Abstract
Regeneration and homeostatic turnover of solid tissues depend on the proliferation of symmetrically dividing adult stem cells, which either remain stem cells or differentiate based on their niche position. Here we demonstrate that in zebrafish lateral line sensory organs, stem and progenitor cell proliferation are independently regulated by two cyclinD genes. Loss of ccnd2a impairs stem cell proliferation during development, while loss of ccndx disrupts hair cell progenitor proliferation but allows normal differentiation. Notably, ccnd2a can functionally replace ccndx, indicating that the respective effects of these Cyclins on proliferation are due to cell type-specific expression. However, even though hair cell progenitors differentiate normally in ccndx mutants, they are mispolarized due to hes2 and Emx2 downregulation. Thus, regulated proliferation ensures that equal numbers of hair cells are polarized in opposite directions. Our study reveals cell type-specific roles for cyclinD genes in regulating the different populations of symmetrically dividing cells governing organ development and regeneration, with implications for regenerative medicine and disease.
Similar content being viewed by others
Introduction
Tissue turnover and regeneration are essential for organismal function and survival and require the proliferation of stem cells. In most solid tissues, this process relies on symmetrically dividing adult stem cells1. For instance, in the epithelia of the intestine, stomach, esophagus and the skin, stem cells divide symmetrically and—depending on their location in the stem cell niche—the daughter cells either maintain their stem cell characteristics, or if displaced from the niche proceed to differentiate2,3,4,5,6,7,8,9. Stem and progenitor cell proliferation need to be tightly controlled, as misregulation of niche signals or uncontrolled proliferation of stem and daughter cells can lead to serious diseases such as cancer. Despite the importance of symmetrically dividing stem cells and their progeny for tissue maintenance, regeneration and disease, whether their proliferation is differentially regulated has not been explored.
The zebrafish sensory lateral line is an excellent model to study the regulation of sensory organ homeostasis and regeneration at the cellular and molecular level within single cells10,11,12. It consists of clusters of 50–80 cells, called neuromasts that are arranged in lines on the head and along the trunk of the fish (Fig. 1A–C). These neuromasts are deposited during embryonic development by migrating primordia13. Each neuromast contains mechanosensory hair cells surrounded by support cells and peripheral mantle cells (Fig. 1C, D). Hair cells possess a long, microtubule based kinocilium and shorter actin rich stereocilia that are sensitive to water motion (Fig. 1B14).
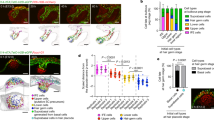
A Representative image of a 5 dpf DAPI-stained zebrafish larva, posterior lateral line neuromast in boxed region. Scale bar = 200 µm. B Scanning electron micrograph of a 5 dpf zebrafish neuromast (dorsal view) with short stereocilia and long kinocilia in purple (adapted from Lush, M.E. and Piotrowski, T. (2014), Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn., 243: 1187-1202. https://doi.org/10.1002/dvdy.24167”10). C Transmission electron micrograph of a transverse section of a 5 dpf neuromast with hair cells in purple and mantle cells in blue. Additional support cells are unlabeled (adapted from Lush, M.E. and Piotrowski, T. (2014), Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn., 243: 1187-1202. https://doi.org/10.1002/dvdy.24167”10). Diagram of a neuromast showing a transverse section (D) and a dorsal view (E). Progenitor cells and hair cells are in purple, amplifying stem cells in the dorsal-ventral poles in pink and mantle cells in blue. F Integrated scRNA-seq UMAP plot of a neuromast regeneration time course (homeostasis, 0 min, 30 min, 1 h, 3 h, 5 h and 10 h after hair cell death; Baek et al., 202212). G–J scRNA-seq Feature Plots (Baek et al., 202212, https://piotrowskilab.shinyapps.io/neuromast_regeneration_scRNAseq_pub_2021/) illustrating gene-specific expression patterns. G pcna labels dividing, differentiating hair cell progenitors (purple arrow) and amplifying stem cells (pink arrow). H atoh1a is expressed in some central cells and marks the lineage from hair cell progenitors to hair cells (purple arrow). I ccndx is expressed in some central cells and along the hair cell lineage and is highest in progenitor cells undergoing differentiating divisions (purple arrow). J ccnd2a is more broadly expressed but is highest in the amplifying cell population (pink arrow) and absent from the hair cell lineage. K Heatmap of scaled gene expression across lateral line cell types during the averaged regeneration time course (Baek et al., 202212). L Heatmap of scaled gene expression during the regeneration time course. All genes are briefly upregulated at 0–30 min but show the largest activation between 3 − 10 h. atoh1a and ccndx show similar expression dynamics, whereas ccnd2a expression peaks slightly later. M Representative images of HCR in situ hybridization of ccndx (yellow) and ccnd2a (magenta) during the regeneration time course. Scale bar = 10 µm. Representative images of HCR in situ hybridization of ccndx (yellow) and atoh1a (magenta) during homeostasis (N) and 5 h after hair cell death (O). Scale bar = 10 µm.
Homeostasis and regeneration of neuromast cells are maintained by two populations of proliferating cells: amplifying support cells and differentiating hair cell progenitors (Fig. 1E, [15]). In the dorsal-ventral (D-V) poles, support cells (Fig. 1E, pink cells) divide and their daughter cells can adopt different fates: remain undifferentiated while in contact with mantle cells, the hypothetical niche, or, be displaced away from the niche and become a hair cell progenitor15,16. Because amplifying cells self-renew and give rise to hair cells, we call them amplifying stem cells. Their degree of plasticity or potency to generate other neuromast cell types remains to be tested. Hair cell progenitors in the center of the organ will in turn divide again and give rise to two hair cells (Fig. 1E, purple cells10,15,17,18,). After hair cell loss progenitors also arise from non-D-V pole support cells, demonstrating plasticity, similar to other epithelial cell lineages19,20.
As in other mechanosensory organs in various species, zebrafish hair cell differentiation in neuromasts is negatively regulated via Notch-dependent lateral inhibition, and loss of Notch signaling causes the development of more hair cells at the expense of support cells15,17,18,21,22. In addition to its function in progenitor fate specification, Notch signaling also inhibits proliferation of differentiating progenitor cells during regeneration15,17. Therefore, during regeneration and immediately after hair cell death, Notch signaling is downregulated, leading to differentiation of progenitor cells, their proliferation and further differentiation into hair cells. The current belief is that cell proliferation is essential for neuromast hair cell regeneration, as cell cycle inhibition with pharmacological inhibitors leads to a failure in regeneration18,23.
Progenitor cell division produces a pair of hair cells with opposing polarity, ensuring the equal generation of hair cells that are sensitive to either rostrad or caudad water flow24. The transcription factor Emx2 determines hair cell polarity in both the lateral line and ear25,26,27,28,29,30. In neuromasts, it is expressed in only one of the two sibling hair cells, where it reverses the cell’s default anterior polarity. Notch-mediated lateral inhibition between the two initially equal progenitors inhibits the expression of Emx2 in one progenitor, and loss of Notch signaling causes both hair cells in the pair to acquire the same polarity25,28,30.
As proliferation is not only essential for life-long regeneration but also for correct hair cell polarity it is essential to elucidate how it is controlled. We previously characterized gene expression dynamics during regeneration in all neuromast cell types using single cell RNA-seq (scRNA-seq)11,12. Here we show that the proliferating amplifying stem cell and progenitor cell populations of the zebrafish lateral line express different cyclinD genes, which are G1-regulators that bind and activate cyclin-dependent kinases (CDK4/6) to initiate the cell cycle31,32. Amplifying cells express ccnd2a and dividing, differentiating progenitor cells express ccndx. Loss of ccndx causes lack of differentiating progenitor divisions while leaving amplifying divisions unaffected. Hair cells still regenerate in lower numbers, demonstrating that progenitor cell proliferation is not required for differentiation and regeneration of hair cells. In contrast, loss of ccnd2a only affects amplifying cell divisions, at least during development. ccnd2a driven by the ccndx promoter rescues progenitor proliferation in ccndx mutants, demonstrating that the effects of these two D-type cyclins are caused by their cell type-specific expression, not because they interact with different targets. Thus, proliferation in amplifying cells and differentiating progenitor cells is mechanistically uncoupled. We also show that Notch signaling inhibits ccndx during homeostasis and that the increase in hair cell progenitor proliferation after Notch downregulation during hair cell regeneration requires ccndx. Lastly, we demonstrate using scRNA-seq and functional analyses that loss of ccndx and progenitor proliferation lead to hair cell polarity defects due to hes2 downregulation and ectopic Emx2 expression. Our findings have important implications for the understanding of how proliferation of symmetrically dividing stem and progenitor cells is controlled during homeostasis and disease.
Results
Lateral line organs possess two populations of dividing cells with unique gene expression profiles
To uncover the mechanisms underlying hair cell regeneration, we set out to identify new regulators of this process. In zebrafish, the antibiotic neomycin induces rapid hair cell death followed by complete regeneration10,33,34. We used this approach in our previous scRNA-seq time course, where we identified all cell types of the regenerating neuromasts (Fig. 1F12,). With regard to dividing cells, two trajectories of proliferating populations were found to be marked by pcna expression (Fig. 1G and Supplementary Fig. 1A, B12,): the dividing hair cell progenitor cells mature into hair cells and are marked by the hair cell-specifying transcription factor atoh1a, whereas amplifying cells are defined as pcna+ cells that do not express atoh1a or hair cell genes (Fig. 1H12).
To identify additional genes specific to either cell population, we queried our scRNA-seq data sets of 5 days post fertilization (dpf) neuromasts11,12. We found the cyclinD family member ccndx to be specifically expressed in differentiating progenitor cells (differentiating divisions, Fig. 1I, K), whereas ccnd2a was enriched in amplifying cells (amplifying divisions, Fig. 1J, K). Both cyclinD genes, as well as atoh1a and pcna were differentially expressed during regeneration with atoh1a and ccndx possessing similar expression dynamics (Fig. 1L and Supplementary Fig. 1C–F12,15,17,33. Upon neomycin treatment, all genes were initially downregulated followed by a strong upregulation between 3–10 h (hrs), when cells started to proliferate (Fig. 1L15). We validated the scRNA-seq time course data with hybridization chain reaction (HCR) mRNA expression analyses. ccndx and ccnd2a were lowly expressed during early stages of regeneration but showed strong upregulation at 5 h post hair cell death (Fig. 1M). As indicated by the scRNA-seq data ccndx was co-expressed with atoh1a in hair cell progenitors during homeostasis and 5 h after neomycin treatment (Fig. 1K, N, O). In contrast to ccndx, ccnd2a was enriched in amplifying cells during homeostasis, with broader expression 5–10 h after hair cell death (Fig. 1M and Supplementary Fig. 1D, H).
