microbial efficiency and its feedback to climate. Nat. Clim. Change 3, 395–398 (2013).
Acknowledgements
J.W. receives supports from the National Natural Science Foundation of China (32241035). S.N. receives supports from the National Natural Science Foundation of China (31988102). M.D. was supported by an Alexander von Humboldt Fellowship. We would like to also thank Mark Bradford and Stefano Manzoni for their constructive comments and suggestions which improved this study.
Author information
Authors and Affiliations
Contributions
K.Y. designed the study with inputs from J.W. and P.G.; K.Y. carried out the analysis with inputs of detrended method from C.A. and data inputs from J.W., L. Y. and M.D.; K.Y., L.H., J.W. and P.G. interpreted results; K.Y. and L.H. developed the model; K.Y. wrote the initial manuscript draft with additional inputs from P.G., L.H., S.N., J.W., M.D., C.A., K.G., J.-s.Y., F.M., L.Y. and T.C.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alice Johnston and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, K., He, L., Niu, S. et al. Nonlinear microbial thermal response and its implications for abrupt soil organic carbon responses to warming. Nat Commun 16, 2763 (2025). https://doi.org/10.1038/s41467-025-57900-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57900-9
This article is cited by
-
Patterns and drivers of soil microbial carbon use efficiency across soil depths in forest ecosystems
Nature Communications (2025)