Abstract
Stem cells hold promise in regenerative medicine as they have the potential to differentiate into a variety of specialized cell types. However, mechanisms underlying stem cell potency and lineage acquisition remain elusive. Epigenetic modifications and genome accessibility prime cellular feedback to signalling cues, influencing lineage differentiation outcomes. Deciphering how this epigenetic code influences the context-dependent response of pluripotent cells to differentiation cues will elucidate how mammalian tissue diversity is established. Using in vitro and in vivo models, we show that lineage-specific epigenetic signatures precede transcriptional activation of germ layer differentiation programs. We provide evidence that while distinct chromatin accessibility and methylome states prime extraembryonic mesodermal fate decisions, it is DNA methylation, and not chromatin accessibility that predetermines the fates of neuroectoderm, definitive endoderm and neuromesodermal lineages. This study establishes that epigenetic machinery fine-tunes epiblast potency, allowing context-specific spatiotemporal responses to promiscuously used signalling cues controlling organogenesis.
Similar content being viewed by others
Introduction
How cells with distinct identities arise in precise locations despite complex signalling landscape remains ambiguous. One explanation is context-dependent mechanism where lineage identities are predetermined by genetic and epigenetic heterogeneity within stem cells. This differential primed nature of stem cells enable them to respond to convergent signalling cues with divergent lineage outcomes. Transcription factors (TFs) with pioneer ability play a key role by guiding chromatin remodelling machinery, triggering genome accessibility to specific sites in response to developmental cues1. Similarly, epigenetic modifications like DNA methylation are also critical for cell fate acquisition, acting as epigenetic barriers that safeguard differentiation and regulate cell potency. DNA methylation fine-tunes differentiation by balancing plasticity (flexibility to environmental stimuli) and canalization (preferential lineage differentiation despite environmental perturbations)1,2. Understanding the molecular mechanisms that establish lineage priming is crucial for decoding how cell type diversity arises.
In mammals, de novo DNA methylation is a hallmark of the post-implantation epiblast state3,4. Site-specific DNA-methylation patterns are established across promoters of lineage genes during the transition from blastocyst to post-implantation3,4. The innate primed nature of post-implantation epiblast cells, their regional identity and association with distinct cell fates, makes them an ideal model to investigate the influence of epigenetic machinery on predetermining lineage identity5,6,7,8,9. While the exact epigenetic differences underlying lineage potential remain unclear, spatial expression of signalling cues and adhesion proteins appears to determine spatial identity, canalizing cells toward distinct developmental paths5,9,10,11. During mouse gastrulation, post-implantation epiblast cells give rise to the three germ layers: ectoderm, mesoderm, and definitive endoderm (DE). Dynamic morphogenetic changes continuously shape the signalling environment driving germ layer differentiation. As gastrulation begins, proximal posterior epiblast cells are the first to ingress into the primitive streak (PS), giving rise to the extraembryonic mesoderm (ExM) lineages in response to high BMP signals12. Fate mapping revealed that those cells are primed for ExM lineages even before PS ingression6,7,8. ExM contributes to the formation of the allantois, yolk sac mesenchyme, and hemato-endothelial progenitors7,8. In contrast, the anterior epiblast cells, characterized by higher levels of adherens and tight junction proteins10, generate the neuroectoderm forming anterior neural tissues13. Meanwhile, distal posterior epiblast cells contribute to the WNT-dependent neuromesodermal progenitors (NMPs), which give rise to the presomitic mesoderm (PSM) and posterior neural tissues5,14.
Mouse epiblast stem cells (EpiSCs), derived from post-implantation epiblast15,16, provide a suitable in vitro model to study priming towards differentiated cell fates9. Like human embryonic stem cells (hESCs), EpiSCs are primed, pluripotent cells that depend on SMAD2/3 and FGF/ERK signalling for self-renewal15. While SMAD2/3 controls the core pluripotent network, FGF/ERK signalling maintains the primed state by inhibiting lineage commitment17,18, although the downstream mechanism remains elusive. Genetic and chemical modulation of the ERK feedback loop has provided insight into downstream mechanisms by which ERK regulates the epigenetic machinery in fixed cell state19,20. However, tracing these effects through the transitory stages of early embryonic development has proven difficult. Since DNA methylation is established in the post-implantation epiblast and reliant on FGF/ERK signalling3,21, EpiSCs offer an ideal model to study ERK-dependent epigenetic effects on germ-layer commitment.
Here, we investigated the primed nature of epiblast cells by evaluating their self-renewal, epigenetic profiles, and responsiveness to WNT (anterior) and BMP (posterior) signalling cues22. We employed a combination of in vitro and in vivo experimental models coupled with single-cell RNAseq (scRNAseq), spatial transcriptomics, chromatin accessibility, and DNA methylation analyses to identify molecular mechanisms influencing epiblast signalling response. By sorting EpiSCs based on the expression of the tight junction protein CLDN6, we compared their transcriptomic profiles with the regional epiblast populations that give rise to neuroectoderm and anterior PS derivatives (APSD): NMPs and DE. We identified ERK activity and differentially expressed ETS proteins as key regulators of distinct epiblast methylomes. These distinct methylomes shape spatiotemporal signalling response and lineage commitment. Using a genetic model with deficiencies in ERK feedback regulation, we demonstrated that distinct epiblast methylomes and chromatin states, modulated by ERK activity, influence BMP-driven ExM lineage specification. Ultimately, our findings highlight the distinct role of DNA methylation and chromatin accessibility as a pretranscriptional guardian of developmental plasticity and lineage fate acquisition, crucial for generating mammalian tissue diversity.
Results
FGF/ERK profiles distinguish anterior and distal posterior epiblast
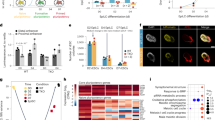
Post-implantation epiblast cells have a predetermined regionalized bias5 towards specific germ layers that is associated with gradients of FGF523 and epithelial markers10, both linked to ERK activity24,25. We hypothesized that epigenetic machinery primes the post-implantation epiblast, influencing cell fate decisions3. Thus, we assessed the relationship of FGF-ERK, regional epiblast identity, epigenetic modifications, and germ layer bias. In order to isolate epiblast cells based on their anterior and distal posterior characteristics, we sought for a surface marker that would enable Fluorescence-activated cell sorting (FACS). Immunofluorescence (IF), scRNAseq10 and high-resolution transcriptional molecular cartography of embryonic day (E) 7.5 embryos showed that the tight junction proteins Cldn6 has an anterior-posterior gradient within epiblast (Fig. 1a, b, Supplementary Fig. 1a). Higher Cldn6 levels colocalized with Atp1b1 in anterior epiblast, while lower amounts with Trh in distal posterior epiblast (Fig. 1a, Supplementary Fig. 1b). Lineages markers also demonstrated the existence of Cldn6 differential expression within early germ layer progenitors (Fig. 1a, b, Supplementary Fig. 1b).
a Spatial transcriptomic analysis showing an anterior (dark yellow arrowhead) and posterior (light yellow arrowhead) gradient of Cldn6 in epiblast of E7.5 mouse embryos. Cldn6 colocalizes with epiblast marker Sox2. Anterior epiblast marker Atp1b1 and distal posterior epiblast marker Trh highlight higher Cldn6 gradient in anterior versus posterior epiblast. High levels of Cldn6 correlate with definitive endoderm (DE) progenitors labelled by Sox17 and Foxa2 markers (pink arrowhead). Cldn6 is absent in primitive streak (PS) identified by T-Bra expression (grey arrowhead) and in NMPs by Tbx6 (blue arrowhead). Cldn6 is not expressed in Hand1pos posterior primitive streak (PPS, grey arrowhead) and extraembryonic mesoderm (red arrowhead) fates. Ep: epiblast, DE: definitive endoderm, NMPs: neuromesodermal progenitors, PPS: posterior primitive streak, ExM: extraembryonic mesoderm. Scale bar: 100 μm. b Cartoon describing the spatial relationship of Cldn6High and Cldn6Low epiblast cells with the emergent PS progenitors. c Representative flow cytometry analysis of CLDN6 in epiblast stem cells (EpiSCs). Pseudocolor plots are gated on live nucleated cells based on CLDN6 levels, showing a CLDN6 gradient. CLDN6Low (light yellow) EpiSCs are sorted from the 20-25% low-level CLDN6-expressing cells. The 10–15% of the highest CLDN6-expressing cells amongst the total EpiSC population are the CLDN6High (dark yellow) EpiSCs. d Heatmap showing differential expression of epithelial-mesenchymal markers, epigenetic machinery and FGF/ERK signalling components in CLDN6Low vs CLDN6High EpiSCs. e GO-term enrichment analysis displays a significant overrepresentation of terms associated with ERK signalling and epithelial signature amongst the differentially expressed genes in CLDN6Low vs CLDN6High EpiSCs (Binomial test with Bonferroni correction). f Flow cytometry analysis of CLDN6 and pERK levels in EpiSCs. CLDN6 levels were assessed across pERKHigh (P4, red) and pERKLow (P5, pink) EpiSCs, with higher CLDN6 levels in pERKHigh (1.9 FC across geometric mean and median of CLDN6 levels). g Illustration describing the FGF/pERK signalling network in CLDN6Low and CLDN6High EpiSCs. For d and e RNAseq data (P ≤ 0.05, fold-change ≥1.5, n = 2 biological replicates). Significance was assessed by DESeq2 using two-sided Wald test with Benjamini–Hochberg adjusted P-values.
Thus, CLDN6 FACS sorting represents a unique strategy to study priming of the regionalized post-implantation epiblast and subsequent differentiation propensity5,6,7,10. We used EpiSCs as an in vitro cellular model because they show a gradient of CLDN6 expression, allowing us to separate them into CLDN6Low and CLDN6High subpopulations (Fig. 1c). Following sorting and reseeding in EpiSC conditions, both populations retain expression of EpiSC pluripotency genes. While the CLDN6High EpiSCs reestablished the original mixed distribution after one passage, CLDN6Low EpiSCs retained their CLDN6Low-state over multiple passages (Supplementary Fig. 1c, d), suggesting this population is more committed to lineage-specific differentiation. This ability of CLDN6Low EpiSCs to self-renew and maintain their restricted potency may be linked to the expression of stemness genes, master regulators of stem cell identity, like Sox9 and Snail226 (Supplementary Fig. 1c). Transcriptional profiling of CLDN6High and CLDN6Low EpiSCs indicated that these in vitro populations reflected anterior and distal posterior epiblast regionalization (Supplementary Data 1). Both populations maintained the expression of core pluripotency-associated genes (Supplementary Fig. 1c, e). CLDN6High EpiSCs exhibited higher expression of epithelial markers, while CLDN6Low EpiSCs showed enrichment in mesenchymal genes (Fig. 1d). Gene ontology (GO) of differentially expressed genes revealed associations with adhesion properties, migratory behaviour, and ERK signalling cascade (Fig. 1e). These data indicated that CLDN6Low and CLDN6High EpiSCs recapitulate the embryonic posterior-anterior and mesenchymal-epithelial identities (Fig. 1d, Supplementary Fig. 1b, c). Consistently, CLDN6High EpiSCs showed higher expression of FGF ligands (Fgf4, Fgf5) and Fgfr2 (Fig. 1d). Dose-sensitive ERK effector TFs from the ETS family showed differential expression between CLDN6Low and CLDN6High EpiSCs. Specifically, Ets1, Etv,1 and Elf427 were upregulated in CLDN6Low EpiSCs, while Ets2 was enriched in CLDN6High EpiSCs (Fig. 1d). Furthermore, GEO sequencing data28,29 revealed enrichment of epithelial-mesenchymal and ETS TFs in CLDN6High EpiSCs and anterior epiblast. In contrast, CLDN6Low EpiSCs resembled distal posterior epiblast cells (Supplementary Fig. 1f). Together, these data suggest the existence of regionalized primed states associated with ERK signalling.
