Abstract
The nucleocapsid N is one of four structural proteins of the coronaviruses. Its essential role in genome encapsidation makes it a critical therapeutic target for COVID-19 and related diseases. However, the inherent disorder of full-length N hampers its structural analysis. Here, we describe a stepwise method using viral-derived RNAs to stabilize SARS-CoV-2 N for EM analysis. We identify pieces of RNA from the SARS-CoV-2 genome that promote the formation of structurally homogeneous N dimers, intermediates of assembly, and filamentous capsid-like structures. Building on these results, we engineer a symmetric RNA to stabilize N protein dimers, the building block of high-order assemblies, for EM studies. We combine domain-specific monoclonal antibodies against N with chemical cross-linking mass spectrometry to validate the spatial arrangement of the N domains within the dimer. Additionally, our cryo-EM analysis reveals novel antigenic sites on the N protein. Our findings provide insights into N protein´s architectural and antigenic principles, which can guide design of pan-coronavirus therapeutics.
Similar content being viewed by others
Introduction
Coronaviruses (CoVs) cause respiratory disease and constitute a major public health issue, as evidenced by the Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), its COVID-19 pandemic, and other epidemics caused by SARS-CoV and MERS-CoV1,2,3. CoVs particles are formed by three structural proteins located in the viral membrane: the glycoprotein or Spike protein (S), Envelope protein (E), and Matrix protein (M)4,5,6,7. A fourth structural protein, the viral nucleocapsid protein (N), is located inside the particle where it packages the viral genome8,9,10.
The N protein is the most abundant viral protein during SARS-CoV-2 infection, and exhibits a high sequence conservation across SARS-CoV-2 variants and the Coronaviridae family11,12. Predominantly localized in the cytoplasm, the N protein is concentrated near double-membrane vesicles (DMVs) where it plays a critical role in encapsidating the newly synthesized viral genome into the capsid as it translocates from the DMVs to the cytoplasm. The viral capsid subsequently migrates to the assembly sites at the endoplasmic reticulum Golgi intermediate compartment (ERGIC) where the N protein interacts to the M protein facilitating the incorporation of the capsid into nascent virions. The critical role of N protein in the viral life cycle underscores its potential as a target for antiviral therapies13,14. However, N proteins have been traditionally overlooked as vaccine candidates because they are “hidden” from the immune system inside infected cells or viral particles. Nevertheless, recent evidence indicates that a subset of the N protein may localize to the cell surface and interact with chemokines involved in leukocyte chemotaxis15. Moreover, N-derived peptides displayed on the cell membrane during infection have been shown to elicit a robust T cell response16,17,18. This response accelerates viral clearance and is associated with less severe COVID-19 disease19,20. In fact, a vaccine based on N peptides that elicit strong CD8+ T cell response is protective in non-human primates21. Furthermore, administering vaccines containing mRNA-N in combination with mRNA-S induces better viral control than administration of mRNA-S vaccines alone22. Due to its high sequence conservation and importance for viral replication and immune modulation, the N protein has become a key target for various therapies, including next-generation COVID-19 vaccines, aimed at generating universal treatments for already known and novel zoonotic CoVs13,14,23,24. Despite the importance of N to the viral life cycle and potential therapeutic approaches, the structure of the full-length N and the mechanisms underlying its oligomerization and encapsidation of the viral genome remain poorly understood for SARS-CoV-2 and other CoVs.
The CoVs capsid is assembled by oligomerization of N along the viral genome, which, at ~30 kb, represents the largest genome among RNA viruses25. The viral capsid has been visualized as flexible filamentous structures when viral particles are treated with detergents to release the encapsidated content26. In situ tomography, inside intact virions, shows the capsid to be constructed of nucleosome-like oligomers or viral ribonucleoproteins (vRNPs) assembled in a beads-on-a-string conformation along the genome27,28. It is estimated that 35–40 vRNPs are distributed along the genome, with each vRNP containing 6–12 copies of N protein and ~800 bp of viral RNA28,29. Dimers of N represent the fundamental repetitive unit of these assemblies by combining two key activities: self-polymerization and RNA binding27,30,31. These vRNPs can be reconstituted in vitro when the N dimers are unphosphorylated32,33 and bind to high-affinity binding RNA sequences such as the 5´ and 3´ UTRs or the packaging signals (PS)34,35,36. The vRNPs are organized into a double-layer cylindrical assembly inside the virus particle37,38. Despite the efforts in elucidating the molecular features of the vRNPs, more knowledge is needed about key aspects including the precise RNA-to-protein stoichiometry, the specific RNA features required for vRNP reconstitution, and the structural details of these macromolecular assemblies.
The limited structural information on the N protein within the capsid, the vRNPs, and dimers is primarily due to its highly dynamic and flexible nature. The SARS-CoV-2 N protomer is ~48 kDa in mass and features a modular organization comprising two rigid domains: an N-terminal RNA-binding Domain (RNA-BD) and a C-terminal Dimerization Domain (DD)39. Crystal structures for both domains have been determined individually27,30. Three intrinsically disordered regions or IDRs separate these two ordered domains: IDRNTD at the N-terminal region, IDRcentral between the RNA-BD and DD, and IDRCTD at the C-terminal region40,41,42. These IDRs lack a sufficiently large hydrophobic core needed for spontaneous folding43. N protein binding to RNA is cooperative and mediated not only by the RNA-BD but also by the three IDRs44 and the DD45. N self-polymerization is mediated by the DD as well as the IDRcentral and IDRCTD46,47. Importantly, the three IDRs in the N protein occupy 45% of the total protein sequence, making the N protein disordered and highly dynamic overall48. However, the normally extended conformation of the N dimer is compacted in the presence of RNA49. The flexibility of the IDRs provides biological advantages that were positively selected during CoVs evolution, including the ability to adopt multiple conformations, bind to RNA, and interact with cellular or viral factors by modulation of post-translational modifications32,50,51,52,53. Although IDRs are functionally important, they represent a significant obstacle to structural analysis of full-length N: no high-resolution structure of a full-length N yet exists for any CoVs.
Indeed, in maps of the CoVs capsid thus far available, the structure of the fundamental dimeric building block of N is unclear. Single-particle analyses produced conflicting models due to the presence of other viral components, structural heterogeneity, or the lack of a pure and homogeneous sample. Therefore, innovative approaches are needed to minimize the mobility of the N IDRs. In this work, we reasoned that an appropriate RNA could capture and perhaps organize the RNA-BDs in the N dimer, stabilize the IDRcentral, and produce a homogeneous conformation amenable to structural analysis. Here, we report the stabilization of N using a panel of viral-derived sequences of RNA, which, when bound to N, promote specific formation of filamentous structures, intermediates of assembly or vRNPs, and dimers. Next, by engineering a symmetrical RNA molecule, we stabilized the fundamental repetitive unit of the viral capsid, the N dimer, to yield a conformation suitable for analysis by negative staining electron microscopy (NS-EM). Although the resolution of the resulting map was insufficient for precise atomic modeling of the N dimer, we validated the spatial arrangement of the N domains using cross-linking mass spectrometry (MS) and domain-specific antibodies raised for this purpose. Characterization of these high-affinity antibodies also illustrates novel antigenic sites of the N protein by cryogenic EM (cryo-EM). Collectively, our findings provide a structural framework for understanding the conformational flexibility of the N protein in its functional contexts, contribute to a better understanding of the antigenic landscape of the N protein, and lay the groundwork for the development of pan-CoVs therapeutic strategies.
Results
The disordered domains are key for SARS-CoV-2 N thermostability
The SARS-CoV-2 Nucleocapsid (N) has been described as disordered, highly flexible, and dynamic40,44,49. Stabilization of the N protein is critical to analyze how each constituent domain contributes to thermostability and protein function. We used Differential Scanning Calorimetry (DSC) to explore the thermal stability of the full-length N protein from different SARS-CoV-2 variants, as well as individual functional domains and truncation mutants of N protein, by heating the samples under controlled conditions and monitoring the folding changes in DSC thermograms.
We first characterized the thermostability of N protein from the Hu-1 (Pango lineage B) and BA.1 (Pango lineage Omicron B.1.1.529) variants (Fig. 1A–C). The unfolding process of the two full-length N proteins fits a two-state model with similar melting temperatures for the two strains (Tm1 = 45.6 °C and Tm2 = 49.3 °C for Hu-1 N vs. Tm1 = 45.4 °C and Tm2 = 49.2 °C for BA.1 N). These Tm values correspond to relatively low thermostability compared to more structured proteins.
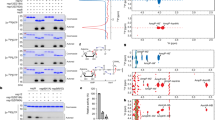
A Schematic diagram of full-length Hu-1 SARS-CoV-2 N and truncation mutants. IDRNTD, intrinsically disordered region located at the NTD; RNA-BD, RNA-binding domain; IDRcentral, intrinsically disordered region connecting the rigid domains RNA-BD and DD; DD, dimerization domain; IDRCTD, intrinsically disordered region located at the CTD. The mutations in BA.1. (Omicron) strains are indicated (BA.1. N). B Specific melting temperatures calculated by Differential Scanning Calorimetry of the constructs shown in A and the type of model used for data processing are indicated. C DSC thermograms of the constructs shown in B. Source data are provided as a Source Data file. D Size exclusion chromatogram of SARS-CoV-2 N (left panel, black). The theoretical mass of the N monomer is 48.6 kDa for reference. The inset shows SDS-PAGE gel analysis of pure SARS-CoV-2 N, along with molecular mass markers (in kDa) for standard proteins. The SEC-MALS chromatogram (right panel, red) shows a molecular weight of ~94 kDa. Native Mass-Spectrometry (Native MS) analysis of the purified sample shows a majority of RNA-free N dimer in the samples. The specific MWs are indicated in the inset table. A representative micrograph of three independent experiments of negative staining electron microscopy (NS-EM) of the flexible dimer is shown (scale bar 100 nm). E Combination of purified SARS-CoV-2 N and purified viral genome of natural SARS-CoV-2 strain virions. Created in BioRender. Saphire, E. (2025) https://BioRender.com/eqzcpd4. Three representative micrographs of three independent experiments of NS-EM showing straight and curved filamentous structures are shown (scale bar 100 nm). 2D averaged classes of these filaments (ovals on right) similar to those found in native capsids of Murine CoVs26 (Supplementary Fig. 1).