During development, ccndx and ccnd2a are both expressed in the 32 h post fertilization (hpf) migrating lateral line primordium (Supplementary Fig. 2A). As in mature neuromasts, ccndx is expressed in atoh1a-positive hair cell progenitor cells in the lateral line primordium and spinal cord neurons as described in the frog and zebrafish35,36. ccnd2a on the other hand is more broadly expressed in primordium cells but excluded from ccndx and atoh1a- expressing progenitor cells (Supplementary Fig. 2A). The exclusive expression of ccndx and ccnd2a is even more obvious in deposited, still developing neuromasts (Supplementary Fig. 2B). The stem cell status of support cells during development still needs to be determined. In conclusion, the two cyclinD genes are expressed in amplifying stem/support cells and proliferating hair cell progenitor cells, respectively, suggesting that proliferation is differentially regulated in stem cells/support cells and their differentiating progeny.
ccndx −/− neuromasts form new hair cells in the absence of proliferation
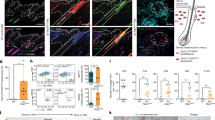
To test the function of ccndx, we generated a mutant using CRISPR/Cas9 (Supplementary Fig. 3A). The mutation introduces a stop codon, but truncated ccndx mRNA is still transcribed in these mutants (Supplementary Fig. 3F-G). 5 dpf ccndx−/− larvae looked grossly normal but lacked an inflated swim bladder and showed abnormal swimming behavior (Supplementary Fig. 3B-C). The mutant larvae failed to inflate their swim bladder and were not viable. Nevertheless, the migrating lateral line primordium deposited neuromasts normally during embryogenesis (Supplementary Fig. 3D-E). Previous morpholino-induced knockdown of ccndx in zebrafish and frogs resulted in defects in motor neuron formation and axonal outgrowth35,36, however anti-acetylated tubulin staining in 5 dpf ccndx−/− larvae showed no obvious differences in motor axon outgrowth (Supplementary Fig. 3H–K). The morpholino-induced phenotype could be more severe than the mutant phenotype as the morpholino also blocks maternal mRNA, or alternatively, it causes off target effects. To test the role of ccndx in hair cell formation, we analyzed a hair cell-specific reporter line (Tg(myo6b:H2B-mScarlet-I)) in ccndx mutants. We observed a reduced number of hair cells in 5 dpf homeostatic ccndx−/− neuromasts and fewer regenerated hair cells 24- and 48 hrs after neomycin-induced hair cell death compared to sibling larvae (Fig. 2A–C). As mutant larvae die soon after, we were not able to assess if mutant hair cell numbers eventually catch up with sibling hair cell numbers.
Time course of hair cell regeneration in sibling (A) and ccndx−/− (B) at homeostasis and 2, 24 or 48 h after hair cell death. Hair cells are labeled by myo6b:H2B-mScarlet-I (magenta). Scale bar = 10 µm. C Hair cell counts in sibling and ccndx−/− at homeostasis and 2, 24 or 48 hrs after hair cell death. Circles represent individual neuromasts. Data are presented as mean values + /− 95% confidence intervals. n = 24 for all. ns = not significant p = 0.99, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. D–F Sibling neuromast expressing sqEt4:GFP/sqEt20:GFP stained with EdU (magenta) after 5 − 6 dpf of homeostasis (D). Scale bar = 10 µm. Spatial analysis of multiple neuromasts showing EdU+ support cells (pink squares), EdU+ hair cells (purple triangles) and surrounding mantle cells (blue X’s, (E). F One-sided binomial analysis of position of EdU+ support cells (pink) and hair cells (purple). ****p < 0.00001, ***p < 0.0001. n = 21. Sibling neuromast 24 h after neomycin treatment expressing sqEt4:GFP/sqEt20:GFP and labeled with EdU (magenta) (G). Scale bar = 10 µm. Spatial analysis of multiple sibling regenerating neuromasts (H) and one-sided binomial analysis of EdU+ cell positions (I). **** p < 0.00001, ns = not significant, p = 0.27. n = 24. J–L ccndx−/− neuromast expressing sqEt4:GFP/sqEt20:GFP labeled with EdU (magenta) after 5 − 6 dpf of homeostasis (J). Scale bar = 10 µm. Spatial analysis of multiple ccndx−/− homeostatic neuromasts (K) and one-sided binomial analysis of EdU+ cell positions, ****p < 0.00001 (L). No EdU+ hair cells are present. n = 21. M–O ccndx−/− neuromast 24 hrs after neomycin treatment expressing sqEt4:GFP/sqEt20:GFP and labeled with EdU (magenta) (M). Scale bar = 10 µm. Spatial analysis of multiple ccndx−/− regenerating neuromasts (N) and one-sided binomial analysis of EdU+ cell positions, ****p < 0.00001 (O). No EdU+ hair cells are present. n = 23. P EdU indexes of sibling and ccndx−/− amplifying, differentiation or total EdU+ cells during homeostasis. Data are presented as mean values + /− 95% confidence intervals. N = 21 for sibling and ccndx−/−. ns = not significant p = 0.35, ***p = 0.0001, *p = 0.0223, two-tailed t-test. (Q) EdU indexes of sibling and ccndx−/− amplifying, differentiation and total EdU+ cells after 24 hrs of regeneration. Data are presented as mean values + /− 95% confidence intervals. n = 24 for sibling and n = 23 for ccndx−/−. ns = not significant p = 0.19, ****p < 0.0001, two-tailed t-test. R Time-lapse analysis of regenerating sibling myo6b:H2B-mScarlet-I+ hair cells from 2 to 42 h after neomycin treatment. Red triangles point to hair cells that survived neomycin. Each new pair of hair cells is marked by differently colored dots. Scale bar = 10 µm. S Time-lapse analysis of regenerating ccndx−/− hair cells from 2 to 42 h after neomycin treatment. Red triangles point to hair cells that survived neomycin. Each new hair cell is marked by differently colored dots. T Quantification of the appearance of the first, second and third hair cell in sibling and ccndx−/− neuromasts over 30 h of regeneration. For siblings, the appearance of each new cell before division was counted. Data are presented as mean values + /− 95% confidence intervals. n = 7 for sibling and n = 6 for ccndx−/−. ns = not significant, p = 1 for first, p = 0.29 for second and p = 0.62 for third myo6b(+) cells, Two-way Anova (genotype, time) with Šidák multiple comparisons test. U atoh1a HCR (magenta) and sqEt4:GFP+ (cyan) hair cells in 5 dpf sibling neuromasts. V A magnified atoh1a+/sqET4:GFP- progenitor cell from (U). Scale bar = 10 µm. W, X atoh1a HCR (magenta) and sqEt4:GFP+ (cyan) hair cells in 5 dpf ccndx−/− neuromasts. X A magnified atoh1a+/sqET4:GFP- progenitor cell from (W). Scale bar = 10 µm. Y Quantification of 5 dpf atoh1a+/sqET4:GFP − progenitor cells showing no difference between sibling and ccndx−/−. Data are presented as mean values + /− 95% confidence intervals. n = 23 for sibling and n = 15 for ccndx−/−. ns = not significant, p = 0.575, two-tailed t-test.
As hair cell regeneration in the zebrafish lateral line is thought to depend on progenitor proliferation18,23, we examined whether proliferation was affected in ccndx−/− neuromasts during homeostasis and regeneration by assessing EdU incorporation for 24 hrs, which labels cells that have entered S-phase of the cell cycle. To determine if the location of amplifying and differentiating divisions was affected by loss of ccndx, we performed spatial analyses of sibling and mutant neuromasts by plotting the location of all EdU+ support and hair cells from multiple neuromasts15,37. EdU+ cells that also express sqEt4:GFP represent differentiating hair cells (purple triangles; Fig. 2E, H, K, N), while EdU+ cells that are not sqEt4GFP+ are amplifying cells, plotted as pink squares. Quiescent sqEt20+ mantle cells are plotted as blue crosses and delineate the neuromast outline15,37.
In homeostatic sibling larvae, amplifying cell divisions were restricted to the D-V poles of neuromasts, while differentiating cell divisions were centrally located (Fig. 1E and Fig. 2D–F15). Upon regeneration, the rate of cell division significantly increased in both cell populations (Fig. 2G–I). In homeostatic and regenerating ccndx−/− larvae, amplifying divisions were also restricted to the D-V poles and EdU indexes were not significantly different from siblings (Fig. 2J–Q). In stark contrast, there were no EdU+ hair cells present in ccndx−/− neuromasts (Fig. 2J–O). Quantification of cell divisions showed that the only dividing cells in ccndx−/− neuromasts were the D-V amplifying cells (Fig. 2P, Q). The decrease in total EdU index in ccndx−/− neuromasts is therefore solely due to the lack of EdU+ hair cells.
To visualize hair cell formation in vivo, we performed time-lapse analyses of regenerating neuromasts expressing myo6b:H2B-mScarlet-I. myo6b:H2B-mScarlet-I started to be expressed early in hair cell differentiation, just before the progenitor divided (Supplementary Movie 1, and Fig. 2R). In contrast, ccndx−/− mutant progenitor cells started to express myo6b:H2B-mScarlet-I but failed to divide (Supplementary Movie 1, and Fig. 2S). Thus, only a single hair cell was generated per progenitor cell compared to two hair cells in siblings. These results challenge the current paradigm that proliferation is required for regeneration and instead demonstrated that neuromasts can regenerate hair cells without progenitor proliferation.
The cell cycle affects differentiation in several cell types38,39,40,41 and we therefore asked if the timing of hair cell differentiation during regeneration differs in the absence of proliferation in ccndx−/− neuromasts. We analyzed time-lapse recordings of the first 30 h of regeneration of myo6b:H2B-mScarlet-I+ hair cells in sibling and ccndx mutant neuromasts. However, the appearances of the first three new hair cells were not significantly different between siblings and ccndx−/− mutant animals (Fig. 2T and Supplementary Movie 1).
As shown in the time lapse movies and EdU experiments, ccndx is required for progenitor proliferation, and we wondered if loss of ccndx might affect how many hair cell progenitors are generated even before they divide. To examine progenitor cell numbers during homeostasis we performed HCRs for atoh1a in neuromasts expressing sqEt4:GFP at 5 dpf. We considered progenitor cells as those without sqEt4:GFP but strongly expressing atoh1a. The number of atoh1a+/sqEt4:GFP - cells in ccndx−/− neuromasts was not significantly different from siblings (Fig. 2U–X), illustrating that progenitor numbers were normal during homeostasis in ccndx−/− larvae (Fig. 2Y). The analysis of hair cell regeneration in ccndx−/− larvae demonstrates that hair cells can regenerate via direct differentiation of progenitor cells in the absence of proliferation. There are fewer ccndx−/− hair cells compared to siblings because mutant progenitor cells generate one hair cell instead of two.
