Abstract
Microbiomes of soil, plants, and the animal gut are pivotal for key life processes such as nutrient cycling, stress resilience, and immunity. While studies have hinted at a shared microbial reservoir connecting these environments, compelling evidence of a soil-plant-gut microbiome axis is scarce. This perspective explores the potential continuum and diversification of microbes along this axis, highlighting specific microorganisms capable of moving from soil to plants to the human gut. A conceptual framework is proposed to better understand the mechanisms driving interactions among these microbiomes. We also examine how soil, plant, and gut microbiomes may co-evolve and influence one another through reciprocal effects. We consider external environmental factors that could strengthen their interconnections, potentially creating beneficial feedback loops that impact ecosystem and human health.
Similar content being viewed by others
Transmission of microorganisms from soil to plant to gut
The composition of the human gut microbiome exhibits a discernible geographic pattern1. This phenomenon is largely influenced by diet and lifestyle, such as food fiber content, urban/rural environments, and exposure to farming2. The effects of these external factors on microbiome composition appear to be stronger than host traits such as relatedness and genetics2. Consequently, the microbiomes present in the soil and plant environment, in particular the edible plant parts, have recently been proposed as significant drivers of the taxonomic and functional diversity of the human gut microbiome3. Hence, the central aim of this perspective is to critically review if there is such a shared microbial reservoir across soil, plants, and the human gut.
The soil microbiome harbors at least 25% of the Earth’s total biodiversity4 and acts as a ‘microbial seed bank’ for the plant microbiome, in particular the roots (i.e., rhizosphere and endosphere)5 but also plant seeds and the phyllosphere, i.e., the aboveground plant parts including flowers and fruits6,7. Several examples have shown that fruit and vegetable-associated bacteria can enter the human gut and may contribute to gut microbial diversity8. In addition to direct transmission of microbes to the human gut via plant-based products, the metabolic content of edible plant products can also indirectly regulate the composition of the human gut microbiome9. Moreover, incidental or deliberate soil consumption (geophagy) has been proposed to provide health benefits, such as enhanced immunological resilience, through the modulation of the human gut microbiome10. Hence, the soil microbiome is regarded as a reservoir or ‘seed bank’ of microorganisms that can enter the human gut directly or indirectly via the plant5. But which microorganisms have been reported to transmit from soil to plant to gut? In turn, which members of the human gut microbiome are introduced back into the soil environment and on/in plant tissues through feces and food/water waste11. To gain a deeper understanding of this putative feedback loop, we will also review current knowledge of the reciprocal selection mechanisms and dynamics of the soil-plant-gut microbiome axis.
Mapping the distribution of microbiota across the soil-plant-human gut axis
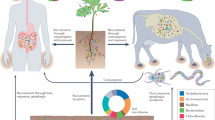
To determine if there is a microbial reservoir along the soil-plant-human gut axis (Fig. 1), we first classified the microbes in these habitats as either specialists or generalists. Specialists are defined here as microbes confined to specific habitats, while generalists are those found across all habitats12. Being classified as habitat specialists does not preclude their existence in other habitats; rather, they are typically found in much lower abundance outside their primary habitat12. Among the generalist microbes, we define potential cross-kingdom microbiota as those with a high abundance across all three habitats13.
Soil, plants, and the human gut are each considered distinct habitats. Within each habitat, specific microbial communities known as habitat specialists are identified. Microbes that are distributed across all habitats are termed here as generalists. Based on their abundance and functions, a group of cross-kingdom microbiota is proposed among the generalist microbes. Created in BioRender. Ma, H. (2025) https://BioRender.com/b51e6if.
Habitat specialists
Most soil microbes interact with plants, yet certain taxa involved in biogeochemical cycling prefer the soil habitat and typically do not rely on nutrient exchange with hosts as seen in human gut commensals or plant symbionts14 (Table 1). Therefore, soil habitat specialists are unlikely to traverse along the soil-plant-human gut microbiome axis. For instance, methanotrophs are soil bacteria that utilize methane as their sole source of carbon and energy15. They are most prevalent in submerged or water-saturated environments, such as wetlands16, but can also be found in termite guts when inhabiting methane-rich environments17. Similarly, Geobacter plays vital roles in soil elemental cycling and can be found in paddy farmland18, soils, and marine sediments, where they engage in syntrophic growth with methanogens19,20. Nitrosomonas, another example of a soil habitat specialist, is a bacterial genus that contributes to ammonia oxidation in aerated soils20. However, not all microbial genera involved in soil elemental cycling are absent in the human gut. For example, sulfate-reducing Desulfovibrio is typically found in soil but also frequently reported in the human gut21 and has been associated with gut microbiome dysbiosis22 and colonic diseases23.
Plant microbiomes typically are formed by recruitment of a subset of the soil microbiome via the spatiotemporal release of exudates from plant seeds, roots, and leaves. Among these are microbial taxa that exhibit an intimate association with plants, here defined as plant habitat specialists. For instance, arbuscular mycorrhizal fungi (AMF) are obligate biotrophs reliant on plants for carbon supply to complete their life cycle24. This symbiotic relationship enhances plant nutrient acquisition (e.g., phosphate and nitrates) from the soil and can also boost disease resistance in plant hosts25. Similarly, N2-fixing rhizobia manifest their symbiosis through the formation of nodules, serving as miniature factories for nitrogen supply to the host plants, predominantly legumes26,27. Similarly, Epichloë, a genus of filamentous fungal endophytes, has co-evolved with cool-season grasses, forming long-term symbiotic associations28. Soil-borne plant pathogens can also be regarded as plant habitat specialists, proliferating upon encountering suitable hosts and entering back into dormancy in soil in the absence of their hosts29. Establishment of these associations involves intricate chemical communication between the plant and the soil microbiome via specific constituents in plant exudates. For example, phenolic acids such as salicylic acid, vanillic acid, can attract saprotrophic fungi like Fusarium species30, while strigolactones play a crucial role in the establishment of symbiotic relationships between plants and AMF31.
The composition of the human gut microbiome is continuously influenced by microbes entering via plant-based food and the environment. Unlike soil and plants, microbes considered as human gut habitat specialists typically possess anaerobic metabolism and adaptability to varying pH levels. The duodenum has a more acidic environment compared to the jejunum and ileum, while the large intestine maintains a relatively neutral pH32. Additionally, human gut habitat specialists should have the ability to utilize mucin, as the gut mucosa forms a protective barrier for epithelial cells and serves as an ecological niche for specific non-harmful members of the gut microbiome2,33. For example, Akkermansia muciniphila is well known for its mucin-degrading capabilities, which not only enable it to thrive in the gut environment but also to contribute to its role in maintaining gut barrier integrity and metabolic health33. A. muciniphila is highly abundant in the human gut34,35, and has been shown to diminish obesity, type 2 and type 1 diabetes mellitus, hepatic steatosis, intestinal inflammation, and various cancers in mice36. Furthermore, bile salts can be abundant in the small intestine, significantly influencing microbial colonization of the human gut37. For instance, the enzymatic preferences of bile salt hydrolases significantly affect the fitness of Lactobacillus and their colonization in humans38. Fermentation by gut microbes is crucial for converting luminal compounds into secondary metabolites and is considered an indicator of a healthy gut microbiome39,40. For example, Faecalibacterium prausnitzii, a major butyrate producer, contributes to gut health by fermenting fibers and producing short-chain fatty acids that have anti-inflammatory effects, which help suppress conditions like irritable bowel syndrome, obesity, and celia disease41. In this context, F. prausnitzii has been proposed as a biomarker for a healthy gut34,35,42. Other beneficial gut habitat specialists include Christensenella minuta, Anaerobutyricum soehngenii, and Oxalobacter formigenes36.