To understand which part of the protein is responsible for the low thermostability of N protein, we designed truncated versions of Hu-1 N (Fig. 1A) that contain the rigid domains in isolation (RNA-BD or DD) or together, connected by their natural IDRcentral (RNA-BD–IDRcentral–DD). The isolated dimerization domain (DD) is the most stable part of the protein (Tm = 52.9 °C), followed by the RNA-BD (Tm = 47.6 °C). In the presence of the connecting IDRcentral, however, the Tm of the combined construct decreases to 45.1 °C. Hence, the presence of IDRcentral decreases overall thermal stability by 2.5–7.8 °C (Fig. 1A–C).
Next, to discriminate the contribution of each terminal IDR, we measured the thermostability of N protein with the deletion of the N-terminal IDR (ΔIDRNTD) or the C-terminal IDR (ΔIDRCTD). Deletion of the N-terminal IDRNTD results in Tm1 = 46.1 °C and Tm2 = 49 °C, which are similar to wild-type, full-length N. In contrast, deletion of the C-terminal IDRCTD results in a single Tm = 46.1 °C (Fig. 1A–C), indicating that deletion of the IDRCTD drives a change in the thermogram from two-state to Gaussian and decreases the overall Tm compared to the full-length N protein. The similarity of Hu-1 and BA.1 N suggests that the thermostability of the full-length N did not change significantly during the evolution of the COVID-19 pandemic. In summary, these results indicate that IDRcentral is the part of the protein that contributes more negatively to the full-length protein thermal stability. Hence, it must be stabilized to permit structural characterization of the CoVs N protein.
SARS-CoV-2 N self-organizes with the viral genome into filamentous viral-like capsids in vitro
We hypothesized that the IDRcentral may undergo structural stabilization by binding to biological partners like the viral genome or specifically, key portions within the viral genome. First, to purify RNA-free N, we expressed and purified full-length Hu-1 SARS-CoV-2 N and carried out size-exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) (Fig. 1D). The <0.7 ratio of 260/280 nm absorbance suggests that there is no RNA in the purified protein sample. Moreover, the molecular weight (MW) calculated based on SEC-MALS measurement is ~94 kDa, consistent with an RNA-free dimer. Native MS used to analyze the oligomeric states of purified N indicated that the majority of the protein was present as RNA-free dimers having an MW of 93.97 kDa (Fig. 1D). We further purified the sample by size exclusion to enrich dimeric N, which has been repeatedly demonstrated to act as the building block for forming higher-oligomeric forms46,54,55,56. The resulting dimeric N, in the absence of RNA, was disordered and lacked any structurally defined conformation when analyzed by electron microscopy (EM) (Fig. 1D).
To examine whether the disordered N organizes into a well-defined conformation upon interaction with RNA, we first asked whether the N protein is stabilized in the presence of the full-length viral genomic RNA. Here, we analyzed N self-polymerization by complexing highly-purified RNA-free Hu-1 N with the ~30 kb genome from virions of highly transmissible variants of SARS-CoV-2 at different protein:RNA ratios (Fig. 1E; Supplementary Fig. 1A, B). We then visualized the complexes by negative staining electron microscopy (NS-EM). Most protein:RNA ratios tested produced heterogeneous clusters of the N protein (Fig. 2A). However, a molar ratio of 100 N protein:1 RNA in 150 mM NaCl and pH 7.5 yielded organized filamentous structures that could be visualized (Fig. 1E). Interestingly, 2D classes and the diameter of the filament (~15 nm) obtained from analysis of these in vitro-assembled filaments are similar to those released from virions of related CoVs26 (Supplementary Fig. 4). Although some filaments had a straight conformation (Fig. 1E) and yielded a NS-EM helical 3D map (Supplementary Fig. 1B; Supplementary Fig. 4), most were curved and flexible (Fig. 1E), thus reducing the likelihood of obtaining high-resolution structural data. Although this flexibility is detrimental for structural purposes, it has been preserved during the evolution of CoVs, and it could explain how their long genome can be packed into an ~80 nm viral particle.
A Genome organization of SARS-CoV-2 with boxes indicating different genes and empty rectangles indicating high-affinity binding RNA sequences. 5’UTR, 5’ untranslated regions; PS, packaging signal, and 3’UTR, 3’ untranslated regions. Representative NS-EM micrographs of three independent experiments of the purified RNA-free N dimer combined to secondary-structure based fragments of the viral PS (scale bar 100 nm). The predicted secondary structure of each RNA fragment (Vienna RNA fold server) is shown above the micrographs. The NS-EM reconstruction maps of the structurally homogeneous complexes are embedded in the corresponding micrographs. B Engineered 24-bp RNA formed by two copies of the high-affinity binding sequence found in PDB 7ACS connected by a 10-bp RNA linker (BLB-RNA). The EM-map of the dimeric N protein bound to the BLB-RNA using a low threshold to show the pseudo-two-fold symmetry is embedded in a representative NS-EM micrograph. C Cartography of the domains of N mapped in the EM reconstruction. D Mapping of cross-linked residues on 3D structure of N dimer. Above: crosslinked residues obtained by mass spectrometry (MS) from three independent experiments are depicted as arcs. Below: Three cross-linking reactions were carried out using the MS-cleavable cross-linker disuccinimidyl sulfoxide (DSSO) that bridges lysines, threonines, or serines with Cα-Cα distance range of 8–27 Å. The chemical structure of the cleavable cross-linker DSSO is shown (center). Representative MS spectra of cross-linked N peptides using DSSO at 5 (green), 10 (purple), and 50 (blue) times the estimated final protein concentration of 10 µM (center). The common residues across all the experiments are shown in the Venn diagram and the tables (right). Mapping (left) of the cross-linked residues in the 3D structure of SARS-CoV-2 N dimer obtained by EM common across all the experiments (black lines) or across at least two replicates (dashed lines) (Supplementary Fig. 5). In this 3D envelope, densities are assigned to the RNA-binding domain (RNA-BD), an intrinsically disordered region (IDRcentral), and the dimerization domain (DD) of each monomer.
This experiment formed the basis for the reconstitution of virus-like capsids and further demonstrates that viral RNA alone can stabilize and allow visualization of the N protein. The filaments purified from full viral genomes, however, still lacked sufficient homogeneity to generate an interpretable map of the repetitive unit from which the assembly is constructed.
Structural rearrangement of N dimers using viral RNA-derived sequences
To identify a conformation of N protein that would be suitable for structural analyses, we truncated the viral RNA genome. We first explored regions of the genome that were previously shown to have high affinity for N, including one of the packaging signals (PS) located at the position 19,785–20,10132 (Fig. 2A). We used sequential, secondary-structure-directed truncations of the SARS-CoV-2 genome around and including these RNA regions to yield a series of viral-derived RNAs of different lengths (Fig. 2A).
We next used this series of viral-derived RNAs in an electrophoretic mobility shift assay to identify which sequences bound N protein, and analyzed the resulting complexes by NS-EM to determine which RNA fragment produced homogeneous conformations of N (Supplementary Fig. 1). In this screening, our standard for considering a sample as structurally homogeneous was if we could obtain 2D class averages with particles that converged and define an initial 3D reconstruction. Most RNA sequences analyzed in this screening platform promoted formation of disorganized clusters (Fig. 2A). This result aligns with preliminary results demonstrating that N binds to RNA in a sequence- and secondary structure-specific manner32,34,48.
We ultimately found, however, that binding of N to certain specific fragments of RNA encoding the PS produced homogeneous structures (Fig. 2) as previously described32,33. Binding of N to one molecule of RNA corresponding to a 318-nt fragment (Fig. 2A; Supplementary Fig. 1C–E) yields an 8-mer or 16-mer complex of N as demonstrated by native MS (Supplementary Fig. 1C; right panel). EM analysis of this RNA-N complex yielded a 3D reconstruction (Fig. 2A; Supplementary Fig. 1D, E) whose volume and diameter (~15 nm) resemble the nucleosome-like structures or vRNPs found in intact virions by in situ tomography29 (Supplementary Fig. 4). The inherent flexibility of the N-318-nt RNA, however, impeded the accurate positioning of the constituent N dimers into the map. We reasoned that shortening the 318-nt sequence of the RNA could produce more simple and interpretable N assemblies. Prediction of the secondary structure of the 318-nt RNA fragment illustrates that it contains three hairpins (Fig. 2A). A panel of secondary-structure-based RNA mutants were created (Fig. 2A), complexed with RNA-free N dimer, and visualized by EM. While most of the RNA fragments derived from the 318-nt RNA yielded disordered clusters, serial truncations of the PS hairpin II (Fig. 2A; blue hairpin) yielded lower-order assembly intermediates that were amenable for EM analysis. 3D maps of the 117-nt and 67-nt derived from this hairpin are shown (Fig. 2A; 117-nt hairpin II, 67-nt hairpin II).