Flow cytometry analyses revealed higher pERK levels in CLDN6High EpiSCs, indicating a positive correlation between pERK dosage and CLDN6 (Fig. 1f), suggesting that epiblast populations with different epithelial and FGF/ERK/ETS signatures mediate differential signalling responses influencing lineage specification (Fig. 1g). Investigating the FGF/ERK molecular mechanism in the primed post-implantation epiblast state has been challenging, as ERK inhibition either induces reversion to the naïve ES-state or promotes differentiation towards neuroectodermal lineages17,21. Indeed, upon ERK inhibition through the sustained administration of 1 μM PD03, EpiSCs spontaneously differentiate, exhibiting decreased expression of CLDN6 and E-CADHERIN (Supplementary Fig. 1g), and increased neuroectodermal and ExM markers (Supplementary Fig. 1h). However, the possibility of sorting epiblast populations based on CLDN6 levels offers the advantage of elucidating the EpiSC intrinsic lineage bias upon differentiation towards ectoderm, mesoderm, and DE. To test the germ layer preprogramming of CLDN6Low and CLDN6High EpiSCs, we established a protocol that leverages high WNT and TGF-β signalling to differentiate EpiSCs towards APSD, which gives rise to DE, NMPs, and cardiac mesoderm. Single cell sequencing (scRNAseq) of these differentiated cultures identified 5 clusters of APSD subpopulations: NMPs, cardiac mesoderm, DE, nascent mesoderm, and PS (Supplementary Fig. 2a, b; Supplementary Data 2)30. Individual APSD subpopulations also have distinct epithelial and mesenchymal transcriptional signatures, with mesoderm lineages showing mesenchymal markers and DE displaying signature of partial EMT phenotype (Supplementary Fig. 2c, Supplementary Data 2)10. We validated the generation of cells having an APSD identity by comparing in vitro differentiated cells at different time points with Eomes-GFP31 isolated from E6.5, E7.0, and E7.75 embryos (Supplementary Fig. 2d, e, Supplementary Data 3). Integration of in vivo (turquoise) and in vitro (light pink) cluster contribution confirmed that our in vitro APSD protocol generates early anterior mesoderm and DE, resembling the in vivo progenitors (Supplementary Fig. 2f). Thus, we developed an efficient APSD differentiation protocol to address the differentiation potency of CLDN6Low and CLDN6High EpiSCs.
Epiblast ERK levels predetermine germ layer lineage decisions
To assess if CLDN6Low and CLDN6High EpiSCs have an innate predisposition towards a specific APSD fate, we sorted and reseeded them directly into APS medium. CLDN6Low EpiSCs differentiated more efficiently into NMPs, generating PSM marked by the expression of TBX6 and Msgn1 (Fig. 2a–c). Conversely, higher FOXA2 and Sox17 suggested that CLDN6High EpiSCs preferentially differentiate into DE (Fig. 2a, b, d). Since both CLDN6Low and CLDN6High EpiSCs can self-renew (Supplementary Fig. 1d, e), we assessed whether they retain the same APSD differentiation potential after reseeding and maintaining them in EpiSC medium. Gross morphology and transcriptional analysis confirmed that CLDN6Low EpiSCs have restricted potency, retaining NMP/PSM-specific potential (Supplementary Fig. 3a, b). Conversely, CLDN6High cells regained the differentiation potential of the unsorted EpiSCs, differentiating into both NMP/PSM and DE (Supplementary Fig. 3a, b), indicating a full reversion to the unsorted EpiSC state.
a Schematics describing the differentiation propensity of CLDN6Low and CLDN6High EpiSCs towards anterior primitive streak derivatives (APSD). Right panel, representative IF staining for FOXA2 (green), TBX6 (red), and DAPI (blue) of CLDN6Low and CLDN6High EpiSCs subjected to APSD-differentiation reveals the higher potential of CLDN6Low to differentiate towards TBX6pos NMP cells, while CLDN6High EpiSCs generate FOXA2pos DE-fated cells with greater efficiency. Scale bar: 50 μm. Each differentiation experiment was performed independently more than three times, and multiple immunofluorescence images were acquired for each experiment. b Illustration of the APSD protocol to differentiate EpiSCs towards NMPs and DE. EpiSCs culture in APS medium for 24 h generated APSD containing NMP and DE progenitors, which are further differentiated toward PSM and DE by administration of PSM medium and DE medium, respectively, for 48 h. c Relative Tbx6 and Msgn1 expression measured by RT-qPCR, revealing high levels of NMP and PSM markers in CLDN6Low EpiSCs differentiated towards APSD. d Relative expression of DE markers, Foxa2 and Sox17, measured by RT-qPCR, showing enriched expression in CLDN6High EpiSCs differentiated towards APSD. Data are mean ± s.e.m. (n = 2, biological replicates) for (c) and (d). e Cartoon describing differentiation of CLDN6Low and CLDN6High EpiSCs towards neuroectoderm. Right panels, representative phase-contrast photomicrographs of the CLDN6Low and CLDN6High anterior neural differentiated cells, showing the formation of neurons (white arrowheads and high magnification of area framed in green boxes, panels I and II) at day (D) 4 and D7 in CLDN6High, while CLDN6Low EpiSCs failed to differentiate towards neuroectoderm. The images are representative of at least three independent differentiations. Scale bars: 100 μm. f Illustration depicting the prospective embryonic contribution of CLDN6Low and CLDN6High epiblast cells to different germ layers. CLDN6High cells generate anterior neural tissues (neuroectoderm) (green) and DE (pink), while distal-posteriorly located CLDN6Low cells specifically contribute to NMPs (blue), giving rise to PSM and posterior neural tissues (spinal cord).
CLDN6High EpiSCs resemble epiblast cells fated to become neuroectoderm based on their anterior epiblast characteristics5,7 (Fig. 1a, d). Indeed, when exposed to neuroectoderm induction medium17, CLDN6High EpiSCs generated neuronal structures (Fig. 2e). However, CLDN6Low EpiSCs failed to differentiate into neuroectoderm (Fig. 2e).
To investigate the role of ERK signalling, we recapitulated low ERK levels by focusing on its upstream kinase MEK, and administered different doses (0.1 μM and 0.5 μM) of the inhibitor PD03. EpiSCs with lower ERK levels exhibited reduced efficiency in differentiating into neuroectoderm, as evidenced by qPCR analysis of Pax6 and Otx2 gene expression (Supplementary Fig. 3c). OTX2 is a marker of anterior neural (forebrain and midbrain) identity. Thus, administering PD03 to EpiSCs before neural differentiation, even at low dosages, biases them towards an Otx2neg hindbrain neural state, indicating that ERK may contribute to priming Otx2pos midbrain and forebrain progenitors in the EpiSCs.
We showed that CLDN6Low EpiSCs (Lower ERK) are biased towards posterior neural (spinal cord) and PSM fates. Similarly, ERK inhibition in PD03-treated EpiSCs shifted APSD differentiation toward Tbx6pos NMPs/PSM rather than Foxa2pos DE progenitor cells (Supplementary Fig. 3d), recapitulating the CLDN6Low and CLDN6High EpiSC biases (Fig. 2a,b; Supplementary Fig. 3a,b), respectively. Thus, CLDN6High EpiSCs (Higher ERK), while pluripotent, are predisposed towards DE and anterior neural (neuroectoderm) fates (Fig. 2f). In contrast, CLDN6Low EpiSCs (Lower ERK) are fate-restricted towards PSM and posterior neural (spinal cord) fates (Fig. 2f). These findings indicate that EpiSCs intrinsically harbour distinct lineage biases linked to ERK signalling.
Epiblast DNA methylation primes WNT-driven germ layer fate
Epigenetic machinery may intrinsically influence the divergent fates of CLDN6Low and CLDN6High EpiSCs under the same differentiation cues. A previous report indicated that WNT signalling presdisposes posterior neural lineage fate segregation22, by establishing a chromatin landscape in posterior epiblast cells that favours NMP differentiation5. Thus, we assessed if different chromatin landscapes underlie the lineage biases of CLDN6Low and CLDN6High EpiSCs. Assay for Transposase Accessible Chromatin using sequencing (ATACseq32) analyses revealed 110 differentially accessible peaks (DAPs) associated with open chromatin in CLDN6High EpiSCs, when compared to CLDN6Low EpiSCs (Fig. 3a, Supplementary Data 4). Surprisingly, none of these DAPs were linked to genes tied to specific germ-layer lineages. Unlike prior work linking WNT signalling to epigenetic heterogeneity and lineage bias5, we did not observe significant difference in chromatin accessiblity between these two populations that could be linked to signalling response. These discrepancies might be linked to the cell system used. Our EpiSCs are cultured under WNT-inhibited conditions, as WNT induces chromatin remodelling and differentiation33. Indeed, only upon WNT stimulation, ATACseq revealed that cis-regulatory elements (CREs)5,33 associated with NMPs become accessible in CLDN6Low APSD (Fig. 3b), coinciding with increased binding of the WNT-effector TF LEF133 (Supplementary Fig. 4a). Thus, DNA accessibility does not explain the alternative cell fate acquisition of CLDN6Low and CLDN6High EpiSCs.
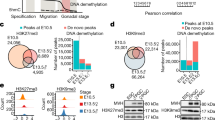
a Volcano plot of ATACseq data showing 110 uniquely differentially accessible peaks (DAPs) (FDR ≤ 0.05, blue) in CLDN6High EpiSCs in comparison to CLDN6Low EpiSCs. b Average density plots (top) and heatmaps (bottom) of chromatin accessibility sites of CLDN6Low and CLDN6High EpiSCs and APSD across neural (anterior and posterior) and PSM putative cis-regulatory elements (CREs). No difference in accessibility across neural CREs is seen in EpiSCs, although there are more accessible CREs in EpiSCs than in APSD. CLDN6Low APSD (NMPs) has more accessible neural CREs than CLDN6High APSD (DE). c Beanplot showing similar CpG methylation levels +/- 5 kb probed across genes in CLDN6Low and CLDN6High EpiSCs and APSD. d Star Wars plot showing intragenic CpG methylation levels (≥10× coverage), highest in CLDN6Low EpiSCs and lowest in CLDN6High APSD. CLDN6High EpiSCs and APSD show similar levels. Means shown as filled circles; whiskers indicate standard error. e The circles represent all the genes in the individual cell types: CLDN6Low and CLDN6High EpiSCs. The overlapping region shows the differentially methylated genes (DMGs) between these two cell types, including hypermethylated and hypomethylated genes. CLDN6Low and CLDN6High EpiSCs have DMGs associated to DE (Foxa2, Sox17, Gsc, Gata6), neuroectoderm (Pax6, Onecut1, Onecut2, Lmx1a, Dbx1) lineages but not NMPs. f WGBS visualization (SeqMonk) of Pax6 genomic region indicating the specific hypermethylated region in CLDN6High EpiSCs (red arrowhead), bearing positional and functional similarities to previously studied PAX6 CRE41, compared to CLDN6Low EpiSCs (white arrowhead). DNA methylation (%) is shown in the bottom bar. g Illustration modelling how DNA methylation primes post-implantation epiblast cells towards WNT-dependent and WNT-independent lineage fates. CLDN6Low EpiSCs, resembling distal posterior epiblast require WNT for EMT and NMP/PSM differentiation5,14,33. Furthermore, the combined activity of pioneer TFs and the WNT-effector LEF1 drives NMPs towards PSM fate33. Conversely, CLDN6High EpiSCs, resembling anterior epiblast, are biased towards DE and neuroectoderm, with DE fate promoted by WNT inhibition after partial EMT10. Both DE and neuroectoderm reside in the WNT-inhibited anterior side of the embryo.
Given that EpiSC maintenance relies on FGF/ERK signalling, which is known to regulate the de novo DNA methylation machinery17,21,34, we performed Whole Genome Bisulfite Sequencing (WGBS). CLDN6Low and CLDN6High EpiSCs and correspondingly differentiated APSD showed comparable overall CpG methylation levels (Fig. 3c), although differences were evident in intragenic DNA methylation, which is implicated in lineage-specific transcriptional regulation35,36,37,38. CLDN6Low EpiSCs showed higher gene body methylation compared to CLDN6High EpiSCs (Fig. 3d), supporting their observed restricted potency (Fig. 2, Supplementary Fig. 3). Reduced intragenic CpG methylation levels between CLDN6Low EpiSCs and WNT-induced CLDN6Low APSD (Fig. 3d) indicate that the addition of WNT triggers epigenetic remodelling and promotes differentiation (Fig. 3b, Supplementary Fig. 4a).
