Abstract
The mammalian brain comprises a vast number of neurons, exhibiting remarkable diversity in both molecular composition and spatial distribution. However, a comprehensive understanding of how these neurons are organized within the brain remains elusive, largely due to the lack of systematic studies providing three-dimensional coverage of molecularly defined neurons across the entire brain. In this study, we utilized transgenic mice and fMOST imaging to map the spatial distribution of glutamatergic, GABAergic, and modulatory neurons at the single-cell level throughout the whole brain. Our approach enabled precise registration of individual cells to the standardized brain coordinate framework, facilitating the construction of whole-brain cell atlases for commonly used Cre recombinase driver lines. Analysis revealed diverse cellular composition patterns across the brain, aligning with the boundaries of known brain regions in some areas while uncovering previously uncharacterized subdivisions in others. Notably, cortical and subcortical nuclei as small as approximately one millimeter in size exhibited intricate three-dimensional organization, suggesting the presence of finer functional zones.
Similar content being viewed by others
Introduction
The mammalian brain harbors the most intricate network identified to date, composed of a vast and diverse array of neurons1,2. During development, neurons of specific numbers and types are generated in appropriate locations and establish proper synaptic connections within circuits3. In recent years, although an increasing variety of cell types have been identified at the molecular level4, a comprehensive understanding of their spatial organization within the brain remains lacking, impeding our comprehension of complex neural networks.
The exploration of cellular brain organization dates back over a century to the era of Cajal. Despite advances, classical histological techniques based on tissue sectioning, developed during that period, remain in use today. On brain sections, it is possible to label specific cell types using chemical and molecular methods, acquiring data about their morphology and spatial distribution, as well as precisely measuring the cell-type composition at specific anatomical area using spatial transcriptomics. Overcoming the limitations of two-dimensional (2D) perspectives requires increasing the number of brain sections. However, current techniques in sectioning, sampling, and alignment still have limitations, resulting in ambiguities and blind spots that hinder comprehensive surveys of cell types.
Over the past decade, advancements in transgenic fluorescent labeling and whole-brain three-dimensional (3D) imaging technologies have provided new methodologies for studying cellular composition5,6,7. For instance, Kim et al. mapped and quantified seven subtypes of GABAergic neurons across the entire mouse brain using the serial two-photon tomography, producing coronal images at 50 μm intervals8. Murakami et al. combined optical clearing with LSFM to analyze the distribution and number of cells throughout the brains of C57 mice of various ages9. We have also employed the fluorescent Micro-Optical Sectioning Tomography (fMOST) series of imaging techniques10 to study the distribution of CRH+, SOM+, and ChAT+ neurons throughout the mouse brain11,12,13. These investigations have significantly deepened the understanding of the mammalian brain, yet there is still a lack of standardized, systematic, and high-quality data accumulation, particularly in terms of integrative analysis. Furthermore, genetically engineered mice that specifically express various marker genes have become vital animal models in neuroscience, although each model comes with inherent limitations and there is generally a lack of prior knowledge about the distribution of neurons expressing these genes throughout the whole brain.
To address these challenges, we have established the Brain-wide Cellular Architectural Deconstruction (BrainCAD) platform specifically for small animal models such as mice. This platform is designed to detect and identify each neuron expressing specific marker genes across the entire mouse brain, accurately mapping these measurements from different individual animals onto the Allen Common Coordinate Framework version 3 (CCFv3)14,15. For future studies requiring higher spatial resolution to resolve single-cell-level structural details, BrainCAD is also designed to support potential adaptation to the recently developed stereotaxic topographic atlas of the mouse brain (STAM)16. This enables precise spatially-located data collection, 3D reconstruction, and information integration. Based on this, we can quantify the number and density of various cell types within areas of interest, thereby measuring the cellular composition at different spatial locations within the brain.
We initially charted a comprehensive cellular atlas of the brain distributions of 19 common types of neurons and one type of glia in mice. The 19 selected neuronal types comprised 8 glutamatergic, 5 GABAergic, and 6 modulatory subtypes, all of which represent cell populations that are widely studied in contemporary neuroscience. Utilizing the resource, we can ascertain the quantity and density of specific cell types and further explore cellular organizational patterns in each brain region, providing crucial data validation and knowledge expansion for traditional studies. Leveraging precise localization, we constructed a high-resolution 3D cell distribution map (10 μm), uncovering previously unknown subdivisions with distinct cellular organization in both cortical and subcortical regions, suggesting the existence of finer functional zones. Specifically, we identified two subregions differentiated by neuronal organizational patterns, which, as anticipated, also exhibit significant functional differences. This highlights the potential of our cell distribution data for exploring brain functional traits on a more refined scale. Data and analysis tools from this study have been collected on the Molecularly defined Cellular Atlas of the Mouse brain (MiCAM), and are available via a web portal (http://atlas.brainsmatics.cn/MiCAM).
Results
Mapping molecularly defined cells in the whole mouse brain
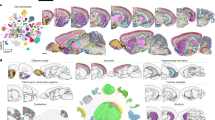
The BrainCAD platform with fMOST is capable of capturing fluorescent signals throughout an entire mouse brain at a voxel resolution of 0.3 μm × 0.3 μm × 1 μm (Methods). This advanced imaging approach surpasses traditional sectioning-based methods by eliminating distortions from slicing, which can impede accurate image registration. The fMOST technology not only enables to distinguish individual cell but also facilitates a comprehensive search across the entire mouse brain, effectively locating all fluorescently labeled cells. Additionally, we acquire the cytoarchitectonic information of the whole brain with propidium iodide (PI)-staining. The platform is strategically designed around the unique capabilities of fMOST, encompassing processes from sample preparation, high-resolution imaging, to post-data processing including cell detection, spatial localization and the final analysis of cell composition (Fig. 1a).
a A pipeline for acquiring and analyzing whole-brain molecularly defined cellular distribution datasets. b Half of a coronal image with GFP-labeled nuclei (green) and PI-stained cytoarchitecture (purple). Enlarged views of nuclei in the orange boxed regions are shown. The image’s position within the whole-brain dataset is indicated in the lower left. c Evaluation of single-cell detection across varying cell densities. The images depict sparse (left) and dense (right) nuclei distributions, with GFP-labeled nuclei and red dots marking automatically detected nuclei centers. Both images are 50 μm maximum projections. The histogram illustrates the precision and recall of cell detection across data blocks with different nuclei densities. d Evaluation of registration across brain regions. The image shows an enlarged view of the registration results, with brain region boundaries (dashed white lines) from CCFv3. This is a 50 μm maximum projection. The box plot displays the Dice score for registration in five brain regions (n, number of biologically independent datasets for evaluating the accuracy of registering certain brain region, n = 20 for each brain region). Constructed based on the interquartile range (IQR), the central line inside the box represents the 50th percentile. The upper and lower boundaries of the box denote the 75th percentile and 25th percentile, respectively. Whiskers extend to 75th percentile + 1.5 × IQR (upper) and 25th percentile − 1.5 × IQR (lower) to truncate outliers, marked with “+”. e Cell quantification of 20 molecularly defined types across 77 brain regions. Nuclei counts per region are color-coded, with row totals representing whole-brain counts (data presented as mean ± SEM, n = 3 mice) shown in the histogram. Circle sizes indicate the proportion of each cell type within a region. Source data are provided as a Source Data file.
We utilized 19 commonly used Cre mouse lines to identify neurons expressing specific marker genes, encompassing various types of glutamatergic, GABAergic, and modulatory neurons. Complete information for all Cre lines is provided in Supplementary Data 6. To specifically label neuronal somata, we crossed LSL-H2B-GFP reporter mice with Cre driver lines. In addition, we used CX3CR1-GFP mice to obtain the whole-brain distribution of microglia, demonstrating that the current platform can be applied not only to mapping neuronal distributions but also to other cell types. The distribution of GAD2+ neurons on a representative coronal plane is depicted in Fig. 1b. The specificity of H2B-GFP labeling for targeted cell types was further validated through immunohistochemical staining (Supplementary Figs. 1 and 2 and Methods).
An automated recognition algorithm was employed to detect cells across the entire brain, determining the spatial positions of individual somata (Methods). In regions with low cell density and minimal adhesion, the algorithm achieved accuracy and recall rates exceeding 95%, while maintaining high performance at approximately 90% in denser cell populations (Fig. 1c and Supplementary Fig. 3). To ensure the quality of cell detection, all automated results were manually reviewed and corrected. For densely packed glutamatergic neurons in specific brain regions with indistinct boundaries, cell counts were estimated based on the fluorescence signal volume (Supplementary Fig. 4). These findings underscore the importance of using section intervals significantly smaller than cellular dimensions to achieve precise localization of all cells.
We implemented an image registration strategy to integrate data from multiple experimental animals into the Allen CCFv3 (Methods). This approach achieves high alignment accuracy across brain regions of varying volumes, with large areas typically achieving Dice scores within the range of 0.95–1.0 (Fig. 1d), and smaller structures scoring predominantly above 0.9 (Supplementary Fig. 5). For smaller brain structures, such as the cerebral aqueduct (AQ), medial habenula (MH), fasciculus retroflexus (fr), and anterior commissure, temporal limb (act), the average registration deviations were remarkably low, ranging from 20 μm to 70 μm (Supplementary Fig. 5). Abbreviations for brain structures are detailed in Supplementary Data 1. To address individual variability that may introduce registration errors, we engaged experienced brain anatomy experts to meticulously review and iteratively refine the registration results, ensuring the highest possible precision.
Using the spatial coordinates of fluorescent protein-labeled cells aligned to the standard mouse brain coordinate framework, we quantified the number, proportion, and density of each neuronal type in distinct regions across the entire brain. The numbers and proportions of 20 cell types were compared across 77 distinct brain areas (Fig. 1e and Supplementary Data 2). Additionally, we developed an interactive dynamic map hosted on a web portal, along with a comprehensive table of the original statistical data (Supplementary Data 3).
To further explore cell composition heterogeneity and account for transitional zones between regions, we divided the entire brain into 3D, equidistant, uniformly sized cubic grids for a more granular analysis (Methods). The grid size is adjustable: with an edge length of 100 μm, the mouse brain is divided into approximately 450,000 units; at 20 μm, this increases to about 28 million units; and at 10 μm, to approximately 227 million units. The distribution patterns of different neuronal types at various grid resolutions are analyzed in subsequent sections, and the results are shared on our web portal.
Organization patterns of glutamatergic neurons
Glutamatergic neurons are a diverse group, comprising multiple subtypes that play distinct roles in transmitting neural signals throughout the brain. These subtypes are heterogeneously distributed, with notable differences between brain regions. For instance, the cortex can be distinguished from the claustrum17 and further subdivided into distinct layers based on the distribution of glutamatergic neurons18.
To explore the distribution patterns of glutamatergic neurons, we analyzed eight neuronal subtypes across the entire mouse brain using soma distribution charts, whole-brain 3D reconstructions, and density heatmaps (Fig. 2a, Supplementary Movie 1 and the online portal). The results revealed significant variability among these neuronal subtypes. While neurons such as Camk2a+, Emx1+, Thy1+, Cart+, and VGLUT2+ were broadly distributed, their spatial patterns varied markedly across the brain. Notable differences were observed in cortical and subcortical regions, including the hippocampal formation (HPF), striatum (STR), thalamus (TH), hypothalamus (HY), midbrain (MB), pons (P), and medulla (MY). In contrast, within the isocortex (ISO), these differences primarily manifested as variations in distribution density across layers and subregions. Neurons such as PlxnD1+, Scnn1a+, and Fezf2+ exhibited more localized distributions, often concentrated in specific brain regions or cortical layers (Fig. 2b).
a The normalized densities of glutamatergic neurons in the whole brain are shown as heatmaps, featuring a dorsal 3D reconstruction and six representative coronal sections. Distances from bregma are labeled in millimeters. Supplementary Movie 1 provides additional data. Scale bar, 2 mm. Regions with high concentrations of PlxnD1+, Scnn1a+, or Fezf2+ nuclei are highlighted. b Hierarchical distribution of eight glutamatergic neuron types in SSp-bfd. Projection thickness is 10 μm for Camk2a+, Emx1+, Thy1+, Cart+, and VGLUT2+, and 100 μm for PlxnD1+, Scnn1a+, and Fezf2+. Scale bar, 300 μm. c A dendrogram-based taxonomy tree of 134 brain regions (left), normalized density fractions of different glutamatergic neuron types across brain regions (middle), and a correlation matrix of glutamatergic neurons in 134 regions (right). Clusters 1–7 (C1–C7) are marked with colored boxes. Brain region or nucleus names are labeled above and to the left of the correlation matrix, with colors representing their parent regions (see Supplementary Data 4). d A 3D reconstruction of the clustering results, displaying only hemispherical regions to enhance the visualization of spatial relationships. e Volume proportions of clusters across 12 anatomical regions. f Heatmap of the densities of 8 glutamatergic neuron types across 7 clusters. g Cubic density analysis of 8 types of glutamatergic neurons. The type of each cube is determined by the maximum density of neurons in the cube. h Volume proportion of cubes in 12 anatomical regions of 8 types. Source data are provided as a Source Data file.