Taken together, these results show that RNA-free N dimers require specific high-affinity binding sites like the hairpin II in the SARS-CoV-2 RNA genome to nucleate and form structurally homogeneous structures, as suggested before by biophysical analysis48. Further, we find that the diameter of the N-based assemblies correlates with the length of the RNA used to form the complex (~15 nm for the 318-nt RNA and ~7 nm for the 67-nt RNA), suggesting that the number of N dimers recruited is lesser with shorter RNAs. These findings contribute to understanding the molecular mechanisms for the assembly of the dimeric Ns into higher-order assemblies, although less flexibility is still needed to generate an interpretable map of the constituent N dimers.
Engineered symmetric RNA stabilizes the N dimer for EM analysis
Based on the successful stabilization of N protein dimers using short sequences of RNA (67-nt hairpin II), we next identified the minimal sequence of RNA that yields a symmetric and interpretable EM map of the N protein dimer (Fig. 2B, C). We hypothesized that by using a symmetric and short RNA to bring RNA-BDs of the dimer in proximity, we can stabilize the IDRcentral by adding a specific cross-linker like DSSO that crosslinks the lysines, threonines, or serines with Cα-Cα distance range of 8–27 Å (Fig. 2D).
First, we designed a synthetic 24-nt RNA fragment that contains two high-affinity binding sequences for N connected by a linker that we named Binding sequence-Linker-Binding sequence- or BLB-RNA (Fig. 2B), and contains as the binding sequence a 7-nt RNA fragment previously crystallized with the monomeric RNA-BD30 (Supplementary Fig. 2A). This 7-nt sequence is repeated five times in the SARS-CoV-2 genome (Supplementary Fig. 2A). By linking two copies of this 7-nt fragment with natural 10-nt sequences that are immediately adjacent in the SARS-CoV-2 genome (Supplementary Fig. 2A) and then adding DSSO, we could purify N protein dimers that yielded interpretable NS-EM maps consistent with a dimeric N (Fig. 2B; Supplementary Fig. 3). We also evaluated the effect of several, longer flanking sequences, but found that the 10-nt sequence gave the most consistent samples (Fig. 2B; Supplementary Fig. 2A).
As we expected, using a short and symmetric piece of RNA produced an EM map of the N dimer stabilized in vitro (Fig. 2B, C). The 2-fold pseudosymmetry and the diameter (~8 nm) obtained from the analysis of these in vitro-assembled N dimers are similar to those released from virions of related CoVs26 (Supplementary Fig. 4). Notably, no symmetry was imposed during the micrograph processing. Processing of 98 K particles obtained by NS-EM resulted in a low-resolution map with distinct domains (Fig. 2B; embedded map, Supplementary Fig. 3). Higher resolution could not be obtained by cryo-EM of this complex or any other yet in the field, likely due to mobility of the molecule. Hence, we set out to use multiple complementary techniques to identify the domain organization of this N assembly.
Integrative structure determination of the SARS-CoV-2 N dimer
To understand the domain organization within the NS-EM map of the N dimer, we first used chemical cross-linking coupled with MS to identify protein residues that are in close spatial proximity within the N dimer. We then translated these data into spatial restraints before building a model of the N dimer that adhered to these distance restraints.
Our strategy to fit the atomic model of SARS-CoV-2 N dimer involved five steps: (i) Calculation of intra-monomer and inter-monomer distances by cross-linking MS; fitting of (ii) DD57 and (iii) RNA-BD30 crystal structures, (iv) fitting of IDRcentral into the EM map using distance restraints determined by cross-linking MS, and (v) experimental validation of the final model by map labeling using domain-specific antibodies.
Calculation of intra-monomer and inter-monomer distances by cross-linking MS
First, to gain complementary information on which residues are in proximity in the assembly, we cross-linked the dimer using increasing concentrations of DSSO as a chemical cross-linker (Fig. 2D; Supplementary Fig. 5). DSSO specifically links Cα-Cα of lysines, threonines, or serines separated by 8–27 Å. Following cross-linking, the samples were denatured, digested and then analyzed by LC-MS/MS on a Thermo Scientific Orbitrap Fusion Tribrid Mass Spectrometer. The resulting fragmentation data were analyzed using Proteome Discoverer with the Xkinkx cross-link data analysis nodes. Both intra- and inter-domain interactions were identified by detecting cross-linked residues appearing in three independent experiments having increasing DSSO concentrations (Fig. 2D; Supplementary Fig. 5).
Fitting the DD crystal structure into the EM map
The intra-DD cross-linked residues were located in the crystal structure (PDB 6WJI) and the real space distances were measured using Pymol visualization software. To discern whether the cross-linked pairs are intra- or inter-monomer, we measured the Cα-Cα distances of the cross-linked residues in both scenarios (Fig. 2D; Supplementary Fig. 5, table in step 5). All the intra-monomer distances are compatible with the DSSO cross-linking length (8–27 Å) (K248-K256 = 13 Å; K248-K257 = 15 Å; and K266-K299 = 11.6 Å) while the inter-monomer lengths are longer (39–48.5 Å). This result is consistent with the fact that N dimers have more frequent intra-monomer contacts than inter-monomer contacts58. Then, we fit the atomic model for the DD into the density of the EM map in which the two asymmetric units of the dimer remained together (Supplementary Fig. 5).
Fitting the central IDR into the EM map
Identification of cross-linked residues between the IDRcentral and the DD (K233–K248, K237–266, K237–249) demonstrated that these domains are in close spatial proximity and are crucial for the correct modeling of this region into the 3D map. In particular, residue K237, which is part of the IDRcentral, is cross-linked with K248, K249, and K266 located in the DD across three independent experiments (Supplementary Fig. 5).
As no experimentally determined atomic model for the IDRcentral region is available to position the cross-linked pairs, we utilized AlphaFold (AF) to generate an initial structural prediction. While AF has limitations in capturing the full conformational range of intrinsically disordered regions (IDRs)59, it provides a valuable starting model for approximating the IDR´s location. This initial model was refined using distance restraints from cross-linked residues and spatial constraints from the EM map (see Supplementary Fig. 5, step 6), resulting in a more accurate representation of the IDRcentral (Fig. 2D). However, the IDRcentral region´s lower symmetry prevents effective use of symmetry-based constraints for fitting60,61 and as a consequence the crosslinked pairs K233-K248, K237-K266, and K237-K249, could not be unambiguously categorized as either intra- or inter-monomer.
Fitting of the RNA-BD-RNA into the EM map
The intra-RNA-BD cross-linked residues are also within the distance range of the DSSO (K61-K102 = 14.6 Å) (Fig. 2D; Supplementary Fig. 5) in the crystal structure (PDB 7ACS) while the potential inter-monomer distance (Supplementary Fig. 5, table in step 5) is not compatible with the DSSO cross-linking length (8–27 Å). This finding is consistent with previous studies using SAXS data which showed that there are no direct contacts between the RNA-BDs and there are more than ~100 Å between them in the context of a dimer58. The RNA-BD-RNA crystal structures do fit into the two lobes of available density separated by more than 27 Å from each other (Fig. 2D; Supplementary Fig. 5).
The general density distribution of the EM map resembles models proposed for the capsid of a related CoV26 (Supplementary Fig. 4) and by other techniques like SAXS58 which showed that the DD holds the dimer together and the RNA-BDs are separated from each other and from the DD. The resulting EM map of the N dimer is also compatible with those previously modeled using viral capsids26. Due to the stabilization of the homodimer using the short and symmetric RNA and the cross-linking with DSSO, we have stabilized the IDRcentral such that it now shows density similar to that obtained for the rigid domains of the protein (Fig. 2B).
As expected, the terminal IDRs (IDRNTD and IDRCTD) are not visible in the EM map. In an attempt to increase the overall resolution of our EM map, we truncated these terminal IDRs (RNA-BD-IDRcentral-DD) of N and complexed the terminally truncated N to BLB-RNA. However, truncation of the terminal IDRs destabilized the homodimer, and no clear 2D classes were found (Supplementary Fig. 2C). This result and the thermostability analysis of these mutants (Fig. 1) indicated that the terminal IDRs, although flexible, are important for the stability of the dimeric N.
mAb discovery platform identifies antibodies that bind to all domains of N
Due to the low resolution of the EM map and the lack of other structural models for full-length N, validation is a priority. We thus developed a panel of anti-N monoclonal antibodies (mAbs) to specifically label each of the five domains of N protein.
First, we immunized mice with purified recombinant N and isolated a panel of 88 novel mAbs using a single B cell-sorting Beacon platform (Berkeley Lights) (Fig. 3A). We characterized the affinity of each mAb by biolayer interferometry assay (BLI) and selected mAbs having nanomolar to picomolar affinities for N protein, which narrowed the panel to 66 mAbs. These 66 mAbs recognize all domains of the N protein: 39 against the RNA-BD, 20 against the DD, and 3, 3, and 1 against the IDRNTD, IDRcentral and IDRCTD, respectively (Fig. 3A, B). We first used the 7 mAbs specific for the IDRs to incubate with N to try to stabilize and label the IDRs in the context of the homodimer N bound to BLB-RNA. We could make 2D classes from the resulting NS-EM of each complex, but none of the 2D class particles yielded a homogenous 3D reconstruction, suggesting that binding of these mAbs to the IDRs triggered conformational change that increased disorder. Hence, to further validate our model, we next screened the 59 mAbs specific for the rigid domains of N. We first sequenced mAbs and classified them phylogenetically (Fig. 3C). We next selected one representative high-affinity antibody from each sequence cluster (10 against the RNA-BD and 7 against the DD) and characterized the association (Ka) and dissociation constant (Kd) for each using quadruplicate BLI assays (Fig. 3D).
