Abstract
How the ubiquitously expressed splicing factors specifically regulate neural crest (NC) development and enhance their vulnerability to splicing perturbations remain poorly understood. Here, we show that NC-specific DLC1, partnering with SF3B1-PHF5A splicing complex, are crucial for determining avian trunk NC cell fate by regulating the splicing of NC specifiers SOX9 and SNAI2 pre-mRNAs rather than their upstream regulators BMP4, WNT1, and PAX7. Mechanistically, SF3B1-PHF5A binds to the intronic branch site (BS) sequences of all factors, while DLC1 interacts with a specific motif near the BS sequences of SOX9 and SNAI2, thereby determining their functional specificity in NC specification. Moreover, DLC1 increases NC cells’ vulnerability to splicing modulator pladienolide B (PB) by reducing the binding capacity of the SF3B1-PHF5A splicing complex to the shorter length of both SOX9 intron 2 and SNAI2 intron 1, which possess weaker polypyrimidine tract 3’ of the BS sequence, resulting in intron retention and loss of NC progenitors. Conversely, somite specific SLU7-SF3B1-PHF5A splicing complex regulates SOX9 and SNAI2 expression and imparts resistance to PB. Our data reveal the cell-type specific splicing complexes with distinct vulnerabilities to PB, highlighting the critical role of the DLC1-SF3B1-PHF5A in determining trunk NC cell fate and enhancing its susceptibility to splicing perturbation.
Similar content being viewed by others
Introduction
In vertebrates, multipotent neural crest cells (NCCs) are specified at the neural plate border between the neural plate and the non-neural ectoderm in the late gastrula stage of chick embryos1. Following neural tube closure, prospective NCCs in the dorsal neural tube undergo epithelial-mesenchymal transition (EMT) to acquire directional migratory behavior toward the periphery, where they give rise to the craniofacial structures and the peripheral nervous system2. Numerous studies have established a gene regulatory network (GRN) in which neural crest (NC) inductive signals (WNTs, BMPs, and FGFs)3 cooperate with neural plate border specifiers (PAX7 and MSX1/2) to activate the transcription of NC specifier genes (SOX9, SNAI2 and FOXD3) that determine NCC fate and EMT features1,4,5,6,7. Notably, SOX9 and SNAI2 are intron-containing genes that require additional underlying regulatory control of their pre-mRNA splicing to generate functional proteins. However, components of the spliceosome complex that regulate the pre-mRNA splicing of these NC specifiers remain largely unknown. In addition, genes encoding the spliceosome components are generally ubiquitously expressed, yet their mutations can lead to craniofacial malformations8, suggesting a specific vulnerability of cranial NCCs to splicing abnormalities. Whether there is a similar susceptibility due to altered spliceosome components in trunk NCCs remains to be determined.
RNA splicing is a multi-step process regulated by the spliceosome, which comprises five small ribonucleoprotein particles (snRNPs) U1, U2, and U4/U5/U6 and their associated proteins unique to each snRNP9. Multi-RNAs and proteins are recruited and assembled on introns and then spliced by the stepwise recognition of the 5’ splice site, the branch site (BS) sequences (CURAY, branch point adenosine [BP-A] underlined), and the 3’ splice site10. These processes produce reactive groups for intron excision and ligation of exons catalyzed by the spliceosome. A core component of U2 snRNP is SF3B1, the largest subunit of the SF3B complex, which plays a crucial role in recognizing BS sequences by base-pairing with the U2 snRNA GUAGUA sequence to form the U2/BS duplex. This duplex contains a bulged adenosine that promotes its stabilization in the early stages of spliceosome assembly11. In mouse embryos, SF3B1 is ubiquitously expressed but is more strongly expressed in the dorsal neural tube along the anterior-posterior axis12. Moreover, SF3B1 null mutants die during preimplantation at around stage 16-3213, indicating SF3B1 has an essential role in embryogenesis. Despite its broad expression, several reports have demonstrated that SF3B1 has specific functions in skeletal patterning13, adult brain neurogenesis14 and regulating the proliferative capacity of hematopoietic stem cells15, indicating that SF3B1 has distinct regulatory roles in different cellular and tissue contexts. Consistently, hypomorphic sf3b1 mutant zebrafish embryos showed a marked reduction in the expression of a subset of trunk NC specifiers (e.g., sox9b, snail1b) partly due to aberrant processing of their pre-mRNAs, which led to defects in NC survival, migration, and lineage differentiation16, suggesting that trunk NCCs are vulnerable to altered splicing machinery, yet the underlying mechanisms are not clear.
The PHD-finger domain protein 5 A (PHF5A), a member of the PHD zinc finger superfamily, is a highly evolutionarily conserved splicing factor ubiquitously expressed in the nucleus of eukaryotes from yeast to humans17. It is intimately associated with SF3B1, SF3B3, and SF3B5, which together form the core of the SF3B complex in U2 snRNA protein18. Moreover, PHF5A serves as a bridge protein to facilitate interactions between U2 snRNA and ATP-dependent helicase in different phases of the splicing process19,20. Structural analysis has revealed that the BP-A is flipped out from the U2/BS duplex and sequestered in a sequence-conserved pocket formed by SF3B1 and PHF5A, a critical step in the pre-mRNA splicing process11,21,22. Several small-molecule antitumor compounds have been identified to act as splicing modulators that bind the BP-A pocket of the SF3B1-PHF5A complex in a competitive manner to disrupt the interaction with BP-A22,23. These compounds affect pre-mRNA splicing patterns in cells by triggering global exon skipping and intron retention depending on the sequence of the inhibited substrate, and the modulator structure, affinity and dose24,25. Apart from its involvement in splicing, PHF5A can function as a transcription factor regulating the expression of connexin43 gene in response to estrogen26, modulating stem cell features in pancreatic cancer cells27, and maintaining the pluripotency of embryonic stem cells (ESCs)28. Given that SF3B1 is required for NC specification via regulating the splicing of NC specifiers and that PHF5A regulates the pluripotency of ESCs, it is plausible that PHF5A is associated with SF3B1 in regulating the formation of multipotent NC stem cells at the transcriptional and/or post-transcriptional level.
Here, we employed the CRISPR-Cas9 genome editing approach in avian embryos to demonstrate the essential role of NC-specific DLC1, in association with the ubiquitously expressed SF3B1-PHF5A complex, in determining the fate of trunk NCCs by regulating the pre-mRNA splicing of NC specifiers SOX9 and SNAI2, rather than their upstream regulators BMP4, WNT1, and PAX7. Global splicing analysis of intronic sequences retained in DLC1 gRNA cells identified a specific DLC1 binding motif near the BS sequence bound by the SF3B1-PHF5A complex within the introns of SOX9 and SNAI2 and a subset of genes associated with NC specification and EMT, thereby determining their functional specificity. Treatment with the splicing modulator pladienolide B (PB) in avian embryos inhibited the recognition of BS sequences by the SF3B1-PHF5A complex, leading to the loss of SOX9 and SNAI2 expression in trunk NCCs. However, the expression of these genes remained unchanged in somites lacking DLC1 expression. Mechanistically, DLC1 weakened the binding capacity of the SF3B1-PHF5A splicing complex to the shorter length of both SOX9 intron 2 and SNAI2 intron 1, which exhibit a weaker Polypyrimidine (Py)-tract than in SOX9 intron 1 and SNAI2 intron 3. This explains the heightened susceptibility of NCCs to PB, resulting in intron retention and loss of NC progenitors. In contrast, the somite-specific complex of SLU7-PHF5A-SF3B1 regulates the splicing of SOX9 and SNAI2 pre-mRNAs and confers resistance to PB. Overall, our findings demonstrate the critical role of the NC-specific DLC1-SF3B1-PHF5A splicing complex in determining trunk NCC fate and increasing its susceptibility to splicing perturbation.
Results
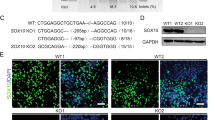
DLC1 is required for pre-mRNA splicing of SOX9 and SNAI2 introns
Our previous studies have shown that DLC1 is expressed in early delaminating and emigrating trunk NCCs from Hamburger and Hamilton (HH) stages (st) 11 to 13 of chick embryo development, consistent with its role in governing directional delamination and migration29. However, a reexamination of DLC1 expression in the trunk region of st 8-12 chick embryos revealed its onset after neural tube closure in the premigratory NC region, where prospective NC progenitors express SOX9 and SNAI2 (Figs. S1a and 1a), suggesting a potential role of DLC1 in NC specification. Expression of DLC1 is downregulated as NCCs migrate further (Fig. S1a). To investigate the role of DLC1 in NC formation, we used CRISPR/Cas9 genome editing to knockout (KO) DLC1 in developing chick neural tubes at st 9-10, coinciding with the onset of DLC1 expression. Two different guide RNA (gRNA) oligos were designed to target DLC1 exon 9 (DLC1 gRNA1 and gRNA2) (Fig. 1b) and a non-targeting gRNA served as a control (Ctrl). Each gRNA or Ctrl gRNA was co-electroporated with a construct encoding nuclear Cas9 and GFP into the right side of the avian trunk neural tubes30 (Fig. 1c). The electroporated embryos were analyzed at 9 h post-transfection (hpt) when the effect of gene KO became evident, as demonstrated by a complete loss of DLC1 protein expression in both gRNAs compared to the Ctrl gRNA (Fig. 1d). We present the results of DLC1 gRNA1 unless otherwise stated, as similar findings were observed with both gRNAs. Electroporation of DLC1 gRNA1 led to cell-autonomous reductions in the expression of SOX9, SNAI2, and FOXD3 in the pre-migratory NC domain compared to the Ctrl gRNA (Fig. 1e). Fluorescence-activated cell sorting of transfected cells for qPCR analysis further confirmed reduction of their mRNA levels (Fig. 1f–h). Importantly, the reintroduction of full-length DLC1 was sufficient to restore the decreased expression of SOX9, SNAI2, and FOXD3 caused by DLC1 gRNA1 (Fig. 1e and Fig. S1b), highlighting the specificity of DLC1 KO. However, domain mapping analysis revealed that neither individual functional domain nor their combinations were able to rescue the loss of NC specifiers expression in DLC1 KO cells (Fig. S1b–d), implying that all domains play a critical role in regulating the expression of these genes. In addition, we observed the nuclear localization of ectopic DLC1 (Fig. 1i), indicating its potential involvement in gene expression regulation. Immunofluorescence staining revealed a loss of SOX9 expression in the dorsal neural tube transfected with DLC1 gRNA1, while the expression of neural stem/progenitor marker SOX2 was upregulated (Fig. 1j, k), suggesting a switch in cell fate from NC to neural lineage. This was accompanied by a significant reduction in migratory NCCs expressing HNK-1 (Fig. 1l). Importantly, the levels of NC inductive signals WNT1, BMP4, and the expression of neural plate border specifier PAX7 were unaffected by DLC1 gRNA1 and gRNA2, indicating that the decrease in expression of NC specifier genes was not due to alterations in the expression of their upstream regulators in the GRN (Fig. S1e–h). In addition, the levels of RNA polymerase II phosphorylation at Ser 5 (PolII-pSer5) and Ser 2 (PolII-pSer2), which are involved in transcription initiation and elongation31, respectively, were not significantly affected by DLC1 gRNA1 and gRNA2 (Fig. 1m). To investigate whether DLC1 regulates transcriptional elongation of NC specifier genes, chromatin immunoprecipitation of PolII-pSer2 was performed at multiple sites along the SOX9 and SNAI2 genomic DNA in embryos treated with DLC1 gRNA1 or Ctrl gRNA. We detected a 4-fold enrichment of PolII-pSer2 at the 500 bp downstream of the SOX9 transcriptional start site in cells transfected DLC1 gRNA1 compared to those transfected with Ctrl gRNA. However, the enrichment was reduced to a level similar to the Ctrl gRNA from 1000 bp onwards to the end of the SOX9 gene body (Fig. S1i). There is no significant difference in the degree of PolII-pSer2 association with SNAI2 genomic DNA at different sites between DLC1 gRNA1 and Ctrl gRNA (Fig. S1j), indicating that transcriptional elongation still occurs in the absence of DLC1. These findings suggest that DLC1 is not directly involved in transcriptional regulation. Instead, intron retention analysis revealed increased levels of SOX9 introns 1 and 2 in DLC1 KO transfected cells compared to the Ctrl gRNA (Fig. 1n, o). Similar results were observed for SNAI2 introns 1 and 3, although intron 2 was too short for detection (Fig. 1p, q). Hybridization chain reaction (HCR) analysis was able to detect the intron-retained SOX9 and SNAI2 mRNAs in the dorsal neural tubes transfected with DLC1 gRNA1 (Fig. 1r). These results suggest that the downregulated expression of SOX9 and SNAI2 in response to DLC1 KO may be attributed to the dysregulated splicing process of their pre-mRNAs. Considering that SOX9 regulates FOXD3 expression6, the loss of FOXD3 expression in DLC1 KO cells may be due to the depletion of SOX9+ NC progenitors. Taken together, these findings suggest that DLC1 is required for NC specification by regulating pre-mRNA splicing of SOX9 and SNAI2.