To further investigate spatial distribution patterns at an anatomical level, we divided the brain into 134 distinct regions based on the Allen CCFv3. The raw data and correlation analyses are provided in Supplementary Data 4. By hierarchically clustering these regions according to the density of the eight glutamatergic neuronal subtypes, we identified seven distinct clusters (Fig. 2c). Whether regions cluster together depends on the presence and relative abundance of the eight types of glutamatergic neurons they contain. Regions with similar compositions tend to cluster into the same group, whereas those with dissimilar compositions are assigned to different clusters. Then clusters were mapped onto the Allen CCFv3 (Fig. 2d and Supplementary Fig. 6a, b), revealing significant spatial correspondence between cluster distribution and anatomical regions. The volume proportions of these clusters within different brain regions underscored region-specific cellular composition patterns (Fig. 2e). Notably, the compositional ratios of glutamatergic neurons varied significantly across clusters (Fig. 2f). For instance, Cluster 1 (C1) exhibited the greatest diversity of glutamatergic neuronal subtypes, encompassing the ISO, HPF, cortical subplate (CTXsp), partial olfactory areas (OLF), and STR regions, thereby covering most anatomical regions of the isocortex and allocortex, which are regarded as homologous structures19.
To compare neuronal heterogeneity across the entire brain and within distinct brain regions, we analyzed density characteristics at a spatial resolution of 100 µm × 100 µm × 100 μm. Distribution maps for each 3D grid were generated (Fig. 2g and Supplementary Fig. 6c), and the volume proportions of grid types for each neuronal subtype within different brain regions were quantified (Fig. 2h). Subregional preferences were observed, characterized by anterior-posterior, laminar, and internal-external spatial features. For example, in the caudoputamen (CP), Thy1+ neurons were most densely distributed dorsolaterally, whereas Camk2a+ neurons were concentrated ventromedially. In the medial amygdalar nucleus (MEA), Thy1+ neurons were denser in the dorsomedial region, while grids in the ventral part were enriched with VGLUT2+, Camk2a+, and Emx1+ neurons. Regions such as the anterior olfactory nucleus (AON), piriform area (PIR), field CA1 (CA1), and prosubiculum (ProS) exhibited laminar distribution patterns. For instance, in CA1, the outer layer predominantly contained Camk2a+ and VGLUT2+ neurons, while the inner layer was mainly composed of Emx1+ neurons. In stratified regions like the superior colliculus (SC), Cart+ neurons were densest in the central fissure. In the periaqueductal gray (PAG), VGLUT2+ neurons were concentrated in the periventricular dorsal part, whereas Thy1+ neurons were distributed in peripheral regions. In the medial vestibular nucleus (MV), Cart+ neurons predominated medially, while Thy1+ neurons were primarily found laterally. These findings highlight subregion-specific differences in the cellular composition of glutamatergic neurons, providing evidence of spatial heterogeneity within anatomical brain areas.
Organization patterns of GABAergic neurons
GABAergic neurons play a pivotal role in regulating neural network activity by inhibiting surrounding neurons. This inhibitory control is essential for maintaining the balance between excitation and inhibition, preventing disruptions that could lead to neurological disorders such as epilepsy and schizophrenia20,21. Understanding the distribution of GABAergic neurons throughout the brain is crucial for advancing our knowledge of the brain’s structural and functional organization, as well as the mechanisms underlying information processing. Moreover, it provides valuable insights into the origins and progression of neuropsychiatric conditions, potentially guiding the development of new preventive and therapeutic strategies.
We analyzed the distribution characteristics of five GABAergic neuron subtypes using original neuron distribution maps (Fig. 3a, Supplementary Movie 2, and the online portal). Among the most extensively studied subtypes, GAD2+ and VGAT+ neurons exhibited consistent distribution patterns throughout the brain. However, regional variations were evident, particularly in the superficial layers of the PIR and the ventral side of the pontine gray (PG) (Fig. 3a and Supplementary Fig. 7a). Regarding the principal subtypes—PV+, SOM+, and VIP+ neurons—our analysis builds upon previous studies detailing their distributions8. We compared our results with previous findings across 12 major brain regions at the whole-brain scale in terms of region volume, cell counts, and cell density (Supplementary Fig. 7b–d and Methods). The Pearson correlation coefficients indicated a high level of consistency between the two datasets, demonstrating the good reproducibility of our current results. We observed notable subtype-specific differences, especially in the ISO. PV+ neurons were most abundant in layers L4–L5, SOM+ neurons predominantly in L5, and VIP+ neurons in layers L2/3 (Fig. 3b). Distinct expression patterns were also evident in subcortical regions. For instance, PV+ neurons were exclusively observed in the ventral posteromedial nucleus of the thalamus (VPM), SOM+ neurons in the PAG, and VIP+ neurons in the medial part of the facial motor nucleus (VII) (Fig. 3a). These results suggest that specific GABAergic neuron subtypes are functionally specialized within particular brain regions.
a The normalized densities of GABAergic neurons in the whole brain are shown as heatmaps, featuring a dorsal 3D reconstruction and six representative coronal sections. Supplementary Movie 2 provides additional data. Scale bar, 2 mm. Some of the brain regions with specific distribution patterns have been highlighted. b Hierarchical distribution of five types of GABAergic neurons in SSp-bfd. The projection thickness is 100 μm. Scale bar, 300 μm. c A dendrogram-based taxonomy tree of 134 brain regions (left), normalized density fractions of different GABAergic neuron types across brain regions (middle), and a correlation matrix of GABAergic neurons in 134 regions (right). Clusters 1–6 (C1–C6) are marked with colored boxes. Brain region or nucleus names are labeled above and to the left of the correlation matrix, with colors representing their parent regions (see Supplementary Data 4). d A 3D reconstruction of the clustering results, displaying only hemispherical regions to enhance the visualization of spatial relationships. e Volume proportions of clusters across 12 anatomical regions. f Heatmap of the densities of 5 GABAergic neuron types across 6 clusters. g Cubic density analysis of 5 types of GABAergic neurons. The type of each cube is determined by the maximum density of neurons in the cube. h Volume proportion of cubes in 12 anatomical regions of 5 types. i Normed Glu/GABA ratios across 134 brain regions. Dashed lines represent the mean normed Glu/GABA ratio (0.34) and the value corresponding to equal densities of glutamatergic and GABAergic neurons (0.00). j Sagittal heatmap showing the cubic distribution of normed Glu/GABA ratios. Source data are provided as a Source Data file.
To examine whole-brain distribution patterns, we performed hierarchical clustering of brain regions based on neuronal composition, identifying six distinct clusters with unique cellular profiles (Fig. 3c and Supplementary Data 4). Here, whether regions cluster together depends on the presence and relative abundance of the five types of GABAergic neurons they contain. Then clusters were spatially mapped onto the mouse brain (Fig. 3d and Supplementary Fig. 7e), and their volume proportions were analyzed across brain regions (Fig. 3e). Similar to glutamatergic neurons, the distribution of GABAergic neurons showed strong correlations with anatomical brain region divisions. The composition of GABAergic neurons varied significantly between clusters (Fig. 3f), while regions within the same cluster exhibited similar composition patterns and anatomical hierarchies. For instance, C3 encompassed the most diverse range of GABAergic neuron subtypes (all five types), spanning the ISO, OLF, HPF and CTXsp, as well as several subcortical regions. This may arise from the requirement of neuronal circuits in these regions to recruit multiple types of GABAergic neurons for the fine-tuned regulation of circuit function. C4, characterized by the highest density of PV+ neurons, was exclusively associated with the cerebellum (CB), in which a large population of Purkinje cells exhibits strong PV expression22, and inhibitory interneurons in the molecular layer also widely express PV23,24.
Further analysis at a 100 µm × 100 µm × 100 µm spatial resolution revealed the predominant GABAergic neuron subtypes within each grid (Fig. 3g and Supplementary Fig. 7f). Volume proportions of these grid types across major brain regions were quantified (Fig. 3h). Subregional preferences were apparent, reflecting heterogeneity within known anatomical areas. For example, in the CP, GAD2+ neurons were densely distributed dorsally, while VGAT+ neurons predominated ventrally. In the CA1 region, stratified layers of VGAT+, GAD2+, and VGAT+ neurons appeared sequentially from the dorsolateral to the ventromedial axis—demonstrating a more intricate layered structure compared to the two-layer division of glutamatergic neurons in CA1 (Fig. 2g). In subcortical regions, distinct patterns emerged. In the rostral part of the lateral septal nucleus (LSr), VGAT+ neurons were denser dorsally, while GAD2+ neurons were more abundant ventrally. In the ventral medial nucleus of the thalamus (VM), SOM+ neurons were densely distributed dorsomedially, whereas VGAT+ neurons dominated ventrolaterally. In the PG and tegmental reticular nucleus (TRN), SOM+ neurons were concentrated in the periphery, with PV+ neurons forming a central core. In the VII, VIP+ neurons were densely distributed centrally, while GAD2+ neurons populated the periphery. Similarly, in the parvicellular reticular nucleus (PARN), SOM+ neurons were most abundant in the anteromedial dorsal area, whereas VGAT+ and GAD2+ neurons predominated posteroventrolaterally. These findings highlight the presence of subregions within anatomical brain areas, each characterized by distinct cellular compositions and potential functional roles. Further experimental validation is warranted to elucidate the underlying mechanisms and functional implications.
Spatial distribution of the ratio of glutamatergic and GABAergic neurons
The balance between glutamatergic (Glu) and GABAergic (GABA) signaling is critical for maintaining synaptic and neural network function within the nervous system25. Disruptions to this balance are implicated in various neurological disorders. For instance, in Alzheimer’s disease, an imbalance skewed towards glutamatergic activity may lead to excessive neuronal excitation, impairing memory and cognitive functions26. Investigating the Glu/GABA ratio across the brain offers valuable insights into its role in regulating cognitive processes and elucidating mechanisms underlying neuropsychiatric disorders, and may provide a basis for future explorations into brain-machine interface design and neuromodulation strategies27,28. However, direct measurement of the Glu/GABA ratio at each synapse across the brain is technically challenging. Here, we indirectly assessed the spatial distribution of the Glu/GABA ratio by comparing the numerical densities of glutamatergic and GABAergic neurons across anatomical regions and localized areas.
To quantify this balance, we defined the neuronal density of glutamatergic or GABAergic neurons in any given brain region as the density of the most abundant subtype within that category. Using this metric, we calculated the normalized Glu/GABA ratio (normed Glu/GABA) as the difference between the densities of glutamatergic and GABAergic neurons, divided by their sum (Methods). A normed Glu/GABA value approaching 1 indicates a glutamatergic bias (Glu-biased), while a value nearing −1 indicates a GABAergic bias (GABA-biased). A value close to 0 represents a balanced distribution. We applied this calculation to 134 anatomical brain regions (Fig. 3i). Our analysis revealed that glutamatergic neurons predominate in most regions, particularly in cortical areas. Conversely, all regions within the CB exhibited a GABAergic bias, primarily attributable to the high density of PV+ neurons. Regions such as the HY, pallidum (PAL), and STR demonstrated a balanced Glu/GABA ratio. In the TH, distinct subregions showed contrasting biases: medial geniculate complex (MG), lateral habenula (LH), and midline group of the dorsal thalamus (MTN) were Glu-biased, while the reticular nucleus (RT) and ventral geniculate group (GENv) were GABA-biased, reflecting their divergent roles in neural network regulation.