Habitat generalists
Habitat generalists should preferably possess specific features such as metabolic flexibility, stress tolerance, and genome plasticity, allowing them to adapt to the different environments43,44. For instance, Clostridium spp. encompasses many habitat generalists such as C. cadaveris and C. senegalense, characterized by its metabolic flexibility, can thrive in diverse environments (Table 1)45. In soil and plants, Clostridium species can exhibit beneficial functions such as biological nitrogen fixation and phosphate solubilization46. In the human gut, Clostridium can ferment carbohydrates to produce short-chain fatty acids, promoting gut health47. C. butyricum, a human gut symbiont known for its butyrate-producing and anti-cancer properties48,49, has also been isolated from soil50. However, certain Clostridium strains, including Clostridium difficile and Clostridium perfringens, produce toxins with neurotoxic, hemolytic, and enterotoxigenic properties47. They were also isolated from the soil and therefore, their transmission along the soil-plant-gut axis warrants attention51. Similarly, Acinetobacter exhibits diverse metabolic functions such as phenol and dibutyl phthalate degradation in soil, as well as phosphate solubilization for plants52,53,54. Acinetobacter species also reside in the human gut, influencing gut epithelium modulation55,56. Several Acinetobacter species, including A. baumannii and A. calcoaceticus, can be found in soil and water, but are also opportunistic human pathogens57,58. Their persistence in these different environments poses a threat to human health and may be attributed to several traits, including their high level of resistance to desiccation59, horizontal gene transfer60, and lipopolysaccharides (LPS) in their outer membrane61. Similarly, Staphylococcus and Streptococcus have been identified as opportunistic human pathogens35, but have also been detected in plant rhizospheres62,63,64,65 and soil environments66. Also, Stenotrophomonas can be considered a habitat generalist, as it inhabits both the plant rhizosphere and endosphere, where it acts as a beneficial microbe by inducing plant growth hormones and chitinases67. In soil, Stenotrophomonas plays a role in nutrient cycling, particularly in nitrogen and sulfur67. In humans, Stenotrophomonas can colonize the gut49,68 and act as a pathogen to immunocompromised and debilitated individuals, leading to severe inflammation and infection69. Its persistence in diverse environments is attributed to its metabolic versatility, ability to form biofilms, production of extracellular enzymes, and resistance to multiple stress conditions69. Also, Pseudomonas and Helicobacter can be considered as habitat generalists. Except for potential pathogenic species, other human gut commensals such as Ruminococcus, which has been identified as one of the most abundant taxa in the human gut by several global studies70,71, have also been reported in the rhizosphere of Antarctic vascular plants72. Therefore, the reservoir of microbial genera along the soil-plant-gut axis may be larger than we currently know. Considering that several competitive traits and their corresponding genes are distinctly different among species of a given genus and even among strains of a given species, comparative meta-analyses will be needed to align so-called metagenome assembled genomes (MAGs) of microbiome members along the axis to further delineate and validate the taxonomic identity and frequency of habitat generalists and cross-kingdom microbiota across these different habitats beyond the genus and species level.
Putative cross-kingdom microbiota along the soil-plant-human gut axis
The core (cross-kingdom) microbiota is traditionally defined by its taxonomic composition, but also its functional roles in relation to the host should be considered73. For instance, Bacillus subtilis, known for its considerable genome diversity, thrives in various environments (Table 1)74. Capable of forming dormant endospores in response to nutrient deprivation and environmental stresses, B. subtilis acts as an important plant growth-promoting rhizobacterium, conferring biotic and abiotic stress tolerance to plants through induced systemic resistance, biofilm formation, and lipopeptide production73,75. Moreover, B. subtilis is widely employed in bioremediation75, but is also extensively studied in humans for the production of vitamins and metabolites with anticancer and antioxidant properties, potentially influencing human longevity and reducing the risk of Parkinson’s disease76. Also, Lactobacillus represents a group of bacteria with beneficial functions for humans, plants, and soil environments. In the human gut, Lactobacillus acts as a probiotic for immune regulation, inhibiting the colonization of enteric pathogens through the production of metabolites such as organic acids, hydrogen peroxide, nitric oxide, short-chain fatty acids, and bacteriocins77. The robust metabolic capabilities of Lactobacillus also contribute to its anti-inflammatory properties78. A recent meta-analysis integrating 168,000 human gut samples globally, reported that Lactobacillus is one of the most abundant gut microbiota, representing a key taxon contributing to the variability observed in gut samples from Europe and North America49. For plants, Lactobacillus has shown growth-promoting effects and has been used as a biocontrol strains in several studies79,80. In soil environments, Lactobacillus has demonstrated the ability to remediate polluting metals81. The generalist nature of Lactobacillus may be attributed to its comprehensive genetic catalog for carbohydrate and protein modification, production of host interaction factors and bacteriocins, and stress response mechanisms82. Another noteworthy genus is Streptomyces, soil-dwelling bacteria renowned for their antibiotic production, plant growth-promoting, and biocontrol traits83. While less prevalent in the human gut, Streptomyces are gaining attention as potential probiotics due to their detoxification of mycotoxins, production of extracellular polymeric substances with anticancer properties, and synthesis of anti-inflammatory polyketides84. Other microbes, such as Lactococcus, are also identified as abundant gut commensals globally49 and have been used as probiotics for treating IBD and Type 2 diabetes85, but also exhibit plant growth-promoting effects attributed to the production of organic acids and siderophores80.
In contrast to these ‘beneficial cross-kingdom microbiota’, the pathogen Salmonella enterica manifests in various disease syndromes, with food contamination as the primary mode of transmission56,86. Studies have shown that Salmonella attaches to and colonizes plants but can even infect them, with the rate of infection contingent upon the activation of the plant immune system, sometimes resulting in plant leaf chlorosis and wilting87. Additionally, soil may serve as a reservoir for Salmonella species, as S. enterica has been found in a sandy loamy soil88. Other human pathogenic bacteria, such as the enteric pathogen Shigella, have been found to colonize the leaves and roots of Arabidopsis through the adoption of type III effectors56,89. However, the effects of Shigella on plants and its survival in soil still require further investigation. Thus, it is imperative to further explore the vast reservoir of beneficial and deleterious cross-kingdom microbial taxa along the soil-plant-human gut axis at genus and preferably at species and strain levels. In this context, identification of the extensive intraspecific diversity of microbial genera is essential to better define their specialist or generalist behavior across the soil-plant-human gut microbiome axis and to identify specific genes or gene clusters associated with that lifestyle90.
Feedback loop and co-evolution between soil, plant and human gut microbiomes
Species originating from distinct lineages can undergo reciprocal evolutionary changes over time, leading to the establishment of interdependent ecological relationships that significantly influence their phenotypes91. Co-evolution is defined as the process whereby a change in a trait of individuals within one population occurs in response to a trait exhibited by individuals in a second population92. In the context of the soil-plant-human microbiome axis, the soil acts as a reservoir for various bacterial taxa such as Helicobacter pylori, which can transfer to the human gut through food contamination or wastewater used for irrigation93,94. Nucleotide sequences of H. pylori in the environment show 96%-100% homology with those in the human gut95. The bacterium’s highly plastic genome, marked by intraspecific recombination, enhances its colonization ability in the human gut, even under antibiotic pressure96,97. In H. pylori, distinct resistance profiles, encompassing single and multidrug resistance, were identified98. Antimicrobial resistance genes induced by medical therapeutics may be reintroduced back into the soil microbiome and impose a subsequent selection pressure on soil-borne H. pylori, which in turn may augment the risk of new infections in humans99. A second example of a bacterium that may exert a reciprocal influence along the soil-plant-human gut microbiome axis includes Pseudomonas. The persistence of Pseudomonas in various habitats may be attributed to its versatile metabolic ability to utilize a wide range of organic and inorganic compounds as energy sources49,100,101, its ability to form protective biofilms to survive environmental stresses102, and the presence of multidrug efflux pumps, which contribute to its antibiotic resistance103. The combination of its genomic plasticity, environmental exposure, and biofilm formation makes horizontal gene transfer particularly prominent in Pseudomonas, enabling it to easily acquire foreign genes and disseminate antibiotic resistance genes, toxins, and other advantageous traits104, eventually mediating functional changes in the different microbiomes along the axis105.