a co-localization of DLC1, SOX9, and SNAI2 mRNA in premigratory NCCs. b Targeting strategy. c Experimental workflow. d Loss of DLC1 expression in both gRNA treatment groups. e Reduced expression of SOX9, SNAI2, and FOXD3 in dorsal neural tubes transfected with DLC1 gRNA (white arrowheads). Black arrows indicate that overexpression of full-length DLC1 restored the expression of SOX9, SNAI2 and FOXD3 in cells transfected with DLC1 gRNA1. Scale bar, 50 µm. f–h Reduced expression levels of SOX9, SNAI2 and FOXD3 in DLC1-depleted cells compared to Ctrl gRNA (n = 5 embryos per treatment). i Nuclear localization of ectopic V5-DLC1 (open arrowheads) in the dorsal neural tube. j Electroporation of DLC1 gRNA1 led to reduced SOX9 expression (white open arrowheads) and upregulated SOX2 expression (white solid arrowheads) in the dorsal neural tube. Dotted lines outline the neural tubes. Scale bar, 50 µm. k Quantification of SOX2 fluorescence intensity. (n = 9 embryos per treatment). l Reduced HNK-1 expression (white open arrowheads) in DLC1 gRNA 1 transfected side. Horizontal white dotted lines indicate the axial level of transverse sections. Scale bar, 100 μm. m Unaltered expression of RNA Pol II-pSer5 and -pSer2 in DLC1 knockout groups. IR-qPCR analysis indicated increased levels of SOX9 introns 1/2 (n) and SNAI2 introns 1/3 (p) in embryos treated with DLC1 gRNA1 (n = 5 embryos per treatment). Arrows indicate primer pairs flanking the BP-A (A) in each intron of SOX9 (o) and SNAI2 (q) pre-mRNA. r Detection of SOX9 introns 1/2 and SNAI2 introns 1/3 in the dorsal neural tubes transfected with DLC1 gRNA1, while no signal was detected in Ctrl gRNA. “+” indicates the transfected side. Scale bar, 50 µm. The minima, maxima, center, percentile values and the exact p-values are listed in the Source data file. Mean ± SD. **P < 0.01, ***P < 0.001. NCCs neural crest cells, Ctrl control, IR intron retention.
DLC1 associates with the PHF5A-SF3B1 splicing complex which is required for pre-mRNA splicing of SOX9 and SNAI2 introns
To elucidate the molecular mechanism by which DLC1 regulates gene splicing, we conducted mass spectrometry (MS) analysis on DLC1-immunoprecipitated lysates from chick neural tubes to identify interacting factors of DLC1. Among the many DLC1-interacting factors involved in diverse molecular functions, one of the major categories is RNA-binding proteins, which mainly consist of splicing factors (Fig. 2a, S2a). This category includes PHF5A and SF3B1, both of which are components of the SF3B complex within the U2 snRNP spliceosome involved in pre-mRNA splicing32 (Fig. 2a, Dataset 1). Immunoprecipitation experiments confirmed the association of endogenous DLC1 with PHF5A and SF3B1 (Fig. 2a). Consistently, MS analysis of PHF5A-immunoprecipitated lysates from chick neural tubes demonstrated the existence of the endogenous PHF5A-SF3B1 complex (Fig. 2b, Dataset 2), and their interaction was further confirmed by immunoprecipitation (Fig. 2b). Additionally, we detected co-localization of the ubiquitously expressed PHF5A-SF3B1 complex with DLC1 in the premigratory NCCs (Fig. S2b). The ability of DLC1 to interact with the PHF5A-SF3B1 splicing complex raises the possibility that PHF5A-SF3B1 could be involved in NC specification. To investigate this hypothesis, we employed a similar CRISPR/Cas9 genome editing approach to KO PHF5A in developing chick neural tubes using two gRNA oligos targeting exons 3 and 4 of PHF5A (gRNA1, gRNA2) (Fig. 2c). After 9 hpt, we observed a significant reduction of PHF5A expression in gRNA-transfected cells compared to the Ctrl gRNA (Fig. 2c), confirming successful PHF5A KO. For subsequent studies, we utilized gRNA1 as similar results were obtained with gRNA2. Electroporation of PHF5A gRNA1 resulted in cell-autonomous downregulation of SOX9, SNAI2, and FOXD3 expression in the premigratory NC region, as confirmed by qPCR analysis of sorted cells when compared to the unaltered expressions observed in Ctrl gRNA and the untransfected side (Fig. 2d–i). Co-expression of PHF5A gRNA1/Cas9 constructs with a vector encoding full-length chick PHF5A cDNA, rather than the PHD domain alone, restored the expression of NC specifier genes (Fig. 2d, f, h and Fig. S2c), demonstrating the specificity of PHF5A KO. Consistent with the downregulation of NC specifiers, the domain of SOX2 expression expanded into the NC territory, where SOX9 was diminished (Fig. 2j, k), indicating a switch in cell fate from NC to neural progenitors. Similar to the results observed with DLC1 KO, PHF5A gRNA did not alter the expression levels of WNT1, BMP4, and PAX7 (Fig. S2d–g). The loss of NC progenitors in PHF5A KO embryos resulted in a lack of migratory NCCs expressing HNK-1 (Fig. 2l). As PHF5A has been implicated in the control of transcriptional elongation of genes that maintain pluripotency in ESCs28, we investigated whether PHF5A associates with PolII to regulate the transcription of NC specifier genes. Immunoprecipitation experiments revealed that PHF5A did not interact with PolII-pSer5 or PolII-pSer2 (Fig. S2h, i)31. Despite the efficient KO of PHF5A, the levels of PolII-pSer5 and PolII-pSer2 remained largely unchanged (Fig. 2m), indicating that the loss of NC specifier gene expression following PHF5A KO is not due to altered levels of the transcriptional machinery. Instead, PHF5A gRNA1 led to increased levels of introns 1 and 2 of SOX9 and introns 1 and 3 of SNAI2, when compared to the Ctrl gRNA (Fig. 2n–q). HCR analysis detected the expression of these introns in PHF5A KO embryos but not in Ctrl gRNA-treated embryos (Fig. 2r). Similarly, SF3B1 KO resulted in a loss of NC progenitors due to intron retention of SOX9 and SNAI2 pre-mRNA without altering the expression of their upstream regulators and transcriptional machinery (Fig. S3a–q). Importantly, PHF5A KO and SF3B1 KO did not affect the expression levels of SF3B1 or PHF5A, respectively (Fig. S3r, s), indicating they do not regulate the expression of each other to determine NCC fate.
a Interactome of DLC1-associated splicing factors. Immunoprecipitation (IP) validated the association of endogenous DLC1 with PHF5A and SF3B1. n = 3 biological replicates, 30 embryos per replicate. b Interactome of PHF5A showing splicing factors in the SF3B complex (green color). IP confirmed the association of endogenous SF3B1 with PHF5A. n = 3 biological replicates, 30 embryos per replicate. c Targeting strategy. Western blot showing loss of PHF5A expression in embryos transfected with PHF5A gRNA1/2. In situ hybridization and qRT-PCR showing reduced expression (white open arrowheads) of SOX9 (d, e), SNAI2 (f, g), and FOXD3 (h, i) in PHF5A-ablated cells compared to Ctrl gRNA. Their expression patterns were restored by pCIG-HA-PHF5A overexpression (black arrows). n = 3 embryos per treatment. Scale bar, 50 μM. j SOX9 expression (white open arrowheads) was reduced in PHF5A-depleted NCCs where SOX2 expression was upregulated (white solid arrowheads). Dotted lines outline the neural tubes. n = 3 embryos per treatment. Scale bar, 50 μM. k Quantification of SOX2 fluorescence intensity. n = 9 embryos per treatment. l HNK-1 expression was reduced (white open arrowheads) in the PHF5A gRNA-transfected side compared to the untransfected side and embryos treated with Ctrl gRNA. n = 3 embryos per treatment. m Expression levels of RNA PolII-pSer5 and PolII-pSer2 remained unaltered in PHF5A KO embryos compared to Ctrl gRNA. IR-qPCR analysis shows increased levels of SOX9 introns 1/2 (n) and SNAI2 introns 1/3 (p) in embryos treated with PHF5A gRNA1 compared to Ctrl gRNA-treated embryos. n = 5 embryos per treatment. Schematic diagrams indicate primer pairs for detecting retained introns of SOX9 (o) and SNAI2 (q). r HCR showing detection of retained introns of SOX9 and SNAI2 in embryos transfected with PHF5A gRNA, while no signal was detected in Ctrl gRNA. n = 5 embryos per treatment. The “+” indicates the transfected side. Scale bar: 50 μM. The minima, maxima, center, percentile values and the exact p-values are listed in the Source data file. Mean ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001.
In contrast, we did not observe a reduction or loss of SOX9 and SNAI2 mRNA expression in cranial NCCs transfected with each gRNA construct (Fig. S4a), indicating that the effects are specific to the trunk.