We further mapped the spatial distribution of glutamatergic and GABAergic neurons across the entire brain at a 100 µm × 100 µm × 100 µm cubic scale (Figs. 2g and 3g). Using this data, we visualized the normed Glu/GABA ratio as a heatmap (Fig. 3j and Supplementary Fig. 8a). This revealed a heterogeneous spatial distribution with anterior-posterior, left-right, inside-out, and laminar preferences. For example, in the CP, the posterior ventral region was more GABA-biased compared to the anterior dorsolateral region. In the MEA, the ventral region showed a glutamatergic bias, while the dorsal region approached a balanced state. The substantia innominata (SI) displayed an anterior dorsolateral band with a stronger GABAergic bias compared to its posterior ventral counterpart. Within the lateral hypothalamic area (LHA), the middle segment along the anterior-posterior axis exhibited a stronger glutamatergic bias than the anterior and posterior segments. The dorsal aspect of the midbrain reticular nucleus (MRN) and the PAG exhibited a stronger glutamatergic bias compared to the ventral aspect. Laminar preferences were evident in the hippocampal CA1 region, where a specific layer exhibited a reduced glutamatergic bias compared to adjacent layers. In the SC, superficial layers were GABA-biased, while deep layers showed a glutamatergic bias. The MY generally displayed an outer glutamatergic bias compared to its inner aspect, with the PARN showing an anterior glutamatergic bias.
To quantitatively depict the normed Glu/GABA distribution, we analyzed the total volume of all 3D grids within intervals of 0.1 across anatomical regions (Supplementary Fig. 8b). Cortical regions, including the ISO, HPF, CTXsp, and parts of the OLF, predominantly exhibited a glutamatergic bias, with nearly all 3D grids in the isocortex falling within normed Glu/GABA > 0. Subcortical regions demonstrated both glutamatergic and GABAergic biases, with the strongest GABAergic preference observed in the CB, where most regions fell within normed Glu/GABA < 0. These findings corroborate prior region-specific analyses and underscore the heterogeneity of Glu/GABA distribution, which extends beyond traditional anatomical boundaries.
Cell organization patterns in the isocortex
The isocortex is organized into numerous subdivisions defined by specialized functions, neuroanatomy, and connectivity patterns. According to the CCFv3, the mouse isocortex comprises 17 major areas further divided into 43 subregions. In cortical information processing, cortical columns—measuring approximately 100 to 380 µm in diameter29—are considered fundamental functional units30,31. The layer-specific variations in cortical subregions partly reflect differences in the distribution of neuron types along the cortical column axis. In this study, we analyzed neuronal organization patterns within anatomical brain regions by integrating cell distribution data along the cortical column direction.
We first examined the cell distribution patterns along the cortical column axis across the 43 cortical subregions by dividing the cortex along its depth and calculating the density distributions of 13 types of glutamatergic and GABAergic neurons (Supplementary Data 5). To accurately capture layer boundaries in the analysis, we adjusted the data based on layer-specific information (Methods). Using Pearson correlation coefficients, we performed hierarchical clustering of the 43 subregions, which identified eight distinct clusters (C1 to C8, Fig. 4a). Whether cortical subregions are clustered together depends on the similarity of the combinatorial distribution patterns of these 13 neuronal types along the cortical columnar axis. Mapping these clusters onto the 3D isocortex revealed spatial patterns: C1 to C5 delineated anterior medial (C5), medial (C2), posterior medial (C4), lateral (C1), and posterior lateral (C3) regions (Fig. 4b), indicating that the distribution of glutamatergic and GABAergic neurons in the isocortex is neither uniform nor random, but instead follows a spatially organized pattern. Due to the spatial folding of the isocortex, a 2D flatmap32 representation was used to fully visualize the clustering results (Fig. 4c). This flatmap, color-coded by clusters, demonstrated spatial correspondence between the clustering patterns and the anatomical atlas. Notably, except for C7 and C8, which comprised isolated subregions, the clusters predominantly included subregions from the same or adjacent brain areas. Regions in C1 and C3 mostly contained layer 4 (black region names in Fig. 4c), while those in C2, C4, C5, and C6 lacked layer 4 (gray region names in Fig. 4c).
a Hierarchical clustering of isocortical subregions based on neuronal density distribution along cortical depth. Left: Taxonomy tree displaying hierarchical clustering results, with a heatmap illustrating the density distribution of various neuron types (columns) across subregions (rows), segmented into 100 equal depth intervals (Methods). Right: Pearson correlation coefficient matrix between subregions, with triangles marking eight clusters (C1–C8) identified in the clustering. b A 3D dorsal view of hierarchically clustered subregions in the hemicerebral isocortex, colored by cluster. c Cortical flatmap of the clustering results with CCFv3 borders. The y-axis represents bregma anterior-posterior (AP) coordinates, while the x-axis shows azimuth coordinates reflecting physical distances traced along the cortical surface on coronal sections. Font color indicates layer 4 presence (black) or absence (gray). d Comparison of neuronal distribution patterns between clusters, using C1 and C5 as examples. Left: Normalized E-I ratios along cortical depth (Methods). Solid gray lines mark hierarchical layer boundaries, and black arrowheads highlight significant differences. Right: Violin plots display the normalized density of 13 neuron types across cortical depths in subregions of C1 (upper) and C5 (lower). e Heatmaps showing neuronal densities with significant differences between C1 and C5. White dashed lines indicate anatomical subregion boundaries. Neuron types and their associated layers are labeled above each flatmap. f Isocortical clustering based on gridded neuronal distribution data. Left: UMAP visualization of gridded data, colored by 17 identified clusters (C1–C17). Right: Clustered isocortical flatmap colored by 17 clusters, with white dashed lines denoting anatomical subregion boundaries. g Heatmaps of neuronal densities showing significant differences among clusters in MOs. Subgraphs include the corresponding 3D subregion distributions and density curves along the AP axis. Neuron types and their layers are labeled above. Each dot represents a nucleus; black arrows indicate MOs locations with significant density changes. Note: Thy1 L6 and Emx1 L6 share the same color bar, as do Cart L5 and Cart L2/3/4. Source data are provided as a Source Data file.
From the clustering results, we derived normalized average density distributions of neuron types along the cortical depth for each cluster (Fig. 4d and Supplementary Fig. 9a). For example, significant heterogeneity was observed between C1 and C5: PlxnD1+ neurons were concentrated in layers 2/3 and 4 in C1 but sparsely distributed in layers 2/3 in C5; Scnn1a+ neurons were prominent near layer 4 in C1 but absent in C5; Fezf2+ neurons were denser in layers 5 and 6 in C5; and PV+ neurons exhibited greater density in layer 5 of C1. Heatmaps of neuronal densities on the cortical flatmap supported these trends (Fig. 4e).
We also analyzed the Glu/GABA ratios across different cortical layers. Consistent with the higher abundance of glutamatergic neurons in the isocortex, the normed Glu/GABA ratio indicated a glutamatergic bias throughout (Fig. 3j and Supplementary Fig. 8). To assess relative differences, we calculated a z-scored normed Glu/GABA ratio (relative normed Glu/GABA) across layers (Supplementary Fig. 9b, Methods). This analysis revealed differential distributions: the lateral association area exhibited lower relative normed Glu/GABA ratios across all layers, while the medial association and visual areas displayed higher ratios. The motor-somatosensory and auditory areas showed a higher glutamatergic bias in deeper layers, whereas the medial prefrontal area had a stronger bias in shallower layers. These findings suggest distinct cell organization patterns across brain areas and layers, reflecting functional specialization within the isocortex.
To investigate neuronal distribution patterns at a finer scale, we applied Leiden clustering33 to the density distributions of 13 neuron types across cortical layers (L1, L2/3/4, L5, L6) in each 100 µm × 100 µm voxel (Fig. 4f and Supplementary Figs. 10, 11). The volume proportions of each cluster within subregions were quantified (Supplementary Fig. 9c). Spatial distributions of these clusters partially aligned with anatomical boundaries, though some subregions from different brain areas were grouped together, and certain subregions were further subdivided. For instance, the secondary motor area (MOs) was split into two clusters along the anteroposterior axis (corresponding to C2 and C12). Heatmaps of neuron densities on the cortical flatmap indicated that this division was primarily driven by density differences in Emx1+ and Thy1+ neurons in layer 6, and Cart+ neurons in layers 2/3/4 and 5 within the MOs (Fig. 4g). This suggests that distinct subregions may exist within the MOs, each potentially subserving different functions34.
Whole-brain distribution of modulatory neurons
Modulatory neurons play a critical role in regulating neuronal signaling by releasing neurotransmitters such as acetylcholine, dopamine, norepinephrine, and serotonin35,36,37. Investigating their distribution provides insights into the mechanisms of information transmission and modulation across brain regions, shedding light on functional states and potential disorders of the nervous system13,38,39,40. A comprehensive understanding of the spatial distribution of modulatory neurons can also facilitate the development of more precise therapeutic strategies41,42,43.
To examine the distribution patterns of modulatory neurons, we utilized neuronal distribution maps, soma distribution charts, whole-brain 3D reconstructions, and density heatmaps for six neuron types across the mouse brain (Supplementary Fig. 12, Supplementary Data 3, Supplementary Movie 3, and the online portal). These neurons exhibited distinct spatial distribution patterns. For example, the MY displayed densities exceeding 4000/mm3, with ChAT+, TH+, and GAL+ neurons densely distributed in the dorsal motor nucleus of the vagus nerve (DMX). In contrast, the nucleus raphe pallidus (RPA) was enriched in CRH+, SERT+, and GAL+ neurons. Other regions, such as the perihypoglossal nuclei (PHY) and the parasolitary nucleus (PAS), displayed distinct distributions of CRH+ and TH+ neurons or ChAT+ and TH+ neurons, respectively. Additionally, specific neuron types showed dense distributions in characteristic regions: ChAT+ neurons in the hypoglossal nucleus (XII), nucleus ambiguus (AMB), VII, and dorsal column nuclei (DCN); CRH+ neurons in the inferior olivary complex (IO); TH+ neurons in the nucleus of the solitary tract (NTS); and SERT+ neurons in the nucleus raphe obscurus (RO).
To explore the interplay between modulatory neurons and glutamatergic or GABAergic populations, we examined their influence on the normed Glu/GABA in 134 brain regions (Supplementary Fig. 12). Using a weighted normed Glu/GABA metric, termed the Modulatory normed Glu/GABA (Methods), we found that ChAT+, TH+, and DAT+ neurons exhibited Modulatory normed Glu/GABA values lower than the normed Glu/GABA average, indicating a preference for GABA-biased regions. Conversely, CRH+, SERT+, and GAL+ neurons showed higher Modulatory normed Glu/GABA values, suggesting a preference for Glu-biased regions. It is important to note that Modulatory normed Glu/GABA reflects the overall distribution of each neuron type across the brain, and their functional roles may vary across specific brain regions and circuits.
To further investigate the relationship between modulatory neurons and normed Glu/GABA ratios, we analyzed the proportional distribution of the six neuron types across 0.1 intervals of normed Glu/GABA within cubic volumes of 100 µm × 100 µm × 100 µm (Supplementary Fig. 13). The analysis revealed distinct distribution trends: ChAT+, TH+, and DAT+ neurons, which preferentially occupy GABA-biased regions, were more abundant in intervals within the “<0” range. In contrast, CRH+, SERT+, and GAL+ neurons, associated with Glu-biased regions, exhibited higher proportions in intervals within the “>0” range. Additionally, TH+ and DAT+ neurons, both dopaminergic, displayed similar distribution patterns across the brain, with DAT+ neurons predominantly located in the OLF, MB, and HY. In comparison, TH+ neurons extended beyond these regions to the ISO, STR, PAL, and MY. These findings provide valuable insights into the potential regulatory roles of modulatory neurons on glutamatergic and GABAergic activity within neural circuits.
