Abstract
Satellite DNA comprises ~11% of the mouse genome and is primarily located in centromeres and pericentromeres. We present comprehensive genomic and epigenomic maps of these regions utilizing Hifiasm assemblies, CUT&RUN-seq, DNA methylation analysis, and RNA-seq alongside recent mouse telomere-to-telomere assembly drafts. We show that 120-mer Minor satellites (MiSats) occupy core centromeres, while MiSat length variants localize at centromere-pericentric junctions. Pericentromeres contain mostly homogeneous Major satellites (MaSats), with divergent MaSats concentrated near pericentric non-satellite repeat islands and pericentric-chromosomal junctions. Most centromeres contain higher-order repeats (HORs), with shorter HORs more common. Centromeres are hypomethylated compared to pericentromeres, and transcripts are detected from a small subset of satellites with lower DNA methylation. CENP-A is highly enriched at 120-mer MiSats but reduced at MiSat length variants. Homogeneous MaSats are enriched with H3K9me3, whereas divergent MaSats are associated with H3K9me3 and H3K27me3. Finally, the density and type of satellite sequence motifs correlate with chromatin signatures.
Similar content being viewed by others
Introduction
Repetitive sequences are a fundamental component of eukaryotic genomes, comprising ~50% of the mammalian genome1. These sequences are crucial for maintaining the structural and functional integrity of the genome, characterized by their high copy number, and include various forms such as transposons, tandem repeats, and satellite DNA2,3,4. Satellite DNA consists of tandemly repeated sequences often located in specific chromosomal regions, such as centromeres, pericentromeres, and telomeres, where they play essential roles in maintaining chromosomal stability, facilitating chromosome segregation during cell division, and protecting chromosome ends2,4,5,6,7,8. Satellite DNA constitutes ~11% of the mouse genome and ~6–8% of the human genome4,9,10. Centromeric satellites are present at the primary constriction, where they assemble specialized nucleosomes in which canonical histone H3 is replaced by its variant called centromeric protein A (CENP-A)11,12. The CENP-A chromatin lays the foundation for the formation of the kinetochore, a protein complex critical for the attachment of chromosomes to spindle microtubules during mitosis and meiosis8,9,13. Pericentric regions, which flank the centromeres, are also rich in satellite sequences and assemble constitutive heterochromatin marked by histone 3 lysine 9 trimethylation (H3K9me3)14,15,16. Pericentric heterochromatin is associated with cohesin, which must be protected from degradation until the onset of anaphase to prevent premature separation of sister chromatids and ensure correct chromosome segregation16,17. Despite their functional importance, assembling these satellite regions has been challenging due to their highly repetitive nature. Recent advances in long-read sequencing technologies have led to significant improvements in genome assemblies, resulting in a complete, fully annotated end-to-end assembly for the human genome.
In humans, centromeres are composed of α-satellite DNA, which consists of tandem repeats of 171 base pairs forming higher-order repeat structures crucial for centromere function8,13,18. These α-satellites can either exist as arrays of monomers, which are inactive and pushed toward pericentric regions, or as higher-order repeats (HORs), in which a set number of monomers (ranging from 2 to 34) are repeated several times and can occur in active or inactive regions of centromeres8,19,20. Human pericentric regions contain various satellite families (e.g., HSATI, HSATII, HSATIII, β-satellites), each distinct in their DNA sequences4,14,21,22. Human centromeric and pericentric regions exhibit high global and local variations, with repeat units in a given satellite class sharing ~60–100% sequence similarity9,13,23. In addition, the human genome contains several chromosome-specific α-satellite sequences and higher-order repeat structures9,23. These sequence variations have allowed the Telomere-to-Telomere (T2T) consortium to assemble a complete, fully annotated, gapless sequence, providing a comprehensive map of human centromeric, pericentric, and telomeric regions9,23.
Unlike humans, where centromeres exhibit high variation in α-satellites and pericentric regions contain multiple satellite families, mouse centromeres are primarily composed of Minor Satellites (MiSats), characterized by 120-base-pair repeat units, and pericentric regions consist of Major Satellites (MaSats), characterized by 234-base-pair repeat units4,10,15,24,25,26,27. Mouse centromeric and pericentric regions are highly homogeneous, and despite being a widely used model organism for genetic and biomedical research, the mouse genome still lacks a fully characterized and annotated map for these repetitive regions. Recently, we characterized the local arrangement of centromeric and pericentric satellites and the enrichment pattern of associated chromatin on PacBio HiFi reads (~15 Kb long)24. We demonstrated the presence of considerable sequence variations within both centromeres and pericentric mouse satellite regions24,28. Similar sequence variations were identified at mouse centromeres at both local and global scales by analyzing short-read sequencing data and MiSat-containing GenBank clones28. Although the extent of these variations is much less than that of human satellites, they can potentially contribute to the assembly of more extended maps for mouse centromere and pericentric regions. More recently, two independent drafts of the mouse T2T assembly from mouse embryonic stem cells derived from the C57BL/6 J strain have become available29,30. These drafts provide timely resources for a comprehensive characterization of mouse satellite regions.
In this study, we created detailed, fully annotated genomic and epigenomic maps for the centromeric, pericentric, and satellite junctions using the recently available mouse T2T assembly drafts in conjunction with our primary representative Hifiasm contigs generated from PacBio HiFi reads obtained from the kidney tissues of the C57BL/6 J strain. We generated fully annotated, high-resolution genomic maps of centromeres and pericentromeres for all mouse chromosomes. We found the evidence for the presence of several HORs on most centromeres. Additionally, we have generated high-resolution chromatin profiling data for CENP-A, sequence-specific centromeric protein CENP-B, H3K9me3, and H3K27me3 chromatin to understand the enrichment patterns in these satellite regions. We also analyzed DNA methylation profiles using kinetics tags of PacBio HiFi reads and found that although mouse centromeres lack clear centromere-depleted regions (CDRs), they exhibit significantly lower DNA methylation levels as compared to the flanking pericentric regions. Furthermore, we generated and analyzed RNA sequencing data from C57BL/6 J mouse tissues to decipher the transcriptional readouts from these regions. Our findings offer critical insights into satellite biology and will play a key role in the development of a fully annotated mouse T2T assembly, similar to the current human T2T assembly.
Results
Fully annotated genomic maps of mouse centromeric and pericentric regions
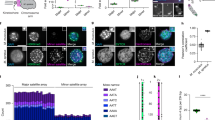
To generate fully annotated genomic maps of mouse centromeres and pericentromeres, we leveraged recent mouse T2T assembly drafts in conjunction with the high accuracy of the minimally processed Hifiasm assemblies that retain the high local sequence accuracy of PacBio HiFi reads31,32, which is critical for mouse satellites that are prone to assembly artifacts due to their high sequence homogeneity and abundance. We analyzed two recent mouse T2T assembly drafts—one from a mouse haploid androgenic embryonic stem cell line (mhaESC)29 and another from mESCs of a C57BL/6 J × CAST/EiJ cross (GCA_964188535.1)30. Both T2T assemblies were generated using a hybrid approach that integrates PacBio HiFi, Oxford Nanopore ultralong (ONT), Illumina short-read, and Hi-C sequencing data, followed by two rounds of assembly polishing29,30. To generate minimally processed contigs, we generated HiFi sequencing data (5 million reads, with an average read length of ~9.5 Kb and 17.6X genome coverage) on the PacBio Revio sequencing platform from the kidney tissue of a male inbred C57BL/6 J mouse. The resulting HiFi consensus reads were assembled using the Hifiasm assembler33. For centromeric analysis, we also generated a Hifiasm assembly from a publicly available female C57BL/6 J HiFi dataset (4 million reads, with an average read length of ~16.4 Kb and 24.6X genome coverage)34. The chromosomal ends of mouse T2T and our Hifiasm contigs spanning satellite regions were annotated using RepeatMasker with the Mus musculus repeat database and NCBI BLAST, employing satellite consensus sequences as databases. We analyzed the distribution and arrangement of centromeric MiSats, pericentric MaSats, centromeric-telomeric junctions, and pericentric-chromosomal junctions in the mouse T2T assembly and our Hifiasm contigs (Fig. 1A, B, Supplementary Figs. 1–5). On average, satellite regions in the mESC T2T assembly were longer than those in the mhaESC, whereas centromeric regions were longer in the mhaESC T2T assembly compared to the mESC T2T assembly (Fig. 1C).
A Chromosome-wise linear maps showing arrangement of satellite and non-satellite repeats at chromosomal termini of mhaESC (Left)29 and mESC (Right, GCA_964188535.1)30 T2T assemblies. B Linear maps of annotated centromeres for each chromosome, including 50 Kb of MaSat sequences from the pericentric side and the full TLC/telomeric repeat containing region from the telomeric side, (C) Length distribution of satellite regions and centromeres displayed as violin plots with overlaid box plots. In the box plots, the center line represents the median, the box bounds correspond to the interquartile range (IQR; 25th to 75th percentile), and the whiskers extend to the most extreme data point within that range. Individual points represent centromere and satellite lengths from each chromosome. The violin shape shows the kernel density estimate of the data distribution. D Pie charts showing the proportional composition of MiSat subtypes within the centromeres, and (E) Pie charts showing the proportion of chromosomes with fragmented centromeres in mhaESC and mESC T2T assemblies. All maps are drawn to scale. Color keys for repeat types are provided at the bottom. ND not detected. Source data are provided with this paper.