The finding that ccndx−/− hair cells differentiate in the absence proliferation contrasts with previous reports that showed pharmacological cell cycle blockers inhibit neuromast hair cell regeneration18,23. To investigate the reason for this discrepancy, we treated 5 dpf control and ccndx−/− larvae with the early S-phase inhibitor aphidicolin and EdU during 48 hrs of regeneration. As published, control larvae treated with aphidicolin showed a severe reduction in regenerated hair cells compared to DMSO controls (Supplementary Fig. 4A, B, E). Aphidicolin-treated ccndx−/− regenerated more hair cells than treated siblings (Supplementary Fig. 4C–E). In aphidicolin-treated siblings, EdU indexes showed a clear reduction in the amplifying and differentiating populations, and therefore total proliferation (Supplementary Fig. 4F–H). EdU+ support cells were also reduced to the same extent by aphidicolin in ccndx−/−, but as ccndx−/− normally do not produce EdU+ hair cells, aphidicolin had no added effect on this population (Supplementary Fig. 4F–H). Aphidicolin-treated siblings often showed enlarged hair cells, as if they were trying to divide but then failed (Supplementary Fig. 4B, yellow asterisk). This result was also seen in time-lapse analyses (Supplementary Movie 2). In sibling neuromasts, hair cells began to differentiate as evidenced by upregulation of myo6b:H2B-mScarlet-I. However, the newly generated hair cells failed to divide, instead enlarged and died. In contrast, aphidicolin-treated ccndx−/− hair cells differentiated and survived over the 42 h of imaging. We conclude that in aphidicolin-treated siblings, new hair cells start to differentiate but eventually die, possibly via activation of cell cycle check-point regulators. As ccndx−/− progenitor cells fail to initiate the cell cycle they escape the toxicity of aphidicolin treatment. Our experiments hence show that proliferation is not required for zebrafish lateral line hair cell regeneration and that previous studies using aphidicolin to stop the cell cycle caused regeneration defects due to drug-induced hair cell death.
ccnd2a drives amplifying cell divisions but substitutes for ccndx if expressed in progenitor cells
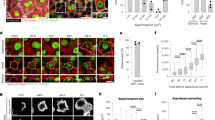
In the amplifying stem cell population, ccnd2a is the most enriched cyclinD gene and proliferation of this cell population is not affected in ccndx−/− mutants (Fig. 2P, Q). To test the function of ccnd2a we generated a CRISPR-Cas12a deletion mutant (Supplementary Fig. 5A). ccnd2a−/− larvae are grossly normal, survive to adulthood and are fertile (Supplementary Fig. 5B, C). However, neuromasts fail to grow between 48-72 hpf after they are deposited by the migrating primordium (Fig. 3A–G, and Supplementary Fig. 5D, E). To test if the smaller neuromasts are caused by a decrease in proliferation during development, we treated ccnd2a+/+ and ccnd2a−/− with EdU for 24 hrs from 2-3 dpf. In ccnd2a−/− neuromasts amplifying divisions were significantly reduced compared to sibling neuromasts and no net change in support cells was observed between 3 – 6 dpf (Fig. 3H, I), whereas hair cell differentiation rates were not significantly different from siblings (Fig. 3H). Thus, ccnd2a−/− neuromasts grow only slightly between 3 dpf and 6 dpf, which is attributed to the addition of new hair cells (Fig. 3J, K).
A–F DAPI stained nuclei of neuromasts from sibling (A - C) or ccnd2a−/− (D - F) at 32, 48 or 72 hpf. Scale bar = 10 µm. G Quantification of total cell number from DAPI stained neuromasts in sibling and ccnd2a−/− at 32, 48 or 72 hpf. ns = not significant p = 0.98, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 4 for 32 hpf sibling, n = 5 for 32 hpf ccnd2a−/−, n = 15 for 48 hpf sibling, n = 18 for 48 hpf ccnd2a-/, n = 16 for 72 hpf sibling and i = 16 for 72 hpf ccnd2a−/−. H Comparison of wildtype and ccnd2a−/− amplifying, differentiation or total EdU indexes between 2 − 3 dpf. ccnd2a−/− has reduced amplification and total EdU indexes. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.55, ***p = 0.0008, **p = 0.0016, two-tailed t-test. n = 16 for ccnd2a+/+ and n = 12 for ccnd2a−/−. Comparisons of wildtype and ccnd2a−/− support- (I) hair cell- (J) and total (K) cell numbers. ns = not significant p = 1 for (I) and p = 0.087 (K), ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 16 for 3dpf wildtype, n = 16 for 6dpf wildtype, n = 12 for 3dpf mutant and n = 20 for 6dpf mutant. L Comparison of sibling and ccndx−/− amplifying, differentiation or total EdU indexes between 2 − 3 dpf. Similarly to 5 − 6 dpf, ccndx−/− neuromasts show reduced differentiation and total EdU indexes and no change in the amplification index. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.25, ****p < 0.0001, *p = 0.0467, two-tailed t-test. n = 18 for both. M– O Comparisons of sibling and ccndx−/− support-, hair cell- and total cell numbers. Data are presented as mean values + /− 95% confidence intervals. ns=not significant p = 0.91, ****p < 0.0001, **p = 0.0019, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. P–S 5 dpf neuromasts from ccndx+/- (P, Q) or ccndx−/− (R, S) expressing sqEt4:GFP without or with ccnd2a-P2A-mScarlet-I driven by the ccndx promoter. Scale bar = 10 µm. T Quantification of sqEt4:GFP+ hair cells in ccndx+/- or ccndx−/− neuromasts at 5 dpf without or with ccnd2a-P2A-mScarlet-I. n = 12 for ccndx+/-/ccnd2a-P2A-mScarlet-I- and n = 16 for all other groups. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.11 and p = 1, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test.
However, ccnd2a is only required for amplifying divisions during development but not regeneration, as after injury, amplifying proliferation still occurred in the D-V poles of ccnd2a−/− neuromasts (Supplementary Fig. 5F–K). The number of regenerated hair cells was lower compared to siblings but the EdU indexes were not different (Supplementary Fig. 5L, M), demonstrating that hair cell proliferation rates were not affected in regenerating ccnd2a−/− neuromasts and the reduction in generated hair cells is due to the overall small size of ccnd2a−/− neuromasts. The finding that ccnd2a−/− mutant support cells are proliferating during regeneration suggests that other expressed cyclinDs, such as ccnd1 may functionally compensate during regeneration (Supplementary Fig. 5N–P). Indeed, ccnd1 is upregulated 3-5 hrs after hair cell death in siblings and ccnd2a−/− mutant neuromasts (Supplementary Fig. 5O, P). We did not observe a difference in ccndx expression between sibling and ccnd2a−/− mutants.
In contrast to ccnd2a−/− developing neuromasts, in 2–3 dpf ccndx−/− neuromasts amplifying proliferation is unaffected and support cell numbers are not significantly different from siblings (Fig. 3L, M). Only differentiating divisions are decreased, leading to fewer hair cells (Fig. 3L, N). Since amplifying divisions are unaffected the neuromasts still grow significantly between 3 dpf and 6 dpf, even though fewer hair cells are added than in sibling neuromasts (Fig. 3N, O). These analyses show that ccndx is required for hair cell progenitor proliferation during development, homeostasis and regeneration, whereas ccnd2a is necessary for amplifying stem cell divisions only during development.
As ccndx and ccnd2a control proliferation of distinct cell populations we wondered if they possess different, cell cycle-independent functions in the two cell types. To test the specificity of these genes we expressed a ccnd2a-P2A-mScarlet-I transgene under the control of ccndx regulatory sequences (described in the following paragraph) in ccndx−/− and sibling larvae. ccnd2a expression does not increase hair cell numbers in ccndx+/- neuromasts but rescues hair cell numbers in ccndx−/− neuromasts (Fig. 3P–T). This demonstrates that ccndx does not possess specific functions in the regulation of hair cell formation. The cell type-specific effects of ccndx are therefore a result of its spatially and temporally restricted expression pattern only in the progenitor cell population.
atoh1a is dispensable for ccndx expression in hair cell progenitors
To better define the sequence of signaling events leading to proliferation of hair cell progenitors we aimed to identify possible upstream regulators of ccndx. To identify regulatory sequences that drive ccndx expression, we compared zebrafish and goldfish genomes42,43, which revealed two conserved non-coding candidate regions upstream of ccndx (Fig. 4A). We generated reporter lines driving EGFP or destabilized EGFP via these elements, cloning a 4.5 kb fragment encompassing both conserved regions. By assaying reporter activity, we found that this 4.5 kb region upstream of ccndx is sufficient to drive H2B-GFP expression in the hindbrain, spinal cord, migrating lateral line primordium and 5 dpf neuromasts (Fig. 4B–E).
A UCSC zebrafish genome browser track showing two regions of conservation with the goldfish genome upstream of ccndx exon 1. The black bar indicates the 4.5 kb region cloned. B, C DAPI stained 32 hpf zebrafish larvae showing H2B-EGFP (cyan) expression in the hindbrain, spinal cord and migrating lateral line primordium driven by the ccndx upstream region with a higher magnification image of the primordium (C). Scale bars = 10 µm and 200 µm, respectively. D, E DAPI stained 5 dpf zebrafish showing H2B-EGFP (cyan) expression in the spinal cord and neuromasts driven by the ccndx upstream region with higher magnification view of a neuromast from a different larva (E). Scale bars =10 µm and 200 µm, respectively. ccndx HCR (yellow) in 5 dpf sibling (F) or atoh1a−/− neuromasts (G). sqEt4:EGFP expression (cyan) shows lack of hair cells in atoh1a−/− larvae. ccndx is expressed in atoh1a−/− neuromasts and shows a broader and more central expression pattern. Scale bar = 10 µm. HCR for ccndx (yellow) and atoh1a (magenta) in heat shocked sibling (H) and hs:atoh1a larvae (I). Scale bar = 10 µm. J ccndx HCR (yellow) in zebrafish expressing the tp1bglobin:egfp Notch reporter (cyan) showing the lack of co-localization. Scale bar = 10 µm. HCR for ccndx (yellow) and egfp (magenta) in ccndx:NLS-d2GFP (cyan) transgenic zebrafish treated with DMSO (K) or LY411575 (L) for 6 hrs. LY411575 treatment induces an increase in ccndx and egfp expression, especially in the central region. Scale bar = 10 µm.Maximum projection still images from time lapses of ccndx:NLS-d2EGFP transgenic zebrafish which were treated with DMSO (M) or LY411575 (N) immediately after neomycin treatment. There is an increase in the number of NLS-d2EGFP+ cells around 10 h after neomycin which decreases in DMSO treated fish but continues to increase with Notch inhibition. Scale bar = 10 µm. HCR for ccndx (yellow) in 5 dpf neuromasts in DMSO treated sibling (O) or atoh1a−/− (P) or LY411575 treated sibling (Q) or atoh1a−/− (R). Scale bar = 10 µm.
One possible regulator of ccndx is atoh1a, as it plays a crucial role in hair cell specification, is co-expressed with ccndx in progenitor cells and exhibits highly similar expression dynamics (Fig. 1N–O). An interaction between the two genes would imply a coupling of hair cell specification and proliferation. We therefore asked if atoh1a impacts ccndx expression by examining atoh1a−/− mutant neuromasts. ccndx expression becomes broader in atoh1a−/− compared to siblings, showing a more central expression pattern (Fig. 4F–G). To test if atoh1a potentially inhibits ccndx we analyzed ccndx expression in heat shocked hs:atoh1a and sibling larvae (Fig. 4H–I). ccndx is still strongly expressed after activation of atoh1a, demonstrating that atoh1a does not repress ccndx. Therefore, atoh1a is neither required for ccndx expression nor does it repress it. Thus, hair cell specification and ccndx expression are uncoupled in the lateral line.
Notch signaling represses ccndx expression in hair cell progenitors
As Notch signaling inhibits hair cell progenitor proliferation during hair cell regeneration15,17, we examined whether Notch signaling inhibits ccndx expression. HCR of the Notch reporter line (tp1bglobin:EGFP) showed that ccndx and egfp are expressed in different cells (Fig. 4J), suggesting that Notch signaling may indeed inhibit ccndx expression. To test if Notch represses ccndx we inhibited Notch signaling with the gamma-secretase inhibitor LY411575 in larvae expressing nuclear localized destabilized GFP driven by the ccndx promoter (ccndx:NLS-d2EGFP). After 6 h of Notch inhibition during homeostasis, both endogenous ccndx and egfp mRNA of the reporter were upregulated compared to untreated neuromasts (Fig. 4K–L). In time-lapse recordings of regenerating ccndx:NLS-d2EGFP/myo6b:H2B-mScarlet-I neuromasts we observed a strong increase in GFP fluorescence after Notch inhibition compared to control larvae (Fig. 4M–N and Supplementary Movie 3). This shows that the 4.5 kb regulatory region of ccndx also responds to Notch inhibition during regeneration. In both DMSO and LY411575 treated ccndx:NLS-d2GFP neuromasts, GFP fluorescence intensity was increased 10 hrs after neomycin, likely in response to the downregulation of Notch signaling that occurs after hair cell death12,15,17. In control larvae, GFP expression then gradually decreased (Fig. 4M and Supplementary Movie 3), whereas in LY411575 treated larvae ccndx:NLS-d2GFP fluorescence intensity continued to increase and stayed strong in support cells to the end of the time-lapse (Fig. 4N and Supplementary Movie 3).