Whole-brain cell organization patterns revealed by unsupervised clustering
Comprehensive and high-resolution cellular distribution data enables the examination of neuronal distribution characteristics and spatial organizational patterns at a level of detail finer than traditional anatomical brain regions. In this study, we utilized a cube size of 100 µm × 100 µm × 100 µm to analyze neuron subtype organization at sub-brain levels, independent of established brain region boundaries (Figs. 2g, 3g, j, 4f and Supplementary Figs. 6c, 7c, 8, 13). These organizational patterns not only strongly correlate with known anatomical regions but also suggest the potential for redefining or further subdividing brain regions based on cellular composition.
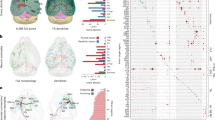
We divided the entire mouse brain into a 3D grid with a resolution of 10 µm × 10 µm × 10 µm (Fig. 5a), where each grid served as a computational unit characterized by a 19-dimensional parameter vector representing the spatial distances of 19 neuron types (Methods). Due to the vast number of grids, we sampled data at 20 µm intervals for clustering and dimensionality reduction analyses (Fig. 5b, c), identifying 18 distinct clusters. Whether 3D grids are grouped into the same cluster depends on the similarity of the distributional composition patterns of the 19 neuronal types within the spatial region centered on each grid. Several clusters aligned with known anatomical regions, revealing the distribution proportions and densities of neuron types within each cluster (Supplementary Fig. 14). When these clusters were mapped onto the CCFv3 (Fig. 5d), a strong correspondence between clustering spatial locations and anatomical boundaries was observed, validating the robustness of our methodology. For instance, in the ISO, layers L1, L2/3, and L5/6 were accurately grouped into C1, C2, and C3, respectively, distinct from the HPF, which corresponded to C7 (Fig. 5e, upper). However, some clustering boundaries deviated from known anatomical regions, such as the division of the motor-related midbrain (MBmot) into two parts (C13 and C14) along the dorsoventral axis (Fig. 5e, middle). Additionally, clustering revealed subdivisions not aligned with known anatomical boundaries, such as the division of the zona incerta (ZI) into two clusters (C9 and C14) (Fig. 5e, bottom).
a Schematic of grid analysis. The CCFv3 3D space was divided into 10 μm cubes; distance parameters for 19 neuron types within each cube served as features (Methods). b UMAP visualization of whole-brain cubic data, downsampled to 20 μm for computation. Each dot represents a cube, colored by its brain region. c The first-stage clustering result of the whole-brain cubic data. d Visualization of clustering results in coronal, horizontal, and sagittal planes. Distances from bregma for coronal planes, from the midline for sagittal planes, and from the dorsal top for horizontal planes of the mouse brain are indicated below, with all dimension units in millimeters. e Enlarged views comparing of clustering results with anatomical boundaries. f Volume proportion of 18 clusters in 12 big brain regions. g 3D views of C6 in the first-stage and coronal visualization of sC13 and sC17 in the secondary-stage. Distances from bregma are shown below (mm). h Heatmaps indicating heterogeneous distributions of Camk2a+, GAD2+ and VGAT+ neurons in CPdl and CPvl. Color-coded distance parameters represent neuron density. i Fiber photometry recording of glutamatergic or GABAergic neurons in CPdl or CPvl. Left, experimental strategy for virus injection with fiber implantation for calcium recording (top) and schematic of open-field test (bottom). Right, average calcium responses of CPdl (blue) and CPvl (red) aligned to locomotion onset (pink line) and end (azure line). Quantification of peak time or area under the curve (AUC) highlights the functional differences between CPdl and CPvl (two-tailed unpaired t-test; mean ± SEM). In Camk2a group, trials = 11, mice n = 7 for CPdl and trials = 5, n = 4 for CPvl; in GAD2/VGAT group, trials = 8, n = 4 for CPdl and trials = 6, n = 4 for CPvl. j Optogenetic stimulation in CPdl and CPvl. Left, behavioral test procedure. Middle, cumulative positions of mice during control and stimulation periods. Right, statistical plots of mean velocity during control (gray) and stimulation (blue) periods (two-tailed paired t-test). In Camk2a group, trials = 6, n = 4 for CPdl, trials = 9, n = 5 for CPvl; in GAD2/VGAT group, trials = 5, n = 5 for CPdl and trials = 6, n = 6 for CPvl. Source data are provided as a Source Data file.
Analyzing the proportions of 3D grid volumes assigned to different clusters within each brain region (Fig. 5f) revealed intriguing patterns. For example, the CB corresponded predominantly to C4, the TH to C10, and the P to C11, while the MY primarily comprised C8 and C16. Although adjacent brain regions generally exhibited higher similarity, exceptions emerged. For instance, while the HY and TH are both adjacent to the PAL, clustering results suggested a higher similarity between HY and PAL, with most of their regions grouped into C9. Additionally, clusters C5 and C7 encompassed nearly all regions of the allocortex (HPF, CTXsp, and OLF), reflecting homogeneity in neuronal composition within these areas.
To refine our understanding of brain region boundaries and uncover potential subregional connections, we performed secondary clustering analysis within each primary cluster, identifying 68 sub-clusters (sCs) across the brain (Supplementary Fig. 15a). These sCs provided finer distinctions, such as the segregation of cortical layers L2/3, L4, L5, and L6 into distinct clusters (Supplementary Fig. 15b, upper left) without clear differentiation across cortical areas, emphasizing greater intra-layer differences than inter-regional variations. Larger regions were further subdivided, such as the polymodal association cortex (DORpm) and sensory-motor cortex (DORsm) related areas in the thalamus, corresponding to sC40 and sC37 (Supplementary Fig. 15b, middle left). These sCs also revealed richer subdivisions than known anatomical boundaries, such as lamination in the main olfactory bulb (MOB), CB, and hippocampal fields CA1 and CA3 (Supplementary Fig. 15b, right panels). Quantitative characterization of the 3D grids within clusters and regions (Supplementary Fig. 15c) indicated that some clusters were primarily driven by the enriched distribution of specific neuron types (Supplementary Fig. 15d).
To validate the biological significance of these newly delineated subregions, we analyzed C6 as a case study. Secondary clustering of C6 identified six sub-clusters, including sC13 and sC17, which divided the posterior-lateral caudoputamen into dorsolateral (CPdl) and ventrolateral (CPvl) regions (Fig. 5g). These subregions exhibited distinct neuronal compositions, with heterogeneous distributions of Camk2a+, GAD2+, and VGAT+ neurons (Fig. 5h). Calcium imaging during mouse movement revealed diametrically opposite response patterns in glutamatergic neurons between CPdl and CPvl, while GABAergic neurons’ responses differed only after movement cessation (Fig. 5i and Supplementary Fig. 16). Optogenetic stimulation further demonstrated functional heterogeneity, with activation of glutamatergic neurons in CPvl or GABAergic neurons in CPdl significantly enhancing locomotor activity, while activation of glutamatergic neurons in CPdl or GABAergic neurons in CPvl showed no significant effect (Fig. 5j). These findings indicate that CPdl and CPvl play distinct roles during mouse locomotion. Previous circuit-based studies have also shown that these two subregions receive inputs from different cortical areas44, suggesting that their functional differences may arise from multiple factors, including cellular composition and circuit architecture. Taken together, our experiments demonstrate that unsupervised clustering can help uncover previously unrecognized structural subregions within established anatomical boundaries, thereby providing important clues for identifying distinct functional domains.
Distinct subregions in subcortical areas
The subcortical regions present a more complex and enigmatic structure and functionality due to the challenges associated with data acquisition. Many boundaries in these areas remain indeterminate or controversial45,46,47. Previous studies have highlighted cellular composition heterogeneity within these regions48,49,50, suggesting that more refined functional specializations exist. However, spatial resolution limitations have hindered accurate subregion boundary delineation. By utilizing foundational data obtained in the present study, we can now investigate the cellular organization in subcortical brain areas at a scale of 10 µm and potentially define subregion boundaries. Here, we conducted preliminary studies on three specific subcortical areas: the basal forebrain (BF), ZI, and SC.
The BF comprises several gray matter structures located medially and ventrally within the telencephalon and diencephalon (Supplementary Fig. 17a), including the SI, magnocellular nucleus (MA), medial septal nucleus (MS), and diagonal band nucleus (NDB)51. We analyzed the distribution density and spatial uniformity of 14 neuron types known to be notably present in the BF (Supplementary Fig. 17b). Both glutamatergic and GABAergic neurons, as well as modulatory types, are found in the BF. These neurons are unevenly distributed (Supplementary Fig. 17c), with significant spatial heterogeneity evident in 3D space (Supplementary Fig. 17d). Using a distance-based density parameter of these 14 neuron types in 10 µm cubic data, we performed unsupervised clustering of the entire BF, resulting in six clusters (Fig. 6a and Supplementary Fig. 17e). UMAP dimensionality reduction and subsequent 2D/3D clustering visualizations revealed that C4 and C5 closely correspond to the MS and NDB subregions, with their boundaries showing a high degree of concordance with known anatomical demarcations defined by differences in neuronal composition (Fig. 6b and Supplementary Fig. 17f). The division is primarily driven by the spatial distribution characteristics of Cart+, PV+, SOM+, and GAL+ neurons (Supplementary Fig. 17g, h). Unlike known anatomical findings, clustering differentiated the ventral part of the pallidum, composed of SI and MA, into four parts (C1, C2, C3, and C6 in Fig. 6a), with no overlap with the boundaries of SI and MA. SI was divided into medial and lateral subareas (C1 and C2), while the medial parts of posterior SI and MA clustered together (C3), and their lateral sides formed C6. This division is mainly due to the distinct distributions of Camk2a+, VGLUT2+, GAD2+, VGAT+, and CRH+ neurons along the medial-lateral axis of SI (Supplementary Fig. 17g, h). Previous studies on cholinergic neuron projections in BF have demonstrated spatial topographic properties52. Our results suggest that such organization may not only underlie its topographic projections but also reflect potential internal heterogeneity. Further biological experiments will be required to validate this hypothesis.
a UMAP dimensionality reduction visualization of all data cubes in BF, colored by anatomical subregions (upper) and clusters (lower) to which they belong. b 3D views and 2D coronal views of clustering results in BF, with borders (black, based on CCFv3) of adjacent anatomical brain regions shown in the 2D views. c UMAP dimensionality reduction visualization of all data cubes in ZI, colored by anatomical subregions (upper) and clusters (lower) to which they belong. d 3D views and 2D coronal views of clustering results in ZI, with borders (black) of adjacent anatomical brain regions shown in the 2D views. e UMAP dimensionality reduction visualization of all data cubes in SC, colored by anatomical subregions (upper) and clusters (lower) to which they belong. f 3D views and 2D coronal views of clustering results of SC, showing the clustering outcomes with clear representations in both perspectives.
The ZI is part of the hypothalamic lateral zone, located ventrally within the mouse brain (Supplementary Fig. 18a), and is involved in a wide range of functions, from motor to social behaviors. Currently, there is no unified standard for the subdivision of ZI in rodents53. Our data show a broad distribution of cell types in ZI (Supplementary Fig. 18b), mostly characterized by spatial unevenness (Supplementary Fig. 18c). We selected eight neuron types with significant numbers (>1000/mm³) and spatial heterogeneity (Supplementary Fig. 18d) for unsupervised clustering within ZI using the same analysis approach as for BF (Fig. 6c and Supplementary Fig. 18e). The results delineated ZI into five clusters, with C5’s boundary almost exactly aligning with the anatomically defined subregion fields of Forel (FF) of ZI in the CCFv3. The other four clusters sequentially divided ZI along the anteroposterior axis into anterior (C1), middle (C2), posterior dorsal (C3), and posterior ventral (C4) sections (Fig. 6d and Supplementary Fig. 18f). The anterior section (C1) was primarily formed by the aggregation of TH+ neurons (Supplementary Fig. 18g), the middle section (C2) by Camk2a+, SOM+, and TH+ neurons, and the posterior dorsal and ventral sections (C3 and C4) by Scnn1a+ and PV+ neurons, respectively (Supplementary Fig. 18h). Notably, SOM+ neurons exhibit a shell-like distribution in the posterior ZI, dividing the anterior part of the posterior section into dorsal and ventral areas, roughly along the boundary between C3 and C4. This previously unreported SOM+ neuron distribution pattern in ZI was confirmed by immunohistochemical staining experiments, showing GFP+ neurons in the middle ZI exhibiting this specific distribution pattern (Supplementary Fig. 19). This result reflects the high degree of neuronal subtype heterogeneity within the ZI, which further partitions the ZI into distinct subregions. Given the complex and diverse functions in which the ZI is involved, these subregions may correspond to specific functional specializations.