Centromeres
The sizes of centromeres ranged from 480 Kb to 2 Mb in the mhaESC T2T assembly, and from 600 Kb to 2.3 Mb in the mESC T2T assembly (Fig. 1B, C). The mESC T2T lacks centromeres on chromosomes 4, 7, and 1830 (Fig. 1B). Some centromeres appeared as three distinct fragments, interrupted by blocks of MaSats (Fig. 1A, B). Cen2 was divided into three regions (Cen2A, Cen2B, and Cen2C) in both T2T assemblies, while Cen9 and Cen16 showed similar fragmentation into three parts each (Cen9A/B/C and Cen16A/B/C), but only in the mhaESC assembly (Fig. 1B). In addition to the predominant 120-mer monomeric MiSats, mouse centromeres contain MiSat length variants arranged as arrays of 112-mers or 112-64-dimers24. While most T2T centromeres were predominantly composed of 120-mer MiSats, we identified two discrete ~224–236 Kb 112-64-dimeric MiSat arrays situated near the centromeric-pericentric junctions. However, these dimeric arrays were placed on different chromosomes in the two assemblies- chromosomes 6 and X in mhaESC, and chromosomes 2 and 5 in mESC (Fig. 1B). Additionally, Cen2B contained low-density 112-mer MiSats distributed across a 730 Kb region in both assemblies (Fig. 1B).
In our Hifiasm assemblies derived from male C57BL/6 J mouse kidney tissue, we identified 14 centromeric contigs that each contained over 5 Kb of MiSats (Supplementary Fig. 1A). Seven of these contigs spanned from a telomeric end to a pericentric end, with centromere lengths ranging from 690 Kb to 850 Kb. In the female C57BL/6 J Hifiasm assembly, we identified 18 centromeric contigs that each contained over 5 Kb of MiSats (Supplementary Fig. 1B), and three of these contigs contained both telomeric and pericentric junctions, with centromere lengths ranging from 712 Kb to 1.02 Mb. Both male and female Hifiasm assemblies included a contig that captured the 236 Kb-long 112-64-dimeric MiSat array flanked by the pericentric MaSats on one side. However, the second dimeric array found in the T2T assembly was missing in these Hifiasm assemblies, likely due to lower sequencing coverage. We also confirmed the presence of the low-density 112-mer MiSat region corresponding to T2T Cen2B in Hifiasm assemblies, but the density of 112-mer MiSats was higher in Hifiasm contigs than in T2T Cen2B (Supplementary Fig. 1A–C and 2). We also generated a Hifiasm assembly using the PacBio HiFi sequencing data (56 million reads, 65X genome coverage and an average read length of ~7.5 Kb) that was used for the mhaESC T2T assembly29. Interestingly, this Hifiasm version showed a higher 112-mer MiSat density than the corresponding Cen2B region in the mhaESC T2T assembly (Supplementary Figs. 1C and 2), indicating that satellite variant density varies across different assembly methods.
In the male C57BL/6 J Hifiasm assembly, we identified a 90 Kb centromeric sequence on a contig with genes known to be present on the centromeric end of Y-chromosomes (Supplementary Fig. 1A). This 90 Kb Y-centromere sequence is similar to the mouse Y-centromere previously mapped to a 90 Kb region using a BAC clone35. Interestingly, the Y-centromere lacked flanking pericentric MaSats on the ~1.2 Mb contig (Supplementary Fig. 1A). The ~166 Kb BAC clone in which the mouse Y-centromere was initially identified also lacked flanking MaSats35. Such a lack of MaSats near Y-centromeres has also been observed during the cytological visualization of mouse male chromosome spreads probed with MaSats35,36.
Centromere-telomere junctions
In mice, the centromere-telomere junctions are occupied by telocentric (TLC) satellite repeats composed of 146 bp monomers and L1 LINE elements30,37,38. These TLC satellites are flanked by an L1 LINE element, which is followed by canonical telomeric simple repeats (TTAGGG)n. These telomeric features were recently confirmed to be present at the ends of most chromosomes in the mESC T2T assembly30. We independently confirmed the presence of TLC satellites, L1 LINE elements, and telomeric simple repeats flanking most mhaESC T2T centromeres and Hifiasm contigs containing centromere-telomere junctions (Fig. 1B, Supplementary Fig. 3A). In the mhaESC T2T assembly, telomeric repeat-containing arrays extending up to ~80 Kb were embedded within the centromeres of four chromosomes (Cen9, 10, 12, and 16) and at one centromeric-pericentric junction (Cen4) (Fig. 1B, Supplementary Fig. 3A–C). Each of these internal telomeric repeat arrays was flanked by TLC satellites and L1 LINE elements, resembling terminal telomeres. However, such interstitial telomeres were absent in the mESC T2T assembly, both male and female C57BL/6 J Hifiasm assemblies, and the mhaESC Hifiasm assembly (Supplementary Figs. 1A–C and 3A–C), indicating possible assembly artifacts in the mhaESC T2T assembly across these regions.
Pericentromeres
The size of pericentric regions ranged from 2.2 Mb on chromosome 4 to 17.5 Mb on chromosome 7 in the mhaESC and from 4 Mb on chromosome 8 to 23 Mb on chromosome 17 in the mESC assembly (Fig. 1A). We have previously shown that ~10% of MaSat-containing HiFi long reads from the C57BL/6 J strain contain divergent MaSat units that share <75% sequence similarity to the MaSat consensus sequence24. In both mhaESCT T2T and Hifiasm assemblies, divergent MaSats were either found as continuous blocks or scattered as monomers within the homogeneous MaSat regions (Fig. 1A, B and Supplementary Fig. 4A–C). Except for mhaESC Cen9C, Cen10, Cen12 and Cen16C, which were located next to a centromeric-pericentric junction with an island of non-satellite repeats, all mhaESC T2T centromeres and Hifiasm MiSat contigs were flanked by homogeneous MaSats (Fig. 1B and Supplementary Fig. 1A–C). In the male C57BL/6 J Hifiasm assembly, we identified 734 continuous MaSat-containing pericentric contigs with MaSat arrays ranging up to ~1.4 Mb (Supplementary Fig. 4A). This large number of contigs is expected because MaSats make up ~10% of the mouse genome4. We analyzed the length distribution of male C57BL/6 J Hifiasm satellite contigs and found that homogeneous MaSat contigs had the shortest average length (Supplementary Fig. 4D). In contrast, pericentric-chromosomal junction contigs were the largest (up to 76 Mb), as expected, because the presence of unique sequences in the chromosomal arms allows for the assembly of longer contigs. Additionally, divergent MaSat and centromeric MiSat Hifiasm contigs were longer than the homogeneous MaSat contigs (Supplementary Fig. 4D), likely reflecting the high sequence identity among the satellite units within the homogeneous parts of the pericentric regions.
Overall, the centromeric and pericentric features are broadly comparable between the mhaESC and mESC T2T assemblies. For subsequent analyses, we focused on the mhaESC assembly for chromosome-level comparisons and the male C57BL/6 J Hifiasm assembly, which includes the CenY, enabling us to generate annotations of all mouse centromeres.
We detected inversion events in satellite regions by examining the directional changes in the satellite sequences in T2T and Hifiasm assemblies. Inversions were highly frequent in centromeric regions, with 19 out of 20 mhaESC T2T centromeres and 9 out of 14 male C57BL/6 J Hifiasm centromeric contigs exhibiting these inversions (Supplementary Fig. 5A, B). In contrast, inversion events were less frequent in pericentric regions, appearing in 10 out of 20 T2T pericentromeres and 14 out of 734 Hifiasm MaSat contigs (Supplementary Fig. 5A, B). While certain pericentric regions, especially divergent MaSat regions, exhibit locally higher numbers of inversions, inversion events are overall more frequent and widespread in centromeres (Supplementary Fig. 5B). The frequent occurrence of inversion events in centromeric regions suggests that they are more prone to chromosomal rearrangements than flanking pericentric regions.
Centromeres and pericentromeres exhibit distinct patterns of sequence identity
We analyzed sequence identity across satellite regions by creating StainedGlass heatmaps39. We found that the highest identity occurs in centromeres, which decreases in pericentromeres, and further decreases in the pericentric-chromosomal arm junctions (Fig. 2A and Supplementary Figs. 6A, B and 7A, B). The satellite sequence identity pattern varied among chromosomes (Fig. 2A). Within centromeres, the 112-64-dimeric MiSat arrays exhibited the highest sequence homology (95–100%), followed by 120-mer MiSat arrays (75–100%), and then the Y centromeres (70-80%) (Fig. 2B, C). In pericentromeres, high-density divergent regions showed low percent identity compared to high-density homogeneous MaSat regions, which were still less homogeneous than centromeric MiSats (Fig. 2C and Supplementary Figs. 6A, B and 7A, B). Interestingly, the most homogeneous MaSats were found on chromosomes 6 and X, which contain the 236 Kb 112-64-dimeric MiSat array adjacent to pericentric regions (Fig. 2A–C and Supplementary Figs. 6A, B and 7A–C).
