Abstract
Different spatial or temporal protein populations, such as cell-surface/intracellular or pre-existing/nascent subpopulations, determine the basal and activity-induced functions of individual synapses within a neuron in vivo. Here, we developed a simple and generalizable platform to image different spatial and temporal subpopulations of endogenous proteins at thousands of synapses in single neurons in the mammalian brain. The platform is based on the development, improvement and integration of CRISPR-Cas9-mediated protein labeling methods, chemical tag labeling techniques, and a semi-automatic analytical pipeline. The combined platform enables whole-cell mapping of total, cell-surface, intracellular, pre-existing, nascent or nascent-and-surface populations of endogenous proteins, such as receptor, scaffold and signaling proteins, at thousands of synapses in individual neurons in living or fixed mouse brain. Our single-cell “synaptome” mapping of endogenous protein subpopulations comprehensively visualizes the spatial representation of synapse diversity in protein localization, trafficking and turnover, providing valuable insights into single-cell organization and computations in the brain.
Similar content being viewed by others
Introduction
Precise mapping of individual proteins with subcellular resolution would improve understanding of cellular processes at the molecular level. In many cases, the same protein has several subpopulations with different biological roles within a cell. For example, transmembrane proteins such as receptors or cell adhesion molecules have spatially different subpopulations: cell-surface populations for signal sensing and transduction and intracellular populations for reserve and recycling pools1,2,3,4. Another example is temporally different subpopulations: pre-existing and nascent protein populations often play distinct roles in basal and activity-induced cellular functions5,6,7. Thus, quantitative, subcellular mapping of different subpopulations of endogenous proteins in the brain is needed to delineate the molecular mechanisms underlying neuronal functions.
Neuronal functions rely on complex protein regulation at synapses to respond to distinct activity in the neural network. A neuron receives a wide range of inputs from other neurons via thousands of synapses8. Individual synapses are highly heterogeneous in their protein composition, trafficking and turnover, which underlies the different structures and functions of each synapse9,10,11,12. This synaptic heterogeneity, revealed through brain-wide “synaptome” analysis, is relevant to behavior, cognitive functions, and brain disorders11,12,13, while brain-wide synaptome analysis conflates variation between cells with synapse variation within a single neuron. Single-cell, quantitative mapping of spatially and temporally different subpopulations of endogenous proteins at thousands of synapses would elucidate the spatial organization of synaptic heterogeneity all along the neuronal dendritic tree, providing an accurate and informative representation of the synaptic landscape in single neurons. Thus, this “single-cell synaptome” analysis would allow for a better understanding of the single-cell organization and computations in the brain. However, there is no simple and generalizable method to differentially visualize these subpopulations of endogenous proteins in single neurons in brain tissue.
We previously developed several methods to label endogenous proteins in single cells in the mammalian brain14,15. These methods, named SLENDR and vSLENDR, are based on precise, homology-directed repair (HDR)-mediated genome editing using the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease Cas9. They provide a high-throughput approach to determine the subcellular localization of endogenous proteins with high resolution, specificity and contrast. In SLENDR and vSLENDR, epitope tags or fluorescent protein tags are genetically fused to specific endogenous proteins. Incorporated epitope tags can be visualized by immunohistochemistry using specific antibodies against the tags. However, the large size of antibodies (~ 150 kDa) hinders their penetration through thick brain tissue, making it challenging to label target proteins within densely-packed protein complexes at the synaptic membrane16. These limitations preclude quantitative spatial measurements of epitope-tagged proteins in brain tissue. Alternatively, fluorescent protein-fused target proteins can be directly imaged without antibody labeling. However, fluorescent proteins’ low brightness and instability provide an insufficient signal-to-noise ratio for quantifying endogenous proteins, especially proteins with low expression levels.
An alternative to epitope tagging and antibody labeling or fluorescent protein tagging is chemical tagging via self-labeling proteins such as HaloTag17, SNAP-tag18, or CLIP-tag19. Chemical tags enable covalent and irreversible labeling with bright and photostable organic fluorophores20,21. The small size (~ 1 kDa) of the organic fluorophore ligands allows fast and efficient tissue penetration to evenly label proteins throughout thick brain tissue22. Notably, the availability of different organic fluorophore ligands in multiple colors and with different cellular permeability permits the detection of spatially restricted subpopulations of tagged proteins. For example, cell-impermeable fluorophore ligands can selectively label cell-surface subpopulations of tagged proteins23. Additionally, pulse-chase labeling with fluorophore ligands of different colors can differentially visualize temporally distinct subpopulations, including pre-existing and nascent proteins7,24. Thus, chemical tag-mediated protein labeling can provide quantitative and spatiotemporal imaging of tagged proteins in tissue.