In this study, EpiSCs were maintained in an FGF-dependent WNT-inhibited state, simulating the hypermethylated post-implantation epiblast3,4. Differentiation of epiblast cells into NMP/PSM requires the combined addition of FGF and WNT14, which rewires lineage-specific DNA methylation loci, rather than altering global DNA methylation levels as previously noted39,40. Thus, our study validates previous findings that DNA methylation influences WNT response, which modulates neuromesodermal differentiation39. We found that CLDN6Low EpiSCs have higher intragenic DNA methylation, suggesting an epigenetic safeguarding mechanism that restricts their differentiation potential. However, subsequent WNT-driven APSD differentiation reduced intragenic DNA methylation in CLDN6Low EpiSCs, potentially creating a more permissive epigenetic landscape to generate NMPs/PSM (Fig. 3d). In the CLDN6High EpiSCs, intragenic DNA methylation remains unchanged upon WNT-induced differentiation towards CLDN6High APSD (DE progenitors) (Fig. 3d). These findings indicate that CLDN6Low and CLDN6High EpiSCs have distinct DNA methylation signatures that differently prime their WNT response, suggesting that epiblast methylomes rather than DNA accessibility are the primary drivers of lineage priming at these stages of development.
To get a deeper insight into distinct epiblast methylomes, we focused on differentially methylated regions associated genes (DMGs) (Fig. 3e, Supplementary Data 5). We identified lineage-specific DMGs associated with anterior neural markers (Pax6, Onecut1, Onecut2, Lmx1a, Dbx1), suggesting a primed epigenetic effect on transcriptionally inactive anterior neural lineage genes in EpiSCs (Fig. 3e). In CLN6Low and CLN6High conditions, the DMGs included both hypermethylated and hypomethylated genes. Notably, these genes were predominantly hypermethylated in the CLN6Low EpiSCs, except for Pax6, which did not follow this pattern. Notably, genes associated with NMPs were not among the DMGs identified in EpiSCs (Fig. 3e). These findings indicate that differences in epiblast methylomes are specifically associated with anterior neural lineage genes. To get a deeper insight into how these DMGs restrict CLDN6Low EpiSCs from differentiating towards neuroectoderm, we focused on the anterior neural master lineage gene Pax6. Pax6 was amongst the DMGs in EpiSCs (Fig. 3e). It was not expressed in either EpiSCs or APSD. Additionally, previous reports indicate that anterior neural competence is lost upon WNT agonism in the epiblast14. We observed increased Pax6 DNA methylation levels across putative cis-regulatory elements (CREs), correlating with increased Pax6 expression and linked to neuronal differentiation41 in CLDN6High EpiSCs (Fig. 3f). This observation corroborates the paradoxical role of DNA methylation in upregulating PAX6 expression41.
To understand if DNA methylation states precede chromatin accessibility changes, we looked at ATACseq signals across the Pax6 locus. Pax6 putative CREs were similarly accessible in both EpiSC populations but not in APSD (Supplementary Fig. 4b), suggesting an association between loss of WNT-induced APSD (NMPs) competence and the inability to form anterior neural tissues5. Similarly to Pax6, the anterior neural differentiation regulators Oncecut2 and Dbx1 showed a similar chromatin accessibility pattern (Supplementary Fig. 4c, d). Conversely, Cdx1 and Cdx2, crucial for posterior neural (NMP) fate acquisition14 were not among the DMGs (Fig. 4e). Their expression in CLDN6Low APSD correlated with increased promoter accessibility (Supplementary Fig. 4e,f). These findings suggest that WNT stimulation remodels the chromatin landscape making it permissive to NMP (posterior neural) fate while inhibiting neuroectoderm (anterior neural) competence5,33. Our results challenge the view that WNT signalling supports the epiblast primed state5, suggesting instead that lineage-specific bias is driven by DNA methylation-mediated priming independent of WNT. Thus, cellular response to WNT signalling divergently influences neural lineage induction by suppressing neuroectoderm and promoting NMP formation.
WNT stimulation induces the differentiation of both NMPs and DE progenitors, which arise after epiblast cells ingress through the PS33. Upon exposure to WNT agonists, CLDN6Low EpiSCs differentiate towards NMPs while CLDN6High EpiSCs form DE (Fig. 2), supporting the existence of different epigenetically primed epiblast populations generating the APSD progenitors (NMPs or DE)10. In EpiSCs, WGBS analyses revealed DMGs associated with DE lineages (Foxa2, Foxa1, Sox17, Gata6, Gsc) but not NMPs (Fig. 3e). Furthermore, in EpiSCs, there were no changes in chromatin accessibility over DE lineage genes (Fig. 3a). Thus, the acquisition of DE fate may also be attributed to the lineage-specific primed methylome of the regionalized epiblast cells. Our data provide further evidence on how these two EpiSC subpopulations are differently primed to respond to WNT signals.
Altogether, we present evidence that DNA methylation patterns may precede WNT-driven chromatin accessibility changes influencing innate priming and lineage acquisition, by safeguarding distal-posterior epiblast cells (CLDN6Low EpiSCs: Lower ERK) from DE and anterior neural differentiation14,33 (Fig. 3g). Spinal cord and somite fate decisions occur during WNT-induced differentiation and rely on the WNT-effector LEF1 and cooperative action of pioneer TFs33, leading to rewiring of the epigenetic landscape. Our findings indicate that epiblast cells with higher ERK, residing near WNT-inhibited regions, may respond differently to the WNT signal by differentiating into ectoderm and endoderm. These results support the hypothesis that DE and Neuroectoderm could originate from common progenitors10,42.
In contrast, epiblast cells with lower ERK, when exposed to WNT stimulation, undergo chromatin changes that promote differentiation towards NMPs.
PBX regulates ERK and epigenetic machinery in epiblast cells
FGF/ERK is known to regulate site-specific DNA methylation in different contexts20,43, but whether it has a similar role in post-implantation epiblast stages remains uncertain. The suppression of the ERK pathway using PD03 has been demonstrated to erode genomic imprints, leading to chromosomal abnormalities and compromising the developmental potential of pluripotent stem cells44,45. This aspect represents a challenge to study FGF/ERK function during the transition from pluripotency towards lineage commitment differentiation, without inducing spontaneous differentiation17,19. The three-amino acid loop extension (TALE) homeodomain proteins (PBX, MEIS, and PREP)46 are alternative candidates as they regulate FGF/ERK signalling in diverse developmental contexts47,48 and cancers49,50. Thus, they are a suitable candidate to study the epigenetic machinery that guides the transition from primed pluripotency to lineage commitment in vitro, while also enabling the observation of developmental phenotypic outputs in vivo.
PBX1 is initially expressed in the late blastocyst stage (Supplementary Fig. 5a, b) and in the post-implantation epiblast (Fig. 4a). Furthermore, epiblast inactivation of Pbx1 with the Sox2Cre allele in a Pbx2-deficient background (Pbx1/2-KO) resulted in stronger developmental defects compared to Pbx1 deletion in PS with the T-BraCre allele (Supplementary Fig. 5c–e), indicating the critical role of Pbx1 in the epiblast. RNAseq analyses of Pbx1-KO ES and EpiSC lines revealed 1,018 differentially expressed genes in Pbx1-KO EpiSCs compared with wild-type (WT) EpiSCs (Fig. 4b and Supplementary Data 6), but no differences in ES cells (Supplementary Fig. 5f). GO analysis of the differentially expressed genes in EpiSCs showed Pbx1 association with the FGF/ERK signalling cascade (Supplementary Fig. 5g). FGF ligands, ETS TFs, and pERK antagonists were amongst differentially expressed genes in Pbx1-KO EpiSCs (Fig. 4b), indicating perturbation of FGF/ERK activity. Immunoblotting analyses confirmed reduced ERK activity in the Pbx1-KO EpiSCs, with no changes in total ERK levels (Fig. 4c, Supplementary Fig. 5h, i). Through PD03-mediated inhibition of ERK, we demonstrated that wild-type EpiSCs treated with 0.5 μM PD03 exhibit CLDN6 levels comparable to those of Pbx1-KO EpiSCs, indicating a similar trend among CLDN6, PBX1, and ERK (Supplementary Fig. 5j).
a Confocal IF of E5.5 post-implantation embryo probed with PBX1 antibody (red), SOX17 (green), and DAPI nuclei counterstaining (grey) revealing PBX1 expression in the epiblast (Ep) and not in primitive endoderm (PrEnd). Scale bar: 50 μm. Images are representative of three independent experiments. b Heatmap displaying differentially expressed genes in WT and Pbx1-KO EpiSCs, showing differences in FGF/ERK signalling components (RNAseq, P ≤ 0.05, fold-change ≥1.5). n = 2 biological replicates. Significance was assessed by DESeq2 on the basis of two-sided Wald test with Benjamini–Hochberg adjusted P-values. c Western blot analysis confirms the absence of PBX1 and reduction of pERK1/2 in PBX1-KO EpiSCs. d Volcano plot of chromatin accessibility data showing 576 DAPs in WT compared with Pbx1-KO EpiSCs, showing enrichment of BMP-responsive genes (Hand1, E2f4, Col26a1, Bmp3, Egr3, and Itga4). e Pie chart depicting all de novo motif analysis results from input DAPs in WT versus Pbx1-KO EpiSCs revealing enrichment of ETS binding hits. f 576 DAPs in WT opposed to Pbx1-KO EpiSCs are enriched for ETS TF motifs (HOMER). g The circles represent all the genes in the individual cell types: Wild type (WT) and Pbx1-KO. The overlapping region corresponds to the differentially methylated genes (DMGs). In WT and Pbx1-KO EpiSCs BMP-responsive genes associated with ExM lineage, like Hand1 and Tbx3, are DMGs. h STRING protein-protein network analysis, showing interactions between ERK/ETS proteins and epigenetic machinery. Unbiased MCL clustering reveals two clusters. Cluster 1 (red) encompasses most ERK/ETS proteins. Cluster 2 (green) shows strong interactions between ETS proteins (Spi1 (PU.1) and ETV2) and de novo DNA methylation and demethylation enzymes. Interactions were experimentally determined from curated databases, text mining, co-expression, and protein homology. i Illustration depicting the mechanism by which PBX1 negatively regulates ERK feedback loop, thereby modulating ERK signalling activity.
To assess if PBX1/ERK regulates the epigenetic machinery in EpiSCs, we performed ATACseq and WGBS comparing WT and Pbx1-KO EpiSCs. ATACseq analyses revealed 576 DAPs (Fig. 4d, Supplementary Data 7), many of which were associated with BMP-responsive genes51,52. HOMER motif enrichment further highlighted ETS TFs as key regulators influencing the epigenetic landscape of these DAPs (Fig. 4e, f). Parallel WGBS corroborated these results by revealing DMGs linked to BMP-responsive ExM lineage genes, including Hand1 and Tbx3 among others (Fig. 4g, Supplementary Data 8). Collectively, these results suggest that the loss of PBX1 leads to pERK reduction, disrupting downstream ETS TF expression, thereby altering the epigenetic regulation of BMP-responsive ExM lineage genes.
We probed how PBX1 regulates ERK activity and epigenetic machinery by doing chromatin immunoprecipitation followed by sequencing (ChIPseq) using the PBX1 antibody. GO of PBX1-bound sites in EpiSCs revealed enrichment of genes associated with FGF/ERK signalling (Supplementary Fig. 5k), with the FGF/ERK negative modulator Spry253 being a strong candidate for direct regulation since it is also upregulated in Pbx1-KO EpiSCs (Fig. 4b). ATACseq revealed that PBX1 binds to two putative CRE in the vicinity of Spry2 (p1 and p2 locus) in EpiSCs (Supplementary Fig. 5l). However, p2 was accessible only in EpiSCs (Supplementary Fig. 5l), indicating an epiblast-specific CRE not accessible in ES cells. Focusing on p2, we identified a PBX-binding sequence within the PBX1 occupancy region. Electrophoretic mobility shift assay (EMSA) with different TALE in vitro-translated proteins demonstrated the preferential assembly of PBX-PREP1, rather than a MEIS2 or HOX-containing complex (Supplementary Fig. 5m). These binding properties suggest a previously unrecognized role for the PBX-TALE complex in regulating FGF/ERK signalling in the epiblast, dissimilar from its pioneer factor activity in NMP-to-PSM differentiation33.