To further confirm the splicing defects in each KO, we conducted RT-PCR with specific primers to examine the levels of unspliced and spliced forms of each intron (Fig. S5a, b). The use of two different gRNAs targeting PHF5A, SF3B1, and DLC1 genes resulted in increased ratios of unspliced transcripts to varying degrees for each intron of SOX9 and SNAI2 when compared to the gRNA Ctrl (Fig. S5c–j). Conversely, the unspliced forms of PAX7, BMP4, and WNT1 were not predominantly expressed in all KO treatments and Ctrl gRNA, except for BMP4 intron 2 which did not show a discernible difference in the ratio of unspliced and spliced counts between each KO and Ctrl gRNA (Fig. S6a–f). These findings are consistent with the unaltered mRNA levels of BMP4, WNT1, and PAX7 when DLC1, PHF5A, or SF3B1 are absent (Figs. S1c–f; S2d–g; S3g–j). Additionally, RT-PCR analysis indicated that there was no exon skipping of SOX9 and SNAI2 in any of the KO treatments (Fig. S7a–d), suggesting that the decreased expression of SOX9 and SNAI2 mRNA in all KO treatments is primarily due to intron retention. In agreement with this, transcriptomic profiling of sorted GFP+ cells from embryos treated with each gRNA confirmed increased intron depth for SOX9 and SNAI2 compared to the Ctrl gRNA (Fig. 3a, Dataset3). Moreover, global splicing analysis revealed that most of the affected genes involved in NC specification and cell-cell adhesion for EMT exhibited intron retention in all KO treatment groups (Fig.3a, S7e, i). For instance, ID133, TFAP2A34, MYC35, SOX1036, LMO437, CDH238, CDH138, CDH1139,40 showed varying levels of intron retention in each KO treatment (Fig. 3a). In addition, some of these factors also manifested defects in other splicing events (Fig. 3b, S7f–i). CDH11 exhibited exon skipping in all treatment groups and 5’ splicing in both DLC1 KO and PHF5A KO, while other factors displayed 5’ and 3’ alternative splicing events depending on the KO treatment (Fig. 3b, S7f–i). Consequently, these factors exhibited aberrant expression levels in each KO group (Fig. 3c–e). In contrast, the levels of retained introns for PAX7, BMP4, and WNT1 were low or comparable in all treatment groups (Fig. 3a). Similarly, no aberration was observed in the splicing of other neural plate border specifiers MYB41, PAX3, ZIC142, the inductive signal FGF143, and the regulators of NC lineages ADAR44, APC245, and MAPK1046 (Fig. 3a). Thereby, expression of these factors remained unaltered in all treatment groups (Fig. 3f–h). Collectively, these findings suggest that nuclear DLC1 associated with the PHF5A-SF3B1 splicing complex has a specific role in NC determination and EMT by regulating the splicing of SOX9 and SNAI2 pre-mRNA and other essential factors involved in these processes.
a List of genes involved in NC specification and EMT with intron retention (IR) observed in DLC1-, PHF5A-, and SF3B1-depleted cells. In contrast, the levels of IR for genes involved in neural plate border specification, NC induction, and NC lineage determinants were either low in all KO treatments or comparable to Ctrl gRNA. The levels of IR were quantified using percent intron retention scores assessed by VAST-TOOLs. b List of genes exhibiting varying degrees of exon skipping (ES), 5’ alternative splicing (5’ AS) and 3’ alternative splicing (3’ AS) in each gRNA treatment. ES events were quantified by percent spliced-in (PSI), while 5’ AS and 3’ AS events were evaluated using percent splice site usage (PSU). qPCR analysis revealed reduced expression levels of the indicated genes in DLC1 gRNA1 (c), PHF5A gRNA1 (d), and SF3B1 gRNA1 (e) compared to Ctrl gRNA. Other genes remained unaffected by these treatments (f–h). The exact p-values are listed in the Source data file. Mean ± SD. ns: no significant difference, **P < 0.01, ***P < 0.001, ****P < 0.0001.
DLC1 determines the functional specificity of the PHF5A-SF3B1 splicing complex in NC specification and EMT by binding to a specific motif within introns related to these processes
Given the ubiquitously expressed SF3B1-PHF5A complex is essential for recognizing BS sequence to create the bulged BP-A pocket during the splicing process11,22,24, the specific splicing function of DLC1 prompted us to examine whether there is a DLC1 binding motif present in the intron of genes essential for NC specification and EMT. By comparing the sequences of introns affected and unaffected by DLC1 KO treatment, we identified a motif (CUCCGGKU), that ranks the highest among others according to their statistically significant predominance in the affected introns compared to the unaffected ones (Fig. S8a, 4a). This motif is located either close to or distant from the BS sequences depending on the introns, with the exception of MYC where the motif falls within its BS sequence (Fig. 4a). Conversely, this motif is absent in the introns that did not exhibit any aberration in DLC1 KO treatment (Figs. 3a, 4a and S8a). To examine the binding of DLC1-SF3B1-PHF5A to the motif and BS sequence within the introns of SOX9 and SNAI2 in chick neural tubes, we performed RNA immunoprecipitation (RIP)-qPCR to evaluate the ability of each endogenous protein to interact with these sequences in each intron. We incubated lysates generated from wild-type HH9-10 embryonic trunks with magnetic beads coated with DLC1, PHF5A, SF3B1, or IgG non-specific control antibodies. The eluted RNA from the beads was analyzed by qRT-PCR using primers flanking their putative binding sequences in each intron (Fig. 4b, f). We found the region encompassing the motif and BS sequence in each intron was significantly enriched by PHF5A, SF3B1, and DLC1 antibodies compared to IgG control (Fig. 4c–e, g–i), confirming their intronic binding capacity. To further determine whether DLC1 is essential for the binding of PHF5A-SF3B1 complex to the BS sequences of SOX9 and SNAI2 introns, RIP-qPCR was performed for PHF5A and SF3B1 in embryos treated with DLC1 gRNA1. The results revealed that DLC1 KO did not disrupt the interaction between SF3B1 and PHF5A1 (Fig. 4j), but significantly reduced the enrichment of PHF5A and SF3B1 at the BS sequences of SOX9 and SNAI2 introns compared to Ctrl gRNA (Fig. 4k–n). These results suggest that DLC1 is necessary for the binding capacity of the PHF5A-SF3B1 complex to these introns, providing a mechanistic explanation for the lack of NC progenitors formation resulting from the loss of DLC1 function due to defective binding of the PHF5A-SF3B1 complex to SOX9 and SNAI2 introns at the BS sequence for splicing initiation. Likewise, DLC1 was found to be associated with its motif located within the introns of CDH2, CDH11, CDH1, ID1, LMO4, MYC, SOX10, and TFAP2A (Fig. S8b), resulting in their abnormal expression in DLC1 KO cells (Fig. 3c).
a DLC1 binding motif (CUCCGGKU), located either near or distant from BP-A, is found in introns of genes affected by DLC1 KO. The motif is ranked the highest among other motifs based on its statistically significant predominance in the affected gene introns compared to the unaffected ones. K represents A/G or U. Motif alignment is highlighted in green, and base pairs (bp) indicate the distance from the motif to BP-A. b, f Schematic diagram showing primer pairs flanking the motif (asterisk) and BP-A of SOX9 (a) and SNAI2 (b) introns for RNA-immunoprecipitation qPCR (RIP-qPCR). Graphs show the binding capacity of endogenous PHF5A (c, g), SF3B1 (d, h), and DLC1 (e, i) to the BP-A of SOX9 introns 1 and 2, as well as SNAI2 introns 1 and 3. n = 3 embryos per treatment. j IP shows that SF3B1 remained associated with PHF5A in the absence of DLC1. k–n RIP-qPCR analysis reveals reduced binding capacity of PHF5A-SF3B1 splicing complex to introns 1 and 2 of SOX9, as well as introns 1 and 3 of SNAI2 in embryos treated with DLC1 gRNA1 compared to Ctrl gRNA. n = 3 embryos per treatment. The exact p-values are listed in the Source data file. Mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We then examined whether PHF5A, SF3B1, and DLC1 could bind to their putative binding sequences within the introns of PAX7, WNT1, and BMP4 pre-mRNAs. While the PHF5A-SF3B1 complex remained strongly associated with the BS sequence of each gene intron (Fig. S9a–f), DLC1 did not bind to any of them, including the introns of ADAR, APC2, FGF1, MAPK10, MYB, PAX3, and ZIC1 (Fig. S9g–i, S8c). This is consistent with the absence of its binding motif in their intronic regions (Fig. 4a, S8a). In agreement with this, DLC1 KO did not affect the binding capacity of the PHF5A-SF3B1 complex to the intronic BS sequences of WNT1, BMP4, or PAX7, and their expression levels were unaffected (Fig. S9j–u, S1e–h). Moreover, the absence of splicing defects in PHF5A KO and SF3B1 KO is unlikely due to a lack of their pre-mRNAs, as they were still expressed in both KO groups (Fig. S2e, g, Fig. 3g, h, Fig. S3i, k and Fig. S6a-d). Analysis of upregulated factors in PHF5A KO and SF3B1 KO from our RNA-seq dataset revealed that CELF3, CELF5, NOVA1, and GPATCH1, all known to play critical roles in pre-mRNA splicing47,48,49, were significantly elevated in both KO groups compared to the Ctrl gRNA (Fig. S10a, b). HCR analysis further confirmed their upregulation in the dorsal NC territory transfected with PHF5A gRNA or SF3B1 gRNA, whereas no expression was detected in the Ctrl gRNA or untransfected side (Fig. S10c). However, epistasis analysis demonstrated that individual KO of these upregulated factors (Fig. S10d) in either PHF5A KO or SF3B1 KO did not alter the mRNA levels of BMP4, WNT1, PAX7 and MYB (Fig. S11a–d, S12a–d). These results suggest that functional redundancy or combined effects among the upregulated splicing factors may compensate for the splicing defects on introns lacking DLC1 binding motifs in PHF5A KO and SF3B1 KO cells.
Altogether, these findings indicate that the ability of DLC1 to associate with its specific intronic motif of SOX9, SNAI2 and other essential factors determines the functional specificity of PHF5A-SF3B1 in NC specification and EMT.
Splicing modulator specifically abolishes NC specifier gene expression
To further investigate the role of the PHF5A-SF3B1 complex in regulating the splicing of SOX9 and SNAI2 pre-mRNAs by binding to BS sequence, we treated HH10 avian trunk neural tubes at the thoracic level with the splicing modulator pladienolide B (PB), which is cytotoxic to cancer cells22,50. PB is known to disrupt the interaction between the PHF5A-SF3B1 complex and BS sequence to form the BP-A binding pocket and stably accommodate the U2/BS duplex, leading to impaired mRNA splicing23,24,50. After a 5-hour treatment with 200 μM PB, we performed in situ hybridization with NC specifier genes on harvested embryos (Fig. 5a). Transverse sections of the stained embryos revealed a complete loss of SOX9, SNAI2, and FOXD3 expression in the premigratory NC region compared to control embryos treated with PBS (Fig. 5b). Intriguingly, expression of SOX9 and SNAI2 in somites remained unchanged (Fig. 5b), suggesting that trunk NCCs are more susceptible to PB treatment. The loss of SOX9 and SNAI2 expression was not due to reduced expression of neural plate border specifier or cell death, as evidenced by the unchanged expression of PAX7 and the absence of Caspase-3 in PB-treated embryos (Fig. 5c, d). Furthermore, PB treatment did not affect the expression levels of DLC1, PHF5A, SF3B1, or PolII (Fig. 5d), nor did it disrupt the interactions between DLC1 and SF3B1 or between PHF5A and SF3B1 (Fig. 5e). The finding supports the proposed mechanism of PB action, which targets the binding capacity of PHF5A-SF3B1 to BS sequence without affecting transcriptional expression and protein-protein interactions. qPCR analysis revealed that PB treatment specifically retained intron 2 of SOX9 and intron 1 of SNAI2 (Fig. 5f–i). This was further confirmed by the detection of the HCR signal for these introns in premigratory and early migratory NCCs. In contrast, there was no HCR signal for intron 1 of SOX9 and intron 3 of SNAI2 (Fig. 5j–n). This difference is likely attributed to the shorter length of SOX9 intron 2 and SNAI2 intron 1, which have a weaker polypyrimidine (Py) tract located at the 3’ end of BP-A compared to SOX9 intron 1 and SNAI2 intron 3 (Table 1, Table S18)51. Consequently, the binding of PHF5A-SF3B1 to BP-A is disrupted upon PB treatment22,24. Indeed, RIP assay revealed that PB treatment reduced the binding capacity of PHF5A-SF3B1 to SOX9 intron 2 and SNAI2 intron 1 but did not alter their ability to bind to SOX9 intron 1 and SNAI2 intron 3 compared to the PBS control (Fig. 5o–r). No exon skipping was detected in SOX9 and SNAI2 after PB treatment (Fig. S6b, d). Overall, these findings demonstrate that PB treatment specifically abolished the expression of NC specifier genes, likely through competitive inhibition of the recognition of BS sequences in SOX9 intron 2 and SNAI2 intron 1 by the PHF5A-SF3B1 complex.