To date, several mechanisms have been proposed that drive these reciprocal effects and co-evolution among soil, plant, and human gut microbiomes. These include molecular mimicry, horizontal gene transfer, cross-feeding, and host selection (Table 2). These mechanisms will be discussed in more detail below.
Molecular mimicry
Molecular mimicry is characterized by analogous structural features shared by molecules originating from distinct genes or their corresponding protein products106. Microorganisms in one microbiome can produce molecules that exhibit structural similarity to essential signaling molecules in the other microbiome (Table 2). This resemblance may give rise to selection pressures between the interconnected microbiomes. For example, lipopolysaccharides (LPS) in the outer membrane of soil-borne Gram-negative bacteria have long been used as markers of soil microbial community structure107. Its structural features also resemble those of host-derived molecules that trigger pattern recognition receptors, such as Toll-like receptors (TLRs) in plants and humans108. In the human gut, this activation subsequently initiates inflammatory responses that not only contribute to tissue immunity but also facilitates coordination with the adaptive immune system109. This structural similarity can result in LPS acting as a pathogen-associated molecular pattern (PAMP), eliciting an immune response that spans plant and human systems110. Another example is peptidoglycan in the bacterial cell wall that is a direct target for innate immune receptors and modulates the accessibility of other PAMPs to additional innate immune receptors111. It has been suggested that this peptidoglycan-mediated host immunity is evolutionary conserved112. Application of eight common clinical antibiotics to soil resulted in significant alterations in gene expression, including the biosynthesis of peptidoglycan113. Hypothetically, these alterations in turn may affect human immunity via exposure to qualitative and quantitative changes in these cellular structures of soil microorganisms when transmitted to the human gut microbiome. Although most examples described so far relate to molecules of pathogenic bacteria, there are a few examples of beneficial molecular mimicry. One such as example is the soil-dwelling butyrate producer Kineothrix alysoides, whose abundance in the gut increased significantly following exposure to high-biodiversity soil10. Butyrate enhances human health by modulating energy metabolism, improving insulin sensitivity, regulating lipid metabolism, and reducing inflammation, primarily through activation of G-protein coupled receptors (GPRs) and epigenetic regulation of gene expression114. Hence, human immune responses may be modulated via the plant115 and food microbiome116, while human activities can affect the biosynthesis and dynamics of these molecules within the soil environment.
Horizontal gene transfer
Horizontal gene transfer is an extensively studied mechanism of interaction between the soil microbiome and the human gut microbiome115,117. Horizontal gene transfer represents mechanisms through which microorganisms acquire foreign DNA from other (micro)organisms. The main mechanisms of horizontal gene transfer within natural microbial communities include transformation, conjugation, transduction, and outer membrane vesicles118. The acquired DNA may confer traits that broaden the ecological niche of a microorganism, alter its interactions with the host or confer a competitive advantage over other microorganisms118. One well-studied example in the context of the soil-plant-human gut microbiome axis is the transfer of antibiotic resistance genes introduced into the soil environment through manure, biosolids, and wastewater derived from human and animal waste119, and even microplastics120. Consequently, antibiotic-resistant bacteria and antibiotic resistance genes present in the soil microbiome can transfer to clinically relevant microbial pathogens or to the human gut microbiota through various routes such as the food chain, drinking water, or environmental exposure117,121,122. During these processes, plant selection of their microbiome and the merger with the manure microbiome, known as community coalescence, have been identified as two critical factors determining the transmission of antibiotic-resistant genes123,124. Accordingly, studies have shown that individuals exposed to specific farms exhibit gut microbiomes with antibiotic resistance gene patterns similar to those found in the environmental samples from the respective farms125. For instance, about 27% of microbes transmitted from environmental soil and dust in swine farms to the human gut harbored at least one antibiotic resistance gene125. This mechanism has the potential to induce significant and rapid co-evolutionary changes across broader phylogenetic lineages of microorganisms, irrespective of their relatedness, extending beyond the confines of solely vertical transmission between the parent and its offspring126. Another group of genes that could be transferred between the soil and human gut microbiomes is the EPS biosynthetic genes. In soil microbes, these genes support various functions, including symbiosis with plants and biofilm formation127. In the human gut, EPS biosynthesis by commensal bacteria benefits the host through immune modulation, reduced gastrointestinal stress, and pathogen inhibition128. Horizontal gene transfer of EPS production genes has been found in various environments, such as food systems129 and bacterial biofilm matrices130. Therefore, it can be hypothesized that the exchange of EPS biosynthesis genes may occur between soil microbes and human gut microbes, likely leading to beneficial reciprocal effects in both habitats along the axis.
Colonization resistance
The concept of the ‘Arms Race’, commonly applied to the co-evolution between hosts and parasites, describes the reciprocal selection that drives adaptation and counter-adaptation of the interacting partners131. Colonization resistance refers to the phenomenon where commensal microorganisms of a host act as a protective barrier against the invasion, colonization, and proliferation of both pathogens and indigenous pathobionts132 (Table 2). This concept has also been employed more broadly to characterize how the resident microbiota hinders the colonization of introduced microorganisms133. In the human gut, colonization resistance becomes evident when antibiotics are used, as several pathogens and invading microbes can only effectively colonize and proliferate in this vacuum134. The mechanisms underlying colonization resistance operate through both direct and indirect means. Indirect mechanisms encompass nutrient competition135, and antagonism by gut microbes producing antimicrobial peptides, short-chain fatty acids or secondary bile acids133. Direct mechanisms of colonization resistance include the mucosal barrier136, oxygen limitation in the gut132, and host antimicrobial peptides and cytokines132. Similarly, colonization resistance in soil can be observed when introduced beneficial microbial strains fail to persist and express specific activities in situ despite demonstrating significant effects under controlled conditions in the lab or greenhouse137. Also, the invasiveness of the human-associated bacterium Escherichia coli O157:H7 in soil was inversely related to the diversity of resident soil microbial communities138. However, microbial invaders can also establish and induce changes in soil bacterial diversity139,140. The variability in the outcome of microbial invasions may be contingent with the taxonomic and functional diversity of the resident microbiome (soil, plant or human). For example, colonization resistance of the human gut is observed under homeostasis, whereas dysbiosis of the gut microbiome compromises colonization resistance132.