In this study, we combined chemical tag labeling with the SLENDR approach to label endogenous proteins in the brain with organic fluorophores. We show that SLENDR and vSLENDR efficiently insert self-labeling tag sequences into a variety of endogenous genes encoding for presynaptic, postsynaptic, and signaling proteins. Multiplexed labeling with different fluorophore ligands enabled single-cell visualization of neuronal protein subpopulations (i.e., surface/intracellular, nascent/pre-existing) in mouse brain tissue. Integrating this approach with a semi-automatic analytical pipeline, we quantified endogenous glutamate receptor subpopulations at the thousands of synapses in single neocortical neurons, providing a whole-cell map of the strength and plasticity of each excitatory synapse (Fig. 1a). Through single-cell synaptome mapping of endogenous protein subpopulations, we reveal the spatial organization of synapse diversity in protein localization, trafficking and turnover in single neurons in vivo.
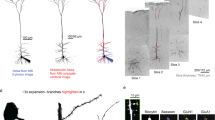
a Schematic of the workflow for single-cell synaptome mapping in the brain. b–k, m–r, Confocal images for HaloTag-fused endogenous proteins (green) including Halo-CaMKIIα (b, c), PSD95-Halo (d, e), Halo-GluN1 (f, g), Halo-GluA1 (h, i), Halo-GluA2 (j, k), Halo-Gephyrin (m, n), and Halo-βCatenin (o–r) in single mEGFP-labeled pyramidal cells (magenta) in the primary somatosensory cortex layer 2/3 at P28. PSD95, a major postsynaptic scaffold at excitaotry synapses; GluN1; a NMDAR subunit; GluA1 and GluA2, AMPAR subunits; Gephyrin, a major postsynaptic scaffold at inhibitory synapses; βCatenin, a multifunctional protein associated with cell adhesion and canonical Wnt signaling. Note selective accumulations of Halo-CaMKIIα (c), PSD95-Halo (e), Halo-GluN1 (g), Halo-GluA1 (i), Halo-GluA2 (k), and Halo-βCatenin (p) at glutamatergic postsynapses formed on dendritic spines, Halo-Gephyrin (m, n) and Halo-βCatenin (q) at GABAergic postsynapses localized on the axon initial segment (arrows in m) and head portion of a few dendritic spines (arrow in n) or immunolabeled for vesicular inhibitory amino acid transporter (VIAAT, yellow) on the somatic surface (arrows in q), and Halo-βCatenin (r) at presynaptic boutons, validating the specificity of our HaloTag labeling. l Structured illumination microscope (SIM) image of the same view as in k, resolving nano-clusters for Halo-GluA2 (arrows). Scale bars, 10 μm (b, d, f, h, j, m, o), 2 μm (c, e, g, i, k, l, n, p–r).