As inhibiting Notch signaling is known to increase atoh1a expression15,17, we wondered whether the increased ccndx expression after Notch inhibition might be due to increased atoh1a levels. We treated atoh1a−/− zebrafish with LY411575 for 6 hrs and performed HCR for ccndx. The central ccndx expression in atoh1a−/− larvae is still increased after LY411575 treatment (Fig. 4O–R) demonstrating that atoh1a is not required for the upregulation of ccndx after Notch inhibition. Together, our results show that Notch signaling inhibits ccndx expression in neuromasts.
Notch inhibition leads to ccndx-dependent progenitor proliferation
The downregulation of Notch signaling during hair cell regeneration increases progenitor cell proliferation (Supplementary Fig. 6A, B15,17,18,25,30). To test if Notch signaling inhibits progenitor proliferation or hair cell specification via its repression of ccndx, we treated sibling and ccndx−/− larvae with LY411575 and EdU for 24 hrs following hair cell death. Spatial analyses of EdU+ cells in sibling neuromasts showed a significant increase in EdU+ hair cells after Notch inhibition as previously described (Fig. 5A, C, I15). The increase in differentiating divisions resulted in an increase in hair cells (Fig. 5J). In ccndx−/− neuromasts treated with DMSO, no EdU+ hair cells are observed, and after Notch inhibition only very few EdU+ hair cells form (Fig. 5E, G). This result is verified by a time-lapse analysis of a LY411575 treated ccndx−/− neuromast which shows no proliferation of hair cell progenitors (Supplementary Movie 4). In contrast to the differentiating divisions, the amplifying divisions occur normally in the D-V poles in the absence of Notch signaling in both siblings and mutants (Fig. 5A–D, E, H, L). Together, these results show that the increased progenitor proliferation induced by Notch inhibition in wildtype neuromasts is strictly dependent on ccndx (Supplementary Fig. 6).
A Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in control DMSO treated sibling neuromasts. B One-sided binomial analysis of EdU+ cell positions. n = 21. ****p < 0.00001, ns=not significant, p = 0.17. C Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in LY411575 treated sibling neuromasts. D One-sided binomial analysis of EdU+ cell positions. n = 27. ****p < 0.00001, **p < 0.02. E Spatial analysis of EdU+ support cells and hair cells after 24 hrs of regeneration in control DMSO treated ccndx−/− neuromasts. F One-sided binomial analysis of EdU+ cell positions. There are no EdU+ hair cells in ccndx−/− neuromasts. n = 18. ****p < 0.00001. G Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in LY411475 treated ccndx−/− neuromasts. H One-sided binomial analysis of EdU+ cell positions. There are few EdU+ hair cells in LY411575 treated ccndx−/− neuromasts. n = 21. ****p < 0.00001, **p < 0.006, ns=not significant, p = 0.11. I EdU index of differentiation divisions after DMSO and LY411575 (LY) treatment in sibling and ccndx−/− regenerating neuromasts. There is no significant difference in differentiating EdU indexes between DMSO or LY411575 treated ccndx−/− neuromasts. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 1, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. J Quantification of sqEt4:EGFP+ hair cells in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. LY411575 induces an increase in regenerated hair cells in both siblings and ccndx−/− neuromasts. ns = not significant p = 0.18, ****p < 0.0001, *p = 0.0113, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. K Quantification of support cell numbers in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. LY411575 decreases support cells in both siblings and ccndx−/− neuromasts. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.067 and p = 0.84, *p < 0.024, ***p = 0.0007, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. L EdU index of amplifying cells in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. ns = not significant p = 0.46 and p = 0.78, *p < 0.018, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−.
Progenitor proliferation and hair cell specification are independently controlled by Notch
In addition to regulating progenitor proliferation, Notch signaling inhibits support cell specification and promotes hair cell specification by a process called lateral inhibition15,17,18,21. This raises the question if the increase in hair cells is caused by the increase in proliferation. We observed that, even though hair cell progenitors do not divide in ccndx−/− neuromasts, Notch inhibition results in the regeneration of more sqEt4:GFP + (but EdU-) hair cells compared to sibling neuromasts (Fig. 5J). This increase in hair cells is accompanied by a decrease in support cell numbers in ccndx−/− neuromasts (Fig. 5K).
Together, these results demonstrate that in the absence of ccndx, Notch inhibition still promotes hair cell fate at the expense of support cell fate, and that therefore lateral inhibition occurs independently of ccndx and proliferation.
ccndx −/− progenitors and hair cells differentiate normally but exhibit polarity defects
Hair cell progenitor divisions produce two hair cells of opposing polarity, resulting in neuromasts with 50% of hair cells possessing a kinocilium on one side and 50% with the kinocilium localized to the opposite side24,44. To examine if hair cell polarity was affected in the absence of progenitor cell divisions we labeled hair cells with the F-actin binding peptide phalloidin. Siblings showed 50% of hair cells of either polarity, while in ccndx−/− neuromasts 70% of hair cells were polarized toward the posterior direction (Fig. 6A–C). This posterior bias in hair cell polarity in ccndx−/− neuromasts was rescued by ccndx-driven ccnd2a expression (Supplementary Fig. 7A–E), showing that the effect on hair cell polarity requires a cyclinD gene in the progenitor cells but is not specific to ccndx. ccnd2a−/− mutant hair cells possess a normal hair cell polarity, as loss of ccnd2a does not affect progenitor proliferation (Supplementary Fig. 7F–H).
Phalloidin staining of sibling (A) and ccndx−/− (B) 5 dpf neuromasts. Scale bar = 10 µm. C Quantification of posterior-polarized hair cells per neuromast. Data are presented as mean values + /− 95% confidence intervals. ****p < 0.0001, two-tailed t-test. n = 18 for sibling and n = 15 for ccndx−/−. Anti-Emx2 immunostaining (magenta) in 5 dpf sqEt4:EGFP (cyan) expressing sibling D and ccndx−/− neuromasts (E). Scale bar = 10 µm. F Quantification of the percentage of anti-Emx2+ hair cells per neuromast during homeostasis and at 24, 48 or 72 h after neomycin treatment. Data are presented as mean values + /− 95% confidence intervals. **p = 0.006, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 30, n = 60, n = 55 and n = 58 for sibling at homeostasis, 24, 48 or 72 hpf respectively, n = 25, n = 61, n = 54 and n = 47 for ccndx−/− at homeostasis, 24, 48 or 72 hpf respectively. G–J Spatial analysis of Emx2+ or Emx2- hair cell nuclei cell position in sibling (G-H) and ccndx−/− (I-J) 72 h post neomycin treatment. H, J One-sided binomial analysis of Emx2+ hair cell positions, n = 18 for each group. K UMAP plot of integrated scRNA-seq datasets of 5 dpf sibling and ccndx−/− lateral line cells with neuromast cell types labeled. L Same UMAP plot as in (K) with sibling cells (cyan) or ccndx−/− cells (red) individually labeled showing a reduction in the number of dividing progenitor cells (arrows) in ccndx−/− larvae. M Dot plot showing percentage of hair cells that express a given known hair cell gene comparing sibling (cyan) and ccndx−/− (red) mature hair cell populations. Magnifications of scRNA-seq UMAP and Feature Plots of differentiating progenitor cells and young hair cells. Individual Feature Plots for atoh1a, hes2.2, and hey2 in sibling (N) and ccndx−/− (O). atoh1a is expressed in progenitor cells and young hair cells. hes2.2 is expressed in a subset of progenitor cells and is greatly reduced in ccndx−/− cells. hey2 expression in young hair cells is reduced in ccndx−/−. P Dot plot of mRNA expression levels of a subset of differentially expressed genes comparing sibling and ccndx−/− young hair cell populations. HCR for atoh1a (yellow) and hes2.2 (magenta) in a 5 dpf neuromast after DMSO (Q) or LY411575 (R) treatment showing co-expression within an individual progenitor cell. With cropped views of a single double-positive cell. Scale bar = 10 µm. Cropped image of hes2.2 Feature Plot from scRNA-Seq regenerating time-course from Baek et al.,202212 in progenitor cells (S) before emx2 expression (T) in young hair cells. Scale bar = 10 µm.
Posteriorly polarized hair cells express the transcription factor Emx2 and loss of Emx2 leads to the absence of posterior polarized hair cells. Emx2 is also sufficient to drive posterior polarization in all hair cells if overexpressed25,28,30,45. Antibody staining for Emx2 during homeostasis and at multiple time points after hair cell death showed that 70% of ccndx−/− hair cells express Emx2 compared to 50% in siblings, matching the phalloidin results (Fig. 6D–F). This higher percentage of Emx2+ hair cells persisted in ccndx−/− neuromasts as more hair cells were added during regeneration (Fig. 6F). Spatial analyses of Emx2+ and Emx2- hair cell nuclei revealed that hair cells are still spatially enriched in their respective halves of ccndx−/− neuromasts rather than being randomly distributed (Fig. 6G–J), showing that ccndx−/− hair cells continue to sense and react to the signals that polarize the field of hair cells across the sensory organ.
To determine if the changes in hair cell polarity are reflected by gene expression changes, we performed scRNA-Seq analyses of FAC-sorted neuromast cells from 5 dpf sibling and ccndx−/− larvae (Fig. 6K, L, Supplementary Data 1). Sibling and ccndx−/− hair cells cluster together suggesting that their transcriptomes are similar. We examined the expression levels of candidate hair cell genes, including mechanotransduction related genes, and did not observe major differences between sibling and ccndx−/− hair cells (Fig. 6M, Supplementary Data 2). We conclude that hair cell differentiation is not affected at a detectable manner by the absence of progenitor proliferation.
In contrast to mature hair cells, we observed more pronounced gene expression changes between sibling and ccndx−/− hair cell progenitors and young hair cells (Fig. 6L, N-O, arrowheads). In siblings, atoh1a-expressing, dividing progenitor cells cluster closer with the amplifying cells, likely due to their expression of cell cycle genes (Fig. 6L, N). In contrast, ccndx−/− hair cell progenitors form a continuous trajectory towards young hair cells (Fig. 6L, O). Examining differentially expressed genes between siblings and ccndx−/− larvae within these cell clusters which could be involved in regulating hair cell polarity, we found members of the protocadherin, dscam and plexin gene families (Fig. 6P). These genes are expressed in either progenitor cells or young hair cells and could be involved in regulating hair cell polarity or hair cell rearrangements that coincide with the establishment of hair cell polarity18,46.