A Mice immunization with purified recombinant N and isolation of a panel of 88 novel mAbs using a single B cell-sorting Beacon platform. B Number of novel high-affinity mAbs distributed across the rigid and the disordered domains of N protein. A, B Created in BioRender. Saphire, E. (2025) https://BioRender.com/6guzz6f. C Above: Phylogenetic tree based on the variable region sequence of the anti-N mAbs. Below: Heat map of mAb binding profile measured by ELISA. Binding domain across N protein and the epitope group of each mAb is indicated. D Iso-affinity kinetic plot (association vs. dissociation constant) grouped by mAb. E Epitope binning clusters (Carterra LSA platform) for the selected mAbs. Two independent antibody binding sites in RNA-BD (RNA-BD I and RNA-BD II) and other two in the DD (DD I and DD II) were identified. 2D class averages illustrate the presence of two independent antibody binding sites in each rigid domain of N. The mAbs NP1-E2 and NP3-D1 were used to complex the RNA-BD. The mAbs NP3-B4 and NP1-E9 were used to complex the DD. Source data are provided as a Source Data file. F NS-EM reconstructions of the RNA-BD (left) or DD (right) bound to mAbs from non-overlapping epitope groups.
The antigenic landscape of N protein reveals at least seven antigenic sites
To identify the number of epitope binding sites in each domain, we carried out high-resolution epitope binning assays on these 17 mAbs using the Carterra LSA platform. Non-competitive and competitive-binding pairs (Fig. 3E) indicate that there are at least two independent antibody binding sites in RNA-BD (RNA-BD I and RNA-BD II), two in the DD (DD I and DD II), and at least one antigenic site across each of the IDRs. We used NS-EM to understand in better detail the spatial disposition of each antigenic site in the rigid domains relative to others. We focused on four mAbs in particular: the RNA-BD binders NP1-E2 and NP3-D1 from RNA-BD groups I and II, respectively, and the DD binders NP3-B4 and NP1-E9 from DD groups I and II, respectively. We determined an NS-EM map of a monomeric RNA-BD simultaneously bound to one copy each of the RNA-BD Fabs NP1-E2 and NP3-D1; and another NS-EM map of the dimeric DD bound to two copies each of the anti-DD Fab NP1-E9 and Fab NP3-B4) (Fig. 3E, F). The footprints of each mAb are non-overlapping, confirming the presence of the non-competitive clusters identified by epitope binning.
Validation of the EM map by labeling N dimer-BLB RNA with domain-specific mAbs
We complexed antibodies representative of the four antigenic sites (RNA-BD I, RNA-BD II, DD I, and DD II), with the dimeric N-BLB RNA complex to try to further confirm domain identity within the NS-EM map of N. However, although we could visualize anti-RNA-BD mAbs with the RNA-BD in isolation, and the anti-DD mAbs with the DD in isolation, within the context of the complete N, most mAbs bound at full occupancy (2 Fabs: 2Ns), they triggered a conformational change that destabilized N-BLB RNA complex such that it was not ordered and not visible by NS-EM. However, one mAb-N complex was ordered and could be visualized, that of the anti-DD NP1-E9 with the N dimer-BLB RNA complex in a 1 Fab: 2 N ratio (Fig. 4A, B). Processing ~1000 micrographs allowed us to produce an NS-EM map of N-BLB RNA-Fab NP1-E9 (Fig. 4B). Structural comparison of this map with the map lacking the Fab revealed an extra density with a volume compatible with a Fab bound to the DD predicted in our atomic model for N dimer. This result experimentally validates the location of the DD within the N-BLB RNA complex and reveals a recognition site in the antigenic landscape of the SARS-CoV-2 N protein.
A Representative SDS-PAGE of three independent experiments of SARS-CoV-2 N bound to BLB-RNA in denaturing conditions (i) or cross-linked with DSSO (iii). SARS-CoV-2 N-BLB RNA bound to Fab NP1-E9 and cross-linked with DSSO (ii). Molecular mass markers (in kDa) of standard proteins are shown. B NS-EM map of SARS-CoV-2 N-BLB RNA bound to Fab NP1-E9. C Molecular model of DD (garnet) bound to Fab NP1-E9 (cyan) and Fab NP3-B4 (grey) fitted into the corresponding cryo-EM map. Magnified views of the DD-Fab interacting interfaces outlined by boxes in the main structure are shown. D Projections of the experimentally determined cryo-EM reconstruction. E Above: Fourier Shell correlation curve (FSC) plot. Below: Euler angle distribution of particles used in the final reconstruction.
To better identify residues involved in the novel interaction with the N-NP1-E9 antibody that targets DD, we solved the DD-Fab NP1-E9-Fab NP3-B4 complex at higher resolution using cryo-EM (Fig. 4C–E; Supplementary Fig. 6). A 3D reconstruction of the 234 kDa DD-two Fab complex reached an overall resolution of 3.7 Å with D1 symmetry imposed (Fig. 4E). The specific location and binding angle of the Fab NP1-E9 to the DD in the context of the N dimer is compatible with that for the isolated DD (Fig. 4B). The residues of the Fab NP1-E9 that interact with DD localize to the CDR1 of the heavy chain (HC) (Y32 of CDR H1 to D348 of DD, and D33 of CDR H1 to Y299 of DD), and to the three CDRs of the light chain (LC) (N32 of CDR L1 to Y355 of DD, S50 of CDR L2 to V350 of DD, and N92 of CDR L3 to T362 of DD) (Fig. 5).
A Binding footprints for Fab NP3-B4 (grey) and Fab NP1-E9 (cyan) from antigenic sites DD-I and DD-II, respectively. Residues of the DD involved in interacting with the corresponding Fab are specified. B Multiple sequence alignment of the DD across variants of concerns of SARS-CoV-2 virus and other highly pathogenic members of the betaCoVs. The interacting residues described in (A) are highlighted. Strictly conserved residues are indicated by stars, conserved residues with strong similar properties are indicated by a colon, and conserved residues with weak similar properties are indicated by the period under the alignment according to ClustalW nomenclature.
The Fab NP3-B4 interacts with DD via residues in the CDR 1 and 3 of the heavy chain (HC) (Y32 of CDR H1 to P364 of DD, Y104 of CDR H3 to N269 of DD, and Y110 of CDR H3 to P364 of DD), and the 3 CDRs of the light chain (LC) (CDR L1 S30-Y268, CDR L2 D49-DD Q283, CDR L3 G90-DD N272, and CDR L3 S91-DD Q272) (Fig. 5; Supplementary Fig. 6). The residues in the DD responsible for interacting with Fab NP3-B4 define the antigenic site DD-I, while those that interact with Fab NP1-E9 define the antigenic site DD-II (Fig. 5). These residues are identical across the Variants of Concern (VOCs) of SARS-CoV-2 and SARS-CoV, and are ~58% conserved with MERS (Fig. 5). This interaction illustrates a new antigenic site for the N protein that both validates the NS-EM map of the N and defines a target for designing universal therapeutics for present and future CoVs diseases.
Discussion
A major gap in knowledge of SARS-CoV-2 assembly is how the basic repetitive unit of the viral capsid, the N dimer, encapsidates its genome, the largest among the RNA virus families, into a viral capsid that can be accommodated in ~80 nm viral particles. In this work, we first reconstituted viral capsids in vitro using purified RNA-free N, plus viral RNA from authentic virions. These capsids are similar in diameter (~15 nm), filamentous shape and density distribution to those released from authentic virions (Supplementary Fig. 4). This side-by-side comparison supports the idea that the filamentous capsids exist both in vitro and upon release from the virions when the space constraints of the viral particle are removed26.
Previous visualization of the viral capsid inside virions by in situ tomography of SARS-CoV-2-infected cells revealed that 3–6 copies of N dimers nucleate into ~15 nm vRNPs similar to encapsidation of eukaryotic DNA mediated by histones to form nucleosomes29. This conformation is called “beads on a string”, and likely assists efficient genome packaging. However, in contrast to nucleosomes, vRNPs are more flexible. This flexibility contributes to the lack of high-resolution structures and in turn, has complicated understanding the precise RNA-to-protein stoichiometry in vRNP formation. To understand these assemblies at a molecular level, we reconstituted them in vitro by combining purified RNA-free N dimers with homogeneous RNA sequences derived from the viral genome. The combination of N with most of the RNAs used in this study yielded heterogeneous clusters suggesting that N protein interacts with the RNA not only by electrostatics but also in a sequence/secondary structure mediated manner. Combination of N with viral RNAs that have high affinity for the N protein34,48,51,62 yielded vRNP-like assemblies32,33 with diameter (~15 nm) and density distribution resembling those extracted from SARS-CoV-2 virions (Supplementary Fig. 4). The homogeneity of the in vitro-reconstituted vRNPs allowed us to perform MS to reveal that each vRNP contains 4 dimers of N and one copy of 318-nt RNA, and that two of these vRNP can further dimerize to yield an assembly of 8 dimers of N and 636-nt RNA (Supplementary Fig. 1C). Although some of these particles converged into clear 2D classes by cryo-EM (Supplementary Fig. 1D), the resulting 3D map did not have sufficient resolution (Supplementary Fig. 1E) to unambiguously assign density for the constituent dimers. However, as the CTD of the N protein, not the NTD, was previously shown to be involved in formation of vRNPs in the presence of RNA46, it is likely that the CTDs constitute the inner rigid core of these in vitro-reconstituted vRNPs.
As the flexibility of the in vitro-reconstituted vRNP (Supplementary Figs. 1 and 4) precluded building of an interpretable EM map, we aimed to stabilize the basic repetitive unit, the N dimer, by using shorter sequences of the 318-nt RNA we used to reconstitute the vRNPs (Fig. 2; Supplementary Fig. 4). We identified a 67-nt RNA that yielded a 3D reconstruction of a dimer (Fig. 2), with a density distribution and a diameter (~8 nm) similar to the N dimer extracted from the capsid of similar CoVs26, although still not rigid enough to permit a high-resolution map. Based on these lessons obtained from the viral-derived RNAs, we decided to engineer an RNA that would: (i) contain a high-affinity sequence of the viral RNA; (ii) be as short as possible to minimize the inherent flexibility of the N protein, (iii) stabilize the simplest order of N assembly, the dimer, so validation of the 3D reconstruction using structural MS is accurate, (iv) is symmetric and presumably yields a symmetric and interpretable 3D reconstruction.