Cross feeding
The fourth mechanism underlying reciprocal effects between soil, plant, and human gut microbiomes involves cross-feeding (Table 2). The cooperative exchange of metabolites as energy and nutrient source among different microbial species or strains141 is largely determined by metabolic dissimilarity and complementarity142,143. Although direct cross-feeding interactions between microbiota along the soil-plant-gut axis have not been widely demonstrated and experimentally validated, shared metabolic functions across these microbiomes suggest potential indirect influences. Bacteria in particular, frequently engage in obligate metabolic mutualisms with other (micro)organisms to expand their ecological niche144. In the human gut, plant fibers containing cellulose and hemicellulose are challenging to digest, necessitating the presence of a diverse array of degrading enzymes such as glycoside hydrolases and polysaccharide lyases. These enzymes are not produced, or are produced only in limited quantities, by mammals themselves145. Interestingly, glycoside hydrolases have been found in soil, plant and human gut microbiomes, with the bacteria producing these enzymes exhibiting conserved functions across these microbiomes146. Soil microorganisms are also capable of producing vitamins, including cobalamin147. Cobalamin (vitamin B12), is crucial in the human gut and many organisms reliant on cobalamin acquisition depend on other species for its provision148. Butyrate-producing bacteria in the gut, which have various beneficial effects on human health, require cobalamin for their growth149. While the human gut does not/seldomly directly feed on soil microbes, the plant microbiome may act as an intermediary by hosting microbes that synthesize or modify essential metabolites150. Through the consumption of plant-based foods, these metabolites, including fibers and vitamins synthesized by soil bacteria, reach the human gut, where gut microbiota further metabolize them150. This interplay along the soil-plant-human gut microbiome highlights the potential for cooperative interactions and the exchange of essential resources, without the need for direct transmission of the involved microorganisms along the axis.
Host selection
Soil type can be a factor influencing the assembly of the plant microbiome; however, its impact is context-dependent. In natural ecosystems, the effects of soil type may be less pronounced compared to agricultural settings151. However, within each soil environmental context, the local proliferation of microbes is influenced by plants, as they actively recruit microbes from the soil into the rhizosphere through selection by root exudation composition, root architecture, and plant litter decomposition152. This subsequently leads to plant species-specific microbiomes, which in turn may influence human gut microbiomes. One illustrative example is the process of plant domestication, where humans transformed wild plant species into highly productive crops for human consumption. These cultivated varieties exhibit traits such as larger fruit size, higher growth rates, and altered chemistry153. Consequently, when compared to their wild relatives, crops exhibit a reduced specific root length, higher root density, altered root exudation, and root-associated microbiomes154. This was further exemplified by microbiome studies of recombinant inbred line populations of a cross between wild relatives and their domesticated crop cultivar that exhibit changes in microbiome compositions associated with specific quantitative trait loci in crop genomes155. To what extent these changes in plant chemistry and microbiome composition affect the human gut microbiome is an emerging topic of research156. A recent study revealed an association between the presence of metabolic pathways in various fruits and vegetables and homeostasis of the human gut microbiome157. This association includes pathways related to the synthesis of glutamate and biotin, as well as numerous carbohydrate-active enzymes156,157.
Microbes, derived from plants (or the diet), along with other host factors such as immune condition and genetic makeup, contribute to the formation of specific enterotypes in the human gut158,159. For example, enterotypes are characterized as Bacteroides, Prevotella, and Ruminococcus types, based on the dominant species160. This leads to functional differentiation among these enterotypes, such as the Bacteroides enterotype, which ferments carbohydrates and possesses a broad saccharolytic potential161. The Prevotella enterotype primarily degrades mucin glycoproteins, while the Ruminococcus enterotype is involved in binding mucins, transporting them, and degrading sugars in the gut161. Conversely, the specificity of enterotypes also influences the future colonization of other microbes, as they are linked to the dysbiosis status of the gut microbiome162 or the disease status of the human host163. This makes it both an important outcome and a deterministic factor in the human host’s selection of their gut microbiome.
Outlook: creating a positive feedback loop in the soil-plant-human gut axis
The reciprocal influences as well as the distribution and proliferation of microbes along the soil-plant-gut axis are subject to various environmental factors. These factors may range from unpredictable climate change events to physiochemical changes in specific micro-habitats. Therefore, managing potential external drivers to create a positive feedback loop along the axis is essential for achieving better “One Health” outcomes.
A healthy plant microbiome can positively influence the human gut microbiome primarily through two mechanisms (Fig. 2). First, the plant phyllosphere and other edible plant parts can transmit soil microorganisms to the human gut. With increased soil microbial diversity, the potential negative impact on the gut microbiome through the plant microbiome may decrease. For instance, enhanced soil microbial diversity correlates with a decreased abundance of antibiotic-resistant genes in the soil164,165,166 and increased antagonistic effects against human pathogens167. Additionally, plant extracts, including phenolic compounds, chlorophyll, and carotenoids, can act as prebiotics for beneficial microbes already present in the human gut168. Secondly, an improved soil microbiome can sustain plants producing food with higher nutrient contents, such as vitamins, polyphenols, and dietary fiber169. For example, biofertilizers have been found to increase the dietary fiber content of onions170 and corn171, the vitamin content of tomatoes172, and the polyphenol content of eggplants173. Subsequently, this higher dietary fiber content enhances the production of short-chain fatty acids by gut bacteria, indirectly contributing to improvement in human health by reducing inflammation, preventing cancer, and supplying energy174. Furthermore, the polyphenols in food promote beneficial strains in the gut, such as A. muciniphila175 and enhance antimicrobial activities against pathogens176.
External drivers, such as crop rotation, diversification, cover cropping, organic compost, and ecological restoration, have been proposed to enhance soil biodiversity and health. A healthy soil ecosystem promotes nutrient uptake and enhances plant disease resistance by fostering the enrichment of beneficial microbes in the plant rhizosphere, phyllosphere and spermosphere. This, in turn, contributes to a robust plant microbiome capable of producing nutrient-rich food, including dietary fiber, polyphenols, and vitamins. The enhanced plant microbiome also has the potential to support microorganisms with probiotic effects while reducing the frequency of human pathogens and antibiotic resistance genes when transferred to the human gut. Furthermore, the soil microbiome can directly interact with the human microbiome. Moreover, a healthy human gut microbiome plays a role in decreasing the frequency of (opportunistic) human pathogens and antibiotic resistance genes reintroduced into the soil through fecal and waste matter, thus completing a positive feedback loop that benefits both soil and human health. Created in BioRender. Ma, H. (2025) https://BioRender.com/b51e6if.
In addition to using plants as a mediator, numerous studies have demonstrated a positive link between soil biodiversity and human health, often referred to as ‘natural immunity’177,178. Particularly in early life, exposure of humans to the natural soil environment aids in the development of a diverse molecular memory, facilitating rapid recognition of harmful organisms179. Furthermore, exposure to the natural environment can introduce beneficial gut microbiota, including butyrate-producing bacteria180 and beneficial fungi181. These microbial components acquired by the gut contribute to activation of the innate immune system. For instance, feeding a soil slurry to mice has been shown to alleviate inflammation178, while exposing inbred laboratory mice to the outdoor environment reduces pathogen infections, increases circulating granulocytes, and promotes differentiation of T cell populations via the colonization of intestinal fungi181. Moreover, higher environmental biodiversity has been associated with an increased proportion of regulatory T cells and elevated plasma TGF-β1 levels in children182. Therefore, exposure to the natural environment can serve as a prophylactic measure in preventing immune-mediated diseases (Fig. 2). In return, a healthy gut microbiome may reduce the presence of antibiotic-resistant genes and human pathogens that could be reintroduced back to the soil environment through feces and waste. Thus, while exposure to the natural environment carries the risk of closer contact with potential human pathogens, a healthy soil microbiome could diminish these chances. In this way, the positive loop along the soil-plant-human gut axis can be reinforced.