Results
A generalizable platform for single-cell, chemical tag labeling of endogenous proteins in the brain
Since HaloTag labeling with organic fluorophore ligands provides bright and photostable fluorescence21,25, we set out to build a toolbox to label endogenous proteins with HaloTag via CRISPR-Cas9-mediated HDR using SLENDR14. However, the original SLENDR technique had low efficiency (< 1%) of knock-in for sequences similar in size to HaloTag or SNAP-tag (~ 600–900 bp). In this study, we utilized a pre-assembled Cas9 protein-guide RNA (gRNA) ribonucleoprotein complex (RNP), which efficiently induces DNA double-strand breaks (DSBs) immediately after the delivery into target cells by in utero electroporation (IUE)26. Moreover, as RNPs are rapidly degraded in cells compared with plasmid vectors, this strategy improves genome editing specificity by decreasing off-target DSBs27,28. The homology donor template was designed with the HaloTag and short linker sequences (0.9 kbp) flanked by ~1 kbp of the genomic sequences upstream and downstream of the HaloTag insertion site. The RNPs and homology donor template were introduced together with a plasmid vector encoding mEGFP as a transfection marker into cortical layer 2/3 pyramidal cells by IUE at embryonic day 14 (E14) or E15 in mice. Brains were fixed at postnatal 3–4 weeks, sectioned, and labeled by incubation with Janelia Fluor (JF) dyes conjugated to HaloTag ligands (JF549-HTL or JF646-HTL) for confocal fluorescent imaging20 (Fig. 1a). We confirmed that brief chemical fixation, matching the duration of our perfusion fixation, allows for spatiotemporally-precise localization of HaloTag signals without any significant signal attenuations (Supplementary Fig. 1a, b)
We first targeted HaloTag to the N-terminus of CaMKIIα by using the same homology arms previously validated for the knock-in of mEGFP14,15. HaloTag-labeled CaMKIIα (Halo-CaMKIIα) was found in the somatodendritic compartment of single pyramidal cells with a peak intensity at dendritic spines, as previously described for endogenous mEGFP-CaMKIIα14,15 (Fig. 1b, c). The RNP-based HaloTag knock-in achieved significantly higher efficiency than the plasmid-based approach14 (Supplementary Fig. 1c, d). This improvement was also observed for other genes, such as βActin and GluA2, indicating the generalizability of our strategy (Supplementary Fig. 1c, d). We confirmed the genome editing specificity by replacing the CaMKIIα-targeting gRNA with a GluA2-targeting gRNA, to find no detectable knock-in signal (Supplementary Fig. 1c). Thus, RNP-based SLENDR achieves specific and efficient HaloTag labeling of endogenous proteins in the brain.
We then applied the HaloTag labeling platform to 10 other endogenous proteins such as postsynaptic scaffolds, receptors, presynaptic release machinery, and intracellular signaling proteins (Fig. 1d–r, Supplementary Fig. 1e–m, and Supplementary Data 1, 2). We confirmed a protein species-specific distribution of HaloTag-fused endogenous proteins in single pyramidal cells. Importantly, we successfully visualized excitatory postsynaptic scaffolds (Fig. 1d, e) and ionotropic glutamate receptors (iGluRs, Fig. 1f–l and Supplementary Fig. 1e), which are difficult to detect by conventional immunohistochemistry due to the poor accessibility of antibodies to densely-packed postsynaptic structures16. Their HaloTag-labeled puncta exclusively overlapped with or were closely apposed to immunohistochemical signals for excitatory presynaptic and postsynaptic markers, demonstrating the specificity of HaloTag signals to excitatory synapses (Supplementary Fig. 2). Moreover, HaloTag-labeled endogenous proteins were further compatible with super resolution imaging by structured illumination microscopy (SIM) (Fig. 1l and Supplementary Fig. 1e) and live monitoring of protein translocation by two-photon microscopy (Supplementary Fig. 1j–n). Therefore, our HDR-mediated protein tagging with HaloTag provides a scalable, single-cell labeling approach for a wide variety of endogenous postsynaptic, presynaptic and cell signaling proteins.
Whole-neuron synaptome mapping of endogenous proteins in thick brain tissue
High brightness, photostability, and permeability of the small-sized fluorogenic HTLs are suitable for quantitative labeling of HaloTag-fused proteins in large tissue22. To confirm this, we performed quantitative, volumetric imaging of endogenous CaMKIIα tandemly tagged with HaloTag and mEGFP (Halo-mEGFP-CaMKIIα) by using vSLENDR in transgenic mice that constitutively expressed SpCas9 (Supplementary Fig. 3a, b)15. In thick (200 µm), fixed and SeeDB2G-cleared brain slices29, fluorescently-labeled HTLs uniformly visualized Halo-mEGFP-CaMKIIα proteins throughout the slice, while antibodies against mEGFP limited the labeled area to the slice surface (Supplementary Fig. 3c–e). The signal intensity for HTLs highly correlated with mEGFP fluorescence even at dendritic spines, where CaMKIIα is enriched at the postsynaptic density30 (Supplementary Fig. 3f). These results show that HTLs enable quantitative measurements of endogenous proteins in thick brain tissue that includes entire individual neurons.