To identify the minimal sequence of RNA that would yield a symmetric and interpretable EM map, we used a 7-nt sequence visualized in the previous structure of the isolated RNA-BD of N30, and which is repeated five times in the genome (Supplementary Fig. 2). By linking two copies of this 7-nt fragment with the natural 10-nt sequence immediately adjacent to it in the SARS-CoV-2 genome, binding to N, and cross-linking the resulting N-RNA complex (Supplementary Fig. 2), we could obtain N-RNA dimers that yielded interpretable EM maps. Importantly, the diameter (~8 nm), the pseudo-two-fold symmetry, and the density distribution of the in vitro reconstituted N dimer is similar to those N dimers extracted from the capsid of similar CoVs26 (Supplementary Fig. 4). Although we successfully captured the N protein in a dimeric conformation using a crosslinker and an engineered RNA, a key limitation of our study lies in the inherent flexibility of the assembly. This structural plasticity prevents achieving higher-resolution maps necessary for constructing an atomic model of the N dimer. Furthermore, it remains unclear whether such symmetric dimers occur in the native viral context, as the flexible nature of the N dimer is critical for supporting its diverse functions in the viral life cycle. Despite the limitations, the stabilization achieved in this study provides valuable insights into the highly dynamic N dimer, shedding light on the spatial rearrangements of its domains. We combined cross-linking MS with domain-specific N mAbs raised specifically for this purpose to reveal the relative positions of the DDs, RNA-BDs, and, interestingly, a central lobe for the IDRcentral. These findings enhance our understanding of the structural adaptability of the N protein, which could be leveraged to engineer novel versions of the N protein with improved stability or immunogenic properties. Such engineered proteins hold potential for use as immunogens in the development of next-generation vaccines against COVID-19 or related CoVs.
In this study, we also produced and characterized 88 novel mAbs against N, including individual mAbs against each of the three IDRs, the RNA-BD, and DD (Fig. 3). SPR-based competition analysis identified at least seven epitope groups across the N protein. Cryo-EM analysis of DD complexed to Fabs NP1-E9 and NP3-E4, yielded a 3.7 Å map that allowed us to build an atomic model (Figs. 4 and 5). In our knowledge, this cryo-EM map reveals two novel antigenic sites of the N protein in the DD, that together with antigenic sites already identified for the RNA-BD (PDB 7STS, 7N3C, 7CR5) expands our knowledge of the antigenic landscape of this immunogenic protein. The discovery of a new panel of anti-N mAbs, including at least one against every ordered domain and IDR, may serve to expand the therapeutic options based on the highly conserved SARS-CoV-2 N or as research tools for the field.
In summary, here, we combined biophysical, biochemical, and structural analysis to shed light on the architectural and antigenic properties of one out of the four structural proteins of CoVs, the N protein. This study sheds light on the complex structural landscape of the N protein, from the viral-like capsids, to intermediate of assemblies or vRNPs, and the building blocks of those assemblies, the N dimers. We identified specific sequences of viral genome RNA that yield defined building blocks and stepwise oligomeric assemblies of N. Engineering shorter RNA sequences stabilized the N dimer and allowed for interpretable 3D reconstructions, revealing key structural features. Novel mAbs targeting N domains were developed, with some permitting high-resolution mapping of antigenic sites. This work expands understanding of the conformational flexibility, and the structural and antigenic properties of the N protein, with implications for vaccine and therapeutic design.
Methods
Our research complies with all relevant ethical regulations, and the study protocol (AP00001239) was approved by the Animal Care Committee at the La Jolla Institute for Immunology.
Protein expression and purification
The coding sequence of the Hu-1 SARS-CoV-2 N protein was synthesized based on the Gene ID NC_045512.2. The sequence was subcloned into the pet46 vector with an N-terminal 6x His tag. The plasmid was sequence-verified and transformed into Rosetta2 cells for expression in E. coli bacteria. The recovered cells were grown in 25 ml LB supplemented with 2500 µg ampicillin and 625 µg chloramphenicol overnight at 37 °C, then diluted into 1 L LB and grown until the OD at 650 nm reached 0.4. The temperature was shifted to 16 °C and when the OD at 650 nm reached 0.8, protein expression was induced by adding IPTG to a final concentration of 0.5 mM and incubating at 16 °C overnight. Then, the cells were pelleted by centrifugation at 8 °C for 25 min and 6000 g. The resulting pellets were resuspended in 50 mM Hepes pH 7.5, 500 mM NaCl, 10% glycerol, 20 mM imidazole and 6 M urea. Benzonase Nuclease (1ul, Millipore 7E1014) was added, and the suspension was lysed using a M-110P microfluidizer (Microfluidics). After clarification, the supernatant was filtered through a 0.22 µm filter and incubated with 14 ml of Ni-NTA bead slurry (Qiagen 30210) for 45 min at RT. After discarding the flowthrough, the beads were washed with a resuspension buffer and the protein was eluted from the beads using a resuspension buffer supplemented with imidazole to a final concentration of 300 mM. The protein was concentrated to a volume of 2 ml using a Vivaspin 10k device and placed in a 10k MWCO snakeskin dialysis bag for overnight dialysis in refolding buffer (50 mM Hepes pH 7.5, 500 mM NaCl and 10% glycerol). The protein was then dialyzed in 50 mM Hepes pH 7.5, 500 mM NaCl to remove the glycerol, and further purified using a s200i size exclusion column (GE Healthcare). Fractions were collected and the purified protein was concentrated to 1–3 mg/ml for use in downstream analyses.
Differential scanning calorimetry (DSC)
The thermal stability of purified RNA-free SARS-CoV-2 Nucleocapsid or truncation mutants was analyzed by Nano DSC (TA instruments). The corresponding protein (100–300 µg) was buffer-exchanged to Phosphate Buffered Saline (PBS) supplemented with 500 mM NaCl and loaded into the sample wells. The temperature ramped from 20 to 100 °C at 1 °C per minute. The resulting thermogram was corrected by subtracting a buffer blank data set and baseline-correcting before fitting to a thermodynamic model to extract the exact melting temperature (Tm).
Size exclusion chromatography-multiangle light scattering (SEC-MALS)
Purified RNA-free SARS-CoV-2 Nucleocapsid (100–300 μg) was applied to a Superdex 200 (GE Healthcare) column in a buffer containing 50 mM Hepes pH 7.5, 500 mM NaCl at a flow rate of 0.5 ml/min. Light-scattering data were collected on a Dawn MiniTreos (Wyatt Technologies) and analyzed with ASTRA (Wyatt Technologies).
RNA synthesis by in vitro transcription
dsDNA gene block(s) including the T3 polymerase promoter sequence followed by the selected sequence were produced by Genewiz (South Plainfield, NJ USA). An in vitro transcription reaction was performed by mixing 50 ug dsDNA gene block as a template, T3 polymerase and RNApol reaction buffer (NEB M0378S), 0.5 mM rNTP Mix (NEB #N0466S), 5 mM DTT, and RNAse inhibitors. The sample was incubated for 3 h at 37 °C. The template DNA was digested using 1 µl DNAse (NEB #B0303S) for 30 min at 37 °C. Then the RNA was purified using TRIzol and precipitated with isopropanol according to the manufacturer’s instructions (Invitrogen 15596026). Using 350 µg DNA as a template ~1.4 mg RNA could be obtained. The 260/280 and 260/230 nm absorbance ratios measured by the NanoDrop were ~2 and ~2.2 respectively, indicating a highly pure RNA sample was obtained.
Formation of N-RNA and N-RNA-Fab complexes
Purified RNA-free SARS-CoV-2 N (700 µg) was added to 100 ug RNA that was synthesized as mentioned above and then 50 mM Hepes pH 7.5 was added to adjust the NaCl concentration to 250 mM before incubation of the complex for 15 minutes at RT in a shaker. Disuccinimidyl sulfoxide (DSSO) cross-linker (Thermo Scientific A33545) was freshly reconstituted with DMSO to produce a 50 mM stock solution. The sample was further incubated at RT for 45 min after the addition of 14 µl DSSO. The cross-linking reaction was quenched with 20 µl 100 mM Tris pH 7.5 and the sample was centrifuged at 10,800 × g for 5 min to check for precipitation. If precipitate was present, the supernatant was moved to a fresh tube and 200 µg of the indicated Fab was added. The sample was incubated for 2 h at RT in a shaker and purified further on a size exclusion s200i to isolate N-RNA-Fab complexes from free Fab and DSSO.
Purification of viral RNA genome from SARS-CoV-2 virions
VeroTMPRSS2 cells (a kind gift from Dr. Michael Diamond, Washington University, St. Louis MO) were grown to 70% confluency in DMEM (Dulbecco’s modified Eagle’s medium) supplemented with 2% heat-inactivated fetal bovine serum (FBS) and penicillin (100 mg/mL), streptomycin sulfate (100 mg/mL), 1 M HEPES (1:100), NEAA (non-essential amino acids 1:100) and 5 µg/mL blasticidin at 37 °C and 5% CO2. Vero TMPRSS2 cells were then infected in a T175 flask at MOI 0.01 with D614G, B.1.617.2 (Delta), or B.1.351 (Beta) SARS-CoV-2 isolates. Viruses were incubated with the cells at 37 °C for 1 h in 15 mL medium. After 1 h, 10 mL of medium was added to each flask and returned to the incubator. After 72 h, the viral supernatant was collected and spun at 4500 × g for 15 minutes at 4 °C. The pellet was discarded, and viral RNA was extracted from 140 µL of viral supernatant using a QIAmp Viral RNA kit (#52904), and quantified on a Nanodrop. All work with infectious SARS-CoV-2 virus was performed in a certified BSL-3 facility at La Jolla Institute for Immunology.