Conclusions
The importance of including the soil and plant microbiomes in human health is increasingly recognized3. In this perspective, we have provided several examples of microorganisms that can transmit from soil or plants to the human gut and exert specific influences. Typical examples highlighted and detailed in the sections above include Bacillus, Pseudomonas and Clostridium species. The question if there are unifying beneficial or detrimental functions in these distinct ecosystems was more difficult to address and the available data and results are still largely circumstantial and hypothetical. To this end, we propose to integrate co-evolutionary concepts, where the soil, plant, and human gut microbiomes may influence each other’s evolutionary trajectories over time, providing a better understanding of the exchange and interactions between the soil microbiome and human gut microbiome. Also identifying habitat specialists, habitat generalists, and potential cross-kingdom microbiota along the soil-plant-human gut microbiome axis at the species and preferably strain level could provide us with a deeper understanding of the microbial fluxes along the axis. In this context, large-scale genomic identification of the extensive intraspecific diversity of microbial genera is essential to identify their life-style associated genes90 and monitor their population dynamics and distribution across the soil-plant-human gut microbiome axis.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Parizadeh, M. & Arrieta, M.-C. The global human gut microbiome: genes, lifestyles, and diet. Trends. Mol. Med 10, 789–801 (2023).
Singh, B. K., Yan, Z.-Z., Whittaker, M., Vargas, R. & Abdelfattah, A. Soil microbiomes must be explicitly included in One Health policy. Nat. Microbiol 8, 1367–1372 (2023).
Sokol, N. W. et al. Life and death in the soil microbiome: how ecological processes influence biogeochemistry. Nat. Rev. Microbiol 20, 415–430 (2022).
Banerjee, S. & Van Der Heijden, M. G. A. Soil microbiomes and one health. Nat. Rev. Microbiol 21, 6–20 (2023).
Sohrabi, R., Paasch, B. C., Liber, J. A. & He, S. Y. Phyllosphere microbiome. Annu Rev. Plant Biol. 74, 539–568 (2023).
Zarraonaindia, I. et al. The soil microbiome influences grapevine-associated microbiota. mBio 6, 10–1128 (2015).
Wicaksono, W. A. et al. The edible plant microbiome: evidence for the occurrence of fruit and vegetable bacteria in the human gut. Gut Microbes 15, 2258565 (2023).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Roslund, M. I., Laitinen, O. H. & Sinkkonen, A. Scoping review on soil microbiome and gut health—Are soil microorganisms missing from the planetary health plate?. People Nat. 6, 1078–1095 (2024).
Blum, W. E. H., Zechmeister-Boltenstern, S. & Keiblinger, K. M. Does soil contribute to the human gut microbiome?. Microorganisms 7, 287 (2019).
Malard, L. A. & Guisan, A. Into the microbial niche. Trends Ecol Evol 38, 936-945 (2023).
Neu, A. T., Allen, E. E. & Roy, K. Defining and quantifying the core microbiome: challenges and prospects. Proc. Natl. Acad. Sci. 118, e2104429118 (2021).
Hooper, L. V. & Gordon, J. I. Commensal host-bacterial relationships in the gut. Science (1979) 292, 1115–1118 (2001).
Hwangbo, M., Shao, Y., Hatzinger, P. B. & Chu, K. Acidophilic methanotrophs: Occurrence, diversity, and possible bioremediation applications. Environ. Microbiol. Rep. 15, 265–281 (2023).
Davamani, V., Parameswari, E. & Arulmani, S. Mitigation of methane gas emissions in flooded paddy soil through the utilization of methanotrophs. Sci. Total Environ. 726, 138570 (2020).
Chiri, E. et al. Termite mounds contain soil-derived methanotroph communities kinetically adapted to elevated methane concentrations. ISME J. 14, 2715–2731 (2020).
Li, T. & Zhou, Q. The key role of Geobacter in regulating emissions and biogeochemical cycling of soil-derived greenhouse gases. Environ. Pollut. 266, 115135 (2020).
Ueki, T. Cytochromes in extracellular electron transfer in Geobacter. Appl. Environ. Microbiol. 87, e03109–e03120 (2021).
Li, Y., Chapman, S. J., Nicol, G. W. & Yao, H. Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301 (2018).
Singh, S. B., Carroll-Portillo, A. & Lin, H. C. Desulfovibrio in the gut: The enemy within?. Microorganisms 11, 1772 (2023).
Figliuolo, V. R., Coutinho-Silva, R. & Coutinho, C. M. L. M. Contribution of sulfate-reducing bacteria to homeostasis disruption during intestinal inflammation. Life Sci. 215, 145–151 (2018).
Sayavedra, L. et al. Desulfovibrio diazotrophica sp. nov., a sulphate reducing bacterium from the human gut capable of nitrogen fixation. Environ Microbiol. 6, 3164–3181 (2021).
Choi, J., Summers, W. & Paszkowski, U. Mechanisms underlying establishment of arbuscular mycorrhizal symbioses. Annu Rev. Phytopathol. 56, 135–160 (2018).
Corradi, N. & Bonfante, P. The arbuscular mycorrhizal symbiosis: origin and evolution of a beneficial plant infection. PLoS Pathog. 8, e1002600 (2012).
Masson-Boivin, C. & Sachs, J. L. Symbiotic nitrogen fixation by rhizobia—the roots of a success story. Curr. Opin. Plant Biol. 44, 7–15 (2018).
Durán, P. The core microbiota across the green lineage. Curr. Opin. Plant Biol. 77, 102487 (2024).
Card, S. D., Bastías, D. A. & Caradus, J. R. Antagonism to plant pathogens by Epichloë fungal endophytes—A review. Plants 10, 1997 (2021).
Ma, M., Taylor, P. W. J., Chen, D., Vaghefi, N. & He, J.-Z. Major soilborne pathogens of field processing tomatoes and management strategies. Microorganisms 11, 263 (2023).
Clocchiatti, A., Hannula, S. E., Van Den Berg, M., Hundscheid, M. P. J. & De Boer, W. Evaluation of phenolic root exudates as stimulants of saptrophic fungi in the rhizosphere. Front Microbiol. 12, 644046 (2021).
Badri, D. V. & Vivanco, J. M. Regulation and function of root exudates. Plant Cell Environ. 32, 666–681 (2009).
Duncan, S. H., Louis, P., Thomson, J. M. & Flint, H. J. The role of pH in determining the species composition of the human colonic microbiota. Environ. Microbiol. 11, 2112–2122 (2009).
Belzer, C. Nutritional strategies for mucosal health: the interplay between microbes and mucin glycans. Trends Microbiol. 30, 13–21 (2022).
Shi, Z. J., Dimitrov, B., Zhao, C., Nayfach, S. & Pollard, K. S. Fast and accurate metagenotyping of the human gut microbiome with GT-Pro. Nat. Biotechnol. 40, 507–516 (2022).
Tap, J. et al. Global branches and local states of the human gut microbiome define associations with environmental and intrinsic factors. Nat. Commun. 14, 3310 (2023).
Cani, P. D., Depommier, C., Derrien, M., Everard, A. & de Vos, W. M. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 19, 625–637 (2022).
Urdaneta, V. & Casadesús, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4, 163 (2017).
Foley, M. H. et al. Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. 118, e2017709118 (2021).
Bäckhed, F. et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 12, 611–622 (2012).
Vieira-Silva, S. et al. Species–function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol 1, 1–8 (2016).
Miquel, S. et al. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261 (2013).
Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J. & Martinez-Medina, M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 11, 841–852 (2017).
Sriswasdi, S., Yang, C. & Iwasaki, W. Generalist species drive microbial dispersion and evolution. Nat. Commun. 8, 1162 (2017).