We therefore applied this approach to quantitatively map α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) in pyramidal cells of layer 2/3 somatosensory cortex. AMPARs are the primary transducer of fast excitatory neurotransmission and have been shown to increase or decrease in response to synaptic plasticity4. Thus, their abundance can be an indicator for synaptic strength. We fused HaloTag to endogenous GluA2 using SLENDR and performed whole-brain labeling of Halo-GluA2 with JF646-HTL in cleared brain slices (300 µm in thickness) (Fig. 2a). Large and high-resolution confocal image stacks captured individual Halo-GluA2 puncta from the entirety of whole neurons (Fig. 2b–d), which likely represented a functional postsynaptic site of excitatory synapses mostly formed on dendritic spines31,32. This was also supported by the immunohistochemical validation for Halo-GluA2 puncta at excitatory synapses (Supplementary Fig. 2e, f). For quantification, we developed a deep learning-based algorithm to detect regions of interest (ROIs) for individual Halo-GluA2 puncta from single neurons in a semi-automatic manner (Supplementary Fig. 3g). Our algorithm simultaneously detected 3016, 1013, and 700 ROIs from three different pyramidal cells within a single imaging volume (Fig. 2e). We measured and mapped the signal intensity for Halo-GluA2 at individual ROIs on each pyramidal cell (Fig. 2f). All three pyramidal cells showed variable signal intensities at each ROI (Fig. 2g). We further reconstructed dendritic branches of the single pyramidal cell with 3016 ROIs by the tree-based method using arrays of ROIs (Supplementary Fig. 3h, see Methods). Based on the reconstructed dendritic branches, we calculated and mapped the density, mean intensity, and coefficient of variation (CV) of Halo-GluA2 puncta on dendritic segments between branching points (Fig. 2h–m). The density of Halo-GluA2 puncta peaked at 25–75 µm distant from the soma (Fig. 2h, k). In contrast, the mean intensity and CV were not significantly different among distinct dendritic segments. However, these findings were not the case for other analyzed neurons with 6311 and 2865 ROIs (Supplementary Fig. 4), suggesting cell-to-cell variation in the spatial distribution of Halo-GluA2 puncta within individual neurons. Taken together, our approach provides quantitative, whole-cell synaptome mapping of endogenous proteins in the brain.
a Schematic of the labeling and analyzing procedure for Halo-GluA2 in single pyramidal cells in the primary somatosensory cortex layer 2/3. b 3D rendering of three pyramidal cells bearing the soma and dendritic branches labeled for Halo-GluA2 in an imaging volume of 424 µm x 424 µm x 257 µm with the lateral and axial resolutions of 0.26 and 0.5 µm/pixel. c Maximum projection image of a pyramidal cell bearing punctate signals for Halo-GluA2 along dendritic branches. d Enlarged image of the boxed area in (c). e Mapping of 3016 (Cell #1, orange), 1013 (#2, green), and 700 (#3, blue) regions of interest (ROIs) for Halo-GluA2-labeled puncta on three single pyramidal cells. f Heat map of Halo-GluA2 intensity at individual ROIs on the whole pyramidal cell (Cell #1). The color bar indicates % of the averaged intensity of all ROIs. g A scatter plot between the distance from soma and Halo-GluA2 intensity at individual ROIs in three single pyramidal cells (Cell #1-3). h–m Heat maps (h–j) and proximal-to-distal distributions (k–m) of the density (h, k), mean intensity (i, l), and coefficient of variation (j, m) of Halo-GluA2-labeled puncta on dendritic segments (n = 156) of the whole pyramidal cell (Cell #1). Bar graph data are presented as mean ± SEM. **, p < 0.01; *, p < 0.05. p = 0.0013 (k, 25–50 vs 100–125); p = 0.0057 (k, 25–50 vs 150–175); p = 0.021 (k, 50–75 vs 100-125); p = 0.0414 (k, 50–75 vs 150–175). One-way ANOVA with Kruskal-Wallis multiple comparison test (k–m). Scale bars, 50 μm (c, e, f, h–j), 10 μm (d). AU indicates arbitrary units. Source data are provided as a Source Data file.