Previous studies have shown that unrestrained FGF/ERK activity due to Spry loss leads to epigenetic deregulation54. Moreover, ERK downstream effectors ETS and JUN/FOS, exibit differential binding sensitivity based on DNA methylation status55, preferring either unmethylated or methylated DNA. ETS TFs directly bind and recruit DNA methylation and demethylation machinery20. Binding of ETS proteins to DNA regulates the epigenetic landscape in primed pluripotent cells19. Furthermore, ETS expression and activity are linked to pERK levels27. Protein-protein interaction analyses using STRING56 revealed a network connecting ERK signalling components, ETS and JUN/FOS proteins, and proteins responsible for de novo DNA methylation and demethylation57. Unbiased Markov Cluster Algorithm (MCL) analysis showed that Cluster 1 (red) primarily includes FGF/ERK components and ETS proteins (Fig. 4h). Strikingly, ETS proteins also directly interacted with de novo DNA methylation and demethylation enzymes in Cluster 2 (green), suggesting ETS ability to directly interact with DNMT3a/b and TET1/2/3 (Fig. 4h, Supplementary Data 9). Thus, our data suggest a systematic network where PBX1 regulation of Spry2 controls differential ERK/ETS activity, which shapes the epigenetic landscape across lineage-specific genes in primed epiblast cells before their transcriptional induction (Fig. 4i). Altogether, we found an ERK-deficient model to explore how FGF/ERK and DNA methylation influence lineage preprogramming.
PBX/ERK prime BMP response towards ExM fates in posterior epiblast
Previous studies demonstrated that endothelial and primitive erythroid cells derive from distinct regionalized proximal posterior epiblast populations8, suggesting differential priming towards BMP-driven ExM lineages in epiblast stages22. Given that BMP-responsive genes were amongst the DAPs and DMGs in Pbx1-KO EpiSCs (Fig. 4d, g), we used the Pbx1-KO model to assess how proximal posterior epiblast populations responds to BMP signals and acquire different ExM fates. IF for the BMP-responsive58 ExM marker HAND130,59 showed that E7.5 Pbx1/2-KO displays severe ExM defects (Fig. 5a, b). We previously established and characterized a protocol to differentiate EpiSCs towards ExM fates via BMP stimulation52. In vitro differentiation of Pbx1-KO EpiSCs towards ExM recapitulated the in vivo ExM phenotype (Fig. 5c–e), with reduced expression of allantois and endothelial markers and upregulation of yolk sac and primitive erythroid genes (Fig. 5d, e). PBX1 regulation of ERK signalling may underpin these lineage biases by modulating the epigenomic landscapes of ExM genes in EpiSCs.
a Illustration depicting of E7.5 ExM lineages: allantois, amnion, and yolk sac influenced by BMP signalling. b Confocal 3D projection of E7.5 control and Pbx1f/f;Pbx2-/-;Sox2Cre/+ embryos stained with the allantois and amnion marker HAND1 (red), DE and node marker FOXA2 (green), and DAPI (grey). N, Node; DE, definitive endoderm; ExM, extraembryonic mesoderm. Images are representative of three independent experiments. WT Scale bar: 100 μm, Pbx1/2-Sox2Cre/+ or 50 μm. c Schematics of WT and Pbx1-KO EpiSCs differentiation protocol52 to PPS and ExM cells. d Heatmap shows lineage-specific DEDG (P ≤ 0.05, fold-change ≥1.5, n = 2 biological replicates) across stages (yellow: EpiSC, grey: PPS, red: ExM). Significance was assessed by DESeq2 on the basis of two-sided Wald test with Benjamini–Hochberg adjusted P-values. e Pbx1-KO EpiSCs show a preferential bias towards primitive erythroid and yolk sac mesenchyme when subjected to ExM differentiation. f Schematic showing DMGs in WT vs Pbx1-KO EpiSCs and their transcriptional changes upon differentiation in PPS and ExM. Yellow, grey, and red circles represent DMGs in EpiSCs, DEGs in PPS, and DEGs in ExM, respectively, illustrating that methylation changes in EpiSCs precede lineage-specific gene activation. g WGBS (SeqMonk) visualization indicating differentially methylated regions across the Hand1 locus in WT and Pbx1-KO EpiSCs, showing hypermethylation in Pbx1-KO EpiSCs. The bar shows CpG methylation levels (%). h UCSC visualization of Hand1 showing increased chromatin accessibility in WT conditions, while the locus remains closed in Pbx1-KO. Accessible loci are indicated with red arrowheads, and reduced accessibility with white arrowheads. i WGBS (SeqMonk) tracks indicating CpG methylation levels across Tbx3 in WT and Pbx1-KO EpiSCs, showing hypermethylation in WT EpiSCs. CpG methylation levels (%) are depicted in the bottom bar. j ATAC-seq tracks of Tbx3 show increased chromatin accessibility in Pbx1-KO vs WT cells. Red arrowheads mark accessible loci; white indicates reduced accessibility.
Epigenomic analyses revealed that many of the DAPs (Fig. 4d, Supplementary Data 7) and DMGs (Figs. 4g, and 5f) in Pbx1-KO EpiSCs overlap with loci transcriptionally regulated during BMP-driven differentiation (Fig. 5d). These findings suggest that PBX1/ERK activity in EpiSCs establishes epigenomic profiles that predetermine ExM lineage outcomes before BMP stimulation.
The expression of the BMP-responsive allantois marker gene Hand1 was significantly disrupted in Pbx1-KO ExM cells during in vitro differentiation (Fig. 5d). In WT EpiSCs, Hand1 exhibits a permissive epigenetic landscape, with hypomethylation and promoter accessibility in EpiSCs, laying the groundwork for successful ExM differentiation towards an allantois fate (Fig. 5g, h). In Pbx1-KO EpiSCs, Hand1 becomes is hypermethylated, and its promoter is inaccessible, resulting in transcriptional repression (Fig. 5 g, h). This restrictive epigenetic state may explain the loss of Hand1 expression during in vitro ExM differentiation (Fig. 5d, e), and the aberrant allantois specification seen in vivo (Fig. 5b). Conversely, the yolk sac lineage gene Tbx330,60 was upregulated in Pbx1-KO in ExM differentiation (Fig. 5d). In WT EpiSCs, Tbx3 is epigenetically silenced with hypermethylation and inaccessible CRE (Fig. 5i, j). In Pbx1-KO EpiSCs, hypomethylation and CRE accessibility at the Tbx3 locus drive its increased expression during differentiation, favoring yolk-sac ExM differentiation (Fig. 5i, j). Altogether, these data indicate that PBX/ERK activity acts upstream of alternative chromatin and methylome signatures modulating BMP-dependent ExM lineage choices (Supplementary Fig. 6). By coordinating these epigenetic changes, PBX1/ERK activity ensures the proper lineage preprogramming of ExM progenitors in response to BMP stimulation.
PBX–ERK primes epiblast epigenome independent of HOX
CLDN6Low EpiSCs, which resemble distal posterior epiblast (Figs. 1, 2; Supplementary Figs. 1, 3) exhibit lower ERK levels, a pattern similar to Pbx1-KO EpiSCs (Supplementary Fig. 5j). Previously, we found that the loss of Pbx1/2 leads to an accumulation of NMPs and reduced PSM differentiation33. We linked this failure to the role of PBX/HOX complex in recruiting LEF1 to activate WNT response on PSM loci, thereby enabling NMP differentiation towards PSM33.
Since Pbx1 is expressed in the epiblast, we investigated whether the NMP accumulation observed in Pbx1/2KO cells is linked to a disruption of the PBX1-ERK- priming mechanism observed in EpiSCs.
In EpiSCs, PBX regulates ERK activity via Spry2 (Fig. 4i), a mechanism independent of Hox genes, as they are not expressed in EpiSCs. EMSA analysis (Supplementary Fig. 5m) further confirmed that the Spry2 locus lacks affinity for the HOX/PBX complex, reinforcing that PBX1 regulates Spry2 in a HOX-independent manner. When Pbx1-KO EpiSCs were differentiated towards APSD, they exhibited increased NMP differentiation with an expected block in PSM specification, as demonstrated by the increased expression of Nkx1.2 and T (NMP genes) and reduced expression of Msgn1 and Tbx6 (PSM genes)33 (Supplementary Fig. 7a–c). Moreover, we observed a NMP bias in CLDN6Low EpiSCs (Fig. 2a), which similar to Pbx1-KO have reduced ERK levels. These findings provide a possible explanation for the accumulation of NMPs, linking PBX1 and ERK levels to the transition of EpiSCs to NMPs and underscore PBX1’s dual role: first, as a priming factor in the epiblast, independent of HOX activity, and second, as a pioneer factor that acts in a HOX-dependent manner during PSM differentiation33.
Additionally, Pbx1-KO EpiSCs exhibited impaired differentiation into DE, with reduced expression of markers such as Foxa2, Cer1, Hhex and Sox17 (Supplementary Fig. 7a, d). Since HOX genes are not expressed in DE61, the role of PBX1 in regulating DE differentiation is also HOX-independent. These findings suggest that PBX1 regulates ERK activity independently of HOX in EpiSCs, which is crucial for balancing subsequent NMP and DE differentiation.
We observed a DE-specific perturbation of FOXA2 expression in E7.5 Pbx1/2-DKO embryos (Fig. 5b). Investigating the underying epigenetic mechanism, WGBS analysis revealed that the Foxa2 locus was hypomethylation in Pbx1-KO EpiSCs. In contrast, in WT EpiSCs, Foxa2 was intragenically hypermethylated (Supplementary Fig. 7e). This observation aligns with prior studies showing that hypermethylation of FOXA2 correlates with high DE differentiation potential62. These findings highlight the role of PBX1-ERK signalling in regulating the epigenetic state of Foxa2. Interestingly, the loss of PBX in the epiblast selectively reduces FOXA2 expression in the definitive endoderm (DE) but does not affect its expression in the node63,64 (Fig. 5b; Supplementary Fig. 7a). Node formation depends on WNT signalling and is disrupted in Foxa2 deficient embryos61,63,64, whereas DE formation occurs independently of WNT. These findings suggests that PBX regulates Foxa2 expression in a lineage-specific manner, potentially by modulating ERK signalling and the associated DNA methylation machinery in the epiblast state. The pleiotropic nature of Foxa2, which is critical for both DE (WNT-independent) and node (WNT-dependent) fates, underscores the importance of this regulation. In ERK-deficient epiblast cells, differential hypomethylation of the Foxa2 locus resulted in disrupted DE formation while sparing node progenitors.
Altogether, loss of Pbx in the epiblast state leads to increased NMP differentiation and reduced DE differentiation, independent of its function as a HOX co-factor. Similarly, PD03 mediated inhibition of ERK in EpiSCs replicates this bias, favoring NMPs while diminishing DE formation (Supplementary Fig. 3d). These data align with the observation that CLDN6High EpiSCs preferentially form DE, while CLDN6Low EpiSCs are biased restricted towards NMPs.
Taken together, these findings show that ERK-mediated epigenetic priming in regionalized epiblast cells determines their divergent responses to the same differentiation signals, ultimately influencing lineage outcomes based on their epigenetic state.
Discussion
In the post-implantation epiblast, de novo DNA methylation plays a major role during the transition from pluripotency towards lineage commitment by targeting lineage-specific genes3. In this work, we identified primed subpopulations of EpiSCs corresponding to distinct regional identities within the epiblast. We reported that ERK signalling and regional identity within the epiblast are associated with divergent epigenetic landscapes that influence how subpopulations of EpiSCs adopt specific lineage programmes in response to differentiation cues. Our results shed light on the interplay between epigenetics and signalling pathways in determining cell fate.