As Emx2 is inhibited by Notch signaling25,30, we searched for gene expression differences in potential Notch pathway genes, as well as genes that are expressed immediately prior to emx2 in progenitor cells. In ccndx−/− neuromasts, the Notch pathway members her4.1, her15.2 and hey2 are downregulated, as are hes2.1, hes2.2, which are expressed before emx2 levels increase (Supplementary Data 2). As an example, in ccndx−/−, hes2.2 is downregulated in hair cell progenitors (Fig. 6N, O, arrows) while hey2 is downregulated in young hair cells (Fig. 6N, O, arrowheads). The downregulation of these genes suggests they could be involved in negatively regulating Emx2. HCR for hes2.2 and atoh1a shows that hes2.2 is expressed in some atoh1a+ cells (Fig. 6Q), which is supported by the scRNA-seq regeneration time course that shows that hes2.2 is transiently expressed in hair cell progenitor cells before they turn on emx2 (Fig. 6S, T12). As hes2.2 is not regulated by Notch signaling in neuromasts because LY411575 does not affect hes2.2 expression (Fig. 6R), the mechanism by which hes2.2 expression is regulated by ccndx is Notch-independent.
ccndx regulates hair cell polarity via activating hes2.2 which inhibits emx2
To test if the loss of hes2.2 expression in ccndx−/− progenitor cells causes hair cell polarity defects, we deleted it with CRISPR-Cas12a. As hes2.1 and hes2.2 are tandem duplicates, we generated an approximately 30 kb deletion spanning both genes. We refer to these double mutants as hes2−/−. hes2−/− neuromasts have a similar phenotype as ccndx−/− neuromasts with greater than 60% of hair cells showing a posterior polarity bias compared to 50% of sibling hair cells (Fig. 7A–C). Accordingly, Emx2+ hair cells are also increased in hes2−/− neuromasts (Fig. 7E–G), even though the total number of hair cells is unaffected (Fig. 7D), demonstrating that hes2 specifically affects hair cell polarity. Hence, hes2 is necessary to establish proper hair cell polarity via negative regulation of Emx2 expression.
Phalloidin staining of 5 dpf neuromasts from hes2+/+ (A) and hes2−/− (B) Scale bar = 10 µm. C Quantification of posterior facing hair cells per neuromast. Data are presented as mean values + /− 95% confidence intervals. ****p < 0.0001, two-tailed t-test. n = 37 for hes2+/+ and n = 47 for hes2−/−. D Quantification of hair cell number in 5 dpf neuromasts from hes2+/+ and hes2−/−. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.12, two-tailed t-test. n = 19 for hes2+/+ and n = 37 for hes2−/−. Anti-Emx2 immunostaining (yellow) in myo6b:H2B-mScarlet-I (magenta) hes2+/+ (E) and hes2-/− (F) 5dpf neuromasts. Scale bar = 10 µm. G Quantification of the percentage of Emx2+ hair cells per neuromast at 5 dpf. Data are presented as mean values + /− 95% confidence intervals. ***p = 0.0009, two-tailed t-test. n = 14 for hes2+/+ and n = 20 for hes2−/−. H–K Phalloidin staining of 5 dpf neuromasts in control or ccndx:hes2.2 expressing, ccndx+/+ or ccndx−/−. Scale bar = 10 µm. L Quantification of posterior facing hair cells per neuromast. Data are presented as mean values + /− 95% confidence intervals. ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 20 for ccndx+/+/hes2-, n = 16 for ccndx+/+/hes2+, n = 24 for ccndx−/−/hes2- and n = 20 for ccndx−/−/hes2+. M–P Anti-Emx2 immunostaining (yellow) in myo6b:H2B-scarlet (magenta) expressing ccndx+/+ or ccndx−/− neuromasts with or without ccndx:hes2.2 expression. Q Quantification of Emx2+ hair cells per neuromast in ccndx+/+ or ccndx−/− mutants with or without ccndx:hes2.2 expression. Data are presented as mean values + /− 95% confidence intervals. ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 20 for ccndx+/+/hes2-, n = 16 for ccndx+/+/hes2+, n = 24 for ccndx−/−/hes2- and n = 20 for ccndx−/−/hes2+. R Quantification of hair cell number in in ccndx+/+ or ccndx−/−mutants with or without ccndx:hes2.2 expression. Data are presented as mean values + /− 95% confidence intervals. **p = 0.0024, ***p = 0.001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 20 for ccndx+/+/hes2-, n = 16 for ccndx+/+/hes2+, n = 24 for ccndx−/−/hes2- and n = 20 for ccndx−/−/hes2+.
To test the sufficiency of hes2.2 in determining hair cell polarity, we generated a transgenic line in which the ccndx upstream region drives hes2.2 expression (called ccndx:hes2.2). We injected this transgene into a ccndx+/- outcross to test potential functional interactions. ccndx:hes2.2 induces an anterior polarity bias in both wildtype and ccndx−/− (Fig. 7H–L). In ccndx−/− all hair cells are anteriorly polarized, whereas in siblings the polarity conversion is less severe. We speculate that additional genes dysregulated in ccndx−/− neuromasts, such as hey2 and emx2 may potentiate the phenotype caused by ccndx:hes2.2 activity.
Matching the polarity phenotype, ccndx:hes2.2 expression decreases Emx2 expression in both wildtype and ccndx−/− (Fig. 7M–Q). Additionally, ccndx:hes2.2 induces a small decrease in hair cell number (Fig. 7R). We conclude that loss of ccndx leads to a reduction in hes2 expression, causing an increase in Emx2, which results in posterior polarized hair cells.
Our combined results demonstrate that proliferation of amplifying stem cells and their progeny is independently regulated by different cyclinD genes. In addition, in the absence of proliferation, hair cells still fully regenerate, but hair cell polarity is affected due to the downregulation of hes2 and emx2.
Discussion
Here, through transcriptional profiling and genetic approaches we show that in the zebrafish lateral line, proliferation of hair cell progenitors and amplifying stem cells is regulated by two different cyclinD family members, ccndx and ccnd2a, respectively. Organ homeostasis and regeneration depend on symmetric proliferation of stem- and progenitor cells to regenerate lost differentiated cells while maintaining both pools. A potential significance of individually regulating proliferation of different stem- and progenitor populations is that it enhances robustness of the regeneration process: Even if proliferation in one population is disrupted, the other still proliferates, leading to a certain level of short-term regeneration, which will, however, eventually come to a halt once the stem cells are depleted.
Our work raises the question if in other regenerating organs, such as the epithelia of the intestine, stomach, esophagus, the skin and the hematopoietic system, proliferation of stem cells and their progeny is also regulated by different CyclinD genes. Some evidence exists that CyclinD genes can have temporal or location-specific functions. For example, mouse developmental and adult neurogenesis depends on different CyclinD genes47,48,49,50,51. Region-specific expression of CyclinD genes also occurs along different segments of the mouse intestine52. However, to our knowledge it is unknown if stem cell and progenitor divisions are regulated by different CyclinD genes during the same neurogenesis stage or if distinct CyclinD genes regulate intestinal crypt or villus cells differentially, which would have intriguing therapeutic implications.
Cyclins can also possess proliferation-independent functions. For example, CyclinD also affects developmental genes independently of association with CDK4/6, e.g. by recruiting histone acetyltransferases or deacetylases41,53,54,55,56. In the mouse retina, CyclinD1 positively regulates Notch1 expression through recruitment of a histone acetyltransferase, thereby affecting Notch signaling57. This raises the question if ccndx and ccnd2a regulate distinct processes in addition to activating proliferation. Here we show that ccnd2a can substitute for ccndx and rescues both the ccndx mutant hair cell number and polarity defects. Therefore, ccndx does not possess any specific functions in hair cell production or polarity that ccnd2a does not possess.
Interestingly, ccndx is present only in anamniotes and has been lost during amniote evolution35,36. ccnd1 and ccnd2 genes and several fgf ligands have maintained synteny during evolution58, which can be observed comparing human and zebrafish ccnd1 and ccnd2 genes (Supplementary Fig. 8A, B). It appears that ccndx evolved from a duplication of this ancestral ccnd-fgf locus, and that amniotes lost ccndx and retained the other syntenic genes (Supplementary Fig. 8A–C). Protein structure prediction shows structural conservation between zebrafish Ccndx, Ccnd1 and Ccnd2a (Supplementary Fig. 8D). We postulate that ccndx was lost in amniotes as other cyclinD genes took over its role. This hypothesis is supported by our finding that ccnd2a rescues ccndx−/− mutant phenotypes.
The expression of cell fate specification genes is often associated with cell cycle regulatory mechanisms, as the cell cycle machinery can affect the epigenome, chromosome architecture and transcriptional programs required for cell fate. CyclinD genes and Cyclin-dependent kinases (CDKs) have been implicated in cell fate decisions in several cell types, and relevant to our study, CCND1/CDK phosphorylates and stabilizes Atoh1 in the mouse cerebellum59. Here we show that, even though atoh1a and ccndx possess highly similar expression dynamics, and contrary to prior dogma, lateral line hair cell differentiation does not require proliferation, even though fewer hair cells regenerate. This finding mirrors what has been described in intestinal organoids where specification occurs prior to proliferation. However, proliferation rates control the abundance of differentiated intestinal cell types after specification. For example, different lineages divide at different rates leading to the generation of substantially more absorptive than secretory cells60. This raises the possibility that in the intestine proliferation is also regulated by different cell cycle genes in various cell lineages as we described for the lateral line system.
In contrast to hair cell differentiation, we show that the establishment of normal hair cell polarity does require progenitor proliferation in the lateral line. The prevailing model for establishment of neuromast hair cell polarity is that progenitor cell division generates two initially equivalent daughter cells, which then initiate Notch signaling between themselves. The posterior-facing hair cells express Emx2, and if overexpressed, Emx2 is sufficient to reverse the polarity of normally anterior-facing hair cells28. It is unknown what induces Emx2, but it is inhibited by Notch signaling25,30. Stochastically, one daughter cell will have less Notch activity, become Emx2+ and posteriorly polarized25,28,30,45. We found that ccndx−/− neuromasts possess more Emx2+ hair cells, and our scRNA-seq analysis revealed that hes2.2 and hey261,62,63,64, among other genes, are downregulated in progenitor cells and young hair cells, respectively (Fig. 6). These findings suggest that these genes could be involved in negatively regulating Emx2 expression, possibly downstream of Notch activation. However, in the mouse ear, Hey2 is not dependent on Notch signaling65,66. Additionally, as in mouse cell lines or the frog retina65,66,67,68, we determined that hes2.2 is not a Notch target in neuromasts. hes2.2 is therefore inhibiting Emx2 expression via a Notch-independent mechanism.
Our hes2 mutation and overexpression experiments strongly suggest that hes2 is acting downstream of ccndx to inhibit Emx2 expression. However, ccndx is expressed earlier and much more broadly than hes2.2, making direct regulation unlikely. On the other hand, hes.2.2 is expressed before progenitor cells undergo cytokinesis but at the same time as mitotic genes such as cdk1. Thus, hes2.2 expression appears linked to the cell cycle, but cell cycle events by themselves are insufficient to trigger hes2.2, as it is not induced in amplifying cells. As hes2 is restricted to the hair cell lineage, it is likely regulated by hair cell progenitor specific genes that still need to be identified. hes2 is also expressed at low levels in the zebrafish and mouse ear69,70,71,72, and it will be interesting to test if Hes2 plays an equally crucial role in setting up hair cell polarity in the inner ear of fishes and other species.
Notch signaling plays multiple functions in the lateral line (Supplementary Fig. 6). Firstly, it controls hair cell versus support cell specification via the process of lateral inhibition15,17,18,21. Secondly, during regeneration, downregulation of Notch signaling also leads to increased differentiation divisions15,17, which we demonstrate here, are entirely dependent on ccndx. The two roles of Notch in cell specification and proliferation are at least partially independent as hair cell differentiation does not depend on ccndx or proliferation (Supplementary Fig. 6). Lastly, Notch signaling affects hair cell polarity by inhibiting Emx2 expression, even though the precise mechanism is not well understood25,30. It is possible that cell proliferation and hair cell polarity are linked, either because loss of progenitor cell division causes defects in the asymmetric distribution of Notch pathway components into the two daughter cells or because ccndx directly regulates Notch pathway genes, as has been described for Ccnd1 in the mouse retina57.