Quantitative profiling of spatially-distinct subpopulations of endogenous AMPARs and NMDARs at individual synapses
HTLs with different cell membrane permeability and colors would allow for multicolor labeling of the surface and intracellular subpopulations of HaloTag-fused transmembrane proteins23. We tested this application to label surface and intracellular iGluRs at individual synapses in single neurons. Surface iGluRs generate excitatory synaptic transmission, while intracellular iGluRs mediate their dynamic trafficking required for synaptic plasticity2,4. Thus, quantitative imaging of surface and intracellular iGluRs, especially AMPAR GluA2 and N-methyl-D-aspartate receptor (NMDAR) GluN1 subunits, would provide a good readout of synaptic strength and plasticity. We generated Halo-GluA2 or Halo-GluN1 using SLENDR in single pyramidal cells in the somatosensory cortex layer 2/3. N-terminal HaloTag fusions of the surface or intracellular receptors result in the HaloTag protein arranged outside or inside of the cell, respectively. To label the extracellular HaloTag fraction, we developed a membrane-impermeable ligand, FLAG2-JF646-HTL, in which JF646 fluorophore was engineered to conjugate two FLAG peptides with high charge and biocompatibility (Supplementary Fig. 5a). FLAG2-JF646-HTL required cell-membrane permeabilization to label intracellular HaloTag proteins in heterologous cell cultures, confirming its membrane impermeability in cellular labeling (Supplementary Fig. 5b, c). Using whole brains fixed at postnatal 3–6 weeks, we sequentially labeled the surface subpopulation (sHalo-GluA2 or -GluN1) with an excess amount of a membrane-impermeable FLAG2-JF646-HTL (1 µM), and then the intracellular subpopulation (iHalo-GluA2 or -GluN1) with a membrane-permeable JF549-HTL (50 nM) (Fig. 3a). JF549-HTL, but not FLAG2-JF646-HTL, clearly labeled the cell bodies, consistent with the contrasting distribution pattern of the surface and intracellular iGluRs in soma (Fig. 3b and Supplementary Fig. 5d)31,32. We also confirmed that FLAG2-JF646-HTL occupied the surface receptors in a dose-dependent manner, and the concentration of 1 µM was high enough for the saturated labeling of the surface receptors (Supplementary Fig. 5e–i). Thus, our sequential labeling specifically visualizes the surface and intracellular subpopulations of endogenous iGluRs.
a Schematic of the dual-labeling for surface and intracellular subpopulations of Halo-GluA2 (sHalo-GluA2 and iHalo-GluA2) or Halo-GluN1 (sHalo-GluN1 and iHalo-GluN1). b Confocal images for sHalo-GluA2 (green) and iHalo-GluA2 (cyan) in a pyramidal cell in the primary somatosensory layer 2/3 at P28. c, g Confocal images for the surface (green) and intracellular (cyan) subpopulations of Halo-GluA2 (c, sHalo-GluA2 and iHalo-GluA2) or Halo-GluN1 (g, sHalo-GluN1 and iHalo-GluN1) in single dendritic branches labeled for mEGFP (magenta) in layer 2/3 pyramidal cells in the primary somatosensory cortex at P28. d–f, h–j, Correlations between the spine-head volume and fluorescent intensity for sHalo-GluA2 (d) or sHalo-GluN1 (h) at individual dendritic spines. Correlations between the spine-head volume and fluorescent intensity for iHalo-GluA2 (e) or iHalo-GluN1 (i) at individual dendritic spines. Correlations between fluorescent intensities for sHalo-GluA2 and iHalo-GluA2 (f) or sHalo-GluN1 and iHalo-GluN1 (j) at individual dendritic spines. These correlations are measured using the same pool of dendritic spines for Halo-GluA2 (n = 215) or Halo-GluN1 (n = 222) obtained from seven dendritic branches of different pyramidal cells in five mice at postnatal 3-6 weeks. k Heat maps of the sHalo-GluA2-to-iHalo-GluA2 (S/I) ratio at individual ROIs on control (left) and SynGAP1-KO (right) pyramidal cells in the primary somatosensory layer 2/3 at P36-38. l–n Cumulative distributions of sHalo-GluA2 intensity (l), iHalo-GluA2 intensity (m), and S/I ratio (n) at individual ROIs in control (blue) and SynGAP1-KO (orange) pyramidal cells. The analyzed ROIs (n = 44,713 and 42,361 for control and SynGAP1-KO, respectively) are obtained from 12 pyramidal cells in four mice for each condition. ***, p < 0.001; ns, not significant. p = 0.8514 (l); p < 0.0001 (m, n). Two-tailed Kolmogorov–Smirnov test (l–n). Scale bars, 10 μm (b, k), 1 μm (c, g). AU indicates arbitrary units. Source data are provided as a Source Data file.