The SC, located dorsally in the midbrain (Supplementary Fig. 20a), is highly conserved across mammals54 and plays a crucial role in various high-level brain functions. A wide distribution of different neuron types is observed in the SC (Supplementary Fig. 20b), with Thy1+, VGLUT2+, Cart+, GAD2+, and VGAT+ neurons being the most densely distributed (>40,000/mm³), followed by SOM+, Camk2a+, and PV+ neurons (>5000/mm³), and less densely distributed neurons including TH+, CRH+, Scnn1a+, GAL+, and VIP+ (<2500/mm³). The density distribution maps reveal that these neuron types are not uniformly distributed (Supplementary Fig. 20c). We first analyzed the proportion of each neuron type across the layers of the SC (Supplementary Fig. 20d), with glutamatergic and modulatory neurons concentrated in the intermediate layers, while GABAergic neurons were predominantly found in the superficial layer, indicating a close relationship between the spatial distribution of neurons and their types within the SC. The 3D distribution further reflects this characteristic (Supplementary Fig. 20e). We clustered the distance-based grid data within the SC, obtaining six clusters (Fig. 6e). These clusters separate the SC in 3D space along dorsoventral and mediolateral axes, forming fine-grained subdivisions along the anteroposterior direction and aligning with anatomical layering directions (Fig. 6f and Supplementary Fig. 20f). To identify the relationship between the clustering location and the spatial distribution of neurons, we analyzed the normalized density of different neuron types within each cluster, which revealed concentrated distributions of one or several neuron types (Supplementary Fig. 20g). C1 mainly includes the zonal layer (SCzo) and superficial gray layer (SCsg) of the SC, with aggregated distribution of GABAergic neurons. C3 is located on the ventral medial side near the midline, with boundary formation determined by CRH+ and Scnn1a+ neurons. C4 is located on the ventral lateral side, primarily formed by Scnn1a+ neurons. C5 is located on the anterior ventral lateral side, mainly formed by SOM+ neurons. C6 is located in the posterior SCzo and SCsg, primarily formed by TH+ neurons. The remaining area forms C2, which spans the medio-lateral and dorsoventral parts, forming divisions along both axes similar to anatomical layering and axis-specific divisions along the mediolateral and anteroposterior axes (Supplementary Fig. 20h). This result is partly consistent with previous findings showing that different ZI subregions project to distinct SC subregions55, suggesting that, beyond the established anatomical lamination, the SC may also exhibit additional medial–lateral and anterior–posterior subdivisions, within which distinct neuronal types are organized to receive and process different streams of input.
These analyses demonstrate that studying the cellular organization in subcortical brain areas or nuclei at a resolution close to individual cells can reveal potential subregion delineations within anatomical boundaries. This approach provides clues for discovering functional subregions within these areas or nuclei and offers data-driven insights for understanding brain functions.
Discussion
The mouse brain contains approximately 70 million neurons, each expressing distinct genes and contributing to neural circuits. While extensive research has mapped the spatial distribution of specific neuronal types8,13,32,56,57,58,59, a comprehensive census of all brain cells has been hindered by technical limitations. Integrating data across studies has been challenging due to inconsistencies60,61,62,63,64.
In this study, we combined transgenic labeling and fMOST imaging to map molecularly defined neurons throughout the entire mouse brain, aiming to create a high-resolution cell atlas. The study involved healthy adult mice (aged over 8 weeks, with at least three individuals per strain) and currently encompasses 20 cell types, with potential for future expansion. Our findings provide insights into the quantity and localization of different neuronal types while exploring cellular organizational patterns and functional mechanisms. The use of genetically engineered mouse models, such as Cre-loxP, allows precise gene manipulation in specific cells, circuits, and time points. We present detailed data on the distribution of neurons in 20 common mouse lines, offering a 3D comparison across the brain, which is currently anchored to the Allen CCFv314 framework to ensure consistent annotation across studies. For future investigations that require higher spatial resolution for single-cell localization in fine structural contexts, these existing neuron distribution datasets could be further leveraged by adapting to the recently developed stereotaxic topographic atlas of the mouse brain (STAM)16. As a reference with isotropic 1-μm resolution and 3D topographies of 916 brain structures, STAM would enable more granular parsing of our cell localization data. These precise spatial data will enable researchers to select brain regions and cell types of interest more accurately, considering potential effects on adjacent circuits and regions, and STAM adaptation could further refine such analyses by linking cellular distributions to finer anatomical subdivisions.
Recent advances in spatial transcriptomics have allowed for the categorization of cell types based on their transcriptomic profiles, providing valuable insights into cell distribution across the brain65,66,67,68,69,70. These studies have subdivided the mouse brain based on cell type distribution65,69,70, partly consistent with our findings. However, current “whole-brain” spatial transcriptomic maps rely on z-sampling data and do not precisely localize individual cells in 3D space. In contrast, our study provides precise measurements at a 10 μm resolution, enhancing our understanding of cellular composition.
We also compared our neuronal distribution results with Allen Mouse Brain in situ hybridization Atlas from the Allen Brain data portal71. In most cases, the results were consistent; however, we found some discrepancies in the distribution of certain neuronal types in specific brain regions (Supplementary Fig. 21). This is likely due to the transient expression of these genes during development in those regions. Additionally, our immunohistochemical experiments show over 90% colocalization of GFP in neurons across the cortical and subcortical areas studied.
Finally, the normed Glu/GABA ratio and Modulatory normed Glu/GABA ratio data we collected reflect glutamatergic and GABAergic neurons distribution characteristics in the brain. While not fully representing synaptic characteristics due to local and long-range connections, these data help researchers consider the local effects between neuron types, including glutamatergic, GABAergic, and modulatory neurons, when selecting brain regions or stimulation targets.
The 19 neuronal types included in this study were selected to encompass, as comprehensively as possible, the most frequently investigated subtypes among glutamatergic, GABAergic, and modulatory neurons, prioritizing those lines most representative of the targeted neuronal populations. For example, the eight glutamatergic neuron types include pan-cortical populations such as Camk2a+, VGLUT2+, and Emx1+ neurons, as well as layer-specific subtypes such as PlxnD1+, Scnn1a+, and Fezf2+ neurons. The five GABAergic types comprise both pan-GABAergic neurons (GAD2+ and VGAT+) and the three major cortical inhibitory subclasses (PV+, SOM+, VIP+). The six modulatory types include major neurotransmitter systems—cholinergic (ChAT+), dopaminergic (TH+ and DAT+), and serotonergic (SERT+)—as well as peptidergic neurons (CRH+ and GAL+). Nevertheless, we acknowledge that our study does not capture the full diversity of neuronal subtypes already identified in the field of neuroscience. The current dataset is presented as an open resource to facilitate further research, with the expectation that it will be expanded in the future to provide a more comprehensive and broadly applicable reference.
An unavoidable issue in our study is the co-expression of neurotransmitters across certain neuronal populations. Previous studies have reported neurons co-expressing glutamate and GABA in regions such as the ventral tegmental area72, the endopiriform nucleus73, and the supramammillary nucleus74, as well as neurons co-expressing TH and GABA in several hypothalamic nuclei75. Such co-expression raises two distinct challenges in our analyses. First, in clustering analyses, co-expression may introduce correlations among the dimensions of the input vectors. To minimize its impact, we employed several strategies. For targeted clustering of specific neuronal subtypes, neuronal densities were log-transformed (Methods), which reduced the dominance of subtypes with considerably high densities and attenuated correlations. We then used correlation coefficients as similarity measures followed by Ward’s hierarchical clustering, thereby focusing on pattern similarity rather than raw Euclidean distances and reducing redundancy across dimensions. For unsupervised clustering encompassing all neuronal populations, we used a reciprocal-nearest-neighbor density metric that incorporated spatial constraints (Methods), applied t-SNE to compress correlated features into lower-dimensional representations, and finally adopted Leiden clustering, a graph-based method that relies on neighborhood structure rather than Euclidean distance. Together, these steps progressively mitigated the distortive effects of correlations, ensuring the robustness of the clustering results. Second, in analyses involving arithmetic operations across neuronal classes, such as calculating the normed Glu/GABA ratio, co-expression presents a greater limitation. For regions where glutamate and GABA co-expression is prevalent, the summed counts of glutamatergic and GABAergic neurons cannot be regarded as fully accurate. Since neurotransmitter co-expression occurs in complex and region-specific patterns, with varying combinations and proportions that are difficult to quantify comprehensively, the precise impact on ratio-based calculations remains uncertain. Future experimental studies will be required to systematically characterize co-expression across the brain and refine such analyses. In summary, although neurotransmitter co-expression imposes limitations on certain aspects of our study, particularly arithmetic-based measures, our clustering framework—by design—was robust against such correlations, thereby preserving the biological validity of the results.
Looking forward, this cell atlas lays a foundational framework for more integrative studies of brain structure and function. By combining our current dataset with additional techniques—such as neural circuit tracing, single-neuron morphological reconstruction, and spatial transcriptomics—future research may generate comprehensive maps of cell types, connectivity, and activity patterns at the whole-brain scale. Moreover, with the ongoing progress in large-scale cellular and connectomic imaging technologies, similar approaches are now being extended to non-human primates, including full-brain high-resolution mapping of neural projections in the monkey brain76,77. These developments pave the way for translating findings from model organisms to humans. The data and methodology presented here can serve as a reference for comparative studies across species, ultimately helping to bridge the gap between basic and clinical neuroscience. Such cross-species comparisons may facilitate a better understanding of the cellular basis of brain functions and disorders at unprecedented resolution.
Methods
Mice
All animal experiments were conducted in accordance with the guidelines approved by the Institutional Animal Ethics Committee of Huazhong University of Science and Technology. Efforts were made to minimize both the number of animals used and their suffering. Animals were housed in a controlled environment at 22 ± 1 °C, with 55 ± 5% humidity, under a 12-h light/dark cycle (lights ON at 06:00, lights OFF at 18:00), with food and water provided ad libitum.
Experiments were performed on adult mice (over 8 weeks old) of both sexes, with three animals per group. Both male and female mice were included in the study, and no sex-specific effects were investigated or observed. Transgenic mouse lines and viral vectors used in this study were sourced from The Jackson Laboratory and The Mutant Mouse Regional Resource Center. To label the nuclei of specific neuron types, Cre driver lines (Camk2a-Cre, Emx1-Cre, Thy1-Cre, Cart-Tg1-Cre, VGLUT2-Cre, PlxnD1-2A-CreER, Scnn1a-Tg2-Cre, Fezf2-2A-CreER, GAD2-Cre, VGAT-Cre, PV-Cre, SOM-Cre, VIP-Cre, ChAT-Cre, CRH-Cre, TH-Cre, SERT-Cre, DAT-Cre, GAL-Cre) were crossed with LSL-H2B-GFP reporter mice. Complete information for all Cre lines is provided in Supplementary Data 6.
For inducible CreER lines (PlxnD1-2A-CreER and Fezf2-2A-CreER), tamoxifen was prepared by dissolving it in corn oil at room temperature with constant rotation overnight. The solution (20 mg/mL) was administered via intraperitoneal injection three times, with injections given every other day. Additionally, three Cx3cr1-GFP mice (12 weeks old) were used to study the whole-brain distribution of microglia.