a Experimental workflow. b PB treatment reduced the expression of NC specifiers (open arrowheads), whereas their expression in NCCs (black arrows) and somites (So) remained unaltered in PBS treatment group. c PB treatment did not affect PAX7 expression (indicated by white arrows) and did not lead to apoptosis, as evidenced by the absence of Caspase-3+ cells. d PB treatment did not affect the expression levels of PHF5A, SF3B1, DLC1, and RNA Pol II, nor induce Caspase 3 expression. e SF3B1 interacts with PHF5A and DLC1 in the presence of PB. Increased levels of SOX9 intron 2 (f) and SNAI2 intron 3 (h) in the PB treatment groups. n = 5 embryos per treatment. Schematic diagrams illustrate intron 2-retained SOX9 pre-mRNA (g) and intron 1-retained SNAI2 (i) pre-mRNA after PB treatment. j Detection of SOX9 intron 2 and SNAI2 intron 1 expression in NCCs of embryos treated with PB. k–n Quantification of fluorescence intensities (6 sections from 6 embryos per treatment). PB treatment reduced the binding of PHF5A and SF3B1 on both SOX9 intron 2, n = 3 embryos per treatment (o, q) and SNAI2 intron 1 (p, r). Scale bar: 50 µm. NT, neural tube. The minima, maxima, center, percentile values, and the exact p-values are listed in the Source data file. Mean ± SD. ns: no significant difference, **P < 0.01, ****P < 0.0001. ISH, in situ hybridization, IF immunofluorescence, IR-qPCR intron retention-qPCR, WB Western blot, IP immunoprecipitation, PB Pladienolide B.
DLC1 determines the increased vulnerability of trunk NCCs to PB treatment
The lack of an effect of PB on SOX9 and SNAI2 expression in somites may be attributed to the absence of NC-specific splicing factors associating with the PHF5A-SF3B1 complex that confer increased susceptibility to trunk NCCs by interfering with BS sequence recognition following PB treatment. Based on the aforementioned results, we hypothesize that DLC1 may serve as an NC-specific splicing factor that determines trunk NCC vulnerability to a splicing modulator, as it was absent in somites and might explain the observed resistance of somites while the increased susceptibility of NCCs to PB treatment (Figs. 1a, S1a, S2b) To test this hypothesis, we electroporated the full-length DLC1 cDNA construct into somites of chick embryos at HH10 and incubated for 8 h before treating with PB or PBS control for 5 h (Fig. 6a). The results showed that ectopic DLC1 did not affect the expression of SOX9 and SNAI2 in the somites of embryos treated with PBS. In contrast, PB treatment led to the loss of SOX9 and SNAI2 expression in DLC1-overexpressing somites in a cell-autonomous manner (Fig. 6b–e). Immunoprecipitation using chick embryo lysates revealed that ectopic DLC1 was still able to interact with endogenous PHF5A and SF3B1 in somites following PB treatment (Fig. 6f), which is consistent with its nuclear localization (Fig. S13a). Intron retention analysis in sorted somites electroporated with DLC1 showed increased levels of SOX9 intron 2 and SNAI2 intron 1 as what was observed in NCCs following PB treatment (Fig. 6g–j). In agreement with this, HCR analysis showed the expression of SOX9 intron 2 and SNAI2 intron 1 in DLC1-transfected somites, whereas the expression of SOX9 intron 1 and SNAI2 intron 3 was not detected (Fig. 6k–o). The RIP assay further showed that electroporation of DLC1 into somites of embryos treated with PB led to a marked reduction in the binding capacity of PHF5A and SF3B1 to both SOX9 intron 2 and SNAI2 intron 1 compared to the PBS control (Fig. 7a–d). This suggests that overexpression of DLC1 specifically weakened the interaction of the PHF5A-SF3B1 complex with the BS sequences of SOX9 intron 2 and SNAI2 intron 1, which have a lower Py-tract score with weaker strength compared to those in SOX9 intron 1 and SNAI2 intron 3, respectively (Table 1, Table S14)51. These findings indicate that NC-specific DLC1 plays a crucial role in conferring increased vulnerability of trunk NCCs to splicing perturbation.
a Experimental strategy. b Ectopic DLC1 expression in somites reduced expression of SOX9 (b, c) and SNAI2 (d, e) in a cell-autonomous manner (open arrowheads) upon PB treatment. Open arrowheads indicate loss of SOX9 and SNAI2 expression in pre-migratory NCCs. There was no alteration of SOX9 (b, c) and SNAI2 (d, e) expression in DLC1-overexpressing somites and in NCCs (black and white arrows) in the PBS treatment group. Scale bar, 50 µm. Yellow dotted boxes outline the magnified view in the rightmost column of each panel. n = 5 embryos per treatment. f IP shows the association of ectopic DLC1 with endogenous PHF5A and SF3B1 in somites of embryos treated with PBS Ctrl and PB. g, i Electroporation of DLC1 in somites resulted in increased levels of SOX9 intron 2 and SNAI2 intron 1 in the presence of PB compared to PBS Ctrl. n = 5 embryos per treatment. h, j Schematic diagrams depict the presence of intron 2-retained SOX9 pre-mRNA (h) and intron 1-retained SNAI2 pre-mRNA (j) in DLC1 overexpressing somites of embryos treated with PB. k The HCR analysis revealed the expression of SOX9 intron 2 and SNAI2 intron 1 in DLC1-overexpressing somites (So) of embryos treated with PB, as compared to the PBS Ctrl. l–o Quantification of fluorescence intensities. 20 sections per treatment were used for the quanitification. Scale bar: 50 µm. The minima, maxima, center, percentile values and the exact p-values are listed in the Source data file. Mean ± SD. ns: no significant difference, ****P < 0.0001. NT neural tube, So somites.
Overexpression of DLC1 in somites of embryos treated with PB resulted in reduced binding capacity of PHF5A to SOX9 intron 2 (a) and SNAI2 intron 1 (b) compared to PBS Ctrl. Overexpression of DLC1 in somites of embryos treated with PB led to a reduction in the binding capacity of SF3B1 to SOX9 intron 2 (c) and SNAI2 intron 1 (d) compared to PBS Ctrl. n = 3 embryos per treatment. Interactomes of splicing factors associated with PHF5A (e) and SF3B1 (f). PHF5A, SF3B1, and SLU7 are highlighted in green. g IP validation of the association of endogenous SLU7 with PHF5A and SF3B1. h Co-localization of SLU7, SOX9 and SNAI2 in somites. Open arrowheads indicate the expression of genes in the dorsal neural tube, somites, and notochord. i RT-qPCR demonstrated reduced expression levels of SLU7 in gRNA treatment compared to Ctrl gRNA, n = 5 embryos per treatment. HCR (j) and RT-qPCR (k) analysis revealed the loss of SOX9 and SNAI2 in SLU7-depleted somites, n = 5 embryos per treatment. IR-qPCR analysis showed increased levels of SOX9 introns 1/2 (l) and SNAI2 introns 1/3 (m) in embryos treated with SLU7 gRNA1 compared to Ctrl gRNA, n = 5 embryos per treatment. n HCR showing unaltered levels of SOX9 and SNAI2 expression in the dorsal neural tubes transfected with SLU7 followed by PB or PBS treatment. Insets indicate the right side of neural tube with SLU7 overexpression. Scale bar, 50 µm. The exact p-values are listed in the Source data file. Mean ± SD. ns: no significant difference. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Somite-specific SLU7-SF3B1-PHF5A splicing complex regulates SOX9 and SNAI2 expression and confers resistance to PB treatment
To identify somite-specific splicing factors that confer resistance to PB, we performed MS on lysates immunoprecipitated with PHF5A and SF3B1 from chick somites. Among all the splicing factors identified (Dataset 4), we selected SLU7 which is present in the interactomes of both PHF5A and SF3B1 (Fig. 7e, f) and has been shown to be crucial for the accurate selection of the 3’ splice site during the second step of the splicing process where SF3B1-PHF5A are essential52. Immunoprecipitation using lysates from somites confirmed the endogenous interaction between SLU7 and PHF5A or SF3B1 (Fig. 7g). HCR analysis revealed that SLU7 is specifically expressed in somites, co-localizing with SOX9 and SNAI2 (Fig. 7h). Electroporation of SLU7 gRNA1 into the somites of chick embryos at st 10 resulted in a loss of SOX9 and SNAI2 expression compared to those treated with Ctrl gRNA (Fig. 7i–k). The absence of their expression was attributed to the retention of SOX9 and SNAI2 introns in somites transfected with SLU7 gRNA1 (Fig. 7l, m). Importantly, electroporation of full-length cDNA encoding chick SLU7 gene into the trunk neural tubes followed by PB treatment did not lead to a loss of SOX9 and SNAI2 expression in the premigratory NC territory (Fig. 7n). Altogether, these findings suggest that the SLU7-SF3B1-PHF5A complex regulates the splicing of SOX9 and SNAI2 pre-mRNAs in somites and imparts resistance to PB treatment.