Redefining pluripotency in an undifferentiated primed state: A single-cell or bulk population property?
Historically, pluripotency has been investigated as a bulk population-based concept. However, bulk transcriptome and lineage differentiation analyses offer only an averaged view of cellular properties, masking inherent differences between subpopulations. Identifying sorted cell populations with common characteristics enables a more precise assessment of the innate lineage potency of EpiSCs9. We challenged the EpiSC differentiation potential using flow cytometry-based sorting and reseeding experiments, and found that EpiSCs consist of a mix of pluripotent and multipotent subpopulations with differing epigenetic landscapes and self-renewal dynamics. These findings suggest a re-examination of bulk population-based studies, which previously suggested that EpiSCs are pluripotent9,15,16.
Furthermore, the existence of subpopulations with distinct epigenetic signatures may provide an explanation for why different hESC lines, which have similar “primed pluripotent” characteristics to EpiSCs, exhibit a canalized tendency to differentiate into specific lineages65. The differentiation potential of these cell lines could be heavily influenced by their intrinsic epigenetic signatures and the conditions in which they are cultured. Thus, understanding how to manipulate the intrinsic epigenetic landscapes of primed pluripotent stem cells is crucial for determining the correct cell lines to use for stem cell-based therapies.
Another caveat working with “primed pluripotent” cells is that they may express early differentiation markers when analysed as a bulk population9,66, raising questions whether the examined population is primed or a mix of differentiated cells. Are “primed differentiated” cells pluripotent? Recent reports have identified a primed mesoderm-fated population expressing primitive streak markers66, further complicating this distinction. These findings show that the WNT-dependent primed state of the “trapped” mesoderm-fated cells is medium-dependent and reversible, as cells regain pluripotency upon switching medium conditions. In our study, the characterized EpiSC populations do not express early mesoderm lineage markers, indicating that they remain in a primed, undifferentiated state without acquiring a primitive streak identity. Moreover, all EpiSC cultures analyzed were maintained and passaged under the same WNT-inhibited medium conditions containing FGF and ACTIVIN, attributing the observed differences to the intrinsic properties of the cells rather than external factors. Thus, is primed pluripotency defined by the innate nature of the cells, or the medium at the time of exposure “trapping” the cells in a transitory differentiated state? Without assessing the self-renewal capacity and differentiation potential of these transitory “trapped” cells at a single-cell or subpopulation level, there is insufficient evidence to conclusively determine whether the entire population is primed and pluripotent. Altogether, these results emphasize the need to refine definitions of primed pluripotency and fosters further discussion on key concepts in stem cell and developmental biology.
Differential ERK activity primes germ layer lineage identity
Using multiple cellular systems, we investigated how differential ERK levels and epigenetic states predetermine the primed states of epiblast subpopulations, influencing cellular response to key germ-layer differentiation signals like WNT and BMP22. Specifically, epiblast response to BMP signalling, crucial for specifying posterior germ-layer lineages including ExM, which give rise to allantois, yolk sac and early vasculature, is influenced by chromatin accessibility and methylome patterns at lineage-determining loci. By examining two BMP-responsive ExM lineage genes58,60, we showed that ERK-deficient epiblast cells are biased towards a Tbx3pos primitive erythroid/yolk sac over a Hand1pos endothelial/allantois fate. Our work provides molecular evidence supporting previous studies reporting that subpopulations of proximal posterior epiblast cells are distinctly primed towards endothelial and primitive erythroid lineages8.
Conversely, WNT signalling response, regulating anterior germ-layer lineage specification (DE, NMPs, neuroectoderm)5,10, appears to be influenced by lineage safeguarding DNA methylation signatures (Fig. 6a). Focusing on anterior versus distal posterior epiblast properties10, we sorted EpiSCs on the levels of the tight junction marker CLDN6. We showed that variations in FGF/ERK levels and lineage-specific DNA methylation signatures precede chromatin accessibility and transcriptional regulatory programmes inducing early neuroectoderm and DE decisions. CLDN6High EpiSCs, are transcriptionally similar to anterior epiblast cells, have higher ERK levels, and can reestablish pluripotency upon self-renewal, giving rise to all three germ layers. However, directly sorting and reseeding these cells showed that CLDN6High EpiSCs are biased towards neuroectodermal and DE fates. This observation aligns with previous studies that support the existence of a common neuroendoderm progenitor within the epiblast10,42. During gastrulation, the neuroectoderm and DE retain epithelial features, unlike their more mesenchymal mesoderm counterpart, which migrates rapidly, fuelling tissue diversity and body expansion in verterbrates10,12,67.
a Illustration of epigenetically driven priming in post-implantation epiblast cells toward neuroectoderm, DE, ExM, and NMP lineages. CLDN6High EpiSCs, with high pERK, resemble anterior epiblast in WNT-inhibited regions and give rise to neuroectoderm without PS ingress. In contrast, CLDN6Low EpiSCs show low pERK, resemble distal posterior epiblast, and exhibit methylation of neuroectoderm genes (Pax6, Onecut2, Dbx1), restricting neuroectodermal potential. Anterior epiblast cells (CLDN6High EpiSCs, high ERK) respond to WNT signalling signalling by forming DE via anterior PS differentiation. In contrast, distal posterior epiblast cells (CLDN6Low EpiSCs, low ERK) possess differential DNA methylation signature over DE genes (Foxa2, Gata6, Sox17), blocking the DE fate but enabling NMP formation upon WNT-induced APS differentiation. DNA methylation safeguards distal posterior epiblast cells from alternative fates, and subsequent chromatin remodelling operated by CDX5 and LEF1-PBX133 pioneer TFs leading to differentiation NMPs generating PSM. Our data indicate that WNT response in primed epiblast cells is shaped by DNA methylation. Proximal posterior epiblasts, with permissive chromatin/methylome, respond to BMP to form ExM tissues. Pbx1-KO EpiSCs have a permissive methylome and chromatin state over yolk sac and primitive erythroid genes (Tbx4, Gata2, Tal1), but repressed states at allantois/endothelial genes (Hand1, Tbx4, Kdr), failing to generate these fates. In contrast, WT EpiSCs, with hypomethylated and accessible allantois/endothelial loci, readily differentiate into these lineages but have reduced yolk sac/erythroid potential. b Revised Waddington’s landscape model illustrating how ERK dosage–dependent DNA methylation regulates cell fate commitment. DNA methylation modulates the balance between canalization and plasticity across germ-layer development, shaping trajectory paths in distinct signalling contexts (yellow, grey, orange, blue landscapes). During epiblast-to-germ-layer transition, cells (coloured balls) roll down lineage paths. DNA methylation (red gates) restricts certain fates, redirecting cells based on methylation signatures (red/blue marks). Dotted lines show fate trajectories shaped by methylation. Shifting signalling landscapes remodel chromatin (grey, blue, orange blocks), with dynamic methylation driving both canalization and plasticity in tissue development. (Adapted from an image by Conrad H. Waddington, licensed under CC BY-SA 4.0 (https://creativecommons.org/licenses/by-sa/4.0/). Source: https://commons.wikimedia.org/wiki/File:Paisagem_epigenetica.jpg. Modifications made by the author).
Conversely, CLDN6Low EpiSCs, indicative of distal posterior epiblast cells, have lower ERK levels, maintain their self-renewal properties with restricted differentiation potential, and show higher capacity to generate NMPs that ultimately will contribute to the somites and spinal cord. By sorting the CLDN6Low EpiSCs, we established the basis of an alternative protocol to enrich NMP-fated populations, which has been challenging to achieve in vitro68,69. Moreover, these NMP-fated EpiSCs are maintained in a WNT-inhibited state, providing an alternative perspective to previous reports that WNT primes posterior neural fates (via NMPs) in the epiblast5. In this paper, we describe an additional system for studying NMP expansion and differentiation.
Furthermore, ERK appears to prime epiblast cells towards anterior neural fates (forebrain and midbrain), whereas the posterior neural fates (hindbrain and spinal cord) require lower ERK (Supplementary Fig. 3c, d). We validated these phenotypic observations in different cellular models, including via dosed ERK inhibition. We found that ERK-dependent differential DNA methylation patterns predetermine neural lineage bias, suggesting a refinement of existing models of neural induction.
Site-specific epigenetic regulation across lineage genes fine-tunes stem cell potency
The global activity of epigenetic machinery has been extensively investigated as pluripotent cells transition towards germ-layer fates57, but how context-providing extracellular signals influence this machinery remains elusive. De novo DNA methylation in post-implantation epiblast cells coincides with FGF/ERK signalling requirements3. Perturbing the ERK cascade or its downstream effector TFs (such as the ETS family) in primed pluripotent cells by RNAi or selective cell-permeable chemical inhibitors induces differentiation19,70 or reversion to a naïve pluripotent state21. To address this limitation, we utilized Pbx1-KO EpiSCs, which have reduced ERK signalling but maintain an undifferentiated state, to investigate ERK-regulated epigenetic mechanisms. Notably, PBX proteins have been implicated in FGF/ERK feedback regulation47,48, and epiblast deletion of Pbx1 results in aberrant lineage acquisition and strong ExM defects (Fig. 5b, d). Probing the molecular mechanisms influencing ExM lineage choice6,7, we observed differential DNA methylation and chromatin accessibility on BMP-responsive ExM lineage genes in Pbx1-KO EpiSCs (Fig. 5g–j). We observed differential expression of ERK downstream cascade effectors such as ETS and JUN/FOS TFs in WT vs Pbx1-KO EpiSCs. Motif enrichment analyses across the differentially accessible regions in WT vs Pbx1-KO EpiSCs highlighted different ETS TFs as putative epigenetic regulators. Altogether, we propose that ERK/ETS drives the recruitment of epigenetic machinery to lineage-specific genes in primed epiblast populations before their transcriptional activation. This hypothesis aligns with the established role of ETS TFs as site-specific recruiters of epigenetic machinery across lineage genes in different developmental contexts20. Thus, the Pbx-KO models provide insight into the ERK-dependent epigenetic mechanisms, providing clues to the origin of cellular specificity to BMP differentiation signals in the selection of ExM lineage choices (Fig. 6a; Supplementary Fig. 6).
Pleiotropic role of TALE proteins in spatiotemporal regulation of embryonic development
We demonstrated that Pbx1-KO EpiSCs recapitulate the immediate germ layer lineage biases observed upon dosed ERK inhibition in EpiSCs, validating the ERK-dependent phenotype. Specifically, we showed that loss of Pbx1 results in increased differentiation towards NMPs over DE (Supplementary Fig. 7a–d). However, unlike CLDN6Low EpiSCs and PD03-treated EpiSCs, where NMPs robustly differentiate into PSM, Pbx1-KO NMPs fail to form PSM. This observation is consistent with the accumulation of NMPs and a block in PSM differentiation in Pbx-KO tailbuds in vivo33. Thus, the role of PBX proteins in the epiblast differs from its previously described MEIS-HOX-dependent function in NMPs, where they regulate WNT-driven PSM formation (Supplementary Fig. 7)33. Our results suggest an alternative PBX-PREP1 complex regulation of ERK activity in the epiblast, independent of its well-established role as a HOX co-factor46. Taken together, these findings shed light on the intricate pleitropic roles of TALE proteins in regulating signalling networks coordinating spatiotemporal aspects of embryonic development.
Insights for cancer and therapeutic applications
The signalling networks and epigenetic machinery surrounding germ layer specification are reactivated in tumorogenesis and metastasis40. Interaction between ERK, BMP and WNT is widely observed in colon cancer71. Despite ERK dose-dependent dynamics in colorectal cancer49,72, therapies still focus on MEK/ERK chemical inhibition rather than on the feedback loop73. Interestingly, ectopic expression of PBX in low metastatic colorectal cancer cells promote migration and invasion by increasing pERK levels, whereas Pbx inhibition lowered pERK dosage, reducing cell migration and invasion49. Our in vitro and in vivo studies provide a biological perspective to PBX1 regulation of ERK driving colorectal cancer. We showed that ERK dosage influences how cells respond to the same signals giving differing outputs. CLDN6Low EpiSCs (Lower ERK) express Ets1, have distinct mesenchymal features, and subsequently differentiate towards mesodermal lineages, which possess migratory properties.