Tissue preparation
Histological procedures were performed as previously described6,7; the specific implementation in this study (consistent with the aforementioned procedures) was as follows: Mice were anesthetized with a 1% sodium pentobarbital solution and subsequently perfused intracardially with 0.01 M phosphate-buffered saline (PBS, Sigma-Aldrich), followed by 4% paraformaldehyde (Sigma-Aldrich) in 0.01 M PBS. Brains were then dissected, fixed in 4% paraformaldehyde for 24 h at 4 °C, and rinsed in PBS for 12 h at 4 °C.
Immunohistochemical staining
For immunofluorescent staining, post-fixed brains were sectioned into 50 μm coronal slices using a vibrating slicer (Leica 1200S, Germany). Sections containing regions of interest were selected to characterize labeled neurons (Supplementary Figs. 1, 2, 7A and 18). The sections were blocked for 1 h at 37 °C in 0.01 M PBS containing 5% (wt/vol) bovine serum albumin (BSA) and 0.3% Triton X-100. Subsequently, sections were incubated with the appropriate primary antibodies (anti:GABA(GAD2), mouse, Sigma-Aldrich, MAB351, 1:200 dilution; anti:GABA(VGAT), rabbit, Sigma-Aldrich, AB5062P, 1:200 dilution; anti:PV, mouse, Sigma-Aldrich, MAB1572, 1:1000 dilution; anti:SOM, mouse, Santa Cruz, sc-7819, 1:200 dilution; anti:ChAT, goat, Sigma-Aldrich, AB144P, 1:500 dilution; anti:TH, rabbit, Sigma-Aldrich, T8700, 1:1000 dilution; anti:TPH2, rabbit, Thermo Fisher, PA1-778, 1:400 dilution) for 12 h at 4 °C, followed by five washes in PBS at room temperature. Next, the sections were incubated with fluorophore-conjugated secondary antibodies (Alexa Fluor™ 594 Donkey anti-mouse lgG (Invitrogen, A-21203, 1:500 dilution); Alexa Fluor™ 594 Donkey anti-goat lgG (Invitrogen, A-11058, 1:500 dilution); Alexa Fluor™ 594 Donkey anti-rabbit lgG (Invitrogen, A-21207, 1:500 dilution)) for 2 h at room temperature. After washing with PBS, DAPI (1 ng/mL) was applied to the stained sections for 5 min. Finally, sections were washed, mounted, and imaged using a confocal microscope (LSM 710, Zeiss, Jena, Germany).
fMOST imaging and preprocessing
Data acquisition was performed using the HD-fMOST system6 as previously described. Briefly, samples were mounted on a high-precision 3D translation stage and immersed in a 2 μg/mL PI solution to counterstain the cytoarchitecture in real time during data acquisition. A two-channel line-illumination modulation (LiMo) microscope was employed to simultaneously image the nuclei and PI-counterstained cytoarchitecture at a voxel resolution of 0.3 μm × 0.3 μm × 1 μm. The HD-fMOST system automated the imaging and sectioning cycles until brain-wide data acquisition was complete. The total data acquisition time for each brain was approximately 120 h, generating a dataset of around 4 terabytes, which included more than 11,000 coronal images.
For the collected high-resolution datasets, image preprocessing, including mosaic stitching and illumination correction, was performed for both the GFP and PI channels as described previously7. Briefly, mosaics of each coronal section were stitched to reconstruct the entire section, ensuring accurate spatial orientation and overlap between adjacent images. Lateral illumination correction was applied section by section. Images from the GFP channel were saved in 16-bit LZW-compressed TIFF format, while those from the PI channel were stored in 8-bit format.
Whole-brain registration
We employed the BrainsMapi scheme15, which leverages anatomical region characteristics, to register whole-brain imaging data to the CCFv3 atlas. To enhance data quality, preprocessing steps were performed, including background removal, uniformization of signal distribution, precise extraction of the brain’s external contour, and initial spatial orientation correction. The preprocessed data were then resampled to achieve the 10-μm isotropic resolution required for registration. Following preprocessing, brain region contours were semi-automatically extracted from the PI channel using the interactive image processing tool Amira. Under the guidance of anatomical experts, 14 brain regions were selected as registration features: the outline, corpus callosum (cc), CP, lateral ventricle (VL), anterior commissure of temporal limb (act), HIP, MH, paraventricular hypothalamic nucleus (PVH), mammillothalamic tract (mtt), fasciculus retroflexus (fr), PG, fourth ventricle (V4), facial nerve (VIIn), and CB. These regions, distributed throughout the brain, were chosen for their anatomical conservation and ease of identification, ensuring registration accuracy and objective feature extraction. Registration parameters were determined by matching the extracted anatomical region features using both linear and nonlinear models implemented in the ANTs tool78. Finally, the deformation parameters were applied to the neuronal distribution channel, aligning it with the standard brain space.
Whole-brain cell counting
The whole-brain cell counting procedure followed a previously described method11; the specific implementation in this study was as follows: The nuclei of neurons were automatically detected using NeuroGPS software79 combined with the TDat data format80 for parallel computation. The programs, written in C++, were executed on computational clusters. The transformation parameters from the registration process were subsequently applied to the centroids of the detected nuclei, mapping them into a unified spatial framework.
For regions with nuclei densities too high to be detected automatically by NeuroGPS (Supplementary Fig. 4a), we developed a pipeline to estimate the number of nuclei. This issue arose in the dentate gyrus (DG) region labeled with VGLUT2+, Emx1+, and Thy1+ nuclei. We assumed that the sizes and intensities of labeled nuclei in the DG were similar to those in other regions and did not account for overlapping nuclei in the images, irrespective of their density. The flowchart for generating a linear model to estimate nuclei counts in regions with considerably dense distributions is shown in Supplementary Fig. 4b. Briefly, data blocks with varying densities of nuclei that could be automatically detected by NeuroGPS were used as original image stacks and binarized using adaptive image thresholding81. After erosion to remove false-positive signals, the stacks served as masks for segmentation, utilizing an active contours region-growing algorithm82. Typical examples of raw data, corresponding detected nuclei, and segmented volumes are shown in Supplementary Fig. 4c. Using the segmented foreground volume and manually verified nuclei counts for each data block, we constructed a linear model (Supplementary Fig. 4d) to estimate nuclei counts for any data blocks with a known volume. This segmentation method was then applied to obtain the foreground volume of the densely labeled DG region, enabling estimation of the total neuron count in this area.
Correlation analysis based on anatomical brain region
We analyzed cellular composition by comparing the neuronal densities of n types across 134 brain regions (n = 8 for Fig. 2c, n = 5 for Fig. 3c). The density of neurons of type i in brain region j is the number of neurons per unit volume in brain region j, defined as \({D}_{{den}}^{{ij}}\). To facilitate further analysis, we transformed \({D}_{{den}}^{{ij}}\) by taking the base-2 logarithm of \(({D}_{{den}}^{{ij}}+1)\), where adding 1 prevents errors when calculating the logarithm of zero-density values. The transformed values were denoted as \({{D}_{{den}}^{{ij}}}^{{\prime} }\). For each brain region, the transformed densities of n neuronal types formed a column vector with n elements. Spearman correlation coefficients were calculated between all pairs of these column vectors, representing the 134 brain regions. Subsequently, Ward’s hierarchical clustering was performed using these correlation coefficients to reveal patterns of similarity among brain regions.
Comparison of GABAergic neuronal subtype distributions with published data
Since the data in ref. 8 are presented in the format of mean ± standard deviation (SD), we also calculated the mean and SD of our data for comparison. For regional volume, we did not measure the volume of each brain region in individual samples but instead used the values provided by the Allen CCFv3; therefore, no SD is available for this dataset. For cell density, the supplementary materials of ref. 8 reported separate mean and SD values for the male group (n = 5) and the female group (n = 5). Thus, the two groups needed to be combined to obtain the mean and SD for a total of 10 samples. Assuming the mean, SD, and sample size of the male group are \({\bar{x}}_{m}\), \({{SD}}_{m}\) and \({n}_{m}\), respectively, and those of the female group are \({\bar{x}}_{f}\), \({{SD}}_{f}\) and \({n}_{f}\), respectively, the combined mean (\(\bar{x}\)) and SD are calculated using the following formulas:
Cortical depths normalization
Using the Allen CCFv3 brain atlas, we normalized cortical depths by solving the 3D Laplace equation, with the isocortex surface potential set to 1 and the deepest layer of the isocortex set to 014. This process generated a cortical depth distribution field ranging from 0 to 1, accurate to two decimal places. The gradient direction of this depth field was interpreted as representing the orientation from the cortical surface toward deeper layers.
To analyze neuronal distributions, we first computed density distributions along the cortical depth axis for 8 glutamatergic and 5 GABAergic neuronal types across various cortical subregions. Let neuronal type i, brain region j, and relative depth d be specified. The density ρ of type i neurons at depth d in region j was determined as: \(\rho=\frac{{N}_{c{ell}}}{{V}_{v{oxel}}\times {N}_{v{oxel}}}\), where \({cell},{voxel}\in {j}_{d}\). Here, \({N}_{c{ell}}\) represents the total number of cells at depth d, \({N}_{v{oxel}}\) denotes the number of voxels, and \({V}_{v{oxel}}\) is the volume of a single voxel within \({j}_{d}\). For each neuronal type in each brain region, this yielded a 100-dimensional density distribution vector. By concatenating the vectors of 13 neuronal types, we obtained a 1300-dimensional feature vector for each brain region. To account for the non-uniform cortical thickness across regions, we established two sets of average depth references for layers L1–L6, based on the presence or absence of layer L4. These cortical subregions were subsequently mapped onto the average depth template using B-spline interpolation. The average depth of a specific layer was computed as the voxel-weighted mean of depth values within that layer. We next calculated the Pearson correlation coefficients for the 43 cortical subregions, generating a correlation matrix to characterize distribution patterns aligned with anatomical delineations. For clustering analysis (Fig. 4a), the density computation was adapted by replacing \({j}_{d}\) with \({j}_{{cd}}\), representing all regions belonging to category c in brain region j at depth d.
Additionally, we divided the cortex into 12,876 cylindrical regions, each with a top surface area of 80 × 80 μm2, referred to as “functional columns.” For each functional column, we computed the relative density distributions of glutamatergic and GABAergic neurons along the depth axis, concatenating them into 1300-dimensional feature vectors. Using these functional columns as the basic clustering units, we performed Leiden clustering33 to generate a cortical delineation based on functional columnar organization (Fig. 4f).
Normed Glu/GABA and relative normed Glu/GABA
To characterize the relative number of glutamatergic and GABAergic neurons within a given brain region or cube, we define their respective densities based on the neuronal type with the highest density in that region or cube. For a specific region or cube j, let \({{Glu}}_{j}\) represent the density of the glutamatergic neuron type with the highest density, and \({{GABA}}_{j}\) represent the density of the GABAergic neuron type with the highest density. The normed Glu/GABA ratio is then defined as: \({{Glu}/{GABA}}_{j}=\frac{{{Glu}}_{j}-{{GABA}}_{j}}{{{Glu}}_{j}+{{GABA}}_{j}}\). This ratio ranges from −1 to 1. A normed Glu/GABA value of −1 indicates that region or cube B contains exclusively GABAergic neurons, with no glutamatergic neurons present. Conversely, a value of 1 signifies the presence of only glutamatergic neurons and no GABAergic neurons. A value of 0 reflects an equal density of glutamatergic and GABAergic neurons. Ratios between −1 and 0 suggest a predominance of GABAergic neurons, whereas values between 0 and 1 indicate a predominance of glutamatergic neurons.