A StainedGlass heatmaps representing the percent identity of self-alignments within the centromeric, pericentric, and pericentric-chromosomal arm junctions of selected mhaESC T2T chromosomes. The heatmap color scale reflecting sequence identity is also shown. StainedGlass heatmaps for the remaining chromosomes are presented in Supplementary Fig. 6A. B StainedGlass heatmaps illustrating identity across the centromeric regions of selected mhaESC T2T chromosomes. StainedGlass heatmaps for the centromeres of the remaining chromosomes are presented in Supplementary Fig. 6B. C Percentage sequence identity among MiSat units (Top) and MaSat units (Bottom) across various mhaESC T2T chromosomes. For chromosomes 2, 6, and X, the percentage identity is calculated separately for 120-mer monomeric (Mon) and 112-64-dimeric (Dim) MiSats in centromeric regions (Top). For chromosomes 2, 4, 6, and X, the percentage identity is calculated separately for homogeneous (Hom) and divergent (Div) MaSats in pericentric regions (Bottom). The Y-centromere sequence is taken from the male C57BL/6 J Hifiasm assembly. Source data are provided with this paper.
The majority of mouse centromeres contain higher-order repeats (HORs)
Previously, the identification of HORs in mouse centromeres was limited to the Y centromere35. We investigated the presence of HORs in all mouse centromeres using CENdetectHOR40. As a proof of concept, we successfully identified known HORs in human centromeres 1, 4, and 13 using CENdetectHOR (Supplementary Fig. 8). Next, using CENdetectHOR, we detected HORs on all T2T centromeres except Cen3 and Cen4 (Fig. 3A). Interestingly, Cen3 and Cen4 are the most homogeneous mouse centromeres (Fig. 2B, C). The HORs are labeled H2 to H45, where the numbers represent the number of monomeric units within the given HOR. We included CenY from Hifiasm assembly in both mhaESC T2T and Hifiasm HOR analysis. CENdetectHOR successfully identified known HORs on the Y-centromere (H7, H13, and H20) and 112-64-dimeric MiSats on Cen6 and CenX (Figs. 1A, B and 3A). As expected due to the highly homogeneous nature of MiSats, mouse centromeric HORs were less abundant than those in humans. Moreover, HORs composed of shorter repeat units, such as dimers (H2) and trimers (H3), were more common than those composed of longer ones (Fig. 3A). Except for CenY HORs and the length variant 112-64-dimeric HORs (LVH2) on Cen6 and CenX, we did not observe any notable chromosome-specific HORs (Fig. 3A). However, we found that certain HORs, such as H3 trimers, were predominantly located at the telomeric ends of the centromeres, whereas H2 dimers were mainly located in the second half of the centromeres away from telomeric ends (Fig. 3A). Similarly, H29 and H15 HORs were located toward telomeric and pericentric ends, respectively. We analyzed nucleotide variations from the consensus MiSat on a few HORs, specifically shorter HORs (H2, LVH2, H3, H4, H5, H6, and H7). We found that, except for 112-64-dimers (LVH2), every HOR contained one or more monomeric units in which bases flanking the CENP-B box were mutated (Fig. 3B). The phylogenetic clustering revealed that CenY HORs were most distantly related to those found in other centromeres (Fig. 3C). Among the non-CenY HORs, the length variant 112-64-dimers (LVHs) were distinctly separated from the remaining HORs, which appeared to cluster into two closely related groups without a clear discernible pattern (Fig. 3D). Similar patterns of HOR formation were validated in the male C57BL/6 J Hifiasm contigs (Supplementary Fig. 9A–C).
A Higher order repeats (HORs) identified by CENdetectHOR in mhaESC T2T centromeres. The Y-centromere sequence is taken from the male C57BL/6 J Hifiasm assembly. B Nucleotide variations in MiSats within specified HORs. The HORs are labeled H2 to H45, where the numbers represent the number of monomeric units within the given HOR. Nucleotide variations are shown for HORs containing 2–7 monomeric units (H2–H7). The blue bidirectional arrow indicates the location of the CENP-B box within the MiSat unit. The red vertical lines denote base changes from the consensus MiSat sequence. C Phylogenetic tree depicting the relationships among various HORs. D Phylogenetic tree depicting the relationships between HORs from different centromeres, excluding the Y-centromere.
Non-satellite repeats exist as individual elements in centromeres and expand into clustered islands across pericentromeres
Mouse centromeric and pericentric satellite regions are interspersed with non-satellite repeat sequences similar to those found in human, Drosophila, and plant satellites24,29,41,42,43,44,45,46,47. We determined the density and arrangement of non-satellite repetitive sequences, including LTR retrotransposons, non-LTR retrotransposons such as LINEs (long-interspersed nuclear elements) and SINEs (short-interspersed nuclear elements), as well as simple repeats in satellite regions in mhaESC T2T and male C57BL/6 J Hifiasm assemblies. Non-satellite repeats appeared either as individual elements or as clusters forming islands in both mhaESC and Hifiasm assemblies (Fig. 4A, B and Supplementary Fig. 10A). Individual elements were found in both centromeric and pericentric regions, while islands were only present in pericentric and flanking regions (Fig. 4A, B). Each island measured up to ~200 Kb in length (Fig. 4A, B and Supplementary Fig. 10A). These islands varied in both number and size by chromosome (Fig. 4C), with chromosome 1 exhibiting the highest abundance (7 islands) and total size (>2 Mb), whereas chromosomes 4, 5, and X showed minimal island content. In the C57BL/6 J Hifiasm assembly, we identified 10 MaSat contigs interspersed with a ~ 100-200 Kb non-satellite islands (Supplementary Fig. 4), as well as an additional 10 MaSat contigs that contained a non-satellite repeat array (<100 Kb) at one end. Additionally, the density of simple repeats, retrotransposons, LINEs, and SINEs increased gradually from centromeric regions to pericentric-chromosomal arm junctions (Fig. 4D and Supplementary Fig. 10B).
A Satellite regions from representative mhaESC T2T chromosomes showing LTRs (long terminal repeats), LINEs (long interspersed nuclear elements), SINEs (short interspersed nuclear elements), and simple repeats as individually inserted elements. B Satellite regions from representative mhaESC T2T chromosomes showing the distribution of LTRs, LINEs, SINEs, and simple repeats clustered as islands. C The number and total size of non-satellite islands on mhaESC T2T satellite regions. D The density of retrotransposons (IAP and non-IAP LTRs), SINEs, LINEs, and simple repeats across satellite regions in the T2T assembly. For A and B panels, maps are drawn to scale, and a zoomed-in view across a smaller region is shown. Source data are provided with this paper.
Divergent MaSats are preferentially localized near non-satellite islands and pericentric-chromosomal arm junctions
We observed large blocks of high-density divergent MaSats near chromosomal arm junctions (Fig. 1A and Supplementary Figs. 4 and 11). We plotted the distribution of divergent MaSats across the entire length of pericentric regions from the centromeric end to the pericentric-chromosomal arm junction for every T2T chromosome (Fig. 5A). We found that the density of divergent MaSats increases from the centromeric-pericentric junctions to the pericentric ends toward the chromosomal arms (Fig. 5A). Furthermore, long stretches of high-density divergent MaSats were also found near pericentric non-satellite repeat islands (Fig. 5A). In male C57BL/6 J Hifiasm contigs, out of the 734 MaSat-containing contigs, 19 spanned pericentric-chromosomal arm junctions, each containing several Mb long chromosomal arm sequences flanking MaSat arrays (Supplementary Figs. 4A and 11). We mapped annotated genes from chromosomal ends of the GRCm39 genome assembly to these contigs, which allowed us to assign 17 of the 19 pericentric-chromosomal junction contigs to specific chromosomes (Supplementary Fig. 11). Among the 19 pericentric-chromosomal junction contigs, 16 contained high-density divergent MaSat sequences localized adjacent to the chromosomal arm regions (Supplementary Fig. 11). Furthermore, the density of divergent MaSat units was highest in the pericentric-chromosomal junction contigs, followed by the pericentric non-satellite repeat island contigs (Fig. 5C).
A Distribution of divergent MaSats and non-satellite repeat islands on mhaESC T2T pericentric regions. Each dot represents the presence of a divergent MaSat unit or island at a specific location. B The density of divergent MaSats in contigs from centromeric-pericentric regions, pericentric regions alone, pericentric regions containing non-satellite islands, and pericentric-chromosomal arm junctions is displayed as the number of divergent MaSat units per 50 Kb. For pericentric regions alone, 15 random contigs are included in the analysis. C Distribution of indicated MaSat-containing contigs based on the density of divergent MaSat units. Source data are provided with this paper.
Pericentric regions exhibit higher DNA methylation than centromeric regions
In humans, centromeric regions contain localized zones of reduced DNA methylation, known as centromere dip regions (CDRs), which are confined to active CENP-A-enriched alpha satellite arrays48. To investigate whether a similar DNA methylation pattern exists in the mouse genome, we assessed CpG DNA methylation in T2T satellite regions and C57BL/6 J Hifiasm satellite contigs by analyzing the kinetic signatures associated with methylated bases in HiFi reads using minimap2 alignments and the modkit49,50. Pericentric MaSats showed the highest DNA methylation across all satellite regions, including pericentric-chromosomal arm junctions in both mhaESC T2T and Hifiasm assemblies (Fig. 6A–E, Supplementary Figs. 12A, B and 13A–13E). Centromeric MiSats exhibited an apparent reduction in DNA methylation (Fig. 6A–E, Supplementary Figs. 12A, B and 13A–E). However, the reduction in DNA methylation in centromeric contigs compared to pericentric regions was less pronounced than the clear dip, with almost no DNA methylation observed in human active centromeres9,48. Interestingly, DNA methylation was higher in the 112-64-dimeric MiSats compared to the 120-mer and 112-mer MiSats (Fig. 6A, B and Supplementary Figs. 12A, B and 13A–E). At pericentromeres, homogeneous MaSats exhibited higher DNA methylation than divergent MaSats (Fig. 6C–E and Supplementary Figs. 12A, B and 13A–E). Non-satellite islands within pericentromeres showed a marked reduction in mCpG density compared to flanking MaSats (Fig. 6A and Supplementary Figs. 12A, B and 13A–E).