Halo-GluA2-labeled pyramidal cells showed a selective distribution of punctate sHalo-GluA2 signals at the spine-head portion, but not in dendritic shafts (Fig. 3c). This suggests that the surface population of endogenous GluA2-containing AMPARs is highly concentrated on the postsynaptic membrane at excitatory synapses, while at very low or undetectable levels on the extrasynaptic membrane of dendritic shafts. The integrated signal intensity of sHalo-GluA2 at the spine-head portion varied from very low to high levels among individual spines and was positively correlated with the spine-head volume measured by the fluorescence intensity of mEGFP (Fig. 3d). Since most AMPARs in pyramidal cells of the hippocampus or neocortex contain GluA233,34, our quantitative imaging is consistent with the functional mapping of AMPAR-mediated postsynaptic responses evoked by single spine stimulations35. We also successfully detected iHalo-GluA2 signals at dendritic spines (Fig. 3c), with its fluorescent peak located between those for sHalo-GluA2 and mEGFP (Supplementary Fig. 5j). The integrated signal intensity of iHalo-GluA2 was variable from spine to spine, and proportional to the spine-head volume (Fig. 3e). Moreover, a tight positive correlation between sHalo-GluA2 and iHalo-GluA2 was noted at individual spines (Fig. 3f). This expression pattern for GluA2 was the case for GluN1. We found accumulations of sHalo-GluN1 signals at the spine-head with the integrated intensity proportional to the spine-head volume (Fig. 3g, h), consistent with functional NMDAR-mediated currents triggered by single-spine stimulations36. We also detected weak iHalo-GluN1 signals at dendritic spines with a proportional relationship to the spine-head volume and sHalo-GluN1 signals (Fig. 3g, i, j). Thus, our in vivo dual labeling for HaloTag-fused endogenous transmembrane proteins with different HTLs is a powerful technique to differentially visualize surface and intracellular subpopulations at individual synapses in single neurons.
Simultaneous, synapse-level quantification of surface and intracellular AMPARs enables precise evaluation of AMPAR-mediated synaptic plasticity. When integrated with single-cell synaptome analysis, this approach offers a comprehensive assessment of synaptic plasticity in individual neurons. Using this methodology, we next performed single-cell synaptome mapping of surface and intracellular GluA2 in SynGAP1 knockout (KO) neurons, a well-established model of neurodevelopmental disorders37,38. While this model is known to impair AMPAR-mediated synaptic plasticity, it remains unclear whether the impairment is widespread across synapses or restricted to a specific subset within neurons. To address this, we used the CRISPR-Cas9 system to simultaneously introduce Halo-GluA2 knock-in and SynGAP1 knockout (Supplementary Fig. 6a, b). We then mapped both sHalo-GluA2 and iHalo-GluA2 at thousands of synapses in control and SynGAP1-KO pyramidal cells of the primary somatosensory cortex (Fig. 3k and Supplementary Fig. 6c). Compared with control neurons, SynGAP1-KO neurons exhibited a global decrease and increase in iHalo-GluA2 and sHalo-GluA2/iHalo-GluA2 (S/I) ratio, respectively, at the single-cell level, indicating a selective and robust reduction in the intracellular pool of GluA2-containing AMPARs at the majority of synapses in SynGAP1-KO neurons (Fig. 3l–n and Supplementary Fig. 6c). These findings suggest that AMPAR-mediated synaptic plasticity is broadly affected rather than confined to specific synaptic populations in SynGAP1-KO neurons. Overall, our single-cell synaptome mapping approach provides a quantitative representation of the synaptic landscape within individual neurons, offering valuable insights into synaptic phenotypes associated with neurodevelopmental disorders.
Single-cell multiplexed synaptome mapping of different endogenous synaptic proteins
Since HaloTag is compatible with other self-labeling protein tags, such as SNAP-tag or CLIP-tag without cross-labeling39, multiplexed chemical tag labeling in the same cell could expand the applicability of the protein labeling toolkit. We confirmed no difference in the signal pattern between HaloTag- and SNAP-tag-mediated protein labeling (Supplementary Fig. 1o–q). To test if SLENDR enabled multiplexed protein tagging with HaloTag and SNAP-tag in the same cells, we simultaneously targeted HaloTag and SNAP-tag into the N-terminals of GluA2 and CaMKIIα, respectively (Supplementary Fig. 7a). We found 3.3% of SNAP-CaMKIIα-positive neurons co-labeled with Halo-GluA2 (n = 61) in cortical slice cultures (Supplementary Fig. 7b), suggesting an efficient multiplexed chemical tag labeling of two different proteins in the same neurons.