Discussion
The formation of NCCs is regulated by a gene regulatory network in various vertebrate species. This network involves signaling pathways, such as WNT1 and BMP4, which activate the expression of transcription factors like PAX7 to establish the neural plate border. These signaling effectors, along with regulators of the neural plate border, then activate the expression of NC specifiers including SOX9, SNAI2, and FOXD3, which determine the fate of multipotent NCC progenitors. However, the factors involved in regulating the splicing of intron-containing SOX9 and SNAI2 genes have not been identified. In this study, we investigated the role of SF3B1-PHF5A spliceosome component in conjunction with NC-specific factor DLC1 in avian trunk NC specification. We demonstrated that these factors are functionally important for NCC formation by regulating the splicing of SOX9 and SNAI2 pre-mRNA, but not their upstream regulators BMP4, WNT1, and PAX7. Mechanistically, the ability of DLC1 to associate with its specific intronic motif of SOX9 and SNAI2 and other essential regulators for specifying NCC fate and EMT determine the functional specificity of SF3B1-PHF5A in these processes. Treatment with the splicing modulator PB in ovo resulted in the loss of SOX9 and SNAI2 expression specifically in trunk NCCs, but not in somites. This differential response is attributed to DLC1, which enhances the vulnerability of trunk NCCs to PB treatment by diminishing the binding capacity of SF3B1-PHF5A to both SOX9 intron 2 and SNAI2 intron 1. These introns characterized by their shorter in length and weaker Py-tract compared to SOX9 intron 1 and SNAI2 intron 3 are more prone to intron retention upon PB treatment. In contrast, the SLU7-SF3B1-PHF5A complex governs the splicing of SOX9 and SNAI2 pre-mRNAs in somites and imparts resistance to PB-induced splicing alterations. Collectively, our results reveal the presence of context dependent-splicing complexes in both NCCs and somites with distinct vulnerabilities to splicing perturbations (Fig. 8a).
a The schematic diagram illustrates the binding of the NC-specific DLC1 to intronic motifs of SOX9 and SNAI2 pre-mRNA, along with the splicing factors SF3B1-PHF5A, which recognize the BS sequence to form a BP-A pocket. The DLC1-SF3B1-PHF5A complex is required for regulating the splicing of SOX9 and SNAI2 pre-mRNA in specifying prospective NCCs. However, this complex also increases the vulnerability of NCCs to the splicing modulator PB, which competitively inhibits the recognition of BS sequence by SF3B1-PHF5A. This inhibition occurs in both SOX9 intron 2 and SNAI2 intron 1, due to their shorter in length and weaker Py tract, resulting in intron retention and loss of NC progenitors. In contrast, the SLU7-SF3B1-PHF5A complex regulates splicing of SOX9 and SNAI2 pre-mRNAs in somites, imparting resistance to splicing perturbation by PB. Py polypyrimidine.
Splicing is considered to be an essential ubiquitous process that involves excision of introns to form mature RNA, which also increases the diversity of proteins generated from the genome53. The spliceosome component SF3B1 is the largest subunit within the SF3B complex of the U2 snRNP. It is mainly involved in the recognition of intronic BS sequence to promote the stable interaction in the initial phase of the splicing process11. Another splicing factor PHF5A closely associated with SF3B1 is important for both spliceosome structural stability, and for linking the spliceosome to histones22,54. Structural analysis has shown that the interface between SF3B1 and PHF5A forms a conserved pocket for BP-A55. We have confirmed the endogenous interactions between SF3B1 and PHF5A, and showed their binding to the BS sequences of SOX9 and SNAI2 introns in chick embryos. Consistently, CRISPR/Cas9-mediated knockout of PHF5A or SF3B1 led to the loss of SOX9 and SNAI2 mRNA expression due to splicing perturbations, resulting in intron retention. Our results are in line with previous studies that showed sf3b1 is required for trunk NCC specification in zebrafish embryos16, suggesting the function of Sf3B1 is conserved between chick and zebrafish embryos. While our global splicing analysis revealed that most of the affected genes exhibited intron retention in all KO treatment groups, sf3b1b406 zebrafish mutants showed increased exon skipping events16. This is consistent with previous studies, which showed that chick exhibits a higher frequency of intron retention than zebrafish56. It is possible that different exon-intron architectures between zebrafish and chick could lead to distinct modes of splicing events.
In addition, the retained introns for SOX9 and SNAI2 likely involve the lack of binding capacity of SF3B1 and PHF5A to BS sequence in PHF5A KO and SF3B1 KO, respectively, which disrupts the assembly and stability of U2-dependent spliceosome for subsequent splicing events, as demonstrated in other studies22,54. Consistent with this notion, overexpression of full-length PHF5A cDNA was able to restore the expression of NC specifiers in PHF5A KO embryos, possibly via the restoration of SF3B1-PHF5A splicing complex formation and binding to BS sequences. Our results support the importance of this complex for gene splicing in vivo.
Although PHF5A has been shown to regulate PolII elongation in genes that are essential for maintaining pluripotency of ESCs28, we found that PHF5A did not associate with PolII-pSer5 and PolII-pSer2, as well as regulate their expression, ruling out a transcriptional role of PHF5A in regulating NC formation. However, previous studies showed that PHF5A functions as a transcription factor regulating genes that contribute to the stemness of cancer cells27,57, which highlights the functional importance of PHF5A in regulating stem cell features in both developmental and pathological contexts. Whether PHF5A functions as a transcription factor or splicing factor in regulating the expression of genes that are essential for the formation of other tissue progenitors and maintaining the other types of cancer stemness remains to be determined.
Our previous studies showed that the asymmetric localization of cytoplasmic DLC1 functions as a RhoGAP to establish differential RHOA activity at the cell rear and front edge that controls trunk NCC polarity for directional delamination and migration29. In contrast, the current study revealed an unprecedented role of nuclear DLC1 in trunk NCC fate determination through the association with SF3B1-PHF5A complex to regulate splicing of SOX9 and SNAI2 pre-mRNA. These studies reveal the spatio-temporal expression and requirement of DLC1 in regulating NC specification and migration, as DLC1 KO at HH9-10 reduced NC formation whereas knockdown in committed NCCs at HH11-12 disrupted their migration directionality29. The underlying mechanisms to controlling DLC1 translocation from the nucleus to the cytoplasm as prospective NCCs undergo delamination remains to be determined. Previously, we showed nuclear DLC1 can associate with FOXK1 transcription factor to cooperatively activate MMP9 expression in promoting melanoma metastasis58. It is conceivable that nuclear DLC1 is an important regulator that controls various developmental and pathological processes at the transcriptional and/or post-transcriptional levels depending on the cellular context. Our study provides evidence of the involvement of DLC1 in the splicing process: 1) nuclear DLC1 can interact with the SF3B1-PHF5A splicing complex, 2) DLC1 binds to a specific intronic motif found in the pre-mRNAs of SOX9, SNAI2 and other essential regulators associated with NC specification and EMT, 3) DLC1 is required for the binding capacity of SF3B1-PHF5A splicing complex to the BS sequences of SOX9 and SNAI2 introns, and 4) DLC1 KO leads to retention of SOX9 and SNAI2 intron transcripts without affecting the expression and function of their transcriptional machinery. In addition, we found this motif is situated either in close proximity to or at a distant from the BS sequence in different introns. The potential impact of the varying positioning of this motif on DLC1 binding and its interaction with the SF3B1-PHF5A splicing complex, which could influence splicing efficiency, requires further investigation.
However, the absence of this motif in the introns of neural plate border specifiers, NC inductive signals and NC lineage determinants, could account for the lack of impact on BS sequence recognition by the SF3B1-PHF5A complex and the subsequent splicing of WNT1, BMP4, and PAX7 introns in the absence of DLC1. These findings suggest that the presence of the DLC1 binding motif together with the BS sequence determines the functional specificity of the DLC1-SF3B1-PHF5A complex in regulating the splicing of genes involved in NC specification and EMT. Based on this notion, it is conceivable that another gene-specific RNA binding proteins associated with SF3B1-PHF5A and other splicing factors are required for regulating the splicing of neural plate border specifiers, NC inductive signals and NC lineage determinants by binding to specific motifs in their pre-mRNAs. While the RNA binding proteins and their motifs are yet to be identified, the threshold requirement for their expression levels may be less stringent compared to DLC1-SF3B1-PHF5A, resulting in distinct responses to the reduced expression of each individual splicing factor. These differences may explain why the KO of PHF5A or SF3B1 only resulted in splicing defects in NC specifier genes rather than their upstream regulators. Alternatively, we revealed an upregulated expression of other splicing factors in the PHF5A KO or SF3B1 KO cells that could play compensatory roles in restoring the splicing of introns without DLC1 binding motifs. Recent studies indicate that there may be feedback mechanisms in cells with PHF5A loss-of-function variants to maintain normal level of SF3B components59. However, these compensatory splicing factors probably do not form stable complexes with DLC1 and SF3B1 or PHF5A on the intronic regions of NC specifiers and genes involved in EMT. Structural analysis to compare their stability on SOX9 and SNAI2 introns warrant further investigations.
The reason why NCCs are more vulnerable to splicing perturbations remains unclear. Previous single-cell transcriptomic studies have indicated that multipotent NC formation is subject to multiple waves of transcriptomic changes, which are likely accompanied by increased levels of alternative splicing and translation to efficiently generate key transcription factors and cellular proteins that are crucial for maintaining its multipotency and initiating EMT, in a short timeframe60,61. As a result, the increased demand for splicing may lead to competition among different unspliced transcripts for access to the limited spliceosomal machinery available. It is tempting to speculate that a subset of genes involved in this process has weaker association with splicing components, allowing for their rapid utilization in the efficient splicing of multiple pre-mRNAs during NC specification. In agreement with this, PB treatment in chick embryos resulted in the loss of SOX9 and SNAI2 expression in NC progenitors, but not in somites. This is due to NC-specific DLC1 that appears to weaken the binding affinity of the SF3B1-PHF5A complex to the BS sequence of SOX9 intron 2 and SNAI2 intron 1. These introns comprise of shorter and weaker Py-tracts compared to those in SOX9 intron 1 and SNAI2 intron 3. Previous findings showed that shorter introns with weaker Py-tracts were more prone to being retained upon inhibition of SF3B1 and PHF5A activity24. This was further supported by the overexpression of DLC1 in somites, which resulted in the loss of SOX9 and SNAI2 expression in the presence of PB due to the retention of SOX9 intron 2 and SNAI2 intron 1. Importantly, we have identified a somite-specific splicing complex SLU7-SF3B1-PHF5A, which regulates the expression of SOX9 and SNAI2 and confers resistance to splicing perturbation by PB. These findings suggest that the presence of cell-type specific splicing factors (DLC1 vs SLU7) in different cellular contexts (NCCs vs somites) determine the susceptibility to splicing perturbation. We speculate that the structure of SLU7-SF3B1-PHF5A splicing complex in association with the intronic regions of SOX9 and SNAI2 pre-mRNAs is more stable than DLC1-SF3B1-PHF5A, resulting in enhanced resistant to PB treatment. Structural studies are required to provide insights into how the conformation and stability of these distinct spliceosomal structures might be altered by PB.
Functional perturbations of several core splicing factors lead to defects in cranial NC formation62. In agreement with this, mutations in the core components of the spliceosome lead to craniofacial disorders8. Most of these defects occur at the levels of NC formation, migration, and differentiation to skeletal lineages. For example, sf3b4 knockdown in Xenopus embryos resulted in the disruption of cranial NC formation, which reduced their migratory capacity and differentiation potency toward skeletal elements in the cranial region63. In contrast, sf3b1b406 zebrafish mutants showed defects in trunk NC formation without affecting cranial NC development16. Consistently, our results in chick revealed that DLC1, SF3B1 and PHF5A are not required for cranial NC development. These findings indicate that trunk NCCs are more susceptible to reduced levels of SF3B1 compared to cranial NCCs in both chick and zebrafish embryos. Whether distinct complexes orchestrate the splicing of genes essential for cranial and trunk NC development in different vertebrate species remains to be elucidated. Nevertheless, our studies suggest that the splicing complexes in cranial NCCs may contain cell-type- or gene-specific factors that contribute to their increased susceptibility to reduced levels, function, or intron binding capacity of splicing factors. Identification of these factors should provide mechanistic insights into how mutations in ubiquitously expressed spliceosome components can lead to craniofacial abnormalities.