Negative stain electron microscopy
SARS-CoV-2 N-RNA, SARS-CoV-2 N-RNA-Fab, DD-Fab NP1-E9-Fab NP3-B4 and RBD- Fab NP1-E2-Fab NP3-D1 complexes were diluted to around 0.02 mg/ml and 4 µl of the complex were deposited on CF400-CU 400 mesh grids (Electron Microscopy Sciences) that were previously glow-discharged for 30 s. For RNA purified from virions, complexes were formed from 10 µg SARS-Cov-2 N and 0.1 µg RNA. The mixture was diluted 1:10 and then 3 µl was added to the grid. When only one Fab was used in complexes with DD and RNA-BD, the mixture was further diluted to around 0.002 mg/ml to allow good separation of the particles on the grid. Excess sample was wicked off the grid, which was then washed with three drops of Milli-Q water. Excess water was blotted between washes using Whatman filter paper. Then, three drops of 0.75% uranyl acetate were added wicking off the excess between each drop. The grid was incubated with the final drop for 2 minutes. Any excess liquid was wicked away and the grid was air-dried before imaging on a FEI Titan Halo 300 kV electron microscope with a K3 direct electron detector at 1.87 pixels/Å and 50 e−/Å2. A total of 200–700 micrographs were collected and image processing and reconstruction was done using cryoSPARC v3.3.163. Micrographs were motion-corrected using patch motion correction, and the CTF parameters were determined using the patch CTF estimation function in cryoSPARC. Particles were picked using Blob Picker and sorted by two-dimensional classification before several rounds of heterogeneous refinement and non-uniform refinement. ChimeraX 1.464,65 was used to prepare figures of the structure.
Native MS
Purified protein (~1 µg/mL) was buffer-exchanged into 500 mM ammonium acetate and the was pH adjusted to 7.5 immediately before analysis on a Thermo Scientific Q Exactive UHMR mass spectrometer equipped with a nanoelectrospray ionization source. Mild collisional activation was used to improve mass measurements by removing extraneous salts and solvent molecules from the gas-phase complexes. Note that no fragmentation of the complexes was observed unless intentionally activated. Calculation of the mass of the intact protein and its complexes with RNA was carried out in Unidec (v4.4). Binding of the N protein to RNA was carried out by mixing 10 µg of protein with 2.1 µg of RNA in a final volume of 10 µL of 220 mM ammonium acetate.
Cross-linking MS
For cross-linking and protein digestion, an Amicon Ultra 0.5 mL centrifugal filter device was used to buffer-exchange purified His-SARS-CoV2-N protein into 50 mM HEPES (pH 7.5) with 500 mM NaCl by filtering with 6 times the original sample volume. The absorbance of the buffer-exchanged protein was measured at 280 nm, and its monomeric concentration was estimated based on the calculated extinction coefficient of 45830 M-1 cm-1 (ExPASy ProtParam). The sample’s 260/280 ratio was greater than 0.5 which suggests nucleic acid contamination and an overestimation of the protein concentration. Multiple cross-linking reactions were carried out using the MS-cleavable cross-linker disuccinimidyl sulfoxide (DSSO) (Cayman Chemical) at 1, 5, 10, and 50 times the estimated final protein concentration of 10 µM (monomer). DMSO required to dissolve DSSO was diluted to 1% in the final reaction mixture that contained 50× DSSO. The cross-linking reaction was allowed to proceed for 90 min at 20 °C and subsequently quenched by addition of Tris-HCl (pH 7.5) for a final concentration of 20 mM. An aliquot of the quenched samples was analyzed by SDS-PAGE and the rest was subjected to in-solution digestion for analysis by OBE (online buffer exchange)-MS, and subjected to in-solution digestion. For in-solution digestion, the samples were denatured with 5 M urea, reduced with dithiothreitol (final concentration: 10 mM) at 37 °C for 20 min, alkylated with iodoacetamide (final concentration: 30 mM) at 37 °C for 20 min, and finally diluted with 100 mM ammonium bicarbonate to reduce the urea concentration to less than 1 M. MS-grade trypsin (Thermo Fisher Scientific) was added for a final protease to protein ratio of 1:50 (w/w) and the proteins were digested at 37 °C overnight. Trifluoroacetic acid (TFA) was used to reduce the pH and inactivate the trypsin. The resulting peptides were desalted using SPEC Pt C18 cartridges (Agilent Technologies). Briefly, the C18 cartridges were equilibrated with 0.1% TFA (3 ×200 μL), the peptide mixture was loaded onto the C18 cartridge, the bound peptides were washed with 0.1% TFA (3 ×200 μL) and eluted with 0.1% TFA, 50% acetonitrile (2 ×200 μL) and 0.1% TFA, 80% acetonitrile (1 ×200 μL), and eluted peptides were dried in a SpeedVac and resuspended in 0.1% TFA. The absorbance of the peptide solutions at 205 nm were used to estimate the peptide concentrations.
Peptides were analyzed by LC−MSn with an UltiMate 3000 RSLCnano coupled to an Orbitrap Fusion Tribrid Mass Spectrometer equipped with an EASY-Spray source. Approximately 1-2 μg of peptide was injected onto an EASY- Spray column (PepMap RSLC C18, 2 μm, 100 A, 75 μm × 25 cm, Thermo Fisher Scientific). The peptides were separated at a flow rate of 0.3 μL min−1 using the following gradient: 0–5 min = 2% B, 5–40 min = to 26% B, 40–62 min = to 40% B, 62–65 min = to 95% B, 65–70 min = to 95% B, where solvent A = H2O with 0.1% formic acid and solvent B = acetonitrile with 0.1% formic acid. Data-dependent CID–MS2 scans (R = 30000, AGC target 5e4,100 ms max injection time) with a normalized collision energy (NCE) of 25%, charge states 3–8, and a dynamic exclusion duration of 30 s were performed on the precursor ions in the full MS1 scan (375–1500 Th, R = 60000, AGC target 4e5, 50 ms max injection time). CID–MS3 scans (NCE 35%) were triggered for the CID–MS2 ions with a mass difference of 31.9721 Da and acquired in the ion trap set to rapid scan rate, AGC target 2e4, and 120 ms max injection time. The MS experiment maximum duty cycle was set to 5 s.
Data were analyzed using Proteome Discoverer v2.4.1.15 and the Xlinkx nodes (Thermo Fisher Scientific). Data were filtered for possible cross-links based on the MS2–MS3 acquisition strategy. The DSSO modification (158.004 Da) was set to be considered for the amino acids K, S, T, and Y. The protein database contained 117 proteins including the SARS-CoV-2N, proteases, enolase, and the common Repository of Adventitious Proteins (cRAP) database. In the XLinkX search, the maximum missed cleavages were 3, minimum peptide length was 5, precursor mass tolerance was 10 ppm, FTMS fragment mass tolerance was 20 ppm, and ITMS fragment mass tolerance was 0.5 Da. Carbamidomethylation of Cys was included as a static modification, and oxidation of Met was included as a dynamic modification. For the cross-linked peptides, an FDR of 1% and a minimum XLinkX score cut-off of 50 was used. The cross-linking results were exported into xiView for further visualization and filtering.
Mouse immunization
Four 8-week-old, female Balb/cJ mice (The Jackson Laboratory) were immunized via subcutaneous injection of each hind flank with 20 µg of viral protein (in 50 mM Tris HCl, 300 mM NaCl) plus AddaVax (InvivoGen). Each mouse received 40 µg viral protein. Mice were boosted 50 days later with the same formulation. The mice were housed under standard conditions with a 12-h light/dark cycle, an ambient temperature of 72 °F +/−3 °F, and relative humidity maintained between 30% and 70%. All animal experiments were approved by the Institutional Animal Care and Use Committee at the La Jolla Institute for Immunology (LJI) and strictly conducted according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Plasma cell isolation
At 6 days post-boost, splenocytes were subject to two-step magnetic cell sorting to isolate plasma cells (PCs). After RBC lysis, non-plasma cells were depleted (Miltenyi Biotec) followed by enrichment of PCs with an EasySep Mouse CD138 Positive Selection kit (Stemcell Technologies).
Beacon-based antibody discovery
Enriched PCs were loaded onto OptoSelect 11 K chips (PhenomeX) and isolated as single cells in nanoliter pens using OEP light cages. Cells were screened in a 30 min time course assay for secretion of antibodies that bound to streptavidin beads (Spherotech) coated with 10 µg/mL biotinylated SARS-CoV-2 N. Secreted antibodies were detected with 1 µg/mL goat anti-human IgG (H + L)-Alexa Fluor 594 (Invitrogen), which was added to the cell culture media used to resuspend the antigen-coated beads.
Synthesis of cDNA from antigen-positive cells was carried out on-chip using the OptoSeq BCR kit (PhenomeX), according to the manufacturer’s directions. First-strand reaction products were exported on mRNA capture beads and deposited into individual wells of a 96-well plate. Total cDNA was amplified using Platinum SuperFi II polymerase (Invitrogen). After enzymatic cleanup, antibody heavy and light chain variable domains were amplified with two rounds of nested PCR using Platinum II Hot-Start polymerase (Invitrogen) using previously published primer sets (Tiller mouse mAbs). The resulting PCR products were assessed using 96w E-gels (Thermo Fisher) and paired wells were Sanger sequenced. Sequences were annotated using the bioinformatics platform PipeBio (PipeBio ApS, Horsens, DK).