In summary, our results demonstrate that proliferation of zebrafish lateral line stem and progenitor cells is independently regulated, with potentially important implications for our understanding of how, in other organs and species, the proliferation of quiescent and activated stem cells and their daughter cells can be manipulated when diseased. We also show that loss of hair cell progenitor proliferation leads to hair cell polarity defects via loss of hes2, which so far has not been implicated in hair cell polarity.
Methods
Zebrafish strains and husbandry
All zebrafish experiments were performed according to the guidelines established by the Stowers Institute for Medical Research IACUC review board.
Zebrafish mutants
atoh1apsi69 12 Primers Fw-GAGCAGAGCGAGTACCCACC and Rv-AGTTTCAGTTCCGACAGCTCG are used for genotyping followed by Sanger sequencing with the forward primer. ccndxpsi76 was generated via CRISPR-Cas9-induced mutagenesis using two single guide RNA (sgRNA) sequences AGCTGTTGTTGTGTCCCTCTGGG and CGGGACAGAGGGTCATTCAGAGG (PAM sites in bold) to generate a deletion from the upstream promoter region through exon one. sgRNA sequences were found using CRISPRscan73. sgRNA guides, without PAM sequences, and Cas9 tracrRNA were ordered from IDT. Guides were diluted to 100 µM in Nuclease-free duplex buffer (IDT). To generate duplexes 1.5 µL of each sgRNA and 3 µL tracrRNA (100 µM) were brought up to 50 µl with dH2O, heated to 95 °C for 5 min and left at room temperature for 20 min and stored at −20 °C until use. For the injection mix 5 µL of duplexed sgRNAs, 1 µl Cas9 protein (1 µg/uL, PNA BIO, CP01-50), 2 µL 0.5% phenol red and 2 µL dH2O were incubated at room temperature for 10 min, then kept on ice until injection. 1-3nL was injected into the cell of one-cell stage zebrafish embryos. Transient mutations were verified at 48 hpf to prove that the sgRNAs worked and remaining embryos were raised to adulthood and screened for germline mutations. For genotyping, primers used were Fw-ATGGCTTTCAGATTGCAACA and Rv-CATTGCTCCAGGTTTTGGTT for the deletion mutation and Rv-CGCACCACAGAGACACAGAC with the same forward primer above for the wildtype PCR.
ccnd2apsi77 was generated via CRISPR-Cas12a induced mutagenesis using single guide RNA sequences TTTAAAATGACAACACAAACTCAC and TTTAGCGGTGGTACCAACAAGAAA to generate a deletion from the upstream promoter region through part of exon two, which generated an approximately 2045b base pair deletion (PAM sites in bold). To find the guide sites, genomic regions were first screened via DeepCpf174 to find scores above 10, then potential guides were BLASTed via the UCSC Genome browser to look for potential off target regions. Cas12a and guide RNA injections were performed based on previously published methods with modifications75. Cas12a guides without PAM sequences (IDT) were diluted to 24 µM in dH2O. 100 uM Lba-Cas12a protein was ordered from New England Biolabs (M0653T). The injection mix consisted of 12 µM guide mix, 20 µM Lba-Cas12a, 0.3 M KCl, 0.05% phenol red and brought up to 5 µL with dH2O then kept for 10 min at 37 °C. 1-3nL of this mix were injected into the cell of one-cell stage zebrafish embryos. Transient mutations were verified at 48 hpf and remaining embryos were raised to adulthood and screened for germline mutations. For genotyping, primers used were Fw-TGACACCAAGAGCATGGGTA and Rv-TGGACCCTTAAAAGCAGTAGGA for the deletion mutation and Rv-CGCATAAAGGGCTGAATGTC with the same forward primer above for the wildtype PCR.
Df(Chr8:hes2.2,hes2.1)psi89 chromosomal deletion was generated via CRISPR-Cas12a-induced mutagenesis using single guide RNA sequences TTTGGGATGAGCTATGTATATTAT and TTTGGTTGAAGCGATCATCAATCAA (PAM sites in bold) located in the 3’ untranslated region of hes2.2 and just upstream of hes2.1, respectively, which generated an approximately 30 kb base pair deletion. sgRNA design and injections were performed as above for ccnd2a. To genotype the mutant allele primers Fw-CGTGGCTTGGTTAATTATTGCG and Rv-AAAATTATTGGCCCCTTTAAGC are used. Primers Fw- AAAGCGCTCATTCTCCCTCT and Rv-CATCATTCTGGAGCTCTGCG are used to genotype the wildtype allele.
Zebrafish transgenics
Previously published zebrafish transgenic lines used
Tg(myo6b:hist2h2l-mScarletI)psi66Tg 76, referred to as myo6b:H2B-mScarlet-I in the text. Tg(Hsp70l:atoh1a)x20TG 77, Tg(she:H2A-mcherry)psi57Tg 76, Tg(tp1bglobin:EGFP)um14Tg 78, Et(krt:EGFP)sqet4ET 79 and Et(krt:EGFP)sqet20ET 79.
Zebrafish transgenic lines generated
Tg(−4.5ccndx:hist2h2l-EGFP)psi78Tg (referred to as ccndx:H2B-GFP in the text) transgenic was generated as follows: a fragment 4.5 kb upstream of ccndx was PCR’d from genomic DNA using primers aactcgagTGGAGGGTTTCTTGAACCTTT (forward) and aaggatccGTCCTGTTGCACGTGTGTCT (reverse) containing XhoI or BamHI sites respectively for cloning into the p5’-entry vector (p5E) from the tol2 kit80. This includes part of the 5’UTR of ccndx. PCR was performed with Phusion High-Fidelity DNA Polymerase (New England Biolabs, E0553) using zebrafish genomic DNA. Primers were ordered from IDT. The p5E-4.5ccndx plasmid was sequence verified via Oxford Nanopore sequencing by Plasmidsaurus. The p5E-4.5ccndx plasmid was used in a standard LR Gateway reaction (Thermo Fisher Scientific, 11791020)using pMiddle entry H2B-EGFP, p3’entry-polyA80 and pToneDest tol1 destination vector (Addgene plasmid #6769181). Tol1 mRNA was generated using the T7 mMessage kit (Thermo Fisher Scientific, AM1344) using linearized pToneTP as the template (Addgene plasmid #6769281). To generate transgenics 1–3 nl of an injection mix consisting of 12.5 ng/µl DNA, 20 ng/µl tol1 mRNA, 0.3 M KCl and 0.05% phenol red, was injected into the cell of a one-cell stage zebrafish embryos from a wildtype Tu in-cross. GFP+ larvae were screened at 48 hpf and raised to adulthood then founders screened for germline transmission of GFP.
For transgenic Tg(−4.5ccndx:NLS-d2EGFP) psi81Tg, the sv40-NLS-d2GFP sequence from the pNSEN-d2 plasmid (Addgene plasmid #5976382) was first cloned into the pMiddle-Entry plasmid. This was then used in a LR Gateway reaction using p5E-4.5ccndx, p3E-polyA and pToneDest. Injection and screening of founders was as described above.
For transgenic Tg(−4.5ccndx:ccnd2a-P2A-mScarlet-I)psi83Tg a ccnd2a expression construct gBlock was ordered (IDT) consisting of zebrafish ccnd2a followed by a P2A-mScarletI sequence. This gBlock was first cloned into the pCR4-topo cloning plasmid (Thermo Fisher Scientific) and sequenced verified. This plasmid was then digested with EcoRI and the subsequent ccnd2a-P2A-mScarlet-I fragment was cloned into the EcoRI site of the pME and sequenced verified. A LR Gateway reaction was performed with p5E-4.5ccndx, pME-ccnd2a-P2A-mScarlet-I, p3E-polyA and the pToneDest Tol1 destination plasmid. 1–3 nl of the injection mix, consisting of 20 ng/µl DNA, 16 ng/µl tol1 mRNA, 0.3 M KCl and 0.05% phenol red, was injected into the cell of a one-cell stage zebrafish embryos from an in cross of ccndxpsi76/sqEt4:EGFP/sqET20:EGFP. mScarlet-I+ larvae were collected at 48 hpf and raised to adulthood. Founders were initially genotyped for presence of the ccndxpsi76 allele, then screened for germline transmission of mScarlet-I fluorescence. Multiple founders were identified, and founders that had a moderate level of ccnd2a-P2A-mScarlet-I expression were characterized.
To generate Tg(−4.5ccndx:v5-hes2.2/cryaa:GFP)psi90Tg, hes2.2 was PCR amplified from zebrafish cDNA using primers to add a V5-tag and a small linker to the N-terminal region and KpnI/NotI cloning sites using primers Fw-aaggtaccATGGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACG GGCTCCGGAATGACTCCCAGCGCCACCGCTG and RV-aagcggccgctCTACCAAGGTCTCCAGACTTC. The PCR product was then cloned into the pMiddle-Entry plasmid and sequenced verified. This Middle-entry plasmid was then used in a LR Gateway reaction using p5E-4.5ccndx, p3E-polyA and pDestTol2pACryGFP (Addgene plasmid #6402283) to allow for screening transgenics by lens GFP expression. Injection and screening of founders was as described above, except injections were into embryos from an out cross of ccndxpsi76/Tg(myo6b:hist2h2l-mScarletI)psi66Tg.
Hair cell ablation and proliferation analysis
To ablate hair cells 5 days post fertilization (dpf) zebrafish were treated with 300 mM neomycin (Sigma-Aldrich, N6386) for 30 min. Neomycin was then extensively washed out and fish were allowed to recover for varying time points depending on the experiment.
To label dividing cells EdU (Carbosynth, 61135-33-9) was added at 3.3 mM with 1% DMSO in 0.5X E2 fish water. Larvae were added to EdU media immediately after neomycin treatment for 24-48 h, depending on the experiment, then fixed in 4% paraformaldehyde overnight at 4 °C. Staining was carried out as described previously11. To visualize GFP in GFP-expressing fish rabbit anti-GFP (1/400, Thermo Fisher Scientific, A11122) immunolabeling was performed. For fish expressing myo6b:H2B-scarlet no antibody staining was needed. Alexa Fluor 594-Azide or Alexa Fluor 647-Azide (Thermos Fisher Scientific, A10270 or A10277) were used during the Click-it reaction. Cell counts, EdU indexes and spatial positioning was performed as described15,37. EdU indexes are calculated by dividing EdU+ cells by the total cell number. Support cells are considered any cell other than a hair cell. Angular positions are represented using rose diagrams generated in R. The enrichment of EdU+ cell types in any quarter between 45°, 135°, 225° and 315° was tested using one-tailed binomial analyses.
Heat shock paradigm
To induce heat shock induction of atoh1a Tg(Hsp70l:atoh1a) transgenic zebrafish were raised to 5 dpf then put at 37 °C for 1 h, followed by 1 hr at 28 °C, then 1 hr at 39 °C, then 1 hr at 28 °C, then 1 hr at 39 °C followed by 1 hr at 28 °C then fixation with 4% paraformaldehyde at 4 °C.