A DNA methylation frequency calculated across 50 Kb bins on satellite-containing regions of mhaESC T2T chromosomes. Below each bar plot, grayscale heatmaps representing DNA methylation frequency are shown, where white indicates low DNA methylation and black indicates high DNA methylation. B DNA methylation frequency and mCpG density calculated across 10 Kb bins on mhaESC T2T centromeres containing 112-64-dimeric MiSat regions. C DNA methylation frequency and mCpG density calculated across 10 Kb bins on mhaESC T2T representative pericentric regions containing high-density divergent MaSats. D Quantification of DNA methylation frequency, and (E) mCpG density across 50 Kb bins on regions containing divergent MaSats, homogeneous MaSats, 120-mer and 112-64-dimeric MiSats. For D and E panels, data are presented as violin plots with embedded box plots. For each box plot, the center line represents the median, the box bounds represent the interquartile range (IQR; 25th to 75th percentile), and the whiskers extend to the most extreme data point within that range. Individual points represent independent data bins. Source data are provided with this paper.
Only a small subset of satellites transcribes into RNA
Both centromeres and pericentromeres have been shown to transcribe in humans and mouse51,52,53,54,55,56,57,58,59. To identify which centromeric and pericentric satellites are transcribed and to compare their expression levels, we performed RNA sequencing (RNA-seq) on total RNA extracted from C57BL/6 J liver tissue and mapped the reads to the mhaESC T2T and Hifiasm assemblies. Our results revealed that only a small subset of MiSat and MaSat was transcribed, and their transcript levels were significantly lower, by several orders of magnitude, compared to the expression of rDNA genes interspersed within satellites (Fig. 7A–D and Supplementary Fig. 14A–C). These findings suggest that satellite non-coding regions are transcribed at very low levels. Notably, none of the pericentric regions with a high density of divergent MaSats showed detectable RNA expression (Fig. 7A and Supplementary Fig. 14A–C). Transcripts from high-density divergent MaSat-containing mhaT2T and Hifiasm regions were restricted to interspersed non-satellite repeats (Fig. 7A and Supplementary Fig. 14A–C). Furthermore, interspersed elements within homogeneous MaSats exhibited higher expression levels than those within high-density divergent MaSats (Fig. 7A–D and Supplementary Fig. 14A–C). We found that satellites with high transcript levels were associated with low CpG methylation density (Fig. 7E and Supplementary Fig. 14D).
A RNA-seq profiles mapped to mhaESC T2T satellite-containing regions and adjacent chromosomal arm junctions. RNA-seq tracks are displayed on a log scale as rendered in IGV. B Transcript levels on mhaESC T2T centromeres (C) Transcript levels on mhaESC T2T pericentromeres. D Transcript levels on regions containing indicated repeats. For panels B, C, and D, bars represent mean ± s.d. from n = 3 independent biological replicates. Each replicate corresponds to an RNA-seq dataset derived from the liver tissue of an individual C57BL/6 J mouse. RNA from each replicate was extracted, processed, and sequenced separately, without pooling. All replicates are biological and derived from three different mice; no technical replicates were used for statistical calculations. Reads were mapped to the mhaESC T2T assembly and normalized to the length of the satellite region. No formal hypothesis testing was performed for (panels B–D); data are descriptive summaries of transcript abundance. E The density of methylated CpG on (Top) and average DNA methylation frequency (Bottom) are plotted against the average transcript levels. Pearson correlation coefficients (r) were calculated using two-sided tests, with exact P values reported. Shaded areas indicate 95% confidence intervals of the regression line. No formal hypothesis testing or adjustments for multiple comparisons were applied, as only these pairwise correlations were tested. Source data are provided with this paper.
Centromeric and pericentric regions and junctions show distinct chromatin enrichment patterns
To explore how the sequence composition and arrangement of satellite variants across different satellite regions influence chromatin organization, we assessed the enrichment of key chromatin marks and centromeric proteins in centromeric and pericentric regions. We generated high-resolution CUT&RUN sequencing data from C57BL/6 J mouse tissue for CENP-A, CENP-B, H3K9me3, H3K27me3, and an IgG control. Sequencing reads were mapped to both unfiltered (Fig. 8A–C, Supplementary Figs. 16A–C and 17A–C) and k-mer-filtered uniquely mappable regions (Supplementary Fig. 15A–C) to rule out potential mapping artifacts. We found that chromatin enrichment patterns on centromeric and pericentric regions were consistent in both mapping approaches (Fig. 8A–C, Supplementary Figs. 15A–C, 16A–C, and 17A–C). Mapping the CENP-A CUT&RUN sequencing reads to mhaESC T2T assembly and C57BL/6 J Hifiasm contigs revealed that regions containing 120-mer MiSats exhibited the highest CENP-A enrichment, while contigs containing 112-mers and 112-64-dimers showed lower enrichment (Fig. 8B and Supplementary Figs. 15B, 16B, and 17A). This observation aligns with our previous observations on CENP-A binding patterns in HiFi reads24 The Y-centromere showed a clear enrichment for CENP-A, a marked depletion of H3K9me3. Given the absence of pericentric MaSats on the Y-chromosome, we did not detect pericentric H3K9me3 heterochromatin in regions flanking the CenY (Supplementary Fig. 16A). CENP-A levels were markedly reduced on TLC satellite sequences and absent on telomeric repeats (Supplementary Fig. 15A–B). Furthermore, IAP elements found within MiSats did not show CENP-A enrichment (Supplementary Fig. 17C), as previously shown24. Interestingly, despite lower CENP-A enrichment, the 112-64-dimer MiSats showed the highest enrichment of CENP-B among all analyzed satellite contigs (Fig. 8B, Supplementary Figs. 15–17). Similarly, although 112-mer-containing contigs had reduced CENP-A levels, their CENP-B enrichment was comparable to that of 120-mer-containing contigs (Fig. 8B, Supplementary Figs. 13–15).
A Chromatin profiles on mhaESC T2T satellite regions showing the log2 enrichment ratio of CENP-A, CENP-B, H3K9me3, and H3K27me3 compared to an IgG control across centromeric and centromeric-pericentric junction. The Y-axis range is consistent across all tracks for a given chromatin mark (-4 to +10 for CENP-A, -3 to +2 for CENP-B, and -6 to +4 for H3K9me3 and H3K27me3). Satellite regions are drawn to scale. B Enrichment signals were quantified by normalizing the number of CENP-A, CENP-B, H3K9me3, and H3K27me3 reads mapped to the indicated mhaESC T2T satellite-containing regions against the IgG control. Data are presented as violin plots with embedded box plots. For each box plot, the centre line represents the median, the box boundaries indicate the interquartile range (IQR; 25th–75th percentile), and whiskers extend to the most extreme data point within that range. Individual points represent independent data values and variability is defined by the distribution shown in the violin plots. C Correlation between CENP-A and CENP-B (Top) and H3K9me3 and H3K27me3 (Bottom) on indicated mhaESC T2T satellites. Pearson correlation coefficients (r) were calculated using two-sided tests, with exact P values reported. No formal hypothesis testing or adjustments for multiple comparisons were applied, as only these pairwise correlations were tested. Source data are provided with this paper.
Next, we analyzed the CENP-A, CENP-B, H3K9me3, H3K27me3, and IgG control CUT&RUN sequencing reads mapped to mhaT2T and Hifiasm pericentric regions. Homogeneous MaSats are consistently enriched with H3K9me3 as continuous domains on contigs, while high-density divergent MaSats show a more discontinuous enrichment of H3K9me3 (Fig. 8A, B). These divergent MaSats, however, were enriched with higher levels of H3K27me3 compared to homogeneous MaSats (Fig. 8B and Supplementary Figs. 15B and 16B). Similarly, non-satellite islands within MaSats displayed reduced H3K9me3 enrichment and increased H3K27me3 levels compared to the surrounding MaSats (Fig. 8A and Supplementary Figs. 15A and 16A). Furthermore, we found a negative correlation between H3K9me3 and H3K27me3 enrichment on pericentric satellites (Fig. 8C). Together, these findings suggest that high-density divergent MaSats and non-satellite islands are associated with reduced H3K9me3 heterochromatin and increased H3K27me3 enrichment, indicating distinct epigenetic regulation in these regions.
Sequence-dependent contribution to centromeric and pericentric chromatin
We determined the distribution of known sequence motifs and their relationship with the chromatin in centromeric and pericentric regions. Centromeric MiSats contains a 17 bp motif called the CENP-B box, shared by MiSats and α-satellites, which binds to CENP-B in a sequence-dependent manner60,61,62,63,64. We determined the density of the canonical (5’-ATTCGTTGGAAACGGGA-3’) and variant CENP-B boxes in T2T centromeres and Hifiasm MiSat contigs (Fig. 9A and Supplementary Fig. 19A). Interestingly, the density of the canonical CENP-B box was highest on centromeres containing 112-64-dimeric MiSats and lowest in 120-mer MiSats interspersed with low-density 112-mers in both mhaESC T2T and Male C57BL/6 J Hifiasm assemblies (Fig. 9A and Supplementary Figs. 18 and 19A). Regions containing 120-mer MiSats alone exhibited an increased density of CENP-B box variants that contained mutations in the bases responsible for CENP-B recognition (Fig. 9B and Supplementary Figs. 19 and 19B). The Y-centromere on our Hifiasm contig lacked CENP-B boxes (Supplementary Fig. 19A), as observed in the human Y-centromere and in ectopic functional centromeres, known as neocentromeres61,65.