Next, we divided each cortical column into 100 equal segments along its depth. At a given cortical depth d, let \({{Glu}}_{d}\) denote the density of the glutamatergic neuron type with the highest density, and \({{GABA}}_{d}\) the density of the GABAergic neuron type with the highest density. The normed Glu/GABA ratio at depth d is calculated as: \({{Glu}/{GABA}}_{d}=\frac{{{Glu}}_{d}-{{GABA}}_{d}}{{{Glu}}_{d}+{{GABA}}_{d}}\). To evaluate the variability across the cortical column, we compute the mean (\(\overline{{Glu}/{GABA}}\)) and standard deviation (\({\sigma }_{{Glu}/{GABA}}\)) of the normed Glu/GABA ratios across the entire depth of the column. The z-score for the relative normed Glu/GABA at depth d, denoted as \({Glu}/{{GABA}}_{d}^{{norm}}\), is defined as: \({Glu}/{{GABA}}_{d}^{{norm}}=\frac{{{Glu}/{GABA}}_{d}-\overline{{Glu}/{GABA}}}{{\sigma }_{{Glu}/{GABA}}}\). This z-score reflects the deviation of the normed Glu/GABA at depth d relative to the average normed Glu/GABA across the full depth of the cortical column.
Calculating the Modulatory normed Glu/GABA
To characterize the relationship between the distribution of modulatory neurons and the normed Glu/GABA ratio within a specific brain region, we introduce the parameter Modulatory normed Glu/GABA. To account for modulatory neurons, we define the density of a specific modulatory neuron type i in a brain region j as \({D}_{{den}}^{{ij}}\), and the maximum density of this neuron type across 134 brain regions as \({D}_{{maxden}}^{i}\). The normalized density of neuron type i in brain region j is calculated as: \({D}_{{normedden}}^{{ij}}={D}_{{den}}^{{ij}}/{D}_{{maxden}}^{i}\). The value of \({D}_{{normedden}}^{{ij}}\) ranges from 0 to 1, where higher values indicate higher relative density. The weighted normed Glu/GABA value for a specific modulatory neuron type i in a brain region j is then calculated by multiplying normed Glu/GABA by \({D}_{{normedden}}^{{ij}}\): \({Weighted\; normed}\frac{G{lu}}{{GABA}}={normed\; Glu}/{GABA}\times {D}_{{normedden}}^{{ij}}\). Finally, summing \({Weighted\; normed}G{lu}/{GABA}\) across all 134 brain regions yields the Modulatory normed Glu/GABA value. This parameter reflects the overall glutamatergic-GABAergic balance associated with the modulatory neuron type across the entire brain.
Cubic distance measuring
The conventional definition of “density” for cell bodies involves dividing the total number of cell bodies within a specific spatial region by the volume of that region. In practice, we partition the spatial domain of the mouse brain into equally sized cubic regions to calculate local cell body densities (Figs. 2–4). This approach effectively uses the average cell density in 3D space to represent the distribution of cell bodies throughout the spatial domain. However, neuronal cell bodies have physical dimensions, typically with diameters ranging from 10 μm to 20 μm. When the size of the cubic region approaches or is smaller than the size of a cell body, many regions yield a local density of 0. Consequently, this density calculation method fails to accurately capture the density of neuronal distributions in 3D space, with the prevalence of zero values introducing significant biases in subsequent analyses.
To address these limitations and enable the analysis of neuronal distribution at finer spatial scales, we introduce a distance-based parameter as an alternative to the traditional density metric (Supplementary Fig. 22). This parameter provides a more robust characterization of neuronal spatial distribution in the mouse brain.
For a given point p within the spatial domain of the mouse brain and a specified neuronal type i, the distance-based parameter is defined as follows: Step 1. Calculate the distances from p to the x nearest neurons of type i, denoted as \({d}_{1}^{{ip}}\), \({d}_{2}^{{ip}}\), …, \({d}_{x}^{{ip}}\). Step 2. Compute the parameter \({D}_{{dist}}^{{ip}}\), defined as the reciprocal of the average distance from to p the x nearest neurons: \({D}_{{dist}}^{{ip}}=\frac{1}{\frac{1}{x}{\sum }_{a=1}^{x}{d}_{a}^{{ip}}}\). When the average distance to the x nearest neurons increases, \({D}_{{dist}}^{{ip}}\) decreases, indicating a sparser local distribution of type i neurons. Conversely, smaller average distances result in higher \({D}_{{dist}}^{{ip}}\), reflecting a denser distribution. To accommodate the dimensions of neuronal cell bodies and the requirements of subsequent analyses, we sampled points every 10 μm throughout the mouse brain and calculated \({D}_{{dist}}^{{ip}}\) for 20 neuronal types with 10 μm precision, where x = 10 (Supplementary Fig. 22a). We then averaged the \({D}_{{dist}}^{{ip}}\) values within 20 μm × 20 μm × 20 μm cubic regions to generate coarser-resolution values, which are accessible via our web portal.
To evaluate the effectiveness of \({D}_{{dist}}\) compared to traditional density parameters \({D}_{{den}}\) in cluster analysis, we constructed a 3D cubic model (Supplementary Fig. 22b). This model, a cubic space with side length 1000 μm, contains 1,000,000 data points with minimum voxel sizes of 10 μm × 10 μm × 10 μm. The model includes three neuronal types (type 1, 2, and 3), each assigned one of two density states: 0 (sparse, 2000 cells/mm³) or 1 (dense, 20,000 cells/mm³). The model is divided into eight spatial regions (500 μm × 500 μm × 500 μm for each), each exhibiting different density patterns represented by binary combinations such as (0,0,1) (0,0,1) (0,0,1) and (1,1,0) (1,1,0) (1,1,0). Neuronal positions within each region were randomly generated to achieve the assigned densities. For each point p in the model, \({D}_{{dist}}\) values were computed at a 10 μm scale, then downsampled to coarser scales of 30 μm, 50 μm, 100 μm, and 150 μm. Similarly, traditional density parameters \({D}_{{den}}\) were calculated by dividing the model into grids of corresponding sizes. At each scale, data points were represented as column vectors containing three elements, corresponding to the \({D}_{{dist}}\) or \({D}_{{den}}\) values for each neuronal type.
We applied UMAP dimensionality reduction to the column vectors, followed by cluster analysis using the Leiden community detection algorithm (Supplementary Fig. 22c). Results showed that at scales of 150 μm and 100 μm, both \({D}_{{den}}\) and \({D}_{{dist}}\) produced satisfactory clustering results. However, at finer scales (50 μm, 30 μm, and 10 μm), \({D}_{{den}}\) failed to distinguish clusters effectively, while \({D}_{{dist}}\) consistently yielded well-defined cluster boundaries.
Our findings underscore the importance of selecting appropriate spatial scales for analyzing neuronal distributions in 3D space. At scales ≥100 μm, both traditional density parameters and \({D}_{{dist}}\) are effective. However, at scales ≤50 μm, traditional density parameters lose efficacy, and distance-based parameters like\({D}_{{dist}}\) should be employed for reliable analysis.
Behavioral test
Virus injection and preparation
For the experiments described in Fig. 5i, all mice were deeply anesthetized via intraperitoneal injection of 100 μl/10 g body weight of a mixed anesthetic solution composed of 2% chloral hydrate and 10% urethane. The mice were then mounted in a stereotaxic apparatus for precise surgical procedures. A total of 150 nl of AAV2/9-EF1α-DIO-GCaMP6s or AAV2/9-EF1α-DIO-ChR2-EGFP (2 × 1012 gc/ml, Brain Case Co., Ltd., China) was injected into either the CPdl (coordinates: bregma −0.83 mm, lateral 3.0 mm, depth 3.1 mm from skull surface) or the CPvl (coordinates: bregma −0.83 mm, lateral 3.1 mm, depth 4.3 mm from skull surface) of Camk2a-Cre, GAD2-Cre, or VGAT-Cre mice to label glutamatergic or GABAergic neurons, respectively. Following virus injection, optical fibers (diameter = 200 μm, NA = 0.37) were implanted into the CPdl or CPvl. The fiber ferrules were secured to the skull using screws and dental cement. After a 3-week recovery period to allow for virus expression, the mice underwent open field testing with either fiber photometry or optogenetic stimulation.
Fiber photometry
During fiber photometry trials, each mouse was allowed to freely explore the open field for 20 min. Calcium signals were recorded using a commercial fiber photometry system (Thinker Tech Nanjing Biotech Co., Ltd.). The fluorescence data from each trial was normalized to the mean baseline fluorescence (F0) using MATLAB-based software provided by Thinker Tech. The transient velocity of the mice was tracked and analyzed using VisuTrack software (Shanghai Xin Ruan Co., Ltd., China). The precision of the velocity data was manually verified. Locomotion bouts were identified in two stages: (1) Peaks in the velocity signal exceeding 8 cm/s were used to confirm locomotion; (2) The start and end points of locomotion were defined as the first and last time points where velocity exceeded 4 cm/s, relative to the peak. Baseline signals preceding locomotion onset or termination (t = 0) were defined as F0, and fluorescence changes (∆F/F) were calculated as \(\left(F-{F}_{0}\right)/{F}_{0}\). Parameters such as peak time and area under the curve (AUC) were extracted during the intervals of interest. Statistical comparisons between CPdl and CPvl groups were performed using a two-tailed unpaired t-test.
Optogenetic stimulation
For optogenetics testing (Fig. 5i), mice explored the open field freely in 6-min trials divided into two phases: (1) Control Period (0–3 min), mice explored the arena without photostimulation. (2) Stimulation Period (3–6 min), photostimulation was applied to activate glutamatergic or GABAergic neurons in the CPdl or CPvl using a 473 nm laser (10 Hz, 15 ms pulse width, 3.5 mW). Stimulation was triggered when the mouse’s transient velocity exceeded 8 cm/s and ceased when velocity dropped below this threshold. The cumulative positions and mean velocities during the control and stimulation periods were calculated using VisuTrack software. Statistical analyses compared these measures across conditions to evaluate the effects of neuronal activation.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Whole-brain spatial location information of 20 distinct types of neurons is available both in Zenodo (https://doi.org/10.5281/zenodo.17180853) and at http://atlas.brainsmatics.cn/MiCAM, a self-built portal. Files of 19-dimensional neuron distance-based parameter vectors for each 10 µm × 10 µm × 10 µm unit in the 3D grid of the entire mouse brain is available at the self-built portal. The quantitative statistical information on the number of 20 distinct types of neurons in various brain regions across the whole brain can be obtained from the Source Data file. Source data are provided with this paper.
Code availability
All custom code supporting the current study is available at http://atlas.brainsmatics.cn/MiCAM.
References
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Herculano-Houzel, S., Catania, K., Manger, P. R. & Kaas, J. H. Mammalian brains are made of these: a dataset of the numbers and densities of neuronal and nonneuronal cells in the brain of glires, primates, scandentia, eulipotyphlans, afrotherians and artiodactyls, and their relationship with body mass. Brain Behav. Evol. 86, 145–163 (2015).
Basu, R., Taylor, M. R. & Williams, M. E. The classic cadherins in synaptic specificity. Cell Adhes. Migr. 9, 193–201 (2015).
Zeng, Z., Miao, N. & Sun, T. Revealing cellular and molecular complexity of the central nervous system using single cell sequencing. Stem Cell Res. Ther. 9, 234 (2018).
Daigle, T. L. et al. A suite of transgenic driver and reporter mouse lines with enhanced brain-cell-type targeting and functionality. Cell 174, 465–480.e22 (2018).
Zhong, Q. et al. High-definition imaging using line-illumination modulation microscopy. Nat. Methods 18, 309–315 (2021).
Gong, H. et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142 (2016).
Kim, Y. et al. Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell 171, 456–469 (2017).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Yang, X. et al. Observing single cells in whole organs with optical imaging. J. Innov. Opt. Health Sci. 16, 2330002 (2023).
Peng, J. et al. A quantitative analysis of the distribution of CRH neurons in whole mouse brain. Front. Neuroanat. 11, 63 (2017).
Zhang, C. et al. A platform for stereological quantitative analysis of the brain-wide distribution of type-specific neurons. Sci. Rep. 7, 14334 (2017).
Li, X. et al. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA 115, 415–420 (2018).
Wang, Q. et al. The Allen mouse brain common coordinate framework: a 3D reference atlas. Cell 181, 936–953 (2020).
Ni, H. et al. A robust image registration interface for large volume brain atlas. Sci. Rep. 10, 2139 (2020).
Feng, Z. et al. A mouse brain stereotaxic topographic atlas with isotropic 1-μm resolution. Nature 645, 448–456 (2025).