Methods
Chick embryos and in ovo electroporation
Fertilized chick Leghorn eggs were purchased by Jinan Poultry Co. China through Tin Hang Technology Ltd and incubated at 38.5 °C in a humidified incubator. Embryos were staged according to Hamburger and Hamiliton (HH) stages30. All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research, The University of Hong Kong (CULATR No: 5969-21). In ovo electroporation was performed as described previously64. Plasmid DNA (3 µg/µL) or gRNAs/Cas9 (Cas9 2 µg/µL + gRNA 1.5 µg/µL) constructs were injected into the lumen of closed neural tubes or the cavity of somites of embryos at HH9-10 using a glass capillary. Electrodes were placed on either side of the neural tube or somites, and electroporation was carried out using a BTX electroporator delivered as five 50 ms pulses, 450 ms interval at 36 V. Electroporated embryos were allowed to develop for 9 h post-transfection (hpt) before being processed for fluorescence-activated cell sorting (FACS) for genotyping analysis, Western blotting, immunofluorescence, whole mount in situ hybridization (ISH), immunoprecipitation, RNA-immunoprecipitation (RIP)-qPCR, intron retention analysis and RNA-sequencing (RNA-seq). Wild-type HH10 or electroporated embryos 8 hpt were treated with PB (200 μM) or PBS before being processed for ISH, hybridization chain reaction (HCR), immunofluorescence (IF), intron retention (IR), and RIP-qPCR analysis.
Plasmids
Full-length chick PHF5A cDNA (NM_001277376.2), PHD domain, DLC1 cDNA29, DLC1 truncated domains and SLU7 cDNA (NM_001006146.2) were inserted at the Xho1 and Cla1 sites upstream of an internal ribosomal entry site (IRES) followed by a tdTomato cDNA in a pCIG expression vector (kind gift of A. McMahon, University of Southern California, USA). Three plasmids for CRISPR/Cas9 KO in chick embryos (U6.3 > gRNA.f + e, #99139, U6.3 > Control.gRNA (GCACTGCTACGATCTACACC). f + e, #99140, pCAGG nls-hCas9-nls-GFP, #99141) were purchased from Addgene. Online program E-CRISP was used for gRNA design. gRNA oligos were synthesized by Integrated DNA technologies (IDT) and cloned into U6.3 gRNA.f + e vector (Table S1).
FACS and genotyping
Electroporated embryos were harvested 8–9 h post-transfection (hpt) and dissociated using 0.05% trypsin. Dissociated cells were sorted using a BD FACS AriaTM SORP based on GFP signal intensity. ~50,000 GFP+ cells were collected for RNA extraction using the TaKaRa MiniBEST Universal RNA Extraction Kit (#9767). cDNA was synthesized from the extracted RNA using the TaKaRa PrimerScriptTM RT Master Mix (#RR036A) and cloned into the plasmid vector (99139_U6.3 gRNA f + e) for genotyping analysis to verify the KO efficiency. Genotyping sequencing was performed using a specific primer (cctataaaaataggcgtatcacg) at the Centre for PanorOmic Sciences, the University of Hong Kong.
RNA-seq
RNA-seq was performed on RNA extracted from sorted GFP+ cells obtained from 30 well-transfected chick embryos. Approximately 50,000 GFP+ cells per treatment were collected for RNA extraction using RNAqueous-Micro Kit (Ambion) and genomic DNA was removed by TURBO DNase treatment for 30 min. RNA quality was assayed using an Agilent 2100 Bioanalyzer. Only samples with an RNA integrity number (RIN) ≥ 8.0 and a 28S/18S ribosomal ratio ≥1.0 were selected for further processing. RNA samples were sent to the Beijing Genomics Institute (BGI) for RNA sequencing. High-quality RNA (~50 ng) was used for RNA amplification and cDNA synthesis. Adapters were ligated to the cDNA followed by amplification to construct single-stranded circular DNA libraries. These circular DNA templates were used to generate DNA nanoballs (DNBs) by rolling circle replication, which enhances fluorescence signals during sequencing. The DNBs were loaded into patterned nanoarrays and sequenced on a BGISEQ-500 platform using 100-bp paired-end sequencing. Biological replicates were included for each KO group to ensure reproducible results.
Global splicing analysis
Aberrant splicing events were analyzed using VAST-TOOLS on the RNA-seq data in FASTA format. VAST-TOOLS maps RNA-seq data to VastDB, quantifying all possible splicing events globally across the genome. Four types of splicing events were assessed: intron retention (IR), exon skipping (ES), 5’ alternative splicing (5’AS), and 3’ alternative splicing (3’AS).
In VAST-TOOLS, IR events were quantified by percent intron retention (PIR), where a higher PIR indicates a greater extent of intron retention. ES events were quantified by percent spliced-in (PSI), representing the proportion of mRNA segments where a specific exon is included, relative to the total number of mRNA segments that can be formed from the intron-exon-intron region. For 5’AS and 3’AS events, the extent of splice site usage was measured by percent splice site usage (PSU), reflecting the frequency of splice site utilization within a given intron.
RNA-seq data from three experimental groups and the control group were analyzed by VAST-TOOLS to quantify all potential aberrant splicing events across the genome. In the case of IR events, those with increased PIR in at least two experimental groups compared to the control group were included. Moreover, the presence of other aberrant splicing events (ES, 5’AS, and 3’AS) from genes showing increased IR was further assessed based on the VAST-TOOLS output.
Motif analysis
To identify the DLC1 binding motif within introns, we selected introns exhibiting increased intron retention within the DLC1 gRNA group out of the 10 genes expressed in the NC. The sequences of these introns were analyzed using the HOMER software to identify any prevalent sequence motifs that could be potentially recognized and bound by DLC1.
In situ hybridization
ISH was performed on HH10 chick embryos with anti-sense DIG-riboprobes for SOX9, SNAI2, FOXD3, WNT1, and BMP4 generated from pBluescript-II KS vector and purified using G-50 column (GE Healthcare). Transfected embryos were fixed in 4% PFA overnight at 4 °C, washed (PBS, 0.1% Tween-20) and dehydrated through 25%, 50%, 75% methanol/PBS, 0.1% Tween-20 then absolute methanol. Embryos can be stored at −20 °C at this point or processed for the rest of the hybridization procedure which took four days. Day 1, embryos were rehydrated through 75%, 50%, 25%, 0 methanol/PBS, 0.1% Tween-20, then bleached in 6% H2O2/PBS, 0.1% Tween-20, incubated in 10 μg/mL proteinase K solution (Invitrogen™, 25530049), post-fixed, pre-hybridized for 1 h and hybridized in solution containing anti-sense probe overnight at 70 °C. Day 2, embryos were extensively washed to remove unbound probe, blocked in 10% sheep serum (Sigma, S3772) for 60–90 min at room temperature then incubated in anti-DIG-AP (Roche, 11093274910) diluted in 1% sheep serum overnight at 4 °C. Day 3, embryos were extensively washed in 1 × TBST 5 times one hour each and incubated in TBST overnight at room temperature. Day 4, embryos were subjected to color development using NBT/BCIP detection kit (1:50, Roche). The duration of color development varied depending on the expression level for each gene. Embryos were fixed (4% PFA overnight at 4 °C) and embedded (Tissue freezing medium, Leica) for cryosectioning.
Whole mount immunofluorescence
Whole mount IF was conducted in transfected embryos which were fixed in 4% PFA overnight at 4 °C and then blocked in blocking buffer (PBS + 1% Triton, 10% FBS, 0.02% NaN3) for an hour. Embryos were then incubated with primary antibodies (1:1000 anti-rabbit GFP, Invitrogen and 1:400 anti-mouse HNK-1, Developmental studies hybridoma bank (DSHB) blocking buffer for 2 days 4 °C with gentle rocking, washed 3 times 30 min each in washing buffer (PBS + 1% Triton) followed by 2 days incubation with secondary antibodies (Alexa Fluor™ 488, Jackson Immuno Research, and Cy™3 AffiniPure donkey anti-mouse IgM) in blocking buffer at 4 °C overnight. The embryos were washed 3 times 30 min each in washing buffer before imaging.
Cryo-sectioning
Immunostained embryos were embedded in Tissue freezing medium (Leica) and cryo-sectioned at 12 μm using a CryoStar NX50 (Thermo Fisher Scientific) for subsequent immunofluorescence imaging. Transfected embryos were freshly dissected, fixed 4% PFA at 4 °C for 2 h. Following fixation, the embryos were dehydrated in 30% sucrose at 4 °C overnight, followed by embedding in Tissue Freezing Medium with silicone molds and snap-frozen at −80 °C for cryo-sectioning.
Immunofluorescence
Transverse sections were washed in (PBS + 0.1% Triton) to remove the OCT (Tissue freezing medium, Leica) and then blocked in blocking buffer (1% BSA in 1 × PBS 0.1% Triton) for 20 min before incubating with primary antibodies (anti-sheep GFP, Bio Rad, anti-rabbit RFP, Rockland, anti-mouse HA tag, Invitrogen, anti-mouse V5 tag, Invitrogen, anti-rabbit SOX9, Merck Millipore, anti-goat SOX2, R&D, anti-rabbit Caspase-3, Cell signaling technology, anti-mouse PAX7, Developmental studies hybridoma bank) at 4 °C overnight, washed (1 × PBS 0.1% Triton) three times and incubated with secondary antibodies (Alexa Fluor™ 488, 594, 647) at room temperature 1–2 h. Stained with DAPI (1:1000 in 1 × PBS) at room temperature for 5 min before mounting (Fluorescence mounting medium, Dako) the slides with cover slips. All incubations were conducted in a humidified chamber.
Imaging
The images of whole mount IF were taken by Leica MZ10F microscopy. Imaging of in situ hybridization, IF on sections and HCR were performed using Zeiss Axiophot microscopy and Zeiss LSM780 confocal microscopy, respectively.
Hybridization chain reaction (HCR)
Electroporated and/or PB-treated embryos were collected at designated time points and placed in a petri dish containing Ringer’s solution. They were then rinsed once before fixation in 4% PFA, at 4 °C for 14 h. Fixed embryos were treated with pre-warmed (37 °C) 10 µg/mL proteinase K solution for 10 min, followed by post-fixation with 4% PFA at room temperature for 20 min. The embryos were then washed in PBST, 50% PBST / 50% 5 × SSCT, 5 × SSCT subsequently. First, the detection procedure involved the following steps: (1) Transfer 1–4 embryos to a 2 mL tube for each sample. (2) Pre-hybridize the embryos in 500 µL of probe hybridization buffer for 30 min at 37 °C. (3) Prepare the probe solution by adding 2 pmol of each probe set (e.g. 2 µL of 1 µM stock) to 500 µL of probe hybridization buffer at 37 °C. (4) Remove the pre-hybridization solution and add the probe solution. (5) Incubate the embryos overnight (>12 h) at 37 °C. (6) Remove excess probes by washing embryos 4 × 15 min with 1 mL of probe wash buffer at 37 °C. (7) Wash the samples 2 × 5 min with 5 × SSCT at room temperature. Second, the amplification procedure involved the following steps: (1) Pre-amplify the embryos with 500 μL of amplification buffer for 5 min at room temperature. (2) Prepare 30 pmol of hairpin h1 and 30 pmol of hairpin h2 separately by snap cooling 10 μL of 3 μM stock (heat at 95 °C for 90 s and cool to room temperature in a dark drawer for 30 min). (3) Prepare the hairpin solution by adding the snap-cooled h1 hairpins and snap-cooled h2 hairpins to 500 μL of amplification buffer at room temperature. (4) Remove the pre-amplification solution and add the hairpin solution. (5) Incubate the embryos overnight (>12 h) in the dark at room temperature. (6) Wash off excess hairpins with 1 mL of 5 × SSCT at room temperature: (a) 2 × 5 min, (b) 2 × 30 min, (c) 1 × 5 min. (7) Samples can be stored at 4 °C protected from light before embedding, cryosectioning and imaging. List of HCR probe sequences for DLC1, PHF5A, SF3B1, SLU7, SOX9, SNAI2, SNAI2 intron 1, SNAI2 intron 3, CELF3, CELF5, NOVA1, GPATHC1 are shown in Table S6–17 respectively.