A Heatmap showing the density of canonical and variant CENP-B box motifs across mhaESC T2T centromeres. The consensus canonical CENP-B box sequence is shown at the top, and single-nucleotide deviations observed in variant CENP-B boxes are indicated in maroon. The color scale indicating motif density per Mb is shown below the heatmap. B Log₂ CENP-A and CENP-B enrichment profiles (Top), DNA methylation profiles (Middle), and stacked bar plots of CENP-B box variant composition across 10 Kb bins (Bottom) on centromeres containing different classes of MiSats (120-mers, 112-mers, and 112-64-dimers). Pie charts summarize the proportion of canonical vs. variant CENP-B box types. The chromatin enrichment values are plotted with a fixed Y-axis range for a given chromatin mark: -2 to +7 for CENP-A and -2 to +2 for CENP-B. C Log₂ H3K9me3 and H3K27me3 enrichment profiles (Top), the density of the MaSat consensus motif (Middle) and StainedGlass heatmaps of sequence identity (Bottom) on representative pericentromeric regions containing either homogeneous or divergent MaSat arrays. The chromatin enrichment values are plotted with a fixed Y-axis range (−4 to +3) for H3K9me3 and H3K27me3 (D) Correlation plots at comparing MaSat motif density with H3K9me3 (Top) and H3K27me3 (Bottom) enrichment on different satellite types. Pearson correlation coefficients (r) were calculated using two-sided tests, with exact P values reported. No formal hypothesis testing or adjustments for multiple comparisons were applied. Source data are provided with this paper.
We then analyzed the relationship between CENP-B box density and the enrichment of CENP-A and CENP-B proteins. Overall, CENP-B box density was positively correlated with both CENP-A and CENP-B enrichment, except in 112-64-dimeric regions (Fig. 8C, Supplementary Figs. 16C and 17A). While these dimers exhibited the highest canonical CENP-B box density and strong CENP-B enrichment, their CENP-A enrichment was lower than that of flanking 120-mer regions. In 112-64-dimers, >90% of CENP-B boxes were canonical (Fig. 9B), whereas in 120-mers, only ~50% were canonical, with the remaining being variant boxes. These observations suggest that the presence of variant CENP-B boxes may promote increased CENP-A binding. Canonical CENP-B boxes contain two CpG sites (5’-ATTCpGTTGGAAACpGGGA-3’), while variants tend to have fewer due to mutations in the CpG sites. Accordingly, we observed higher mCpG density in 112-64-dimers relative to surrounding 120-mers, suggesting that CpG mutations in variant CENP-B boxes reduce DNA methylation and thereby increase CENP-A enrichment (Fig. 9B and Supplementary Figs. 18 and 19B–C).
Next, we determined the contribution of DNA sequence in chromatin enrichment at pericentric regions. Recently, we have characterized a 12-bp motif, called the MaSat motif (5’- GAAAACTGAAAA -3’), within MaSats24. While the CENP-B boxes are essential for recruiting and binding to the centromeric protein CENP-B, the function of the MaSat motif is not explored. We found that the density of the Major motif was inversely proportional to the density of divergent MaSats in both mhaESC T2T and Hifiasm assemblies (Fig. 9C). Furthermore, H3K9me3 enrichment showed a positive correlation with MaSat motif density, while H3K27me3 exhibited an inverse relationship (Fig. 9D). These findings indicate that homogeneous MaSat regions with high MaSat motif density favor H3K9me3 enrichment, while divergent MaSat regions with reduced MaSat motif density, especially near non-satellite repeat islands or arm junctions, exhibit decreased H3K9me3 and increased H3K27me3, suggesting a sequence-dependent chromatin transition.
Discussion
Our analyses of mouse centromeres and pericentromeres using the recently available mouse T2T assembly drafts and Hifiasm long-read assembly suggest that mouse centromeres vary in length from 90 Kb to ~1.2 Mb, similar to humans, where centromeres range from 500 Kb to 5 Mb9. A few centromeres are fragmented and interspersed within pericentric MaSats. Most centromeres contain 120-mer MiSats, while large blocks containing high-density length variants, 112-mers and 112-64-dimer MiSats, are adjacent to pericentric regions of a few chromosomes. These variants are present in both male and female C57BL/6 J animals, indicating that they are not specific to the Y chromosome. Multiple mhaESC T2T centromeres contained interstitial telomeres that were not detected in the mESC T2T30 or Hifiasm assemblies, suggesting possible assembly artifacts.
Interestingly, we confirmed that the Y-centromere lacks flanking pericentric MaSats, which agrees with previous findings suggesting that the majority of the mouse Y chromosome is euchromatic66,67. Heterochromatin protein 1 (HP1), which binds to pericentric H3K9me3 heterochromatin, plays a crucial role in recruiting and maintaining the cohesin complex, essential for ensuring proper sister chromatid cohesion and preventing premature separation prior to anaphase17,68,69. Since the mouse Y chromosome lacks detectable pericentric MaSats, and H3K9me3 heterochromatin, future studies investigating the loss rate of the mouse Y chromosome may shed light on whether the absence of pericentric MaSats reduces the chromosome segregation efficiency of the mouse Y chromosome.
Our results also show that most pericentric regions, including those spanning centromeric-pericentric junctions, are populated mainly by homogeneous MaSats, while divergent MaSats preferentially localize near non-satellite islands within pericentric regions or the pericentric-chromosomal junctions, suggesting that, as in humans, sequence divergence of pericentric satellites increases with distance from the core centromere. Additionally, while centromeric regions contain rare short, interspersed transposons (a few Kb long), pericentric regions contain both short transposons (a few Kb long) and long islands of non-satellite sequences (ranging from ~100−~200 Kb), indicating that pericentric regions are more permissive to non-satellite sequence insertions compared to centromeric regions likely due to their proximity to chromosomal arms. Furthermore, while inversions are present in centromeric and pericentric regions, they are more frequent within centromeric regions. In contrast, human active alpha satellite arrays, especially those within well-defined HOR domains, largely lack inversions48. This contrast likely reflects differences in satellite sequence homogeneity and evolutionary pressures between species. Human centromeres, being more divergent, may limit the potential for sequence-based inversions. In contrast, the high sequence homogeneity of mouse centromeric MiSats, facilitates inversions via non-allelic homologous recombination. In younger, more homogeneous centromeres such as those in mice, inversion events may have evolutionary significance, particularly by generating structural variations that drive further divergence. As centromeres accumulate more abundant HOR structures and sequence variations, as seen in humans, inversion frequency may decline. Together, we speculate that the inversion frequency within satellites is influenced by sequence homogeneity and centromere maturation, providing valuable insights into the evolutionary trajectory of centromeres.
Our study shows the progressive increase in non-satellite repeat density from centromeres to pericentromeres to chromosomal arm junctions. While non-satellite repeats are present as individual, isolated elements at centromeric and homogeneous pericentric regions, they have expanded into clustered islands within the divergent parts of pericentromeres. This observation suggests that as MaSats divergence increases, non-satellite insertions increase and lead to the formation of islands. Similar non-satellite repeats have been observed in centromeres of other species. For example, Drosophila centromeres contain several satellite and non-satellite repeat families45,70,71,72. However, the majority of functional CENP-A enriched regions are located on the islands of retrotransposons45. Similarly, retrotransposons interspersed within plant centromeric satellites, which are more divergent than those of mouse centromeres, are part of functional centromeres41,42,44,73,74. Given our observation that rare LTRs within homogeneous MiSats are devoid of CENP-A, retrotransposons likely adopt centromeric function when satellites become more divergent and non-satellite repeats cluster into islands during evolution.
We detected HORs in 19 out of 21 mouse centromeres. The two mhaESC T2T centromeres, Cen3 and Cen4, which were the most homogeneous among all mouse centromeres, did not contain HORs. We found that most mouse centromeres lack chromosome-specific HORs. However, certain HORs tend to preferentially localize to either the ends or the middle of centromeres. Additionally, Cen 5, 14, 15, 17, 18, and 19, which share identical lengths and exhibit a highly similar sequence identity pattern, contain highly similar HOR patterns, suggesting that these centromeres may have undergone recent duplication or homogenization. However, further investigation is required to rule out duplication due to possible assembly artifacts due to highly homogeneous satellite sequences.
Among all satellite and DNA repeats analyzed, we found that homogeneous pericentric regions exhibit the highest levels of DNA methylation. In humans, the centromeric core exhibits a sharp dip in DNA methylation, with almost no signals on active CENP-A-enriched HORs9,48. While we observed a significant decrease in DNA methylation within mouse centromeric regions compared to pericentric regions, we did not observe well-defined CDRs seen in human active HORs9,48. Human inactive HORs, typically found near pericentric satellites, are hypermethylated and lack CDRs48. Similarly, we observed increased DNA methylation on dimeric MiSats that are present at centromeric-pericentric junctions. Our findings suggest that the modest decrease in DNA methylation in younger, homogeneous satellites may represent an early step toward the development of CDRs in more evolutionarily divergent satellite centromeres, such as those found in humans. Additionally, we found that only a small subset of MaSat and MiSat sequences were transcribed into RNA. Within homogeneous MaSats, RNA expression was associated with regions of lower mCpG density, similar to transcription observed in regions of hypomethylation within human neocentromeres75.