Unique VH and VL domains were cloned into linearized human antibody expression vectors (human IgG1 and kappa light chain) using Gibson assembly (NEB) according to the manufacturer’s directions. Ligation reactions were transformed into 5-alpha F’Iq competent E. coli cells (NEB). QIAprep 96 Turbo Miniprep kits (Qiagen) were used to isolate plasmids according to the manufacturer’s instructions. Briefly, S block wells containing 1.1 mL LB media spiked with antibiotic were inoculated with single colonies and incubated overnight at 37 °C with agitation. DNA extraction was carried out with Qiagen buffer solutions and protocols. Plasmids were sequenced to ensure that the genes were in-frame, and that the cloned heavy and light chain variable domains matched PCR sequences.
ELISA-based binding analysis
Test expressions of each isolated antibody were carried out in 2.5 mL cultures of ExpiCHO cells cultured in 24-well blocks. ExpiCHO cells were transiently transfected and antibodies were allowed to express for 5 days. Cell supernatants were clarified and used directly in ELISA screens as previously described. Briefly, half-area ELISA plates were coated with 2 µg/mL of full-length N, the RNA-BD or DD. After blocking with 3% BSA/PBS, a 1–10 dilution of expression supernatant was added to each well, in duplicate. Bound mAbs were detected with anti-human secondary antibody (Southern Biotech).
High-throughput SPR binding kinetics
Binding kinetics measurements were performed on the Carterra LSA platform using HC30M sensor chips (Carterra) at 25 °C. The chip was activated with a freshly prepared solution of 130 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) (Pierce PG82079) and 33 mM N-hydroxysulfosuccinimide (Sulfo-NHS) (ThermoFisher Scientific 24510) in 0.1 M MES pH 5.5 using the single flow cell (SFC). Antibodies were diluted to 10 µg/mL in 10 mM sodium acetate (pH 4.5)/0.01% Tween and immobilized in quadruplicate using the 96PH for 15 minutes. Unreactive esters were quenched with a 7-minute injection of 1 M ethanolamine-HCl (pH 8.5).
A two-fold dilution series of the N RNA-BD or DD was prepared in 1xHBSTE-BSA buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20, supplemented with 0.5 mg/ml BSA). Protein was injected onto the chip surface using the SFC, from the lowest to the highest concentration, without regeneration in between. Five to six injections of buffer before the lowest non-zero concentration were used for signal stabilization. For each concentration, baseline data were collected for 120 s, association data for 300 s and dissociation data for 600 s. After the titration of each analyte, the chip surface was regenerated with two pulses (17 s per pulse) of 10 mM Glycine, pH 2.0. The running buffer for all kinetic steps was 1xHBSTE-BSA.
Titration data were processed with the Kinetics software package (Carterra), including reference subtraction, buffer subtraction and data smoothing. Analyte binding time courses for each antibody were fitted to a 1:1 Langmuir model to derive ka, kd and KD values.
High-throughput SPR epitope binning
Epitope binning was performed on the Carterra LSA® HT-SPR with the same array as used for kinetic analysis in HBSTE-BSA. A classical sandwich assay was used for competition analysis for the RNA-BD as previously described66. Briefly, 50 nM of the RND-BD was injected in each cycle for 4 min, followed immediately by a 4-min injection of the analyte antibody at 200 nM. Epitope binning for the N dimerization domain (N-DD) was performed with a premix assay format. A mixture of 50 nM of N-DD and 200 nM of each analyte antibody was incubated for at least 30 min before injecting over the array for 4 min. The sample injection for every sixth cycle was N-DD only instead of antibody-N-DD mixture. In both cases, the surface was regenerated each cycle with double pulses (30 s per pulse) of 10 mM Glycine pH 2.0. Data was processed and analyzed with Epitope® software (Carterra). Data was referenced using unprinted locations (nearest reference spots) on the array for both experimental set-ups. For the sandwich assays, the binding level of the analyte antibody just after the end of the injection was compared to that of a buffer only injection. For pre-mixed assays the response of each injection cycle was normalized to the response level in N-DD only cycles. Competition results were visualized as a heat map that depicts blocking relationships of analyte/ligand pairs. Clones that suffered from severe loss of activity or lack of complete dissociation from either N domain when used as ligands were excluded from analysis. Antibodies with similar patterns of competition are clustered together in a dendrogram and are assigned to shared communities.
Antibody expression
ExpiCHO cells were grown to a density of 8–12 × 106 cells/ml and then diluted to 6–7z106 cells/ml in a 25 ml culture on the day of transfection. Plasmids encoding HC and LC (10 µg each) were mixed with 80 µl Expifectamine, allowed to sit for 1–2 minutes and added to cells dropwise over 5 min. The flask containing the transfected cells was allowed to shake at 120 rpm in an incubator maintained at 37 °C, 8% CO2. The cells were fed 18–22 h after transfection with a mixture of 6 ml cold feed and 150 µl cold enhancer and moved to a 32 °C incubator with 5% CO2 and incubated with agitation at 120 rpm.
Antibody purification
The cells were harvested 7-8 days post transfection by centrifugation at 4000 × g for 30 min. The supernatant was filtered using a 0.22 µm filter and the pH was adjusted to 7.5. Packed protein A beads (2 ml; #Praesto AP Purolite Life Sciences PR00300-310) were washed with TBS and incubated with the supernatant for between 30 min and 2 h at RT. The mixture was passed through a gravity column and the beads were washed with 20 CV TBS to remove unbound proteins; IgG was eluted using 6 ml elution buffer (100 mM glycine pH 2.2). Neutralization buffer (900 µl, 1 M Tris pH 8) was added to bring the pH to neutral. IgG was dialyzed into TBS pH 7.5 overnight at 4 °C. Aliquots (1 mg/ml) were made and stored at −20 °C.
Digestion of IgG to Fabs
Each antibody IgG (5 mg) was incubated with Ides (Promega) 5% w/w for 2 h at 37 °C. L-cysteine (Sigma) was added to a final concentration of 15 mM and incubated for 1 h at 37 °C. Digestion was quenched with 50 mM iodoacetamide. The Fc portion was removed by passing the mixture over a column containing protein A beads. Pure Fab was recovered from the flowthrough by buffer exchanging with TBS using a Vivaspin 10k concentrator.
Cryo-EM data processing and model building
DD (200 µg) was complexed with 100 µg of Fab NP1-E9 and 100 µg Fab NP3-B4 and allowed to incubate overnight at 4 °C. The complexes were purified using an S75 size exclusion column. The peak fractions were collected and concentrated to 0.15 mg/ml. The sample (3 µl) was deposited on CF-2/1-3Cu-T50 grids and flash frozen using the FEI Vitrobot Mark IV (Thermo Fisher Scientific) in 100% humidity and 4 °C using a 10-s blot time. A dataset was collected on a 300-keV Titan Krios electron microscope with a K3 direct electron detector at 1.1 Å/pixel and a total dose of 50 e−/Å2 at the cryo-EM facility of La Jolla Institute for Immunology. A total of 7858 movie stacks were motion-corrected using the patch motion correction, and the CTF parameters were determined using the patch CTF estimation in cryoSPARC 72. TOPAZ neural network picker was used to select 1,520,588 particles. After two-dimensional classification and several rounds of hetero-refinement and non-uniform refinement (Supplementary Fig. 7), a set of 145,884 selected particles was used to produce a medium-resolution 3D reconstruction. To further improve the map quality, a new set of 632,077 particles were picked with TOPAZ. Subsequent ab initio reconstruction, heterogeneous refinement, and NU refinement yielded a 3.7 Å cryo-EM structure with D1 symmetry (Supplementary Figs. 6, 7). Model building was performed with Coot 0.9.8.767 and guided by the crystal structure PDB 6WJI57. Model refinement and validation was performed in Phenix 1.2068. For visualization purposes the map was processed by DeepEMhancer69 and ChimeraX 1.464,65 was used to prepare representations of the structure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and the cryo-EM map of the SARS-CoV-2 Nucleocapsid DD bound to Fab NP1-E9 and Fab NP3-B4 have been deposited to the RCSB Protein Data Bank and the Electron microscopy Data Bank respectively with accession numbers 9C2H and EMD-45157 (https://www.rcsb.org/structure/9C2H). Negative stain EM map of the SARS-CoV-2 N dimer complexed to 24 bp RNA is deposited to the EMDB, with accession number EMD-45158. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org/) via the PRIDE partner repository70 with the dataset identifier PXD052584 and 10.6019/PXD052584. Source data are provided with this paper.
References
Telenti, A. et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 596, 495–504 (2021).
Peck, K. M., Burch, C. L., Heise, M. T. & Baric, R. S. Coronavirus host range expansion and middle east respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Ann. Rev. Virol. 2, 95–117 (2015).
Machhi, J. et al. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 15, 359–386 (2020).
Wang, X., Yang, Y., Sun, Z. & Zhou, X. Crystal structure of the membrane (M) protein from a bat betacoronavirus. PNAS Nexus 2, pgad021 (2023).
Zhang, Z. et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat Commun 13, 4399 (2022).
Mandala, V. S. et al. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 27, 1202–1208 (2020).
Hsieh, C. L. et al. Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge. Nat Commun 15, 1553 (2024).
Wang, Y. T. et al. Spiking pandemic potential: structural and immunological aspects of SARS-CoV-2. Trends Microbiol. 28, 605–618 (2020).
Hardenbrook, N. J. & Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 52, 123–134 (2022).
Sola, I., Almazán, F., Zúñiga, S. & Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Ann. Rev. Viro. 2, 265–288 (2015).
Wu, W., Cheng, Y., Zhou, H., Sun, C. & Zhang, S. The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J. 20, (2023).