Xu, Q. et al. Microbial generalists and specialists differently contribute to the community diversity in farmland soils. J. Adv. Res. 40, 17–27 (2022).
Pahalagedara, A. S. N. W. et al. Culture and genome-based analysis of four soil Clostridium isolates reveal their potential for antimicrobial production. BMC Genomics 22, 1–14 (2021).
Figueiredo, G. G. O., Lopes, V. R., Romano, T. & Camara, M. C. Clostridium, Beneficial Microbes in Agro-Ecology, 477–491 (Academic, Cambridge 2020).
Guo, P., Zhang, K., Ma, X. & He, P. Clostridium species as probiotics: potentials and challenges. J. Anim. Sci. Biotechnol. 11, 1–10 (2020).
Chen, D. et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467 (2020).
Abdill, R. J. et al. Integration of 168,000 samples reveals global patterns of the human gut microbiome. Cell 188, 1–19 (2025).
ROSS, D. J. Influence of Media on the Counts of Clostridium butyricum in Soils. Nature 181, 1142–1143 (1958).
Lim, S. C., Knight, D. R. & Riley, T. V. Clostridium difficile and one health. Clin. Microbiol. Infect. 26, 857–863 (2020).
Liu, Y. et al. Phenol biodegradation by Acinetobacter radioresistens APH1 and its application in soil bioremediation. Appl. Microbiol. Biotechnol. 104, 427–437 (2020).
Sharma, N., Kumar, V., Maitra, S. S., Lakkaboyana, S. K. & Khantong, S. DBP biodegradation kinetics by Acinetobacter sp. 33 F in pristine agricultural soil. Environ. Technol. Innov. 21, 101240 (2021).
He, D. & Wan, W. Phosphate-solubilizing bacterium Acinetobacter pittii gp-1 affects rhizosphere bacterial community to alleviate soil phosphorus limitation for growth of soybean (Glycine max). Front. Microbiol. 12, 737116 (2021).
Glover, J. S., Browning, B. D., Ticer, T. D., Engevik, A. C. & Engevik, M. A. Acinetobacter calcoaceticus is well adapted to withstand intestinal stressors and modulate the gut epithelium. Front. Physiol. 13, 880024 (2022).
Organization, W. H. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance, to Guide Research, Development, and Strategies to Prevent and Control Antimicrobial Resistance (World Health Organization, 2024).
Pal, R. B. & Kale, V. V. Acinetobacter calcoaceticus-an opportunistic pathogen. J. Postgrad. Med. 27, 218–221 (1981).
Castillo-Ramírez, S. The importance of Acinetobacter baumannii from non-human sources. Lancet Microbe 4, e761–e762 (2023).
Jawad, A., Snelling, A. M., Heritage, J. & Hawkey, P. M. Exceptional desiccation tolerance of Acinetobacter radioresistens. J. Hosp. Infect. 39, 235–240 (1998).
Cooper, R. M., Tsimring, L. & Hasty, J. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. Elife 6, e25950 (2017).
Moffatt, J. H. et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977 (2010).
Oliva, G. et al. Counteracting action of Bacillus stratosphericus and Staphylococcus succinus strains against deleterious salt effects on Zea mays L. Front. Microbiol 14, 1171980 (2023).
Al-Quwaie, D. A. H. Bacterial community dynamics with rhizosphere of Calotropis procera and Senna alexandrina desert plants in Saudi Arabia. Bioinformation 16, 567 (2020).
Mundt, J. O., Coggin, J. H. Jr & Johnson, L. F. Growth of Streptococcus faecalis var. liquefaciens on plants. Appl. Microbiol 10, 552–555 (1962).
Holguin, G., Guzman, M. A. & Bashan, Y. Two new nitrogen-fixing bacteria from the rhizosphere of mangrove trees: Their isolation, identification and in vitro interaction with rhizosphere Staphylococcus sp. FEMS Microbiol. Lett. 101, 207–216 (1992).
Singh, R. P. & Kumari, K. Genome Sequence of Environmental Isolate Staphylococcus aureus OS-6. Isolated from a soil sample. Microbiol Resour. Announc 12, e00238–23 (2023).
Ryan, R. P. et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525 (2009).
Hellmig, S. et al. Life-threatening chronic enteritis due to colonization of the small bowel with Stenotrophomonas maltophilia. Gastroenterology 129, 706–712 (2005).
An, S. & Berg, G. Stenotrophomonas maltophilia. Trends Microbiol 26, 637–638 (2018).
Costea, P. I. et al. Subspecies in the global human gut microbiome. Mol. Syst. Biol. 13, 960 (2017).
Lee, S. et al. Global compositional and functional states of the human gut microbiome in health and disease. Genome Res. 34, 967–978 (2024).
Teixeira, L. C. R. S. et al. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 4, 989–1001 (2010).
Lemanceau, P., Blouin, M., Muller, D. & Moënne-Loccoz, Y. Let the core microbiota be functional. Trends Plant Sci. 22, 583–595 (2017).
Earl, A. M., Losick, R. & Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275 (2008).
Mahapatra, S., Yadav, R. & Ramakrishna, W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J. Appl Microbiol 132, 3543–3562 (2022).
Goya, M. E. et al. Probiotic Bacillus subtilis protects against α-synuclein aggregation in C. elegans. Cell Rep. 30, 367–380 (2020).
Rastogi, S. & Singh, A. Gut microbiome and human health: Exploring how the probiotic genus Lactobacillus modulate immune responses. Front. Pharm. 13, 1042189 (2022).
Aghamohammad, S. et al. Anti-inflammatory and immunomodulatory effects of Lactobacillus spp. as a preservative and therapeutic agent for IBD control. Immun. Inflamm. Dis. 10, e635 (2022).
Strafella, S. et al. Comparative genomics and in vitro plant growth promotion and biocontrol traits of lactic acid bacteria from the wheat rhizosphere. Microorganisms 9, 78 (2020).
Jaffar, N. S., Jawan, R. & Chong, K. P. The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production - A mini review. Front. Plant Sci. 13, 1047945 (2023).
Zhang, S. et al. Lactic acid bacteria promoted soil quality and enhanced phytoextraction of Cd and Zn by mustard: A trial for bioengineering of toxic metal contaminated mining soils. Environ. Res. 216, 114646 (2023).
Sun, Z. et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6, 8322 (2015).
Gopalakrishnan, S., Srinivas, V. & Prasanna, S. L. Streptomyces. In Beneficial Microbes in Agro-Ecology 55–71 (Elsevier, 2020).
Cuozzo, S. et al. Streptomyces genus as a source of probiotics and its potential for its use in health. Microbiol. Res. 266, 127248 (2023).
Bermúdez-Humarán, L. G. et al. Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol 16, 278–283 (2013).
Aljahdali, N. H., Sanad, Y. M., Han, J. & Foley, S. L. Current knowledge and perspectives of potential impacts of Salmonella enterica on the profile of the gut microbiota. BMC Microbiol. 20, 1–15 (2020).
Schikora, A., Garcia, A. V. & Hirt, H. Plants as alternative hosts for Salmonella. Trends Plant Sci. 17, 245–249 (2012).
Zheng, J. et al. Colonization and internalization of Salmonella enterica in tomato plants. Appl. Environ. Microbiol. 79, 2494–2502 (2013).
Jo, S. H. et al. A human pathogenic bacterium Shigella proliferates in plants through adoption of type III effectors for shigellosis. Plant Cell Environ. 42, 2962–2978 (2019).