CENP-B binding strongly correlates positively with CENP-A enrichment, except for the 112-64 dimers, which show reduced CENP-A enrichment compared to the 120-mer MiSat regions, despite having higher CENP-B enrichment. Interestingly, these 112-64-dimers display a higher density of canonical CENP-B boxes and a corresponding increase in CENP-B enrichment compared to the 120-mer MiSats, despite their lower levels of CENP-A. DNA methylation is negatively correlated with CENP-A enrichment and has been shown to inhibit CENP-B binding9,48,76. Notably, canonical CENP-B boxes contain two CpG sites, whereas many CENP-B box variants we analyzed have mutations in these sites. Accordingly, we observed higher mCpG density in 112-64-dimers compared to 120-mers, indicating that DNA methylation might inhibit CENP-A binding more strongly than CENP-B, or that other elements may impact CENP-A and CENP-B binding on 112-64 dimers at the centromeric-pericentric junctions. Since these variants exhibit higher DNA methylation and lower CENP-A levels compared to the 120-mers, resembling the features of inactive HORs in humans, the 112-64-dimers are probably evolving into elements similar to human inactive HORs. The reason why these variants retain high CENP-B enrichment requires further investigation. While the role of CENP-B in promoting CENP-A chromatin formation is well established, recent studies also suggest that CENP-B functions as a molecular switch depending on the chromatin context by facilitating recruitment of either CENP-A assembly factors (e.g., ASH1L) or heterochromatin-promoting proteins (e.g., Suv39h1, HP1)77. Together, our observations combined with previous studies support a dual regulatory model for CENP-B’s influence on CENP-A binding: a sequence-dependent increase in CENP-A binding by CENP-B on 120-mer MiSats, and a methylation-dependent regulation that reduces CENP-A enrichment without a proportional decrease in CENP-B binding on 112-64-dimers in the centromere-pericentric transition zones.
We also found that homogeneous MaSats, predominantly localized near centromeres and in the core regions of the pericentromeres, were more enriched with H3K9me3, a marker of transcriptionally silent constitutive heterochromatin. In contrast, divergent MaSats, concentrated at the pericentric-chromosomal arm transitions, exhibited lower levels of H3K9me3. We found that homogeneous MaSats, which occupy the majority of mouse pericentric regions, contain denser MaSat motifs than divergent MaSats, which are localized near pericentric non-satellite islands and pericentric-chromosomal arm junctions, suggesting that the presence of the MaSat motif may influence H3K9me3 heterochromatin enrichment. Mice HMGA1 and Drosophila D1, AT-hook proteins, bind to AT-rich pericentric satellites and suppress position-effect variegation (PEV), indicating their role in H3K9me3-mediated silencing78. Given that the MaSat motif also contains poly A/T tracts, it may facilitate the binding of HMGA1 and other AT-hook proteins by binding to the narrow minor groove of poly A/T tracts79, potentially contributing to the recruitment or stabilization of the H3K9me3 chromatin state at homogeneous MaSats. Interestingly, we observed the opposite pattern for the enrichment of facultative H3K27me3 heterochromatin on pericentric regions. High levels of constitutive H3K9me3 heterochromatin are preferentially associated with highly homogeneous, MaSat motif-rich pericentric MaSats. In contrast, more divergent pericentric satellites exhibit reduced H3K9me3 enrichment, accompanied by a corresponding increase in facultative H3K27me3 heterochromatin. Furthermore, non-satellite islands embedded within MaSat regions are largely devoid of H3K9me3 and instead show preferential enrichment for H3K27me3, indicating a shift toward facultative heterochromatin.
In conclusion, we utilized recent mouse T2T assembly drafts29,30 and shorter minimally processed Hifiasm assemblies, combined with epigenomic and transcriptomic profiling, to provide comprehensive genomic and epigenomic maps of mouse centromeres and pericentromeres (summarized in Fig. 10). This work will enable the complete annotation of the mouse T2T assembly, similar to the current human T2T assembly, and facilitate future studies aimed at gaining mechanistic insights into mouse centromeres and pericentromeres.
Generalized models describing five major types of configurations of mouse satellite regions observed in T2T and Hifiasm assemblies. Created in BioRender. Thakur, J. (2025) https://BioRender.com/4dj7bs9.
Methods
This study was conducted in compliance with all relevant institutional regulations.
Animals
Animals were maintained at Emory University per the protocol approved by the Institutional IACUC under protocol number PROTO202100038. Experiments were performed using inbred Mus musculus (C57BL/6 J) male mice (Jackson Laboratory #000664). Male mice (aged 15–20 weeks at the time of tissue collection) were chosen to enable characterization of the Y chromosome. To evaluate whether sex-specific differences might influence our findings, we also analyzed a publicly available dataset generated from female mice. Except for expected Y chromosome–specific features present only in male-derived samples, no substantial differences were detected between our male data and the female dataset (Supplementary Fig. 1). Disaggregation by sex does not apply to these datasets.
Nuclei preparation
Nuclei were prepared from C57BL/6 J kidney (for genomic DNA) and liver (for CUT&RUN profiling) tissues. First, the flash-frozen tissue was weighed and ground into a fine powder using liquid nitrogen in a prechilled mortar and pestle. The powdered tissue was then resuspended in 4 mL of ice-cold extraction Buffer (0.32 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, and 1X protease inhibitor) per gram of tissue. The suspension was homogenized using a Tekmar tissuemizer for 15 pulses of ~5 s each, with 20 s intervals on ice. Finally, the cell suspension was filtered through a 100 μm cell strainer to remove large tissue chunks. The filtered homogenate suspension was centrifuged at 6000 x g for 10 min at 4 °C. The pellet was resuspended in 4 mL (per gram of tissue) of ice-cold Extraction buffer supplemented with 0.2% IGEPAL, and the samples were incubated on ice for 10 min. The nuclei suspension was layered on top of ice-cold sucrose buffer (1.2 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, and 1X protease inhibitor) to create a sucrose cushion. Using prechilled rotors, the mixture was centrifuged at 10,000 x g for 20 min at 4 °C. The nuclei pellet was resuspended in a buffer suitable for downstream processing.
PacBio sequencing
High molecular weight genomic DNA was isolated from the kidneys of C57BL/6 J mice using the Monarch High Molecular Weight DNA Extraction Kit, following the manufacturer’s instructions. The nuclei pellet was incubated with 600 μL Monarch Lysis Buffer and 20 μL Proteinase K and incubated at 55 °C with gentle agitation at 700 rpm for 45 min. Following lysis, 10 µL of RNase A was added to the lysate and incubated at 55 °C with gentle agitation at 700 rpm for 45 min for 10 min. Subsequently, 300 μL of Monarch Protein separation Buffer was added to the lysate, mixed vigorously for 1 min by inverting the tube, and centrifuged at 16,000 x g for 10 min to pellet the precipitated proteins.
The DNA supernatant was transferred to a clean tube containing 500 μL isopropanol and two Monarch DNA capture beads. The solution was mixed by vertically rotating the tube at 10 rpm for 5 min to attach DNA to the beads. The supernatant was discarded, beads were washed twice with 500 µl Monarch gDNA wash buffer, and DNA was eluted from beads with 200 µL of Monarch DNA Elution Buffer. The purified high molecular weight genomic DNA was sequenced using PacBio’s Revio platform with single-molecule, real-time (SMRT) sequencing technology to generate raw HiFi long reads. The consensus HiFi reads were used for downstream analyses, including satellite assembly and methylation analysis. This comprehensive approach ensured the generation of long, high-quality reads suitable for in-depth genomic studies.
CUT&RUN sequencing and data analysis
The pelleted nuclei from liver tissue were resuspended in CUT&RUN wash buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, and 0.5 mM spermidine). Concanavalin beads were prepared by washing the bead slurry with three times the volume of binding buffer (20 mM HEPES, 10 mM KCl, 1 mM CaCl2, and 1 mM MnCl2) and resuspended in an equal volume of binding buffer as the initial slurry volume. Approximately one million nuclei were used for each CUT&RUN reaction. To each CUT&RUN reaction, 20 µL of beads per million nuclei were added and the mixture was incubated at room temperature for 15 min on a rotator. The beads were washed, resuspended in 200 µL of antibody buffer containing 4 µL of antibodies (anti-CENP-A antibody from Cell Signaling Technologies - #C51A7, anti-CENP-B antibody from Santa Cruz - # sc-376392, anti-H3K9me3 antibody from abcam - #ab8898, anti-H3K27me3 antibody from Cell Signaling Technologies - # 9733, and anti-IgG antibody from abcam - # ab46540) per reaction, and incubated at 4 °C overnight. The beads were washed in 500 µL of cold Dig-Wash buffer (20 mM HEPES (pH 7.5), 150 mM NaCl, 0.5 mM spermidine, and 0.05% digitonin) twice, resuspended in 200 µL of Dig-Wash buffer containing 1 µL pA-MN, and incubated for 1 h at room temperature on a rotator. The beads were then washed twice with cold Dig-Wash buffer, resuspended in 200 µL of Dig-Wash buffer with 3 mM CaCl2, and incubated at 0 °C for 30 min. After 30 min, the reaction was stopped by adding 200 µL of 2X DRSTOP buffer (550 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% digitonin, 40 µg/mL glycogen, 50 mg/mL RNase A and 0.0025 ng/µL yeast spike-in DNA) and the mixture was incubated for 10 min at 37 °C. The samples were centrifuged for 5 min at room temperature, and the supernatant was collected. DNA was extracted from the supernatant and libraries were prepared using the KAPA HyperPrep kit. Amplified libraries were sequenced using NextSeq 500/550 instrument. Reads were aligned to the mouse T2T and Hifiasm assemblies using Bowtie2 with the multimapping parameter -k 10, and alignments were filtered to retain only those with a mapping quality (MAPQ) score ≥10. Additionally, to restrict mapping to uniquely mappable regions, K50-mers were generated using Meryl80, and the assemblies were masked using these K50-mers. Reads were then aligned to these K50-mer-filtered assemblies using Bowtie2 with -k 10, followed by the same MAPQ ≥ 10 filtering.