CLDN6High EpiSCs (Higher ERK) express Ets2, exhibit more epithelial features, and preferentially differentiate towards neuroectoderm and DE, which have epithelial features10. Our observations align with the reported ERK-driven opposing migratory functional roles of Ets1 and Ets2 in different cancer cell lines27,74. Based on these findings, we propose reconsidering MEK inhibition therapies that target lineage switching in colorectal cancer75,76,77,78. A potentially effective approach could involve targeting uniquely expressed ETS79,80,81,82 or PBX proteins49, which modulate ERK levels and influence the oncogenic epigenome.
Proposed model of lineage determination
In summary, our study offers detailed understanding of the epigenetic signatures that precede spatiotemporal lineage fate acquisition during gastrulation. We propose a model where DNA methylation and chromatin accessibility balance canalization and plasticity2, enabling context-specific responses to differentiation signals such as BMP and WNT (Fig. 6b). These mechanisms drive the formation of diverse germ-layer tissues and offer important insights into both developmental biology and disease.
Methods
Cell lines
EpiSC lines derived from E6.5 mouse embryos (129S6/SvEvTac background) were a kind gift from Dr. M.M. Rothová (The Novo Nordisk Foundation Centre for Stem Cell Biology, University of Copenhagen, Denmark). The lines were derived in EpiSC medium as previously described83. EpiSC lines were maintained in 6-well cell-culture plates (Corning, 3506) pre-coated with 15 µg/ml human Fibronectin (Merck Millipore, FC01015) in N2B27 medium, prepared as described in Ying et al.84, supplemented with 20 ng/ml Activin A, 10 ng/ml bFGF and 1 µM XAV939 (EpiSC medium). Chemical inhibition of ERK signalling via PD03 was tested at different dosages (0.1 µM, 0.5 µM and 1 µM), and treated over 2–3 passages where applicable. Mouse ES lines were cultured in 0.1% gelatin-coated 6-well cell-culture plates. Mouse ES cells were cultured in standard conditions in either serum/LIF or 2i/LIF conditions with CHIR-99021, PD03, and LIF (made in-house). All lines used in this study were karyotyped and routinely tested for Mycoplasma.
Generation of Pbx1 ES/EpiSC lines using the CRISPR/Cas9 system
The strategy to inactivate Pbx1 in ES cells and EpiSCs was the same. Pbx1-KO lines were generated by inserting a floxed CMV-hygromycin-Poly (A) cassette between Exon 2 and Intron 2 of Pbx1 to disrupt Pbx1 gene expression, employing the CRISPR Nickase system. Guide RNAs (gRNAs) were designed using the CRISPR/Cas9 design tool (crispr.mit.edu) and cloned into pX335 (a gift from Prof. F. Zhang; Addgene, 42335). Cells were co-transfected with a pair of gRNA-pX335 plasmids (gRNA 1: GTCCAGGCTCTGGTCCGTGA; gRNA 2: CCAAGTGAGTGCCCCCACCC; 100 ng each vector) and 1 µg linearized targeting vector using Lipofectamine 2000 (Thermo Fisher Scientific, 11668030), seeded at low density and selected for hygromycin resistance for 8–10 days. Single colonies were screened by diagnostic PCR, Southern blot, and Western blot analyses. Pbx1 gRNA used: GGCCATCCCGACCCCAGCGT.
Flow cytometry analysis and Fluorescence-Activated Cell Sorting (FACS)
EpiSCs were seeded at ~6 x 103 cells/cm2 in 6-well cell-culture plates pre-coated with 15 µg/ml human Fibronectin. Cells were dissociated using Accutase for 3 min at 37 °C and counted. 150 x 106 cells per condition were incubated with the CLDN6 Alexa Fluor 488-conjugated antibody (R&D Systems, FAB3656G, 1:200) and Ghost Dye™ Red 780 (Tonbo, 13-0865, 1:1000) for 20 min on ice, washed with ice-cold FACS buffer (10% (v/v) FBS in PBS) and resuspended into 1 ml. Cells were sorted into MACSQuant Tyto Running Buffer using the MACSQuant Tyto sorter (Miltenyi Biotec) or into ice-cold FACS buffer using the SONY SH800S cell sorter (Sony Biotechnology) at the DanStem Flow Cytometry Platform (University of Copenhagen, Copenhagen, Denmark). All reseeding experiments were performed using the MACSQuant Tyto sorter (Miltenyi Biotec), taking advantage of high-speed, multi-parameter flow sorting in the safety of a fully enclosed cartridge to prevent contamination. Post-sorting, cells were reseeded into Fibronectin pre-coated 6-well cell culture plates at around 10.5 x 103 cells/cm2.
EpiSC differentiation to germ-layer lineages
All differentiation was conducted in serum-free, feeder-free, and monolayer conditions in chemically-defined N2B27-based media. EpiSCs were seeded at ~10.5 x 103 cells/cm2 in 6-well cell-culture plates pre-coated with 15 µg/ml human Fibronectin and cultured in EpiSC medium for 24 h. EpiSCs were differentiated towards either APSD using 30 ng/ml Activin A, 4 µM CHIR-99021, 20 ng/ml bFGF, and 100 nM PIK-90 (APS medium) for 24 h. For NMPs/PSM differentiation, EpiSCs were first differentiated towards APSD, and then to NMPs/PSM using 1 µM A-83-01, 3 µM CHIR-99021, 250 nM LDN193189 and 20 ng/ml bFGF for 48-72 h (PSM medium). For DE differentiation, EpiSCs were first differentiated towards APSD in N2B27-basal medium over 24 h, and then to DE using 30 ng/ml Activin A, 4 µM CHIR-99021, and 100 nM PIK-90 in RPMI(Glutamax) + B27 (without insulin) basal medium (DE medium) for 48 h. EpiSCs were differentiated towards neuroectoderm based on known media requirements17. Differentiation of EpiSCs towards PPS and ExM was done as previously described52. A complete list of cytokines and signalling pathway inhibitors used for cell maintenance and differentiation is provided in Supplementary Table 1.
Mice
The Tg(Eomes::GFP) BAC transgenic strain is described by Kwon31. The Pbx mutant alleles used in this study were previously described33. The Tg(T-Cre)1Lwd allele85 used for inactivating Pbx1 in the primitive streak and its lineage derivatives, was purchased from MRC Harwell Institute under MTA agreement (#NCI, MTA 47362-20). All animals used in this study were maintained in laboratory animal housing facilities at macroenvironmental temperature and humidity ranges of 20–24 °C and 45–65%, respectively, with a 12/12 h light/dark cycle. All animal work was carried out under European legislation. All work was authorized and carried out under the Project License 2017-15-0201-01255 issued by the Danish Regulatory Authority.
Immunofluorescence staining of adherent cells
EpiSCs were seeded at ~4 x 103 cells/cm2 in EpiSC medium in 8-well microscopy slides (Ibidi, 80826) pre-coated with 15 µg/ml human Fibronectin, and differentiated to PS and derivatives as described above. At desired time points, cells were briefly washed twice in PBS (Sigma, D8537) and fixed in 4% (w/v) paraformaldehyde diluted in PBS from 16% methanol-free formaldehyde stock (Pierce Biotechnologies, 28906) for 15 min at RT. Fixed cells were then permeabilized in PBS containing 0.3% (v/v) Triton X-100 (PBST) for 10 min at RT and blocked in 3% (v/v) Donkey serum in PBST containing 1% (w/v) BSA (blocking solution) for 15 min at RT. Primary and secondary antibody incubations were performed in the blocking solution overnight at 4 °C and for 2 h at RT, respectively. Antibody incubations were followed by three quick washes in PBST, followed by two washes in PBST for 15 min at RT. After the final incubation, cells were stored in PBST containing 1 µg/ml DAPI at 4 °C in the dark and packed in Parafilm to prevent evaporation. Confocal imaging was performed using a Leica TCS SP8 or Zeiss LSM 770 microscope, and images were processed using the ImarisTM software with default settings. The antibodies used are listed in Supplementary Table 2.
Immunofluorescence staining of embryos
Embryos were dissected in cold PBS and fixed in 4% (w/v) paraformaldehyde for 30 min at RT and processed as described in Mariani et al.33 Images were acquired using a Leica TCS SP8 confocal microscope. Image processing and 3D reconstruction were performed using the ImarisTM software. Antibodies are listed in Supplementary Table 3.
Imaging of embryo morphology
Embryos were dissected in cold PBS, fixed in 4% (w/v) paraformaldehyde for 30 min at RT, and washed twice in cold PBS. Images were acquired using a Zeiss AxioZoom.V16 microscope at the Core Facility for Integrated Microscopy (University of Copenhagen, Copenhagen, Denmark).
Electrophoretic Mobility Shift Assay (EMSA)
EMSAs were performed as described33, using in vitro-translated proteins or nuclear extracts purified from cells. Sequences of the oligonucleotides spanning the PBX binding site on the Spry2 regulatory region used in EMSA are described in Supplementary Fig. 7a. The uncropped image can be found in the source data file.
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from 0.5-1 x 106 cells using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen: ID: 74104). First-strand synthesis was performed on 1 μg of total RNA using random hexamers (Thermo Fisher Scientific, N8080127) and SuperScript III reverse transcriptase (Thermo Fisher Scientific, 18080-044) according to the manufacturer’s instructions. A mix of concentrated cDNA from all samples was used to generate standard curves. Amplification was detected using the SYBR Green master mix (Roche, 04707516001) or the Universal Probe Library system on a LightCycler480 Real-Time PCR System (Roche). The measures were normalized to Tbp, Sdha, and Pgk1 reference genes unless mentioned otherwise. All reactions were performed in duplicates. See Supplementary Table 4 for a list of primers and probes used.
Western Blot
Immunoblottings were performed as described33. The following primary antibodies were used: polyclonal Rabbit anti-PBX1 (CST, 4342, 1:1000), Rabbit anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) XP® (CST, 4370, 1:1000), Rabbit anti-phospho-MEK1/2 (Ser217/221) (41G9) (CST, 9154, 1:1000), Rabbit anti-p44/42 MAPK (total Erk1/2) (137F5) (CST, 4695, 1:1000) and monoclonal Mouse anti-H3 (Abcam, ab10799, 1:30,000). The secondary antibodies used include Donkey anti-Rabbit HRP (Thermo Fisher Scientific, A16023, 1:10,000) and Donkey anti-Mouse HRP (Thermo Fisher Scientific, A16011, 1:10,000). For chemiluminescence detection, the membrane was incubated with Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare, RPN2236) and imaged on a ChemiDoc MP (Bio-Rad). For reprobing with different primary antibodies, the membrane was soaked in Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific, 46430) 2 times for 10 min at RT to inactivate horseradish peroxidase (HRP).To ensure comparable densitometry results, gel electrophoresis, protein transfer, and antibody staining were performed in parallel for all samples used to generate each Western blot figure panel. All the uncropped images are found in the source data file.