Hybridization chain reaction (HCR) in situ hybridization
HCR probes and amplifiers were purchased from Molecular Instruments. Zebrafish were fixed at least two days in 4% paraformaldehyde at 4 °C, dehydrated in a graded series of methanol, then stored in 100% methanol at −20 °C until staining. HCR was performed according to the manufacture’s protocol (Molecular Instruments) with minor changes as previously described84. Zebrafish were permeabilized with 100% acetone at −20 °C for 20 min for 5-6 dpf zebrafish or 10 min for 32 hpf zebrafish. HCR Probes used were ccndx-B1, ccndx-B4, atoh1a-B2, atoh1a-B5, pcna-B4, ccnd2a-B2, ccnd1-B4, d2EGFP-B2, and hes2.2-B5. For ccndx and atoh1a double HCR only the ccndx-B4 and atoh1a-B5 combination worked. Amplifiers used were conjugated with either Alexa-647, Alexa-546 or Alexa-488.
Phalloidin staining and Immunohistochemistry
Phalloidin staining was performed as described with either Alexa Fluor 647, Alexa Fluor 568 or Alexa Fluor 488 phalloidin (Thermo Fisher Scientific, A22287, A12380, A12379)85. For anti-acetylated tubulin staining zebrafish were fixed in 2% paraformaldehyde/1% Trichloroacetic acid for 10 min at room temperature, then washed in PBS/0.8% Tween-20 (PBSTw) followed by 5 min wash in dH2O, 7 min in acetone at −20 °C, then washed again in PBSTw. Larvae were then blocked at least 1 h in 10% normal goat serum (GEMINI BIO-PRODUCTS, #100-109) in PBSTw. Anti-acetylated tubulin (Sigma-Aldrich, T6793) was diluted to 1/1000 in blocking buffer and incubated overnight at 4 °C. Larvae were then washed in PBSTw and incubated in secondary antibody, goat anti-mouse Alexa-568 (Thermo Fisher Scientific, A11004) 1/1000 in blocking buffer, overnight at 4 °C. The samples were then extensively washed.
Immunohistochemistry against Emx2 (TransGenic, K0609) was performed as described in ref. 29. Secondary antibody goat anti-mouse Alexa Fluor 568 (Thermo Fisher Scientific, A11004) was used at 1/1000. In some cases, this was done simultaneously with rabbit anti-GFP (Thermo Fisher Scientific, A11122) at 1/400 dilution overnight. DAPI (1/1000, Thermo Fisher Scientific, D1306) was used to visualize nuclei, followed by Alexa Fluor 647 phalloidin staining at 1/40 dilution (Thermo Fisher Scientific, A22287).
Protein analysis
Ccnd protein structure comparison was performed with Robetta protein structure prediction (https://robetta.bakerlab.org/)86, using zebrafish Ccnd1, Ccnd2a and Ccndx amino acid sequences downloaded from Ensembl43. The protein structures were visualized and superimposed using Mol* 3D Viewer, https://www.rcsb.org/3d-view87.
Imaging
Images were taken on either a Zeiss LSM 780 confocal microscope or Nikon Ti2 Eclipse equipped with a CSU-W1 Yokogawa spinning disk. Post-processing of images was performed with ImageJ (Fiji)88.
Time-lapse imaging
Hair cells were first killed with neomycin as described above before mounting. Zebrafish anesthesia and mounting in low-melt agarose were performed as described37. Time-lapse imaging was performed with a Nikon Ti2 Eclipse with a Yokogawa CSU-W1 spinning disk head with a Hamamatsu Flash 4.0 sCMOS with a temperature-controlled stage set to 28.5 °C. During each time-lapse experiment the L2 and L3 neuromasts from two fish were imaged. Z-stacks were taken every 5 min for up to 42 h. For quantification of myo6b:H2B-mScarlet-I hair cells the time when fluorescence signal could first be observed was recorded.
Drug treatments
The gamma-secretase inhibitor LY411575 (Selleckchem, S2714), dissolved in DMSO, was diluted to a final concentration of 50µM. Aphidicolin (Sigma-Aldrich, A0781), dissolved in DMSO, was used at 100µM for EdU analysis and at 60µM for time lapse experiments. DMSO only was used as a negative control for both experiments.
Statistical analysis
All statistics and graphs were generated with GraphPad Prism 9 (version 9.4.1). For comparing two groups unpaired t-test was used. For experiments with multiple groups a Two-way ANOVA with Šidák multiple comparisons test was used to test significance. A p-value smaller than 0.05 was considered significant.
Single cell RNA-sequencing
Embryo dissociation and FACS
scRNA-seq was performed in duplicate using 5 dpf homeostatic ccndx sibling and ccndx−/− fish. Siblings were collected from a cross between ccndxpsi76/Tg(she:H2A-mcherry)psi57Tg and Tg(she:H2A-mcherry)psi57Tg zebrafish and sorted for mCherry expression at 4 dpf. Mutant larvae were collected from an incross of ccndxpsi76/Tg(she:H2A-mcherry)psi57Tg and sorted for mCherry+ fish lacking an inflated swim bladder at 4 dpf. These larvae were then allowed to recover until 5 dpf, then processed for FACS. Approximately 400–500 larvae were collected for each replicate.
Embryo dissociation and FACS were performed similarly to previously published protocols with modifications12. 5 dpf larvae were anesthetized with MS-222, then split between two Falcon 70 µm cell strainers (Corning, 352350) and placed in 0.25% Trypsin-EDTA (Thermo Fisher Scientific, 25200056) on ice. The larvae from each strainer were then added to a 5 ml round bottom tube (Corning, 352063) and the tissue was broken up by slowly pipetting up and down using a pipette man and a glass Pasteur pipette, keeping the tubes on ice. The total time between placing larvae in trypsin to finishing pipetting was approximately 8 min. The dissociated tissue was then filtered through a 70 µm Filcon filter (BD Biosciences, 340634), pelleted for 5 min at 2000rpm at 4 °C and the pellet was resuspended in 1xDPBS (Thermo Fisher Scientific,14190)/0.04%BSA (Sigma-Aldrich, A1595). The pelleting and resuspension in 1xDPBS/0.04%BSA were repeated two more times. After the final wash, the pellet was resuspended in 0.5 mL 1xDPBS/0.04%BSA. The cells were combined and filtered through another 70 µm Filcon filter into a 5 ml round bottom tube. Cells were then incubated in Draq5 (1/200 dilution, Thermo Fisher Scientific, 65-0880-96) for 5 min on ice. Draq5+/mCherry+ cells were sorted by the Stowers Institute Cytometry Core Facility using a BD Influx Cell Sorter (BD Biosciences, San Jose, CA. USA), with cells collected in tubes containing 10µL 1XDPBS/0.04% BSA. Sorted cells were then spun down at 350rcf at 4 °C for 2 min in a swinging bucket centrifuge with slow deceleration. Most of the supernatant was removed and the cells were resuspended in approximately remaining 100 µL.
10X Chromium scRNAseq library construction
The cells were then given to the Sequencing and Discovery Genomics Department (Stowers Institute for Medical Research) for sequencing. Dissociated, sorted cells were assessed for concentration and viability using a Luna-FL cell counter (Logos Biosystems). Cells deemed to be at least 80% viable were loaded based on live cell concentration onto a Chromium X Series instrument (10x Genomics) and libraries prepared using the Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 (10x Genomics) according to manufacturer’s directions. Resulting cDNA and short fragment libraries were checked for quality and quantity using a 2100 Bioanalyzer (Agilent Technologies) and Qubit Fluorometer (Thermo Fisher Scientific). With cells captured estimated at ~6800–9600 cells per sample, libraries were pooled and sequenced to a depth necessary to achieve at least 30,000 mean reads per cell on an Illumina NextSeq 2000 instrument utilizing RTA and instrument software versions current at the time of processing with the following paired read lengths: 28 × 10 × 10 × 90 bp.
Single cell RNA-seq data analysis
Raw reads were processed using 10X Genomics software Cellranger v7.0. Reads were demultiplexed into Fastq file format using cellranger mkfastq with default parameters. Genome index was built under zebrafish danRer11, Ensembl version 102 and used cellranger count function for cell gene count matrix generation.
Feature count matrix generated by cellranger were then loaded into R package Seurat (v4.3). Here we have two replicates for both sibling and mutant populations. We first processed each sample individually for quality control. The cells with less than 500 UMI counts or more than 10% of mitochondrial expression were filtered out. Then top 2000 variable genes were selected to run PCA analysis for nearest neighbor graph and clustering (PC = 40, res = 0.8), as well as the UMAP visualization. Then all replicates and samples were merged based on Seurat’s CCA scRNA-seq integration method for all downstream analysis. Finally, Seurat’s FindAllMarkers function (ROC test) was used to generate marker genes for every cluster and annotated based on ZFIN database knowledge.
Cell State analyses
We applied two RNA Velocity based pseudo time analyses, scVelo and partition-based graph abstraction (PAGA), to infer the single cell states. First, we ran the Velocyto pipeline to obtain unspliced/spliced mRNA reads for each single cell. Then, we applied the generalized dynamical model to solve the full transcriptional dynamics using the Python package scVelo. Finally, the PAGA graph was generated based on the latent time calculated by scVelo.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw data from images used in this manuscript can be accessed from the Stowers Institute for Medical Research Original Data Repository at http://www.stowers.org/research/publications/libpb-2516. The scRNA-seq data generated in this study, including sorted BAM files and count matrices produced by CellRanger, has been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE268255. Source data are provided with this paper.
References
Beumer, J. & Clevers, H. Hallmarks of stemness in mammalian tissues. Cell Stem Cell 31, 7–24 (2024).
Doupe, D. P., Klein, A. M., Simons, B. D. & Jones, P. H. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell 18, 317–323 (2010).
Doupe, D. P. et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337, 1091–1093 (2012).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330, 822–825 (2010).
Leushacke, M., Ng, A., Galle, J., Loeffler, M. & Barker, N. Lgr5(+) gastric stem cells divide symmetrically to effect epithelial homeostasis in the pylorus. Cell Rep. 5, 349–356 (2013).
Fullgrabe, A. et al. Dynamics of Lgr6(+) Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Rep. 5, 843–855 (2015).
Rompolas, P. et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016).
Kulukian, A. & Fuchs, E. Spindle orientation and epidermal morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130016 (2013).
Lush, M. E. & Piotrowski, T. Sensory hair cell regeneration in the zebrafish lateral line. Dev. Dyn. 243, 1187–1202 (2014).
Lush, M. E. et al. scRNA-Seq reveals distinct stem cell populations that drive hair cell regeneration after loss of Fgf and Notch signaling. eLife 8, e44431 (2019).
Baek, S. et al. Single-cell transcriptome analysis reveals three sequential phases of gene expression during zebrafish sensory hair cell regeneration. Dev. Cell 57, 799–819.e796 (2022).
Ghysen, A. & Dambly-Chaudiere, C. The lateral line microcosmos. Genes Dev. 21, 2118–2130 (2007).
Kindt, K. S., Finch, G. & Nicolson, T. Kinocilia mediate mechanosensitivity in developing zebrafish hair cells. Dev. Cell 23, 329–341 (2012).
Romero-Carvajal, A. et al. Regeneration of Sensory Hair Cells Requires Localized Interactions between the Notch and Wnt Pathways. Dev. Cell 34, 267–282 (2015).
Thomas, E. D. & Raible, D. W. Distinct progenitor populations mediate regeneration in the zebrafish lateral line. eLife 8, e43736 (2019).
Ma, E. Y., Rubel, E. W. & Raible, D. W. Notch signaling regulates the extent of hair cell regeneration in the zebrafish lateral line. J. Neurosci. 28, 2261–2273 (2008).