Guerrero-Egido, G. et al. bacLIFE: a user-friendly computational workflow for genome analysis and prediction of lifestyle-associated genes in bacteria. Nat. Commun. 15, 2072 (2024).
Darwin, C. R. On the various contrivances by which British and foreign orchids are fertilised by insects, and on the good effects of intercrossing (John Murray, London, 1862).
Janzen, D. H. When is it coevolution. Evolution 34, 611–612 (1980).
Duan, M. et al. Transmission routes and patterns of helicobacter pylori. Helicobacter 28, e12945 (2023).
Kawaguchi, K., Matsuo, J., Osaki, T., Kamiya, S. & Yamaguchi, H. Prevalence of Helicobacter and Acanthamoeba in natural environment. Lett. Appl. Microbiol. 48, 465–471 (2009).
Sasaki, K. et al. Helicobacter pylori in the natural environment. Scand. J. Infect. Dis. 31, 275–279 (1999).
Nguyen, A. N. T. et al. Recombination resolves the cost of horizontal gene transfer in experimental populations of Helicobacter pylori. Proc. Natl. Acad. Sci. 119, e2119010119 (2022).
Boyanova, L., Hadzhiyski, P., Gergova, R. & Markovska, R. Evolution of Helicobacter pylori resistance to antibiotics: a topic of increasing concern. Antibiotics 12, 332 (2023).
Tshibangu-Kabamba, E. & Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—from biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 18, 613–629 (2021).
Li, H.-Z. et al. Active antibiotic resistome in soils unraveled by single-cell isotope probing and targeted metagenomics. Proc. Natl. Acad. Sci. 119, e2201473119 (2022).
Rojo, F. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34, 658–684 (2010).
Shaffer, J. P. et al. Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nat. Microbiol. 7, 2128–2150 (2022).
Drenkard, E. & Ausubel, F. M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743 (2002).
Lorusso, A. B., Carrara, J. A., Barroso, C. D. N., Tuon, F. F. & Faoro, H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int J. Mol. Sci. 23, 15779 (2022).
Freschi, L. et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol. Evol. 11, 109–120 (2019).
Hacquard, S. et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe 17, 603–616 (2015).
Oldstone, M. B. A. Molecular mimicry, microbial infection, and autoimmune disease: evolution of the concept. Mol. Mimicry: Infect.-Induc. Autoimmune Dis. 296, 1–17 (2005).
Zelles, L., Bai, Q. Y., Beck, T. & Beese, F. Signature fatty acids in phospholipids and lipopolysaccharides as indicators of microbial biomass and community structure in agricultural soils. Soil Biol. Biochem 24, 317–323 (1992).
Pieterse, C. M. J. et al. Induced systemic resistance by beneficial microbes. Annu Rev. Phytopathol. 52, 347–375 (2014).
Graham, D. B. & Xavier, R. J. Conditioning of the immune system by the microbiome. Trends Immunol. 44, 499–511 (2023).
Gauthier, A. E., Rotjan, R. D. & Kagan, J. C. Lipopolysaccharide detection by the innate immune system may be an uncommon defence strategy used in nature. Open Biol. 12, 220146 (2022).
Wolf, A. J. & Underhill, D. M. Peptidoglycan recognition by the innate immune system. Nat. Rev. Immunol. 18, 243–254 (2018).
Cloud-Hansen, K. A. et al. Breaching the great wall: peptidoglycan and microbial interactions. Nat. Rev. Microbiol. 4, 710–716 (2006).
Pino-Otín, M. R. et al. Impact of eight widely consumed antibiotics on the growth and physiological profile of natural soil microbial communities. Chemosphere 305, 135473 (2022).
Zhang, L., Liu, C., Jiang, Q. & Yin, Y. Butyrate in Energy Metabolism: There Is Still More To Learn. Trends Endocrinol. Metab. 32, 159–169 (2021).
Chen, Q.-L., Cui, H.-L., Su, J.-Q., Penuelas, J. & Zhu, Y.-G. Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 24, 530–541 (2019).
De Filippis, F., Pasolli, E. & Ercolini, D. The food-gut axis: lactic acid bacteria and their link to food, the gut microbiome and human health. FEMS Microbiol. Rev. 44, 454–489 (2020).
Scaccia, N., Vaz-Moreira, I. & Manaia, C. M. The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 26, 1213–1226 (2021).
Brito, I. L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 19, 442–453 (2021).
Jia, W.-L. et al. Antibiotics in soil and water: Occurrence, fate, and risk. Curr. Opin. Environ. Sci. Health 32, 100437 (2023).
Yu, X. et al. Microplastics exacerbate co-occurrence and horizontal transfer of antibiotic resistance genes. J. Hazard Mater. 451, 131130 (2023).
Wang, F. et al. Antibiotic resistance in the soil ecosystem: A One Health perspective. Curr. Opin. Environ. Sci. Health 20, 100230 (2021).
Thanner, S., Drissner, D. & Walsh, F. Antimicrobial resistance in agriculture. mBio 7, 10–1128 (2016).
Wen, X. et al. Community coalescence and plant host filtering determine the spread of tetracycline resistance genes from pig manure into the microbiome continuum of the soil–plant system. Microbiol. Res. 284, 127734 (2024).
Custer, G. F., Bresciani, L. & Dini-Andreote, F. Toward an integrative framework for microbial community coalescence. Trends Microbiol. 32, 241–251 (2024).
Sun, J. et al. Environmental remodeling of human gut microbiota and antibiotic resistome in livestock farms. Nat. Commun. 11, 1427 (2020).
Hall, J. P. J., Brockhurst, M. A. & Harrison, E. Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philos. Trans. R. Soc. B: Biol. Sci. 372, 20160424 (2017).
Costa, O. Y. A., Raaijmakers, J. M. & Kuramae, E. E. Microbial extracellular polymeric substances: ecological function and impact on soil aggregation. Front. Microbiol. 9, https://doi.org/10.3389/fmicb.2018.01636 (2018).
Hidalgo-Cantabrana, C. et al. Genomic overview and biological functions of Exopolysaccharide Biosynthesis in Bifidobacterium spp. Appl Environ. Microbiol. 80, 9–18 (2014).
Rossi, F., Rizzotti, L., Felis, G. E. & Torriani, S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 42, 232–243 (2014).
Vandana & Das, S. Genetic regulation, biosynthesis and applications of extracellular polysaccharides of the biofilm matrix of bacteria. Carbohydr. Polym. 291, 119536 (2022).
Laanto, E., Hoikkala, V., Ravantti, J. & Sundberg, L.-R. Long-term genomic coevolution of host-parasite interaction in the natural environment. Nat. Commun. 8, 111 (2017).
Caballero-Flores, G., Pickard, J. M. & Núñez, G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol. 21, 347–360 (2023).
Mullineaux-Sanders, C., Suez, J., Elinav, E. & Frankel, G. Sieving through gut models of colonization resistance. Nat. Microbiol. 3, 132–140 (2018).
Lawley, T. D. & Walker, A. W. Intestinal colonization resistance. Immunology 138, 1–11 (2013).
Spragge, F. et al. Microbiome diversity protects against pathogens by nutrient blocking. Science (1979) 382, eadj3502 (2023).
Karita, Y., Limmer, D. T. & Hallatschek, O. Scale-dependent tipping points of bacterial colonization resistance. Proc. Natl. Acad. Sci. 119, e2115496119 (2022).
de Boer, M. et al. Control of Fusarium wilt of radish by combining Pseudomonas putida strains that have different disease-suppressive mechanisms. Phytopathology 93, 626–632 (2003).