Erwin, S. R. et al. Spatially patterned excitatory neuron subtypes and projections of the claustrum. eLife 10, e68967 (2021).
Costa, M. R. & Müller, U. Specification of excitatory neurons in the developing cerebral cortex: progenitor diversity and environmental influences. Front. Cell. Neurosci. 8, 449 (2015).
Molnár, Z. Evolution of cerebral cortical development. Brain Behav. Evol. 78, 94–107 (2011).
Righes Marafiga, J., Vendramin Pasquetti, M. & Calcagnotto, M. E. GABAergic interneurons in epilepsy: more than a simple change in inhibition. Epilepsy Behav. 121, 106935 (2021).
Xu, M. & Wong, A. H. C. GABAergic inhibitory neurons as therapeutic targets for cognitive impairment in schizophrenia. Acta Pharmacol. Sin. 39, 733–753 (2018).
Brandenburg, C. et al. Parvalbumin subtypes of cerebellar Purkinje cells contribute to differential intrinsic firing properties. Mol. Cell. Neurosci. 115, 103650 (2021).
Bastianelli, E. Distribution of calcium-binding proteins in the cerebellum. Cerebellum 2, 242–262 (2003).
Wang, W. X., Qiao, J. & Lefebvre, J. L. PV-IRES-Cre mouse line targets excitatory granule neurons in the cerebellum. Mol. Brain 15, 85 (2022).
El-Ansary, A. & Al-Ayadhi, L. GABAergic/glutamatergic imbalance relative to excessive neuroinflammation in autism spectrum disorders. J. Neuroinflamm. 11, 189 (2014).
Palop, J. J. et al. Aberrant excitatory neuronal activity and compensatory remodeling of inhibitory hippocampal circuits in mouse models of Alzheimer’s disease. Neuron 55, 697–711 (2007).
Lopatina, O. L. et al. Oxytocin and excitation/inhibition balance in social recognition. Neuropeptides 72, 1–11 (2018).
Mitani, A., Dong, M. & Komiyama, T. Brain-computer interface with inhibitory neurons reveals subtype-specific strategies. Curr. Biol. 28, 77–83.e4 (2018).
Woolsey, T. A. & Van der Loos, H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain Res. 17, 205–242 (1970).
Jones, E. G. Microcolumns in the cerebral cortex. Proc. Natl. Acad. Sci. USA 97, 5019–5021 (2000).
Haueis, P. The life of the cortical column: opening the domain of functional architecture of the cortex (1955–1981). Hist. Philos. Life Sci. 38, 2 (2016).
Wu, Y. et al. Quantitative relationship between cerebrovascular network and neuronal cell types in mice. Cell Rep. 39, 110978 (2022).
Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Yang, J. H. & Kwan, A. C. Secondary motor cortex: broadcasting and biasing animal’s decisions through long-range circuits. Int. Rev. Neurobiol. 158, 443–470 (2021).
Briand, L. A., Gritton, H., Howe, W. M., Young, D. A. & Sarter, M. Modulators in concert for cognition: modulator interactions in the prefrontal cortex. Prog. Neurobiol. 83, 69–91 (2007).
Tully, K. & Bolshakov, V. Y. Emotional enhancement of memory: how norepinephrine enables synaptic plasticity. Mol. Brain 3, 15 (2010).
Thiele, A. & Bellgrove, M. A. Neuromodulation of attention. Neuron 97, 769–785 (2018).
Beier, K. T. et al. Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell 162, 622–634 (2015).
Sun, P. et al. Sex-related differential whole-brain input atlas of locus coeruleus noradrenaline neurons. Front. Neural Circuits 14, 53 (2020).
Lin, W. et al. Whole-brain mapping of histaminergic projections in mouse brain. Proc. Natl. Acad. Sci. USA 120, e2216231120 (2023).
Durand-de Cuttoli, R. et al. Manipulating midbrain dopamine neurons and reward-related behaviors with light-controllable nicotinic acetylcholine receptors. eLife 7, e37487 (2018).
Carson, R. P. & Robertson, D. Genetic manipulation of noradrenergic neurons. J. Pharmacol. Exp. Ther. 301, 410–417 (2002).
Nishitani, N. et al. Manipulation of dorsal raphe serotonergic neurons modulates active coping to inescapable stress and anxiety-related behaviors in mice and rats. Neuropsychopharmacology 44, 721–732 (2019).
Hintiryan, H. et al. The mouse cortico-striatal projectome. Nat. Neurosci. 19, 1100–1114 (2016).
Krishnaswamy, P. et al. Sparsity enables estimation of both subcortical and cortical activity from MEG and EEG. Proc. Natl. Acad. Sci. USA 114, E10465–E10474 (2017).
Alkemade, A., Keuken, M. C. & Forstmann, B. U. A perspective on terra incognita: uncovering the neuroanatomy of the human subcortex. Front. Neuroanat. 7, 40 (2013).
Forstmann, B. U., de Hollander, G., van Maanen, L., Alkemade, A. & Keuken, M. C. Towards a mechanistic understanding of the human subcortex. Nat. Rev. Neurosci. 18, 57–65 (2017).
Cheng, S. et al. Vision-dependent specification of cell types and function in the developing cortex. Cell 185, 311–327.e24 (2022).
Chen, A. et al. Global spatial transcriptome of macaque brain at single-cell resolution. Cell 186, 3726–3743.e24 (2023).
Hara, Y. et al. Spatial transcriptome of developmental mouse brain reveals temporal dynamics of gene expressions and heterogeneity of the claustrum. bioRxiv 2023.04.12.536360 https://doi.org/10.1101/2023.04.12.536360 (2023).
Eickhoff, S. et al. The basal forebrain cholinergic nuclei and their relevance to schizophrenia and other psychotic disorders. Front. Psychiatry 13, 909961 (2022).
Bloem, B. et al. Topographic mapping between basal forebrain cholinergic neurons and the medial prefrontal cortex in mice. J. Neurosci. 34, 16234–16246 (2014).
Arena, G. et al. Disentangling the identity of the zona incerta: a review of the known connections and latest implications. Ageing Res. Rev. 93, 102140 (2024).
Wheatcroft, T., Saleem, A. B. & Solomon, S. G. Functional organisation of the mouse superior colliculus. Front. Neural Circuits 16, 792959 (2022).
Yang, Y. et al. Whole-brain connectome of GABAergic neurons in the mouse Zona Incerta. Neurosci. Bull. 38, 1315–1329 (2022).
Roostalu, U. et al. Quantitative whole-brain 3D imaging of tyrosine hydroxylase-labeled neuron architecture in the mouse MPTP model of Parkinson’s disease. Dis. Model. Mech. 12, dmm042200 (2019).
Newmaster, K. T. et al. Quantitative cellular-resolution map of the oxytocin receptor in postnatally developing mouse brains. Nat. Commun. 11, 1885 (2020).
Bjerke, I. E. et al. Densities and numbers of calbindin and parvalbumin positive neurons across the rat and mouse brain. iScience 24, 101906 (2021).
Carrasco, A., Oorschot, D. E., Barzaghi, P. & Wickens, J. R. Three-dimensional spatial analyses of cholinergic neuronal distributions across the mouse septum, nucleus basalis, globus pallidus, nucleus accumbens, and caudate-putamen. Neuroinform 20, 1121–1136 (2022).
Rodarie, D. et al. A method to estimate the cellular composition of the mouse brain from heterogeneous datasets. PLoS Comput. Biol. 18, e1010739 (2022).
Bjerke, I. E., Yates, S. C., Carey, H., Bjaalie, J. G. & Leergaard, T. B. Scaling up cell-counting efforts in neuroscience through semi-automated methods. iScience 26, 107562 (2023).
Attili, S. M., Moradi, K., Wheeler, D. W. & Ascoli, G. A. Quantification of neuron types in the rodent hippocampal formation by data mining and numerical optimization. Eur. J. Neurosci. 55, 1724–1741 (2022).
Keller, D., Erö, C. & Markram, H. Cell densities in the mouse brain: a systematic review. Front. Neuroanat. 12, 83 (2018).
Erö, C., Gewaltig, M.-O., Keller, D. & Markram, H. A cell atlas for the mouse brain. Front. Neuroinform. 12, 84 (2018).
Shi, H. et al. Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622, 552–561 (2023).
Bhattacherjee, A. et al. Spatial transcriptomics reveals the distinct organization of mouse prefrontal cortex and neuronal subtypes regulating chronic pain. Nat. Neurosci. 26, 1880–1893 (2023).
Fang, R. et al. Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science 377, 56–62 (2022).
Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021).
Zhang, M. et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354 (2023).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Ng, L. et al. An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362 (2009).
Root, D. H. et al. Single rodent mesohabenular axons release glutamate and GABA. Nat. Neurosci. 17, 1543–1551 (2014).
Shabel, S. J., Proulx, C. D., Piriz, J. & Malinow, R. GABA/glutamate co-release controls habenula output and is modified by antidepressant treatment. Science 345, 1494–1498 (2014).
Root, D. H. et al. Selective brain distribution and distinctive synaptic architecture of dual glutamatergic-GABAergic neurons. Cell Rep. 23, 3465–3479 (2018).
Negishi, K. et al. Distributions of hypothalamic neuron populations coexpressing tyrosine hydroxylase and the vesicular GABA transporter in the mouse. J. Comp. Neurol. 528, 1833–1855 (2020).
Gou, L. et al. Single-neuron projectomes of macaque prefrontal cortex reveal refined axon targeting and arborization. Cell 188, 3806–3822.e24 (2025).
Zhou, C. et al. Continuous subcellular resolution three-dimensional imaging on intact macaque brain. Sci. Bull. 67, 85–96 (2022).
Klein, A. et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46, 786–802 (2009).
Quan, T. et al. NeuroGPS: automated localization of neurons for brain circuits using L1 minimization model. Sci. Rep. 3, 1414 (2013).
Li, Y. et al. TDat: an efficient platform for processing petabyte-scale whole-brain volumetric images. Front. Neural Circuits 11, 51 (2017).
Bradley, D. & Roth, G. Adaptive thresholding using the integral image. J. Graph. Tools 12, 13–21 (2007).
Chan, T. F. & Vese, L. A. Active contours without edges. IEEE Trans. Image Process. 10, 266–277 (2001).
Acknowledgements
The authors are grateful to Dr. Mu-ming Poo, Dr. Hongwei Dong, Dr. Tao Yang, Dr. Pengcheng Li and Dr. Ben Long for their critical comments on this work; Dr. Siqi Chen, Dr. Mei Yao, Dr. Ayizuohere Tudi, Tingting Sun and Jiamei Qi for assistance with animal experiments; Shuxin Wang, Hao Dong, Ziming Chen and Yunyi Gu for the data analysis; Dr. Chi Xiao and Hui Han for their advice and help with data preprocessing; Xiaowei Chen for the portal website. We would like to thank the MOST group and all staff from HUST-Suzhou Institute for Brainsmatics. This study was supported by STI 2030-Major Projects (2021ZD0201002, led by A.L.; 2021ZD0201001, led by H.G.), the Innovational Fund for Scientific and Technological Personnel of Hainan Province (KJRC2023A03) led by Q.L. and the National Natural Science Foundation of China Grants (T2122015), led by A.L.
Author information
Authors and Affiliations
Contributions
Q.L. and H.G. conceived and organized the study; H.G., A.L. and X.L. designed the research; A.L., X.L., X.Y. and J.Y. supervised and organized the imaging and analysis; M.R. performed hybridization and resin-embedding experiments; T.J. contributed to the fMOST system design, maintenance, and data capturing under the supervision of X.Y. and J.Y.; M.Z., Z.F. and X.J. performed the image preprocessing and format transformation; Z.M. and F.T. performed the function-related experiments and data analysis; M.Z., J.Z., C.D., L.C. and S.B. performed data analysis and modified the methods; M.Z., J.Z. and A.L. wrote the manuscript with comments from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yongsoo Kim and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, M., Zhou, J., Jiang, T. et al. Molecularly defined cellular atlas of the entire mouse brain with isotropic single-cell resolution. Nat Commun 16, 10167 (2025). https://doi.org/10.1038/s41467-025-65238-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65238-5