Sample preparation for mass spectrometry
Endogenous DLC1 and its associated proteins were immunoprecipitated from lysates of chick embryos (30 embryos per sample) using a DLC1-specific antibody, with IgG serving as a control. The immunoprecipitated proteins were separated by SDS-PAGE and visualized using QC Colloidal Coomassie Stain (BioRad, #1610803). Distinct and unique protein bands were observed in the DLC1 antibody samples but not in the IgG control, confirming the specificity of the pull-down. The samples were electrophoresed through the stacking gel until reaching the boundary between the stacking and separating gels. The Coomassie blue-stained bands were excised from the gel and subjected to reduction and alkylation by 10 mM TCEP and 55 mM 2-chloroacetamide, respectively. Protein digestion was performed by incubating samples with trypsin (1 ng/μl) overnight at 37 °C. Subsequently, tryptic peptides were extracted from the gel sequentially with 50% acetonitrile (ACN) containing 5% formic acid (FA) and then with 100% ACN. The peptide extracts were pooled and dried using a SpeedVac. The peptides were desalted using C18 StageTips prior to LC-MS/MS analysis at the Centre for PanorOmic Sciences, Li Ka Shing Faculty of Medicine, the University of Hong Kong. The same approach was applied to identify PHF5A-associated proteins in neural tubes as well as PHF5A- and SF3B1-interacting factors in somites.
Mass spectrometry data acquisition
Eluted peptides were analyzed using a Dionex Ultimate3000 nanoRSLC system coupled to Thermo Fisher Orbitrap Fusion Tribid Lumos mass spectrometer. Separation was performed on a commercial C18 column (75 μm i.d. × 50 cm length × 2μm particle size) connected to a NanoTrap column (75 μm i.d. × 2 cm length × 3 μm particle size) (Thermo Fisher). Peptides were separated using a linear gradient of increasing buffer B (80% ACN, 0.1% FA) and decreasing buffer A (0.1% FA) at a flow rate of 300 nL/min. The gradient program was as follows: buffer B was increased to 30% over 90 min, then ramped to 45% over the next 15 min, followed by a rapid increased to 95% and held for 5 min. Subsequently, the gradient was returned to 5% B for column re-equilibration. Mass spectrometer was operated in positive polarity mode with a capillary temperature of 300 °C. Full MS survey scan was acquired at a resolution of 120,000, with an automatic gain control (AGC) target of 5 × 104, a maximum ion injection time (IT) of 30 ms, and a scan range of 350–1500 m/z. A data-dependent top 10 method was employed, utilizing higher-energy collisional dissociation (HCD) for fragmentation. MS2 spectra were acquired at a resolution of 30,000, with a normalized AGC target of 200%, a maximum ion injection time (IT) of 100 ms, an isolation width of 1.6 m/z, and a normalized collisional energy of 30 s. Precursor ions selected for HCD were dynamically excluded for 22 s.
Mass spectrometry data analysis
The mass spectrometry raw files were processed and searched against the Uniprot Gallus gallus reference proteome (updated as of 2023.08.19, containing 43,710 entries) and the Uniprot Homo sapiens reference proteome (updated as of 2023.04.15, containing 38,977 entries) using MaxQuant version 1.6.14.0. The digestion was set to specific, with trypsin/P selected. Variable modifications included oxidation of methionine and N-terminal acetylation of peptides, while Carbamidomethyl of cysteine was set as a fixed modification. The peptide length was limited to 7–25 amino acids, with up to 2 missed cleavages allowed. The precursor mass tolerance was set to 20 ppm for the initial MS/MS search and 4.5 ppm after mass recalibration for the main search. Fragment ion mass tolerance was set to 20 ppm. Protein identifications were made using a target-decoy approach with a reversed database, and both peptide and protein false discovery rates were controlled at 1% (0.01). Following the MaxQuant analysis, the generated proteinGroups.txt file was imported into Perseus version 1.6.13.0 to extract the iBAQ values for each sample for quantitative comparison. Differentially interacting proteins were identified based on statistical significance determined by t-a test (q < 0.05) and a predetermined log2 fold change threshold.
The STRING database was used to predict protein interactions with our bait targets. The resulting PPI network was imported into Cytoscape software version 3.10.0 for visualization and analysis of functional modules. GO databases were used to analyze the functions of proteins interacting with DLC1, PHF5A, and SF3B1. Enrichment of GO terms was assessed using the Hypergeometric test, with p < 0.05 considered statistically significant.
Immunoprecipitation and RNA-immunoprecipitation (RIP)-qPCR
Wild-type chick embryos or electroporated chick embryos were dissociated in IP lysis buffer supplemented with protease and phosphatase inhibitors (Invitrogen™ Halt™ Protease and Phosphatase inhibitor cocktail, 100 ×, 78442) or ribonuclease inhibitors (Invitrogen™ RNaseOUT™, 10777019). 10% of the lysate from each treatment was reserved as the input control and stored at −80 °C for further analysis. The remaining lysates from each treatment were incubated with antibodies in tubes on a rotating wheel overnight at 4 °C to form the immune complexex. 25 µL beads (Thermo Scientific 90409) were added to each treatment and incubated for 4 h with rocking at 4 °C. The samples were then washed, and the bead-immune complexes were collected using a magnetic stand. Subsequently, the immunoprecipitated lysates from each sample together with 10% of the inputs were mixed with 1 × loading dye and boiled for 10 min at 95 °C to dissociate the immunocomplexes from the beads. The resulting samples were analyzed by Western blot or RIP-qPCR using primers listed in Table S2.
Chromatin immunoprecipitation
1 × 106 sorted GFP cells were crosslinked with 1% formaldehyde (Thermo Scientific, 28908) at room temperature for 10 min in a chemical fume hood. The crosslinking reaction was quenched by incubating the cells with glycine solution for 5 min under the same conditions. Formaldehyde-containing waste was properly disposed of according to safety guidelines. The cells were washed twice with ice-cold PBS to remove residual formaldehyde and glycine. The membrane and cytosol were lysed, and the chromatin was digested with micrococcal nuclease (MNase, Thermo Scientific, 26157). The chromatin was further fragmented by sonication using a Diagenode Bioruptor Pico ultrasonication system. The fragmented chromatin was incubated with either anti-RNA Pol II pSer2 or anti-RNA Pol II pSer5 (Cell signaling technology) at 4 °C for 4 h. Subsequently, 25 µL of ChIP grade protein A/G magnetic beads were added per reaction, and the mixture was incubated overnight at 4 °C. The bead-antibody-chromatin complex was collected, washed to remove non-specific interactions, and subjected to elution. The immunoprecipitated complex was treated with proteinase K at 65 °C for 40 min to recover DNA which was further purified and analyzed by ChIP-qPCR using primers listed in Table S5.
Western blot
Electroporated chick embryos were dissociated in RIPA buffer (0.1% SDS, 1% NP-40, 1% sodium deoxycholate, 150 mM NaCl, 25 mM Tris-HCl pH 7.6), lysed on ice for 40 min with vortexing every 10 min, then centrifuged for 15 min at 16000 g at 4 °C to collect the supernatants. The concentration of lysates was determined using the BCA assay (Thermo Scientific, 23228) with the establishment of a standard curve. Samples could be stored at −80 °C for further analysis or processed immediately. To assess the expression levels of endogenous protein or protein-protein interaction, protein lysates were separated on SDS-PAGE with varying percentages based on the molecular weight of protein of interest. The transfer duration was adjusted and varied based on the protein mass. Protein bands were visualized on films (Fujifilm, 47410 19274) using chemiluminescent substrates (Advansta, K-12045-D50; Carestream dental GBX, 661 0091) following incubation with primary antibodies (anti-rabbit PHF5A, Proteintech, anti-mouse SF3B1 MBL, anti-rabbit DLC1 BD Biosciences, anti-SLU7 Abcam, anti-rabbit cleaved caspase-3 Cell Signaling Technology, anti-mouse RNA Pol II Cell Signaling Technology, anti-rabbit RNA Pol II Ser5 Cell Signaling Technology, anti-rabbit RNA Pol II Ser2 Cell Signaling Technology) and secondary antibodies (anti-mouse IgG HRP, anti-rabbit IgG HRP, Thermo Fisher Scientific). Full scan blots are provided in the Source Data Files.
Reverse transcription polymerase chain reaction, qPCR, intron retention and statistical analysis
Semi-quantitative RT-PCRs were performed in embryos treated with different gRNA treatments using Vazyme, 2 × Rapid Taq Master Mix (P222-02) to analyze the levels of unspliced and spliced transcripts, as well as exon skipping. Quantification of DNA band intensity was conducted using Adobe Photoshop 2021. qPCR was performed with cDNAs obtained from sorted cells using TaKaRa, TB Green™ Premix Ex Taq ™ II (RR820A) to analyze the transcript levels of various genes or with cDNAs generated from RNAs for the intron retention analysis (Table S2). The list of primers used for semi-quantitative RT-PCR, qPCR, and ChIP-qPCR is shown in Tables S3, S4, and S5, respectively.
Data from at least three separate experiments were presented as mean ± SD. The quantitative data from qPCR were analyzed using t-test (GraphPad Prism 10). Benjamini-Hochberg correction was applied for multiple comparisons. Significant differences were shown as *: p < 0.05, **: p < 0.01, ***: p < 0.001, ****: p < 0.0001.
Data availability
All data supporting the conclusions of this study are presented in the manuscript, supplementary materials, and the source data files. The RNA-seq data generated in this study were deposited into the Gene Expression Omnibus database under the accession number GSE299465 and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE299465. Mass spectrometry data were deposited to ProteomeXchange database under the ID PXD064804 and are available at the following URL: https://www.iprox.cn/page/project.html?id=IPX0012214000. The differentially interacting factors of DLC1 and PHF5A in NCCs, as well as those of PHF5A and SF3B1 in somites, are available in Dataset 1, Dataset 2 and Dataset 4, respectively. In addition, the global splicing and motif analysis of the RNA sequencing data are available in Dataset 3. Source data are provided with this paper.
References
Basch, M. L., Bronner-Fraser, M. & Garcia-Castro, M. I. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218–222 (2006).
Liu, J. A. & Cheung, M. Neural crest stem cells and their potential therapeutic applications. Dev. Biol. 419, 199–216 (2016).
Stuhlmiller, T. J. & Garcia-Castro, M. I. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol. Life Sci. 69, 3715–3737 (2012).
Monsoro-Burq, A. H., Wang, E. & Harland, R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Dev. Cell 8, 167–178 (2005).
Simoes-Costa, M., Stone, M. & Bronner, M. E. Axud1 integrates Wnt signaling and transcriptional inputs to drive neural crest formation. Dev. Cell 34, 544–554 (2015).