scRNAseq procedure and analysis
ScRNAseq was performed using the Massively Parallel Single-Cell RNA-sequencing technology (MARS-seq). For in vitro experiments, EpiSCs were seeded at ~6 x 103 cells/cm2 in 6-well cell-culture plates pre-coated with 15 µg/ml human Fibronectin and differentiated to APSD as described above. At desired time points, cells were dissociated using Accutase (Thermo Fisher Scientific, 00-4555-56) for 3 min at 37 °C and counted. 500,000 cells per condition were washed with ice-cold FACS buffer (10% (v/v) FBS in PBS) and resuspended in 1 ml ice-cold FACS buffer containing 1 µg/ml DAPI. For in vivo experiments, E6.5/E7.0/E7.75 Tg(Eomes::GFP) BAC transgenic embryos were individually dissociated into 300 μl warmed Trypsin 0.05% (w/v) EDTA for 10 min at 37 °C. Digestion was stopped by adding 600 μl ice-cold FACS buffer and centrifugation at 350 x g for 3 min. Cells were washed with 500 μl ice-cold FACS buffer, centrifuged again at 350 x g for 3 min, and resuspended into 250 μl ice-cold FACS buffer containing 1 µg/ml DAPI. Single cells from either in vitro-differentiated cells or transgenic embryos were sorted into Eppendorf Polypropylene U-shaped 384-well Twin Tec PCR Microplates (Thermo Fisher Scientific, 10573035), containing 2 μl of lysis solution (0.2% (v/v) Triton X-100) supplemented with 0.4 U/μl RNasin Ribonuclease Inhibitor (Promega, N2515) and 400 nM indexed RT primer from group 1 (1-96 barcodes) or group 2 (97-192 barcodes) as described86. Additionally, 71 WT EpiSCs were sorted into each plate as a spike-in control for batch-effect correction. Capture plates were prepared on the Bravo automated liquid handling robot station (Agilent) using 384-filtered tips (Axygen, 302-82-101). Index sorting was performed using either a FACS Aria III cell sorter (BD Biosciences) or a SONY SH800S cell sorter (Sony Biotechnology) at the DanStem/reNEW Flow Cytometry Platform (University of Copenhagen, Denmark), gating in SSC-A versus FSC-A to collect live cells, and then in FSC-W versus FSC-A to sort only singlets. For in vivo experiments, only GFPpos cells were sorted to capture Eomes-expressing cell types. Immediately after sorting, plates were spun down, snap-frozen on dry ice, and stored at −80 °C until further processing. Semi-automated library preparation was performed as described in Jaitin et al., 2014, using 10-12 cycles of PCR amplification and AMPure XP beads (Beckman Coulter™Agencourt: A63881) for purification. DNA concentration was measured with a Qubit Fluorometer (Thermo Fisher Scientific, Q32854), and fragment size was determined with a Fragment analyzer (Advanced Analytical). Libraries were paired-end sequenced on a NextSeq 500 Sequencer (Illumina) at the DanStem/reNEW Genomics Platform (University of Copenhagen, Denmark). Between 1146 and 1528 cells were sequenced per lane.
R1 and R2 fastq files were generated using bcl2fastq (v2.19.1), and the pooling and well information was extracted from the sequence using umis (v1.0.3) [https://github.com/vals/umis] into a unique fastq file. The reads were then filtered based on the pooling barcodes with 1 mismatch allowed. The poly-Ts at the end of the reads were trimmed using Cutadapt (v1.18)87. The reads were mapped to the mouse genome (GRCm38/mm10 together with ERCC92) using HISAT2 (v2.1.0), the alignments were processed with Samtools (v1.7)88, and the reads were counted with featureCounts (Subread (v1.5.3))89 using Ensembl v93, and the UMIs using UMI_tools (v1.0.0)90. Expression data were analyzed using Seurat (v3.1)91. Data filtering, normalization, and scaling were performed using the standard pre-processing workflow91. Integration of different datasets was performed as described92. Spiked-in EpiSCs were used as a reference to correct the batch effect between integrated datasets. Marker genes of each cell cluster were outputted for GO-term analysis to define the cell type.
RNAseq procedure and analysis
Cells were lysed, and total RNA was extracted using the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen). RNA concentration was quantified by NanoDrop, and quality was verified with a Fragment analyzer. For all conditions, two biological replicate samples were collected from independent experiments. Library preparation was carried out using 0.5 μg of RNA with NEBNext Ultra II RNA Library Prep Kit (New England BioLabs, E7770S) as per the manufacturer’s instructions. Libraries were amplified for 5 PCR cycles and purified with AMPure XP beads. DNA concentration was measured with a Qubit Fluorometer, and fragment size was determined with a Fragment Analyzer. All samples were sequenced on a NextSeq 500 Sequencer (Illumina) at the DanStem/reNEW Genomics Platform (University of Copenhagen, Denmark). RNAseq was performed, mapped, and analyzed as previously described33.
ATACseq procedure and analysis
ATACseq was performed, mapped, and analyzed as previously described33. For all conditions, two biological replicate samples were collected from independent experiments. Library quality was assessed using the Fragment Analyzer and quantified by Qubit assay. The libraries were sequenced with 50 bp paired-end reads on a NextSeq 500 Sequencer (Illumina) at the DanStem/reNEW Genomics Platform (University of Copenhagen, Denmark). Prediction of cis-regulatory elements (CREs) and gene annotation was made using rGREAT (v4.0.4)93,94 or HOMER (v4.7.2)95.
ChIPseq procedure and analysis
ChIPseq was performed, mapped, and analyzed using a previously detailed protocol33. All samples were sequenced on a NextSeq 500 Sequencer (Illumina) at the DanStem/reNEW Genomics Platform (University of Copenhagen, Copenhagen, Denmark). Prediction of cis-regulatory elements (CREs) and gene annotation was made using rGREAT (v4.0.4)93,94.
Spatial transcriptomics
Embryos were collected at E7.5 and fixed accordingly to the protocol of Resolve BioSciences’ MC technology96,97, with PAXgene fixative sample preparation (Qiagen: ID: 765312) for 1 h, followed by PAXgene stabilizer (Qiagen, ID: 765512) for 2 h, transferred to 30% sucrose in PBS for 30 min and finally included in OCT (Sakura) and snap-frozen in liquid nitrogen for cryo-sectioning. The samples were cryo-sectioned into 10 μm sections using an HM560 cryostat onto Resolve coverslips. Resolve BioSciences’ MC technology performed the slide hybridization according to the manufacturer’s instructions (protocol 3.0). The probes for 29 genes were designed using Resolve’s proprietary design algorithm. Imaging and signal decoding were performed by BioSciences’ MC technology according to the company’s standard procedures. Final visualization was performed using the ImageJ plugins from Resolve BioSciences (Polylux). The spatial information and colour attribution to RNA were attributed by the count matrix format provided by Resolve BioSciences and ImageJ. Representative images are from E7.5 embryos (n = 4 biological replicates).
GEO-sequencing
Published RNAseq data from iTranscriptome (https://www.picb.ac.cn/hanlab/itranscriptome/Home/) based on genes expressed in the epiblast of the gastrulating mouse embryo (E7.0-7.5)28,29.
Whole Genome Bisulfite Sequencing (WGBS) and analysis
DNA was extracted from sample pellets using the Qiagen DNeasy Blood & Tissue Kit (Qiagen ID: 69506) following the manufacturer’s instructions. 1 μg of sample gDNA was used to prepare the libraries. We utilized the EZ DNA Methylation-Lightning Kit (Zymo Research, #D5031) for bisulfite conversion. CLDN6Low versus CLDN6High WGBS library preparation was done using NEBNext Ultra II DNA Library Prep Kit (E7645S), enzymatic fragmentation using NEBNext dsDNA Fragmentase (M0348S), and sensitive adaptors NEBNext Multiplex Oligos (Methylated Adaptor, E7535S), as per manufacturer’s instructions. Samples were sequenced on NextSeq2000 (200 cycles kit), averaging 256M reads per sample. WT versus Pbx1-KO WGBS libraries were prepared via the post-bisulfite library construction method using Illumina’s TruSeq DNA Methylation Kit (EGMK81312) with sensitive adaptors (EGIDX81312) following the manufacturer’s instructions. Samples were sequenced three times on the NextSeq500 (150 cycles kit), and the three runs were concatenated to get an average of 256M reads per sample.
Adapter trimming, sequence alignment, and methylation calls were performed using Bismark (v0.18.1)98. Differential methylation analyses were performed using methylKit (v1.25.0)99, applying Fisher’s exact test for comparing the fraction of methylated Cs in differentially probed samples, with a cutoff for the absolute value of methylation percentage change (25%) and qvalue of differential methylation statistic (0.01). Differentially methylated Cs were annotated with rGREAT (v4.0.4) using a single nearest gene approach93 and defined as differentially methylated regions near genes (DMGs). The coverage cutoff was set at 10 in the CLDN6 sorted dataset and 4 in WT versus Pbx1 dataset, respectively. The output from Bismark was also reported into SeqMonk (v1.46.0; Babraham Bioinformatics) for parallel DSS-based100 methylation validation analyses and WGBS data visualization over key genes of interest. On the visualization track, FirstEF101 shows predicted CREs. Hypo- and Hyper-methylated regions were not separately probed since multiple studies determined that it does not correlate directly with gene expression as these regions could be bound by repressors, especially in the early lineage transcription factors of interest in this study41,62.
STRING
Protein-protein interaction networks were analyzed using STRING (v11.5)56 based on known and predicted associations. Unbiased MCL clustering of networks was done based on the simulation of stochastic flow. The inflation parameter was set as default (3), and edges between clusters are denoted by dotted lines.
Gene ontology and Motif enrichment analysis
Biological pathway enrichments were determined using Panther (v15.0)102. Statistical significance was assessed using the Binomial test with Bonferroni correction for multiple testing (P ≤ 0.05). We performed known motif enrichment analysis using HOMER (v4.7.2)95.
Statistics and reproducibility
The sample size was estimated based on previous preliminary experiments. No statistical method was used to pre-determine the sample size. Experiments and quantifications were not done in a blinded fashion. Each differentiation (and subsequent RT-qPCR analysis) was performed at least three or more times independently with similar results. Samples used for RT-qPCR and RNA-seq come from different differentiation batches, ensuring multiple biological replicates and reproducibility. Each immunoblot and EMSA experiment was repeated at least three times independently. Unless otherwise noted, each immunofluorescence staining and phase-contrast photomicrographs were repeated at least three times independently with similar results. Statistics were derived using GraphPad Prism (v8.4.3) (GraphPad Software, San Diego, CA, USA). The raw data underlying all reported averages in graphs and charts are provided in the Source data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated in this study are included in this article and its Supplementary Information files. NGS data supporting the findings have been deposited in the ArrayExpress database at EMBL-EBI under the following accession numbers: E-MTAB-13407, E-MTAB-13574, E-MTAB-13618, E-MTAB-13660. Further data from published papers: (scRNA-seq of embryonic tailbuds, E-MTAB-9785; scRNA-seq of in vitro cells, ATAC-seq, E-MTAB-9776, ChIP-seq of LEF1 in NMPs, E-MTAB-9775)33 [https://doi.org/10.1038/s41467-021-25370-4]; (ATAC-seq - E-MTAB-6337)5 [https://doi.org/10.1016/j.cell.2018.09.040]; (Stereo-sequencing: 3D Transcriptome of the mouse embryo at gastrulation, GSE65924) [https://doi.org/10.1016/j.devcel.2016.02.020. Source data are provided with this paper.
References
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Pujadas, E. & Feinberg, A. P. Regulated noise in the epigenetic landscape of development and disease. Cell 148, 1123–1131 (2012).
Borgel, J. et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 42, 1093–1100 (2010).
Auclair, G., Guibert, S., Bender, A. & Weber, M. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 15, 545 (2014).
Metzis, V. et al. Nervous system regionalization entails axial allocation before neural differentiation. Cell 175, 1105–1118 e17 (2018).
Harland, L. T. G. et al. The T-box transcription factor Eomesodermin governs haemogenic competence of yolk sac mesodermal progenitors. Nat. Cell Biol. 23, 61–74 (2021).
Kinder, S. J. et al. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126, 4691–4701 (1999).
Padron-Barthe, L. et al. Clonal analysis identifies hemogenic endothelium as the source of the blood-endothelial common lineage in the mouse embryo. Blood 124, 2523–2532 (2014).
Kojima, Y. et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14, 107–120 (2014).
Scheibner, K. et al. Epithelial cell plasticity drives endoderm formation during gastrulation. Nat. Cell Biol. 23, 692–703 (2021).
Osteil, P. et al. Dynamics of Wnt activity on the acquisition of ectoderm potency in epiblast stem cells. Development 146 (2019).
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Quinlan, G. A., Williams, E. A., Tan, S. S. & Tam, P. P. Neuroectodermal fate of epiblast cells in the distal region of the mouse egg cylinder: implication for body plan organization during early embryogenesis. Development 121, 87–98 (1995).