Zhao, H. et al. Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein. Preprint at https://doi.org/10.1101/2022.02.08.479556 (2022).
Royster, A. et al. SARS-CoV-2 nucleocapsid protein is a potential therapeutic target for anticoronavirus drug discovery. Microbiol. Spectr. 11, e0118623 (2023).
Peng, Y. et al. Structures of the SARS -CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 39, e105938 (2020).
Domingo López-Muñoz, A., Kosik, I., Holly, J. & Yewdell, J. W. Cell Surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci. Adv, 8, eabp9770 (2022).
Lineburg, K. E. et al. CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity 54, 1055–1065.e5 (2021).
Sette, A. & Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184, 861–880 (2021).
Grifoni, A. et al. SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host and Microbe 29, 1076–1092 (2021).
Taus, E. et al. Dominant CD8+ T cell nucleocapsid targeting in SARS-CoV-2 infection and broad spike targeting from vaccination. Front. Immunol. 13, 835830 (2022).
Eser, T. M. et al. Nucleocapsid-specific T cell responses associate with control of SARS-CoV-2 in the upper airways before seroconversion. Nat. Commun. 14, 2952 (2023).
Harris, P. E. et al. A synthetic peptide ctl vaccine targeting nucleocapsid confers protection from sars-cov-2 challenge in rhesus macaques. Vaccines 9, 520 (2021).
Hajnik, R. L. et al. Dual spike and nucleocapsid MRNA vaccination confer protection against SARS-CoV-2 Omicron and Delta variants in preclinical models. Sci. Transl. Med. 14, https://www.science.org (2022).
Saxena, A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J. Biosci. 45, https://doi.org/10.1007/s12038-020-00067-w (2020).
Iacob, S. & Iacob, D. G. SARS-CoV-2 treatment approaches: numerous options, no certainty for a versatile virus. Front. Pharmacol. 11, https://doi.org/10.3389/fphar.2020.01224 (2020).
Enjuanes, L. et al. Molecular basis of coronavirus virulence and vaccine development. In Advances in Virus Research vol. 96, 245–286 (Academic Press Inc., 2016).
Gui, M. et al. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell 8, 219–224 (2017).
Ye, Q., West, A. M. V., Silletti, S. & Corbett, K. D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 29, 1890–1901 (2020).
Klein, S. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun 11, 5885 (2020).
Yao, H. et al. Molecular architecture of the SARS-CoV-2 virus. Cell 183, 730–738.e13 (2020).
Dinesh, D. C. et al. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 16, e1009100 (2020).
Tayeb-Fligelman, E. et al. Low complexity domains of the nucleocapsid protein of SARS-CoV-2 form amyloid fibrils. Nat. Commun. 14, 2379 (2023).
Carlson, C. R. et al. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell 80, 1092–1103.e4 (2020).
Carlson, C. R. et al. Reconstitution of the SARS-CoV-2 ribonucleosome provides insights into genomic RNA packaging and regulation by phosphorylation. J. Biol. Chem. 298, 102560 (2022).
Korn, S. M., Dhamotharan, K., Jeffries, C. M. & Schlundt, A. The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5’-genomic RNA elements. Nat. Commun. 14, 3331 (2023).
Iserman, C. et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell 80, 1078–1091.e6 (2020).
Savastano, A., Ibáñez de Opakua, A., Rankovic, M. & Zweckstetter, M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 11, 6041 (2020).
Calder, L. J., Calcraft, T., Hussain, S., Harvey, R. & Rosenthal, P. B. Electron cryotomography of SARS-CoV-2 virions reveals cylinder-shaped particles with a double layer RNP assembly. Commun. Biol. 5, 1210 (2022).
Fukuhara, H. et al. Unprecedented spike flexibility revealed by BSL3 Cryo-ET of active SARS-CoV-2 virions. Preprint at https://doi.org/10.1101/2023.10.10.561643 (2023).
Morse, M., Sefcikova, J., Rouzina, I., Beuning, P. J. & Williams, M. C. Structural domains of SARS-CoV-2 nucleocapsid protein coordinate to compact long nucleic acid substrates. Nucleic Acids Res. 51, 290–303 (2023).
Zachrdla, M., Savastano, A., Ibáñez de Opakua, A., Cima-Omori, M. S. & Zweckstetter, M. Contributions of the N-terminal intrinsically disordered region of the severe acute respiratory syndrome coronavirus 2 nucleocapsid protein to RNA-induced phase separation. Protein Sci. 31, e4409 (2022).
Tenchov, R. & Zhou, Q. A. Intrinsically disordered proteins: perspective on COVID-19 infection and drug discovery. ACS Infect. Dis. 8, 422–432 (2022).
Schiavina, M., Pontoriero, L., Tagliaferro, G., Pierattelli, R. & Felli, I. C. The role of disordered regions in orchestrating the properties of multidomain proteins: The SARS-CoV-2 nucleocapsid protein and its interaction with enoxaparin. Biomolecules 12, 1302 (2022).
Basu, S. & Bahadur, R. P. A structural perspective of RNA recognition by intrinsically disordered proteins. Cell. Mol. Life Sci. 73, 4075–4084 (2016).
Chang, C.-K. et al. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 83, 2255–2264 (2009).
Takeda, M. et al. Solution structure of the C-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 380, 608–622 (2008).
Chen, C. Y. et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 368, 1075–1086 (2007).
Cong, Y., Kriegenburg, F., De Haan, C. A. M. & Reggiori, F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 7, 5740 (2017).
Cubuk, J. et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 12, 1936 (2021).
Ribeiro-Filho, H. V. et al. Structural dynamics of SARS-CoV-2 nucleocapsid protein induced by RNA binding. PLoS Comput Biol. 18, e1010121 (2022).
Wright, P. E. & Dyson, H. J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 (1999).
Tugaeva, K. V. et al. The mechanism of SARS-CoV-2 nucleocapsid protein recognition by the human 14-3-3 proteins:SARS-CoV-2 N association with host 14-3-3 proteins. J. Mol. Biol. 433, 166875 (2021).
Cheng, N. et al. Protein post-translational modification in SARS-CoV-2 and host interaction. Front. Immunol. 13, 13 (2023).
Perdikari, T. M. et al. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J 39, e106478 (2020).
Yu, I. M., Oldham, M. L., Zhang, J. & Chen, J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteriviridae. J. Biol. Chem. 281, 17134–17139 (2006).
Van Nguyen, T. H. et al. Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. Acta Crystallogr. D. Struct. Biol. 75, 8–15 (2019).
Jayaram, H. et al. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80, 6612–6620 (2006).
Vandervaart, J. P. et al. Serodominant SARS-CoV-2 nucleocapsid peptides map to unstructured protein regions. Microbiol Spectr. 11, e0032423 (2023).
Różycki, B. & Boura, E. Conformational ensemble of the full-length SARS-CoV-2 nucleocapsid (N) protein based on molecular simulations and SAXS data. Biophys. Chem. 288, 106843 (2022).
Reid Alderson, T., Pritišanac, I., Kolaric, D., Moses, A. M. & Forman-Kay, J. D. Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2. Proc. Natl Acad. Sci. USA 120, e2304302120 (2023).
Vreven, T. et al. Integrating cross-linking experiments with ab initio protein–protein docking. J. Mol. Biol. 430, 1814–1828 (2018).
Maheshwari, S. & Brylinski, M. Predicted binding site information improves model ranking in protein docking using experimental and computer-generated target structures. BMC Struct. Biol. 15, 23 (2015).
Wang, Y. et al. Modular characterization of SARS-CoV-2 nucleocapsid protein domain functions in nucleocapsid-like assembly. Mol. Biomed.4, 16 (2023).
Kang, S. et al. A SARS-CoV-2 antibody curbs viral nucleocapsid protein-induced complement hyperactivation. Nat. Commun. 12, 2697 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Pettersen, E. F. et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Yu, X. et al. The evolution and determinants of neutralization of potent head-binding antibodies against Ebola virus. Cell Rep. 42, 113366 (2023).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acid Res. 50, D543–D552 (2021).
Acknowledgements
We thank Dr. Ruben Diaz-Avalos for excellently managing the cryo-EM facility at La Jolla Institute for Immunology. We wish to thank Dr. Laurence Cagnon for oversight of and assistance in the La Jolla Institute for Immunology BSL-3 facility. We also thank the GHR Foundation for support of the K3 detector with which these images were collected. We also thank Dr. Sharon L. Schendel for editing this manuscript. The technical assistance of Dipti Parekh is gratefully acknowledged. We thank NIH R21 AI178427-01 and CDRF Global grant DAA3-20-66949-1 for financial support (E.O.S. and S.L.B.). This project was also funded by the COVID-19 Advancement by Postdoctoral Research, kindly supported by Walter Green and Lisa Liguori (E.O.S. and S.L.B.). S.L.B. is funded by a RYC2023-044971-I and CIGE/2023/169.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.L.B., Methodology: S.L.B., C.H., Investigation: S.L.B., C.H., R.D.A., A.S.N., D.T.S., K.M.H., S.H., M.Z., R.R.R., E.O., R.M., Supervision: S.L.B., S.S., V.H.W., E.O.S., Writing original draft: S.L.B., E.O.S., Writing—review & editing: S.L.B., C.H., R.D.A., A.S.N., D.T.S., K.M.H., S.H., M.Z., R.R.R., E.O., R.M., S.S., V.H.W., E.O.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alexander Leitner, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Landeras-Bueno, S., Hariharan, C., Avalos, R.D. et al. Structural stabilization of the intrinsically disordered SARS-CoV-2 N by binding to RNA sequences engineered from the viral genome fragment. Nat Commun 16, 6521 (2025). https://doi.org/10.1038/s41467-025-61861-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61861-4