Cheung, M. & Briscoe, J. Neural crest development is regulated by the transcription factor Sox9. Development 130, 5681–5693 (2003).
Cheung, M. et al. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell 8, 179–192 (2005).
Beauchamp, M. C., Alam, S. S., Kumar, S. & Jerome-Majewska, L. A. Spliceosomopathies and neurocristopathies: Two sides of the same coin?. Dev. Dyn. 249, 924–945 (2020).
Kambach, C., Walke, S. & Nagai, K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9, 222–230 (1999).
Wahl, M. C., Will, C. L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Lardelli, R. M., Thompson, J. X., Yates, J. R. 3rd & Stevens, S. W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16, 516–528 (2010).
Isono, K. et al. Molecular cloning, genetic mapping, and expression of the mouse Sf3b1 (SAP155) gene for the U2 snRNP component of spliceosome. Mamm. Genome 12, 192–198 (2001).
Isono, K., Mizutani-Koseki, Y., Komori, T., Schmidt-Zachmann, M. S. & Koseki, H. Mammalian polycomb-mediated repression of Hox genes requires the essential spliceosomal protein Sf3b1. Genes Dev. 19, 536–541 (2005).
Lim, D. A. et al. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol. Cell Neurosci. 31, 131–148 (2006).
Wang, C. et al. Depletion of Sf3b1 impairs proliferative capacity of hematopoietic stem cells but is not sufficient to induce myelodysplasia. Blood 123, 3336–3343 (2014).
An, M. & Henion, P. D. The zebrafish sf3b1b460 mutant reveals differential requirements for the sf3b1 pre-mRNA processing gene during neural crest development. Int J. Dev. Biol. 56, 223–237 (2012).
Trappe, R. et al. Identification and characterization of a novel murine multigene family containing a PHD-finger-like motif. Biochem Biophys. Res. Commun. 293, 816–826 (2002).
Yan, C., Wan, R., Bai, R., Huang, G. & Shi, Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 353, 904–911 (2016).
Rzymski, T., Grzmil, P., Meinhardt, A., Wolf, S. & Burfeind, P. PHF5A represents a bridge protein between splicing proteins and ATP-dependent helicases and is differentially expressed during mouse spermatogenesis. Cytogenet. Genome Res. 121, 232–244 (2008).
Cordin, O. & Beggs, J. D. RNA helicases in splicing. RNA Biol. 10, 83–95 (2013).
Finci, L. I. et al. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 32, 309–320 (2018).
Teng, T. et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex. Nat. Commun. 8, 15522 (2017).
Cretu, C. et al. Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors. Nat. Commun. 12, 4491 (2021).
Butt, H. et al. Overlapping roles of spliceosomal components SF3B1 and PHF5A in rice splicing regulation. Commun. Biol. 4, 529 (2021).
Vigevani, L., Gohr, A., Webb, T., Irimia, M. & Valcarcel, J. Molecular basis of differential 3’ splice site sensitivity to anti-tumor drugs targeting U2 snRNP. Nat. Commun. 8, 2100 (2017).
Oltra, E., Pfeifer, I. & Werner, R. Ini, a small nuclear protein that enhances the response of the connexin43 gene to estrogen. Endocrinology 144, 3148–3158 (2003).
Karmakar, S. et al. RNA polymerase II-associated factor 1 regulates stem cell features of pancreatic cancer cells, independently of the PAF1 complex, via interactions with PHF5A and DDX3. Gastroenterology 159, 1898–1915 e1896 (2020).
Strikoudis, A. et al. Regulation of transcriptional elongation in pluripotency and cell differentiation by the PHD-finger protein Phf5a. Nat. Cell Biol. 18, 1127–1138 (2016).
Liu, J. A. et al. Asymmetric localization of DLC1 defines avian trunk neural crest polarity for directional delamination and migration. Nat. Commun. 8, 1185 (2017).
Hamburger, V. & Hamilton, H. L. A series of normal stages in the development of the chick embryo. Dev. Dyn. 195, 231–272 (1992).
Shilatifard, A., Conaway, R. C. & Conaway, J. W. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 72, 693–715 (2003).
Golas, M. M., Sander, B., Will, C. L., Luhrmann, R. & Stark, H. Molecular architecture of the multiprotein splicing factor SF3b. Science 300, 980–984 (2003).
Light, W., Vernon, A. E., Lasorella, A., Iavarone, A. & LaBonne, C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development 132, 1831–1841 (2005).
Luo, T., Lee, Y. H., Saint-Jeannet, J. P. & Sargent, T. D. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. USA 100, 532–537 (2003).
Kerosuo, L. & Bronner, M. E. cMyc regulates the size of the premigratory neural crest stem cell pool. Cell Rep. 17, 2648–2659 (2016).
McKeown, S. J., Lee, V. M., Bronner-Fraser, M., Newgreen, D. F. & Farlie, P. G. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev. Dyn. 233, 430–444 (2005).
Ferronha, T. et al. LMO4 is an essential cofactor in the Snail2-mediated epithelial-to-mesenchymal transition of neuroblastoma and neural crest cells. J. Neurosci. 33, 2773–2783 (2013).
Rogers, C. D., Sorrells, L. K. & Bronner, M. E. A catenin-dependent balance between N-cadherin and E-cadherin controls neuroectodermal cell fate choices. Mech. Dev. 152, 44–56 (2018).
Taneyhill, L. A. & Schiffmacher, A. T. Should I stay or should I go? Cadherin function and regulation in the neural crest. Genesis 55, e23028 (2017).
Manohar, S., Camacho-Magallanes, A., Echeverria, C. Jr. & Rogers, C. D. Cadherin-11 is required for neural crest specification and survival. Front. Physiol. 11, 563372 (2020).
Betancur, P., Simoes-Costa, M., Sauka-Spengler, T. & Bronner, M. E. Expression and function of transcription factor cMyb during cranial neural crest development. Mech. Dev. 132, 38–43 (2014).
Plouhinec, J. L. et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev. Biol. 386, 461–472 (2014).
Copeland, J. & Simoes-Costa, M. Post-transcriptional tuning of FGF signaling mediates neural crest induction. Proc. Natl. Acad. Sci. USA 117, 33305–33316 (2020).
Gacem, N. et al. ADAR1 mediated regulation of neural crest derived melanocytes and Schwann cell development. Nat. Commun. 11, 198 (2020).
Hasegawa, S. et al. Apoptosis in neural crest cells by functional loss of APC tumor suppressor gene. Proc. Natl. Acad. Sci. USA 99, 297–302 (2002).
Dinsmore, C. J. & Soriano, P. MAPK and PI3K signaling: at the crossroads of neural crest development. Dev. Biol. 444, S79–S97 (2018).
Peng, S. et al. The role of CELF family in neurodevelopment and neurodevelopmental disorders. Neurobiol. Dis. 197, 106525 (2024).
Jensen, K. B. et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371 (2000).
Sales-Lee, J. et al. Coupling of spliceosome complexity to intron diversity. Curr. Biol. 31, 4898–4910 e4894 (2021).
Sciarrillo, R. et al. Splicing modulation as novel therapeutic strategy against diffuse malignant peritoneal mesothelioma. EBioMedicine 39, 215–225 (2019).
Corvelo, A., Hallegger, M., Smith, C. W. & Eyras, E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 6, e1001016 (2010).
Frank, D. & Guthrie, C. An essential splicing factor, SLU7, mediates 3’ splice site choice in yeast. Genes Dev. 6, 2112–2124 (1992).
Marasco, L. E. & Kornblihtt, A. R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 24, 242–254 (2022).
Zheng, Y. Z. et al. PHF5A epigenetically inhibits apoptosis to promote breast cancer progression. Cancer Res. 78, 3190–3206 (2018).
Cretu, C. et al. Structural basis of splicing modulation by antitumor macrolide compounds. Mol. Cell 70, 265–273 e268 (2018).
Grau-Bove, X., Ruiz-Trillo, I. & Irimia, M. Origin of exon skipping-rich transcriptomes in animals driven by evolution of gene architecture. Genome Biol. 19, 135 (2018).
Yang, Y. et al. PHF5A contributes to the maintenance of the cancer stem-like phenotype in non-small cell lung cancer by regulating histone deacetylase 8. Ann. Clin. Lab. Sci. 52, 439–451 (2022).
Yang, X. et al. Nuclear DLC1 exerts oncogenic function through association with FOXK1 for cooperative activation of MMP9 expression in melanoma. Oncogene 39, 4061–4076 (2020).
Harms, F. L. et al. De novo PHF5A variants are associated with craniofacial abnormalities, developmental delay, and hypospadias. Genet. Med. 25, 100964 (2023).
Berube-Simard, F. A. & Pilon, N. Molecular dissection of CHARGE syndrome highlights the vulnerability of neural crest cells to problems with alternative splicing and other transcription-related processes. Transcription 10, 21–28 (2019).
Soldatov, R. et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536 (2019).
Park, B. Y., Tachi-Duprat, M., Ihewulezi, C., Devotta, A. & Saint-Jeannet, J. P. The core splicing factors EFTUD2, SNRPB and TXNL4A are essential for neural crest and craniofacial development. J. Dev. Biol. 10, 29 (2022).
Devotta, A., Juraver-Geslin, H., Gonzalez, J. A., Hong, C. S. & Saint-Jeannet, J. P. Sf3b4-depleted Xenopus embryos: a model to study the pathogenesis of craniofacial defects in Nager syndrome. Dev. Biol. 415, 371–382 (2016).
Liu, J. A. et al. Fbxo9 functions downstream of Sox10 to determine neuron-glial fate choice in the dorsal root ganglia through Neurog2 destabilization. Proc. Natl. Acad. Sci. USA 117, 4199–4210 (2020).
Acknowledgements
We thank the members of M. Cheung’s laboratory, R. Fässler, S.C. Kwon, T. Cheung, Z.J. Zhou, Y.W. Huang and B. Gao for their helpful discussions. We are also grateful to the members of M.E. Bronner’s laboratory for their advice on the generation of the CRISPR-Cas9/gRNA constructs. Furthermore, we extend our appreciation to the technical staff of the Centre for PanorOmic Sciences for their expertise in confocal imaging and FACS analysis. The work was supported by Hong Kong Research Grants Council General Research Fund (Project no. 17101622, 17102420) and Seed Fund for Basic Research, HKU (Project no. 202011159132, 201910159225) to M.C. Z.F.Z. was supported by the postgraduate scholarship HKU and General Research Fund.
Author information
Authors and Affiliations
Contributions
Conceptualization: Z.F.Z., J.A.I.J., and M.C. Investigation: Z.F.Z., S.S.G., H.Y.T., Y.X.R., and M.N.H. Methodology: Z.F.Z., S.S.G., J.K.W., Y.X.R., A.W.L.L., K.K.W.W., R.S., and M.P.L.C. Visualization: Z.F.Z., S.S.G., H.Y.T., and M.N.H. Funding acquisition: Z.F.Z. and M.C. Supervision: M.C. Writing (original draft): Z.F.Z. and M.C. Writing (review & editing): Z.F.Z. and M.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, Z., Guo, S., Tam, H.Y. et al. Determination of trunk neural crest cell fate and susceptibility to splicing perturbation by the DLC1-SF3B1-PHF5A splicing complex. Nat Commun 16, 6718 (2025). https://doi.org/10.1038/s41467-025-62003-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62003-6