Wibowo, I., Pinto-Teixeira, F., Satou, C., Higashijima, S. & Lopez-Schier, H. Compartmentalized Notch signaling sustains epithelial mirror symmetry. Development 138, 1143–1152 (2011).
Ge, Y. et al. Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell 169, 636–650.e614 (2017).
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466–471 (2012).
Brown, R. & Groves, A. K. Hear, Hear for Notch: Control of Cell Fates in the Inner Ear by Notch Signaling. Biomolecules 10, 370 (2020).
Neves, J., Abello, G., Petrovic, J. & Giraldez, F. Patterning and cell fate in the inner ear: a case for Notch in the chicken embryo. Dev. Growth Differ. 55, 96–112 (2013).
Mackenzie, S. M. & Raible, D. W. Proliferative regeneration of zebrafish lateral line hair cells after different ototoxic insults. PLoS One 7, e47257 (2012).
Lopez-Schier, H. & Hudspeth, A. J. A two-step mechanism underlies the planar polarization of regenerating sensory hair cells. Proc. Natl. Acad. Sci. USA 103, 18615–18620 (2006).
Jacobo, A., Dasgupta, A., Erzberger, A., Siletti, K. & Hudspeth, A. J. Notch-Mediated Determination of Hair-Bundle Polarity in Mechanosensory Hair Cells of the Zebrafish Lateral Line. Curr. Biol. 29, 3579–3587.e3577 (2019).
Ji, Y. R. et al. Function of bidirectional sensitivity in the otolith organs established by transcription factor Emx2. Nat. Commun. 13, 6330 (2022).
Jia S. et al. The dark kinase STK32A regulates hair cell planar polarity opposite of EMX2 in the developing mouse inner ear. eLife 12, eLife.84910 (2023).
Jiang, T., Kindt, K. & Wu, D. K. Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife 6, e23661 (2017).
Kindt, K. S. et al. EMX2-GPR156-Galphai reverses hair cell orientation in mechanosensory epithelia. Nat. Commun. 12, 2861 (2021).
Kozak, E. L. et al. Epithelial Planar Bipolarity Emerges from Notch-Mediated Asymmetric Inhibition of Emx2. Curr. Biol. 30, 1142–1151.e1146 (2020).
Malumbres, M. & Barbacid, M. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009).
Malumbres, M. Cyclin-dependent kinases. Genome Biol. 15, 122 (2014).
Harris, J. A. et al. Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio). J. Assoc. Res Otolaryngol. 4, 219–234 (2003).
Thomas, E. D., Cruz, I. A., Hailey, D. W. & Raible, D. W. There and back again: development and regeneration of the zebrafish lateral line system. Wiley Interdiscip. Rev. Dev. Biol. 4, 1–16 (2015).
Lien, H. W. et al. Zebrafish cyclin Dx is required for development of motor neuron progenitors, and its expression is regulated by hypoxia-inducible factor 2alpha. Sci. Rep. 6, 28297 (2016).
Chen, J. A., Chu, S. T. & Amaya, E. Maintenance of motor neuron progenitors in Xenopus requires a novel localized cyclin. EMBO Rep. 8, 287–292 (2007).
Venero Galanternik, M., Navajas Acedo, J., Romero-Carvajal, A. & Piotrowski, T. Imaging collective cell migration and hair cell regeneration in the sensory lateral line. Methods Cell Biol. 134, 211–256 (2016).
Liu, L., Michowski, W., Kolodziejczyk, A. & Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 21, 1060–1067 (2019).
Muhr, J. & Hagey, D. W. The cell cycle and differentiation as integrated processes: Cyclins and CDKs reciprocally regulate Sox and Notch to balance stem cell maintenance. Bioessays 43, e2000285 (2021).
Soufi, A. & Dalton, S. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143, 4301–4311 (2016).
Hydbring, P., Malumbres, M. & Sicinski, P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat. Rev. Mol. Cell Biol. 17, 280–292 (2016).
Chen, Z. et al. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 5, eaav0547 (2019).
Martin, F. J. et al. Ensembl 2023. Nucleic Acids Res. 51, D933–D941 (2023).
Lopez-Schier, H., Starr, C. J., Kappler, J. A., Kollmar, R. & Hudspeth, A. J. Directional cell migration establishes the axes of planar polarity in the posterior lateral-line organ of the zebrafish. Dev. Cell 7, 401–412 (2004).
Ohta, S., Ji, Y. R., Martin, D. & Wu, D. K. Emx2 regulates hair cell rearrangement but not positional identity within neuromasts. eLife 9, e60432 (2020).
Mirkovic, I., Pylawka, S. & Hudspeth, A. J. Rearrangements between differentiating hair cells coordinate planar polarity and the establishment of mirror symmetry in lateral-line neuromasts. Biol. Open 1, 498–505 (2012).
Obernier, K. et al. Adult Neurogenesis Is Sustained by Symmetric Self-Renewal and Differentiation. Cell Stem Cell 22, 221–234.e228 (2018).
Kowalczyk, A. et al. The critical role of cyclin D2 in adult neurogenesis. J. Cell Biol. 167, 209–213 (2004).
Ansorg, A., Witte, O. W. & Urbach, A. Age-dependent kinetics of dentate gyrus neurogenesis in the absence of cyclin D2. BMC Neurosci. 13, 46 (2012).
Obernier, K. & Alvarez-Buylla, A. Neural stem cells: origin, heterogeneity and regulation in the adult mammalian brain. Development 146, dev156059 (2019).
Pastor-Alonso, O. et al. Generation of adult hippocampal neural stem cells occurs in the early postnatal dentate gyrus and depends on cyclin D2. EMBO J. 43, 317–338 (2024).
Yang, R., Bie, W., Haegebarth, A. & Tyner, A. L. Differential regulation of D-type cyclins in the mouse intestine. Cell Cycle 5, 180–183 (2006).
McMahon, C., Suthiphongchai, T., DiRenzo, J. & Ewen, M. E. P/CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc. Natl. Acad. Sci. USA 96, 5382–5387 (1999).
Fu, M. et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J. Biol. Chem. 280, 29728–29742 (2005).
Ratineau, C., Petry, M. W., Mutoh, H. & Leiter, A. B. Cyclin D1 represses the basic helix-loop-helix transcription factor, BETA2/NeuroD. J. Biol. Chem. 277, 8847–8853 (2002).
Lin, H. M., Zhao, L. & Cheng, S. Y. Cyclin D1 Is a Ligand-independent Co-repressor for Thyroid Hormone Receptors. J. Biol. Chem. 277, 28733–28741 (2002).
Bienvenu, F. et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 463, 374–378 (2010).
Itoh, N. & Ornitz, D. M. Evolution of the Fgf and Fgfr gene families. Trends Genet. 20, 563–569 (2004).
Miyashita, S. et al. Cyclin D1 controls development of cerebellar granule cell progenitors through phosphorylation and stabilization of ATOH1. EMBO J. 42, e113613 (2023).
Zheng, X. et al. Organoid cell fate dynamics in space and time. Sci. Adv. 9, eadd6480 (2023).
Kageyama, R., Ohtsuka, T. & Kobayashi, T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 (2007).
Iso, T. et al. HERP, a new primary target of Notch regulated by ligand binding. Mol. Cell Biol. 21, 6071–6079 (2001).
Iso, T., Kedes, L. & Hamamori, Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194, 237–255 (2003).
Nakagawa, O. et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. USA 97, 13655–13660 (2000).
Benito-Gonzalez, A. & Doetzlhofer, A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J. Neurosci. 34, 12865–12876 (2014).
Basch, M. L., Ohyama, T., Segil, N. & Groves, A. K. Canonical Notch signaling is not necessary for prosensory induction in the mouse cochlea: insights from a conditional mutant of RBPjkappa. J. Neurosci. 31, 8046–8058 (2011).
Nishimura, M. et al. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49, 69–75 (1998).
Solter, M. et al. Characterization and function of the bHLH-O protein XHes2: insight into the mechanisms controlling retinal cell fate decision. Development 133, 4097–4108 (2006).
Jimenez, E. et al. A regulatory network of Sox and Six transcription factors initiate a cell fate transformation during hearing regeneration in adult zebrafish. Cell Genom 2, 100170 (2022).
Orvis, J. et al. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat. Methods 18, 843–844 (2021).
Shen, J., Scheffer, D. I., Kwan, K. Y. & Corey, D. P. SHIELD: an integrative gene expression database for inner ear research. Database (Oxf.) 2015, bav071 (2015).
Scheffer, D. I., Shen, J., Corey, D. P. & Chen, Z. Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. J. Neurosci. 35, 6366–6380 (2015).
Moreno-Mateos, M. A. et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015).
Kim, H. K. et al. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol. 36, 239–241 (2018).
Fernandez, J. P., Vejnar, C. E., Giraldez, A. J., Rouet, R. & Moreno-Mateos, M. A. Optimized CRISPR-Cpf1 system for genome editing in zebrafish. Methods 150, 11–18 (2018).
Peloggia, J. et al. Adaptive cell invasion maintains lateral line organ homeostasis in response to environmental changes. Dev. Cell 56, 1296–1312.e1297 (2021).
Millimaki, B. B., Sweet, E. M. & Riley, B. B. Sox2 is required for maintenance and regeneration, but not initial development, of hair cells in the zebrafish inner ear. Dev. Biol. 338, 262–269 (2010).
Parsons, M. J. et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009).
Parinov, S., Kondrichin, I., Korzh, V. & Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 231, 449–459 (2004).
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007).
Shin, M. et al. Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development 143, 3785–3795 (2016).
Yoo, A. S., Staahl, B. T., Chen, L. & Crabtree, G. R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646 (2009).
Berger, J. & Currie, P. D. 503unc, a small and muscle-specific zebrafish promoter. Genesis 51, 443–447 (2013).
Peloggia J, Lush ME, Tsai YY, Wood C, Piotrowski T. Environmental and molecular control of tissue-specific ionocyte differentiation in zebrafish. bioRxiv, (2024).
Navajas Acedo, J. et al. PCP and Wnt pathway components act in parallel during zebrafish mechanosensory hair cell orientation. Nat. Commun. 10, 3993 (2019).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Sehnal, D. et al. Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 49, W431–W437 (2021).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank Dr. Nina Könecke for initial scRNA-seq analyses of the lateral line. We also thank members of the Stowers Institute Core facilities including Allison Scott, Kaitlyn Petentler and Anoja Perera from the Sequencing and Discovery Genomics Core, and Fang Liu and Jeff Haug from the Cytometry Core for expert technical assistance. We thank all members of the Stowers Institute Aquatics Facility for excellent zebrafish husbandry. We thank Drs. Robb Krumlauf and Aurelie Hintermann for comments on the manuscript and all members of the Piotrowski laboratory for helpful comments throughout this project. This research was supported by NIH (NIDCD) award 1R01DC015488-01A1 to T.P., funding from the Hearing Health Foundation, Award #991977, to T.P., and institutional support from the Stowers Institute for Medical Research to T.P.
Author information
Authors and Affiliations
Contributions
M.E.L. and T.P. conceived the project, designed experiments and interpreted results. M.E.L., Y.T., D.M., J.P. and J.E.S. performed experiments and analyzed experimental data. S.C. performed computational analysis. M.E.L. and T.P. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks David Raible who co-reviewed with Selina Baeza-Loya; Shawn Burgess and Angelike Stathopoulos for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lush, M.E., Tsai, YY., Chen, S. et al. Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes. Nat Commun 16, 5913 (2025). https://doi.org/10.1038/s41467-025-60251-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60251-0