Van Elsas, J. D. et al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. 109, 1159–1164 (2012).
Mawarda, P. C., Lakke, S. L., van Elsas, J. D. & Salles, J. F. Temporal dynamics of the soil bacterial community following Bacillus invasion. iScience 25, 104185 (2022).
Xing, J. et al. The legacy of bacterial invasions on soil native communities. Environ. Microbiol 23, 669–681 (2021).
Culp, E. J. & Goodman, A. L. Cross-feeding in the gut microbiome: Ecology and mechanisms. Cell Host Microbe 31, 485–499 (2023).
Giri, S. et al. Metabolic dissimilarity determines the establishment of cross-feeding interactions in bacteria. Curr. Biol. 31, 5547–5557 (2021).
Schäfer, M. et al. Metabolic interaction models recapitulate leaf microbiota ecology. Science (1979) 381, eadf5121 (2023).
Oña, L. et al. Obligate cross-feeding expands the metabolic niche of bacteria. Nat. Ecol. Evol. 5, 1224–1232 (2021).
Martínez-Romero, E. et al. We and herbivores eat endophytes. Micro Biotechnol. 14, 1282–1299 (2021).
Berlemont, R. & Martiny, A. C. Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 12, e1005300 (2016).
Lu, X., Heal, K. R., Ingalls, A. E., Doxey, A. C. & Neufeld, J. D. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 14, 53–66 (2020).
Sokolovskaya, O. M., Shelton, A. N. & Taga, M. E. Sharing vitamins: Cobamides unveil microbial interactions. Science (1979) 369, eaba0165 (2020).
Louis, P. & Flint, H. J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8 (2009).
Carmody, R. N., Varady, K. & Turnbaugh, P. J. Digesting the complex metabolic effects of diet on the host and microbiome. Cell 187, 3857–3876 (2024).
Philippot, L., Raaijmakers, J. M., Lemanceau, P. & Van Der Putten, W. H. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol 11, 789–799 (2013).
Cordovez, V., Dini-Andreote, F., Carrión, V. J. & Raaijmakers, J. M. Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol 73, 69–88 (2019).
Fernandez, A. R., Sáez, A., Quintero, C., Gleiser, G. & Aizen, M. A. Intentional and unintentional selection during plant domestication: herbivore damage, plant defensive traits and nutritional quality of fruit and seed crops. N. Phytologist 231, 1586–1598 (2021).
Pérez-Jaramillo, J. E., Carrión, V. J., de Hollander, M. & Raaijmakers, J. M. The wild side of plant microbiomes. Microbiome 6, 1–6 (2018).
Oyserman, B. O. et al. Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nat. Commun. 13, 3228 (2022).
Serrano, K. & Bezrutcyzk, M. Genome to gut: crop engineering for human microbiomes. Nat. Rev. Microbiol. 21, 132 (2023).
Soto-Giron, M. J. et al. The edible plant microbiome represents a diverse genetic reservoir with functional potential in the human host. Sci. Rep. 11, 24017 (2021).
Costea, P. I. et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16 (2018).
Larzul, C. et al. Driving gut microbiota enterotypes through host genetics. Microbiome 12, 116 (2024).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Siezen, R. J. & Kleerebezem, M. The human gut microbiome: are we our enterotypes?. Micro Biotechnol. 4, 550 (2011).
Frioux, C. et al. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe 31, 1111–1125 (2023).
Rao, X., Shao, Y. & Wu, H. Fishing for obesity-related gut microbiome enterotype. Cell Host Microbe 32, 1209–1211 (2024).
Chen, Q.-L. et al. Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol. Lett. 1, 3–13 (2019).
Han, B. et al. The source, fate and prospect of antibiotic resistance genes in soil: A review. Front Microbiol 13, 976657 (2022).
Yang, X. et al. High microbiome diversity constricts the prevalence of human and animal pathogens in the plant rhizosphere worldwide. One Earth. 7, 1301–1312 (2024).
Brennan, F. P. et al. Harnessing agricultural microbiomes for human pathogen control. ISME Commun. 2, 44 (2022).
Holkem, A. T., Silva, M. P. & Favaro-Trindade, C. S. Probiotics and plant extracts: A promising synergy and delivery systems. Crit. Rev. Food Sci. Nutr. 63, 9561–9579 (2023).
Etalo, D. W., Jeon, J.-S. & Raaijmakers, J. M. Modulation of plant chemistry by beneficial root microbiota. Nat. Prod. Rep. 35, 398–409 (2018).
Petrovic, B., Sękara, A. & Pokluda, R. Biofertilizers enhance quality of onion. Agronomy 10, 1937 (2020).
de Matos Nascimento, A. et al. Biofertilizer application on corn (Zea mays) increases the productivity and quality of the crop without causing environmental damage. Water Air Soil Pollut. 231, 1–10 (2020).
Molla, A. H., Manjurul Haque, M., Amdadul Haque, M. & Ilias, G. N. M. Trichoderma-enriched biofertilizer enhances production and nutritional quality of tomato (Lycopersicon esculentum Mill.) and minimizes NPK fertilizer use. Agric. Res. 1, 265–272 (2012).
Sharma, M. et al. AMF and PSB applications modulated the biochemical and mineral content of the eggplants. J. Basic Microbiol. 62, 1371–1378 (2022).
Ye, S. et al. A critical review on interplay between dietary fibers and gut microbiota. Trends Food Sci. Technol. 124, 237–249 (2022).
Kumari, M. et al. Fostering next-generation probiotics in human gut by targeted dietary modulation: An emerging perspective. Food Res. Int. 150, 110716 (2021).
Wan, M. L. Y., Co, V. A. & El-Nezami, H. Dietary polyphenol impact on gut health and microbiota. Crit. Rev. Food Sci. Nutr. 61, 690–711 (2021).
Von Hertzen, L., Hanski, I. & Haahtela, T. Natural immunity: biodiversity loss and inflammatory diseases are two global megatrends that might be related. EMBO Rep. 12, 1089–1093 (2011).
Ottman, N. et al. Soil exposure modifies the gut microbiota and supports immune tolerance in a mouse model. J. Allergy Clin. Immunol. 143, 1198–1206 (2019).
Rook, G. A. Regulation of the immune system by biodiversity from the natural environment: an ecosystem service essential to health. Proc. Natl. Acad. Sci. 110, 18360–18367 (2013).
Brame, J. E., Liddicoat, C., Abbott, C. A. & Breed, M. F. The potential of outdoor environments to supply beneficial butyrate-producing bacteria to humans. Sci. Total Environ. 777, 146063 (2021).
Yeung, F. et al. Altered immunity of laboratory mice in the natural environment is associated with fungal colonization. Cell Host Microbe 27, 809–822 (2020).
Roslund, M. I. et al. Biodiversity intervention enhances immune regulation and health-associated commensal microbiota among daycare children. Sci. Adv. 6, eaba2578 (2020).
Acknowledgements
The contribution of H.M. was funded by the National Natural Science Foundation of China (32201402) and Chinese Scholarship Council (202206205006). The contributions of D.C. and J.M.R were funded by the Dutch Research Council (NWO/OCW), as part of the MiCRop Consortium program, Harnessing the second genome of plants (Grant number 024.004.014).
Author information
Authors and Affiliations
Contributions
All authors contributed to the manuscript’s conception. H.M. led the writing, with valuable revisions and input from J.M.R. and D.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, H., Cornadó, D. & Raaijmakers, J.M. The soil-plant-human gut microbiome axis into perspective. Nat Commun 16, 7748 (2025). https://doi.org/10.1038/s41467-025-62989-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62989-z