Gouti, M. et al. A gene regulatory network balances neural and mesoderm specification during vertebrate trunk development. Dev. Cell 41, 243–261.e7 (2017).
Brons, I. G. et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 (2007).
Tesar, P. J. et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 (2007).
Greber, B. et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215–226 (2010).
Perera, M. & Brickman, J. M. Common modes of ERK induction resolve into context-specific signalling via emergent networks and cell-type-specific transcriptional repression. Development 151 (2024).
Goke, J., Chan, Y. S., Yan, J., Vingron, M. & Ng, H. H. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol. Cell 50, 844–855 (2013).
de la Rica, L. et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 (2013).
Ficz, G. et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell 13, 351–359 (2013).
Martyn, I., Kanno, T. Y., Ruzo, A., Siggia, E. D. & Brivanlou, A. H. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 558, 132–135 (2018).
Khoa le, T. P. et al. Visualization of the Epiblast and visceral endodermal cells using Fgf5-P2A-Venus BAC transgenic mice and epiblast. Stem Cells PLoS One 11, e0159246 (2016).
Ciruna, B. & Rossant, J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell 1, 37–49 (2001).
Rajalingam, K. et al. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat. Cell Biol. 7, 837–843 (2005).
Guo, W. et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 (2012).
Plotnik, J. P., Budka, J. A., Ferris, M. W. & Hollenhorst, P. C. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res 42, 11928–11940 (2014).
Chen, J. et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 12, 566–580 (2017).
Peng, G. et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell 36, 681–697 (2016).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Kwon, G. S. & Hadjantonakis, A. K. Eomes::GFP-a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis 45, 208–217 (2007).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Mariani, L. et al. A TALE/HOX code unlocks WNT signalling response towards paraxial mesoderm. Nat. Commun. 12, 5136 (2021).
Shirane, K. et al. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell 39, 87–103 (2016).
Deaton, A. M. et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 21, 1074–1086 (2011).
Maunakea, A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Ronnerblad, M. et al. Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood 123, e79–e89 (2014).
Lorincz, M. C., Dickerson, D. R., Schmitt, M. & Groudine, M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 11, 1068–1075 (2004).
Li, X. et al. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. USA 113, E8267–E8276 (2016).
Smith, Z. D. et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 549, 543–547 (2017).
Ziller, M. J. et al. Dissecting the functional consequences of De Novo DNA methylation dynamics in human motor neuron differentiation and physiology. Cell Stem Cell 22, 559–574.e9 (2018).
Miura, S. & Mishina, Y. The DVE changes distal epiblast fate from definitive endoderm to neurectoderm by antagonizing nodal signaling. Dev. Dyn. 236, 1602–1610 (2007).
Staber, P. B. et al. Sustained PU.1 levels balance cell-cycle regulators to prevent exhaustion of adult hematopoietic stem cells. Mol. Cell 49, 934–946 (2013).
Choi, J. et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature 548, 219–223 (2017).
Di Stefano, B. et al. Reduced MEK inhibition preserves genomic stability in naive human embryonic stem cells. Nat. Methods 15, 732–740 (2018).
Selleri, L., Zappavigna, V. & Ferretti, E. ‘Building a perfect body’: control of vertebrate organogenesis by PBX-dependent regulatory networks. Genes Dev. 33, 258–275 (2019).
Delgado, I. et al. Control of mouse limb initiation and antero-posterior patterning by Meis transcription factors. Nat. Commun. 12, 3086 (2021).
Osterwalder, M. et al. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev. Cell 31, 345–357 (2014).
Han, H. B. et al. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J. Gastroenterol. 20, 18260–18270 (2014).
Lamprecht, S. et al. PBX3 is part of an EMT regulatory network and indicates poor outcome in colorectal cancer. Clin. Cancer Res 24, 1974–1986 (2018).
Argelaguet, R. et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019).
Drozd, A. M. et al. Progesterone receptor modulates extraembryonic mesoderm and cardiac progenitor specification during mouse gastrulation. Int. J. Mol. Sci. 23 (2022).
Hanafusa, H., Torii, S., Yasunaga, T. & Nishida, E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 4, 850–858 (2002).
Nabet, B. et al. Deregulation of the Ras-Erk signaling axis modulates the enhancer landscape. Cell Rep. 12, 1300–1313 (2015).
Gustems, M. et al. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 42, 3059–3072 (2014).
Szklarczyk, D. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Grosswendt, S. et al. Epigenetic regulator function through mouse gastrulation. Nature 584, 102–108 (2020).
Zheng, M., Erhardt, S., Ai, D. & Wang, J. BMP signaling regulates Hand1 in a dose-dependent manner during heart development. Int. J. Mol. Sci. 22 (2021).
Morikawa, Y. & Cserjesi, P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development 131, 2195–2204 (2004).
Davenport, T. G., Jerome-Majewska, L. A. & Papaioannou, V. E. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development 130, 2263–2273 (2003).
Beck, F., Erler, T., Russell, A. & James, R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219–227 (1995).
Bahar Halpern, K., Vana, T. & Walker, M. D. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. J. Biol. Chem. 289, 23882–23892 (2014).
Ang, S. L. & Rossant, J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78, 561–574 (1994).
Kitajima, K., Oki, S., Ohkawa, Y., Sumi, T. & Meno, C. Wnt signaling regulates left-right axis formation in the node of mouse embryos. Dev. Biol. 380, 222–232 (2013).
Wu, H. et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc. Natl. Acad. Sci. 104, 13821–13826 (2007).
Stavish, D. et al. Generation and trapping of a mesoderm biased state of human pluripotency. Nat. Commun. 11, 4989 (2020).
Ferretti, E. & Hadjantonakis, A. K. Mesoderm specification and diversification: from single cells to emergent tissues. Curr. Opin. Cell Biol. 61, 110–116 (2019).
Edri, S., Hayward, P., Baillie-Johnson, P., Steventon, B. J. & Martinez Arias, A. An epiblast stem cell-derived multipotent progenitor population for axial extension. Development 146 (2019).
Sambasivan, R. & Steventon, B. Neuromesodermal progenitors: a basis for robust axial patterning in development and evolution. Front Cell Dev. Biol. 8, 607516 (2020).
Sterneckert, J. et al. Neural induction intermediates exhibit distinct roles of Fgf signaling. Stem Cells 28, 1772–1781 (2010).
Kim, D., Rath, O., Kolch, W. & Cho, K. H. A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene 26, 4571–4579 (2007).
Brandt, R. et al. Cell type-dependent differential activation of ERK by oncogenic KRAS in colon cancer and intestinal epithelium. Nat. Commun. 10, 2919 (2019).
Sclafani, F. MEK and PD-L1 inhibition in colorectal cancer: a burning blaze turning into a flash in the pan. Lancet Oncol. 20, 752–753 (2019).
Neri, F. et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017).
Korkaya, H. & Wicha, M. S. Cancer stem cells: nature versus nurture. Nat. Cell Biol. 12, 419–421 (2010).
Tse, J. W. T., Jenkins, L. J., Chionh, F. & Mariadason, J. M. Aberrant DNA methylation in colorectal cancer: what should we target?. Trends Cancer 3, 698–712 (2017).
Harmston, N. et al. Widespread repression of gene expression in cancer by a Wnt/beta-Catenin/MAPK Pathway. Cancer Res 81, 464–475 (2021).
Heide, T. et al. The co-evolution of the genome and epigenome in colorectal cancer. Nature 611, 733–743 (2022).
Deves, C. et al. Analysis of select members of the E26 (ETS) transcription factors family in colorectal cancer. Virchows Arch. 458, 421–430 (2011).
Eskandari, E., Mahjoubi, F. & Motalebzadeh, J. An integrated study on TFs and miRNAs in colorectal cancer metastasis and evaluation of three co-regulated candidate genes as prognostic markers. Gene 679, 150–159 (2018).
Jiang, W., Xu, Y., Chen, X., Pan, S. & Zhu, X. E26 transformation-specific variant 4 as a tumor promotor in human cancers through specific molecular mechanisms. Mol. Ther. Oncolytics. 22, 518–527 (2021).
Cheng, X. et al. ETS variant 5 promotes colorectal cancer angiogenesis by targeting platelet-derived growth factor BB. Int J. Cancer 145, 179–191 (2019).
Guo, G. et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 (2009).
Ying, Q. L. & Smith, A. G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327–341 (2003).
Perantoni, A. O. et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132, 3859–3871 (2005).
Jaitin, D. A. et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014).
M, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12 (2011).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive Integration of Single cell data. Cell 177, 1888–1902.e21 (2019).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Tanigawa, Y., Dyer, E. S. & Bejerano, G. WhichTF is functionally important in your open chromatin data?. PLoS Comput. Biol. 18, e1010378 (2022).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Rothova, M. M. et al. Identification of the central intermediate in the extra-embryonic to embryonic endoderm transition through single-cell transcriptomics. Nat. Cell Biol. 24, 833–844 (2022).
Silvia Groiss, D. P. et al. Highly resolved spatial transcriptomics for detection of rare events in cells. bioRxiv (2021).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012).
Wu, H. et al. Detection of differentially methylated regions from whole-genome bisulfite sequencing data without replicates. Nucleic Acids Res. 43, e141 (2015).
Davuluri, R. V., Grosse, I. & Zhang, M. Q. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29, 412–417 (2001).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Acknowledgements
We thank the staff of the DanStem Core Facilities: M. Michaut, H.M. Neil, H. Wollmann, G. De la Cruz, P. van Dieken, J.M. Bulkescher, A. Shrestha, M. Paulsen, and E. Fernandez-Rebollo. We thank Anna-Katerina Hadjantonakis for the Eomes-GFP mice. We are grateful to M.M. Rothová for sharing the wild-type 129 EpiSC line, L. Selleri for the Pbx mutant alleles, and the Core Facility for Transgenic Mice at the University of Copenhagen, especially J. Martin Gonzalez, for mouse line derivation. Special mention to Benjamin Claass and Olof Berggren from Miltenyi Biotec Norden AB for their support with the MACS Tyto sorters, which were crucial for the sorting and reseeding experiments. We thank Anna Maria Drozd, Jan Zylicz and Anna-Katerina Hadjantonakis for critically reading the manuscript. The Novo Nordisk Foundation Centre for Stem Cell Medicine, reNEW, is funded by the Novo Nordisk Foundation (NNF21CC0073729). Data analysis performed using the Danish National Life Science Supercomputing Centre, Computerome. This study is supported by the Novo Nordisk Foundation grant number NNF17CC0027852 and the Danish Council for Independent Research project grant 4183-00516A. Work supported by Marie Curie Reintegration Fellowship (H2020-MSCA-IF-2014_RI) to E.F.; and Lundbeckfonden Ph.D. Fellowship (grant number R180-2014-2980) to X.G.
Author information
Authors and Affiliations
Contributions
N.A.M. and E.F. designed and conceived the study. N.A.M. and K.J.P. performed the CLDN6 flow cytometry analyses, sorting, reseeding, and differentiation experiments. L.M. and X.G. performed and analyzed the scRNAseq data. N.A.M. performed the bulk NGS experiments. N.A.M. and A.K. analyzed the NGS data. X.G. generated the Pbx1-KO EpiSC cell lines and performed the BF and confocal imaging of the PBX1/2-KO embryos. K.F. did the PBX1 IF of E5.5 embryos. K.J.P. did the CLDN6 E7.5 IF. L.M. and K.S. did the PBX1 ChIPseq. V.C. did the Western blots. E.F. supervised the work. C.M.S., A.P., J.M.B., and J.S. helped perform critical EpiSCs PD03 treatment experiments and provided critical insights during the manuscript revision. N.A.M. and E.F. prepared the figures and wrote and edited the manuscript with input from other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Menezes, N.A., Peterson, K.J., Guo, X. et al. Cell-context response to germ layer differentiation signals is predetermined by the epigenome in regionalized epiblast populations. Nat Commun 16, 5000 (2025). https://doi.org/10.1038/s41467-025-60348-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60348-6