Generation and annotation of satellite contigs
Assembled contigs were generated from PacBio HiFi reads using Hifiasm software with default parameters. Satellite-containing contigs were then filtered from the total contig pool using NCBI-BLAST against satellite consensus sequences. Specific satellite types, including 120 mer MiSats, 112 mer MiSats, 112-64-mer MiSats, TLC-Sats, homogeneous MaSats, and divergent MaSats, were annotated on these satellite contigs using NCBI-BLAST. Non-satellite repeat sequences (LINE, SINE, LTR, simple repeats, rDNA, and tRNA) were annotated using RepeatMasker on the Mus musculus repeat database81.
Generation of identity heatmaps
We utilized the StainedGlass39 software package to generate colored identity dotplots for satellite regions of the T2T mouse genome assembly. We used default settings for StainedGlass, which fragments the input sequence into non-overlapping 1 Kb windows and computes pairwise alignments between all fragments using minimap2, and calculates the sequence identity for each alignment (percent Identity = 100(M/M + X + I + D), where M is the number of matches, X is the number of mismatches, I is the number of insertions, and D is the number of deletions). For broader genome-scale visualization, we utilized HiGlass, a web-based genome interaction viewer82, in cooler format.
Detection of higher-order repeats
We utilized CENdetectHOR40 to identify the HORs within mouse T2T centromeres and Hifiasm centromeric contigs by employing the consensus sequences for minor satellite. CENdetectHOR generated phylogenetic trees of the monomers, yielding a set of HOR calls for each region. As we move from the root to the leaves, the distance utilized to cluster monomers into families diminishes. The output trees from CENdetectHOR were visualized in PhyloTreeGUI to assess the composition of each HOR, including monomeric families and their frequency in the analyzed sequence. The locations and types of HORs were exported into bed files for visualization in IGV.
Annotations of genes in pericentric-chromosome arm junctions
We identified 2-5 Kb sequences from the extreme pericentric end of each chromosome as annotated in the GRCm39 genome. These sequences were subsequently used in NCBI-BLAST searches against Hifiasm contigs to identify contigs containing the respective sequences. Next, genes and transcribed pseudogenes located within the first 5 Mb (if the contig is >5 Mb) of all autosomes except chromosomes 2, 3, and 10, as well as chromosome X scaffold NT_165789.3, were identified using the NCBI genome annotation associated with the mouse GRCm39 genome. Genes specific to chromosome Y near the centromere were similarly identified.
The corresponding RNA transcripts were searched for matches against the Hifiasm contigs using NCBI BLAST. BLAST hits with >99% identity were selected for further analysis. For each chromosome, the selected hits were further restricted to the previously identified contig containing the pericentric junction. This contig was converted to a BED file and loaded into IGV for visual comparison to GRCm39 (viewed using the NCBI Genome Data Viewer). Non-specific BLAST hits were identified by comparison to GRCm39 and removed. The resulting cleaned BED file was then processed to combine all exon BED records into a single BED record per gene.
DNA Methylation Analysis
To analyze DNA methylation, BAM files generated from PacBio HiFi reads were converted to FASTQ format using samtools fastq -T ‘*’, which retains MM/ML tags corresponding to base modifications. Reads were then aligned to the mouse T2T and Hifiasm assemblies using minimap2 with the -y option to preserve modification annotations and the map-hifi preset for HiFi reads. Multimapping was allowed by setting the -N 10 parameter. DNA methylation calls were extracted using modkit pileup, generating output in the bedMethyl format. To ensure high-confidence methylation calls, only sites with a posterior probability ≥ 0.8 were retained. Average methylation frequency across defined genomic regions was calculated using bedtools map, taking the mean of site-specific methylation percentages. mCpG density was calculated as the number of methylated CpG sites per region, normalized by the region length.
RNA-sequencing and analysis
Total RNA was extracted from the liver tissue of C57BL/6 J using the Trizol method and sequenced using an Illumina platform by Azenta Life Science. Raw reads were assessed for quality using FastQC (v0.11.9), and adapter sequences along with low-quality bases were trimmed using Trimmomatic83. Resulting high-quality reads were aligned to the T2T and Hifiasm assemblies using STAR (v2.7.11b), with parameters optimized for retaining multi-mapping reads commonly found in repetitive regions. Specifically, reads mapping to up to 1,000 genomic loci were retained (--outFilterMultimapNmax 1000), and anchor windows were extended to improve mapping sensitivity in repetitive sequences (--winAnchorMultimapNmax 1000). Unique alignment was preserved in the final BAM file using --outSAMmultNmax 1 to support downstream quantification and visualization. Mapped reads were quantified using bedtools coverage84, and alignment output was used to determine transcriptional readout across annotated and repetitive genomic regions.
Statistics & Reproducibility
No statistical method was used to predetermine sample size. Sample sizes were chosen based on common practice in the field and the availability of biological material. Replicates are biological replicates, defined as samples derived from different animals. Technical replicates were not used for statistical analyses. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. For descriptive analyses (e.g., transcript levels, DNA methylation frequency, enrichment signals), mean values are reported with error bars representing standard deviation (s.d.) across biological replicates, unless otherwise specified in figure legends. Correlation analyses were conducted using Pearson’s correlation coefficient (two-sided), with exact r values and p values reported in figure legends. All attempts at replication were successful. Replicates are explicitly defined in figure legends, and all raw data underlying figures and statistical analyses are provided in the Source Data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. Raw PacBio long-read sequencing, RNA-sequencing, and CUT&RUN-sequencing data are deposited in the NCBI Sequence Read Archive under Bioproject number PRJNA1173940 and can be accessed at the following link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1173940?reviewer=1frnl4j5o23ukda2nn8l02ovg1. The mhaESC T2T assembly29 was accessed from the following link: https://github.com/yulab-ql/mhaESC_genome/releases. The mESC T2T30 was accessed using GenBank accession number GCA_964188535.1. PacBio HiFi dataset from the mhaESC cell line was accessed from the NCBI BioProject PRJNA109700029. PacBio HiFi data for the female C57BL/6 J mouse was accessed from the NCBI Sequence Read Archive accession number SRR1160687034.
Code availability
All bioinformatic tools used in this study are open source and freely available. Hifiasm33 was used to generate assemblies from PacBio HiFi data. RepeatMasker81 was used to identify non-satellite repeats. NCBI BLAST85 was used for sequence similarity searches, StainedGlass39 was used to create sequence identity heatmaps, HiGlass82 was used to visualize sequence identity maps, and CENdetectHOR40 was utilized for HOR detection. RNA sequencing data were aligned using the STAR aligner86, while chromatin profiling data were aligned using Bowtie287. Meryl80 was used for k-mer filtering. PacBio HiFi reads were mapped to T2T and Hifiasm assemblies using minimap249, and DNA methylation calling was performed using modkit50. Figure 10 was created with BioRender.com under an academic license, in compliance with the BioRender publication guidelines. The citation to use Fig. 10: Thakur, J. (2025) https://BioRender.com/4dj7bs9. Hifiasm assemblies and annotation files for genomic maps of satellite regions in mouse T2T and Hifiasm assemblies can be accessed using the following link. https://github.com/nacpatil/Repeats.
References
de Koning, A. P., Gu, W., Castoe, T. A., Batzer, M. A. & Pollock, D. D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7, e1002384 (2011).
Biscotti, M. A., Olmo, E. & Heslop-Harrison, J. S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 23, 415–420 (2015).
Feschotte, C. & Pritham, E. J. DNA transposons and the evolution of eukaryotic genomes. Annu Rev. Genet 41, 331–368 (2007).
Thakur, J., Packiaraj, J. & Henikoff, S. Sequence, chromatin and evolution of satellite DNA. Int. J. Mol. Sci. 22, 4309 (2021).
Garrido-Ramos, M. A. Satellite DNA: an evolving topic. Genes (Basel) 8, 230 (2017).
Plohl, M., Meštrović, N. & Mravinac, B. Satellite DNA evolution. Genome Dyn. 7, 126–152 (2012).
Rudd, M. K., Wray, G. A. & Willard, H. F. The evolutionary dynamics of alpha-satellite. Genome Res. 16, 88–96 (2006).
Alexandrov, I., Kazakov, A., Tumeneva, I., Shepelev, V. & Yurov, Y. Alpha-satellite DNA of primates: old and new families. Chromosoma 110, 253–266 (2001).
Altemose, N. et al. Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022).
Komissarov, A. S., Gavrilova, E. V., Demin, S. J., Ishov, A. M. & Podgornaya, O. I. Tandemly repeated DNA families in the mouse genome. BMC Genomics 12, 531 (2011).
Ando, S., Yang, H., Nozaki, N., Okazaki, T. & Yoda, K. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell Biol. 22, 2229–2241 (2002).
Henikoff, J. G., Thakur, J., Kasinathan, S. & Henikoff, S. A unique chromatin complex occupies young α-satellite arrays of human centromeres. Sci. Adv. 1, e1400234 (2015).
Aldrup-Macdonald, M. E. & Sullivan, B. A. The past, present, and future of human centromere genomics. Genes (Basel) 5, 33–50 (2014).
Altemose, N. A classical revival: Human satellite DNAs enter the genomics era. Semin Cell Dev. Biol. 128, 2–14 (2022).
Guenatri, M., Bailly, D., Maison, C. & Almouzni, G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 (2004).
Hahn, M. et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev. 27, 859–872 (2013).
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001).
Rudd, M. K. & Willard, H. F. Analysis of the centromeric regions of the human genome assembly. Trends Genet 20, 529–533 (2004).
Waye, J. S. et al. Chromosome-specific alpha satellite DNA from human chromosome 1: hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics 1, 43–51 (1987).
Hartley, G. & O’Neill, R. J. Centromere repeats: hidden gems of the genome. Genes (Basel) 10, 223 (2019).
Greig, G. M. & Willard, H. F. Beta satellite DNA: characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics 12, 573–580 (1992).
Waye, J. S. & Willard, H. F. Human beta satellite DNA: genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl Acad. Sci. USA 86, 6250–6254 (1989).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Packiaraj, J. & Thakur, J. DNA satellite and chromatin organization at mouse centromeres and pericentromeres. Genome Biol. 25, 52 (2024).
Hayashi, T. et al. A variant family of mouse minor satellite located on the centromeric region of chromosome 2. Genomics 17, 490–492 (1993).
Jones, K. W. Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature 225, 912–915 (1970).
Kipling, D., Ackford, H. E., Taylor, B. A. & Cooke, H. J. Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics 11, 235–241 (1991).
Rice, W. R. Centromeric repeats of the Western European house mouse I: high sequence diversity among monomers at local and global spatial scales. bioRxiv https://doi.org/10.1101/2020.08.28.272245 (2020).
Liu, J. et al. The complete telomere-to-telomere sequence of a mouse genome. Science 386, 1141–1146 (2024).
Francis, B. et al. The structural diversity of telomeres and centromeres across mouse subspecies revealed by complete assemblies. bioRxiv https://doi.org/10.1101/2024.10.24.619615 (2024).
Lang, D. et al. Comparison of the two up-to-date sequencing technologies for genome assembly: HiFi reads of Pacific biosciences sequel ii system and ultralong reads of Oxford nanopore. Gigascience 9, giaa123 (2020).
Cosma, B. M. et al. Evaluating long-read de novo assembly tools for eukaryotic genomes: insights and considerations. Gigascience 12, giad100 (2022).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Hon, T. et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data 7, 399 (2020).
Pertile, M. D., Graham, A. N., Choo, K. H. & Kalitsis, P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 19, 2202–2213 (2009).
Falconer, E. et al. Identification of sister chromatids by DNA template strand sequences. Nature 463, 93–97 (2010).
Cazaux, B. et al. Evolution of the structure and composition of house mouse satellite DNA sequences in the subgenus Mus (Rodentia: Muridea): a cytogenomic approach. Chromosoma 122, 209–220 (2013).
Kalitsis, P., Griffiths, B. & Choo, K. H. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc. Natl; Acad. Sci. USA 103, 8786–8791 (2006).
Vollger, M. R., Kerpedjiev, P., Phillippy, A. M. & Eichler, E. E. StainedGlass: interactive visualization of massive tandem repeat structures with identity heatmaps. Bioinformatics 38, 2049–2051 (2022).
Daponte, A. et al. CENdetectHOR: a comprehensive tool for CENtromere profiling and HOR detection. bioRxiv https://doi.org/10.1101/2025.01.07.631657 (2025).
Ananiev, E. V., Phillips, R. L. & Rines, H. W. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078 (1998).
Cheng, Z. et al. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14, 1691–1704 (2002).
Meštrović, N. et al. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 23, 583–596 (2015).
Zhong, C. X. et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825–2836 (2002).
Chang, C. H. et al. Islands of retroelements are major components of Drosophila centromeres. PLoS Biol. 17, e3000241 (2019).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Scott, K. C., Merrett, S. L. & Willard, H. F. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16, 119–129 (2006).
Gershman, A. et al. Epigenetic patterns in a complete human genome. Science 376, eabj5089 (2022).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
ONT, O. N. T. Nanoporetech/Modkit. https://github.com/nanoporetech/modkit (2025).
Velazquez Camacho, O. et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife 6, e25293 (2017).
McNulty, S. M., Sullivan, L. L. & Sullivan, B. A. Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 42, 226–240.e226 (2017).
Heyt, K. M. & Thakur, J. Regulation of epigenetic processes by non-coding RNAs. Nucleus 64, 285–301 (2021).
Bobkov, G. O. M., Gilbert, N. & Heun, P. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 217, 1957–1972 (2018).
Chan, F. L. et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl Acad. Sci. USA 109, 1979–1984 (2012).
Chen, E. S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008).
Probst, A. V. et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell 19, 625–638 (2010).
Ferri, F., Bouzinba-Segard, H., Velasco, G., Hubé, F. & Francastel, C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37, 5071–5080 (2009).
Chen, Y. L. et al. Determinants of minor satellite RNA function in chromosome segregation in mouse embryonic stem cells. J. Cell Biol. 223, e202309027 (2024).
Fachinetti, D. et al. DNA Sequence-Specific Binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327 (2015).
Masumoto, H., Masukata, H., Muro, Y., Nozaki, N. & Okazaki, T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 (1989).
Meštrović, N. et al. Conserved DNA Motifs, including the CENP-B box-like, are possible promoters of satellite DNA array rearrangements in nematodes. PLoS One 8, e67328 (2013).
Ohzeki, J., Nakano, M., Okada, T. & Masumoto, H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775 (2002).
Okada, T. et al. CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300 (2007).
Alonso, A. et al. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8, R148 (2007).
Morgan, A. P. & Pardo-Manuel de Villena, F. Sequence and structural diversity of mouse Y chromosomes. Mol. Biol. Evol. 34, 3186–3204 (2017).
Soh, Y. Q. et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 (2014).
Yi, Q. et al. HP1 links centromeric heterochromatin to centromere cohesion in mammals. EMBO Rep. 19, e45484 (2018).
Shimura, M. et al. Epigenetic displacement of HP1 from heterochromatin by HIV-1 Vpr causes premature sister chromatid separation. J. Cell Biol. 194, 721–735 (2011).
Garavís, M. et al. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 5, 13307 (2015).
Sun, X., Le, H. D., Wahlstrom, J. M. & Karpen, G. H. Sequence analysis of a functional Drosophila centromere. Genome Res. 13, 182–194 (2003).
Talbert, P. B., Kasinathan, S. & Henikoff, S. Simple and Complex Centromeric Satellites in. Genetics 208, 977–990 (2018).
Naish, M. & Henderson, I. R. The structure, function, and evolution of plant centromeres. Genome Res. 34, 161–178 (2024).
Houben, A. et al. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116, 275-–283 (2007).
Wong, N. C. et al. Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet 2, e17 (2006).
Tanaka, Y., Kurumizaka, H. & Yokoyama, S. CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS J. 272, 282–289 (2005).
Otake, K. et al. CENP-B creates alternative epigenetic chromatin states permissive for CENP-A or heterochromatin assembly. J. Cell Sci. 133, jcs243303 (2020).
Jagannathan, M., Cummings, R. & Yamashita, Y. M. A conserved function for pericentromeric satellite DNA. Elife 7, e34122 (2018).
Reeves, R. & Nissen, M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265, 8573–8582 (1990).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 245 (2020).
Sit, A., Hubley, R & Green, P. RepeatMasker http://www.repeatmasker.org (2013-2015).
Kerpedjiev, P. et al. HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 19, 125 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Acknowledgements
This work was supported by the National Institutes of Health (NIH) grant 1R35GM147558-01 awarded to J.T. J.C. and J.P. were supported by the Summer Undergraduate Research Experience (SURE) program at the Emory College of Arts and Sciences. We thank members of the Thakur lab and Roger Deal for their valuable discussions and input on the manuscript. Illumina sequencing of CUT&RUN samples was performed at the Emory Integrated Genomics Core. PacBio sequencing and RNA sequencing were performed at Azenta Inc.
Author information
Authors and Affiliations
Contributions
G.C., J.C., and J.T. conceived and designed the study. G.C., J.C., L.S., S.B., J.P., X.L., and J.T. performed analyses. J.T. and S.B. performed experiments. G.C., J.C., and J.T. wrote the manuscript. G.C., J.C., L.S., and S.B. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Peer review
Peer review information
Nature Communications thanks Nicolas Altemose, Damian Dudka and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chaudhry, G., Chen, J., Snipes, L. et al. Genomic and epigenomic maps of mouse centromeres and pericentromeres. Nat Commun 16, 9688 (2025). https://doi.org/10.1038/s41467-025-64689-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64689-0