The AMPA/NMDA ratio is known to be highly correlated with synaptic plasticity, but it is not feasible to estimate the AMPA/NMDA ratio in single excitatory synapses. To quantitatively image both AMPARs and NMDARs in the same dendritic spines, we next targeted HaloTag and SNAP-tag to GluN1 and GluA2 in pyramidal cells of the somatosensory cortex layer 2/3 (Fig. 4a). We found dually-labeled pyramidal cells for Halo-GluN1 and SNAP-GluA2 with different color ligands (100 nM JF646-HTL and 10 nM TMR-Star SNAP-tag ligand (TMR-STL)) in fixed brain slices at postnatal day 27 or 35 (P27 or P35) (Supplementary Fig. 7c). Both Halo-GluN1 and SNAP-GluA2 accumulated at the same dendritic spines labeled for mEGFP (Fig. 4b). Consistent with Fig. 3, Halo-GluN1 and SNAP-GluA2 integrated intensities were proportional to the spine-head volume (Supplementary Fig. 7d, e), and positively correlated with each other at individual spines (Fig. 4c). We noted a low correlation between the Halo-GluN1/SNAP-GluA2 ratio and spine-head volume (Fig. 4d). This low correlation may be due to differences in the distribution of this ratio between small and large dendritic spines. Indeed, the CV for the SNAP-GluA2/Halo-GluN1 ratio is higher at small dendritic spines with the bottom 20% of mEGFP intensity, compared to large dendritic spines with the top 20% of mEGFP intensity (CV = 0.68 vs 0.44, 40 spines from three mice for each), suggesting greater variability in the ratio among smaller spines. Intriguingly, some small dendritic spines accumulated Halo-GluN1 signals but not SNAP-GluA2 signals (Fig. 4b, arrows, d), which may represent “silent” synapses that contain NMDARs but no AMPARs40.
a Schematic of Halo-GluN1 and SNAP-GluA2 dual-labeling. b Confocal images for Halo-GluN1 (green) and SNAP-GluA2 (cyan) in an mEGFP-labeled dendritic branch (magenta) of the double knock-in pyramidal cell in the layer 2/3 in the primary somatosensory cortex at P35. Arrows represent a small spine that shows signals for Halo-GluN1 but not SNAP-GluA2. c A correlation between the fluorescent intensities for Halo-GluN1 and SNAP-GluA2 at individual dendritic spines (n = 192) obtained from three pyramidal cells of three mice at P27 or P35. d A correlation between the spine-head volume and fluorescent ratio of SNAP-GluA2 to Halo-GluN1 at individual dendritic spines. The pool of dendritic spines analyzed is same as that in (c). e Schematic of the dual-labeling of surface Halo-GluA2 (sHalo-GluA2) and PSD95-SNAP. f Confocal images for sHalo-GluA2 (green) and PSD95-SNAP (cyan) in an mEGFP-labeled dendritic branch (magenta) of a double knock-in pyramidal cell in layer 2/3 primary somatosensory cortex at P16. Arrows represent a dendritic spine labeled for PSD95-SNAP but not sHalo-GluA2. g–i Heat maps of the synaptic ROI size (g), PSD95-SNAP density (Intensity/ROI size) (h), and sHalo-GluA2 density (i) at individual ROIs (n = 1273) on a single pyramidal cell (Cell #1). The color bar indicates % of the averaged ROI size (g), PSD95-SNAP density (h), or sHalo-GluA2 density (i) of all ROIs. j–m Correlations of the synaptic ROI size (j, k) or distance from the soma (l, m) with the density of PSD95-SNAP (l, j) or sHalo-GluA2 (k, m) at individual synaptic ROIs on a single pyramidal cell. n, Mapping of synaptic ROIs with high (top 5%, magenta) and low (bottom 5%, cyan) sHalo-GluA2 density on a single pyramidal cell. o, Difference in the synaptic ROI size among the three groups of synaptic ROIs with high (n = 64), medium (n = 1147), and low (n = 64) sHalo-GluA2 density in the Cell #1. Bar graph data are presented as mean ± SEM. p, Comparison of the averaged synaptic ROI size among the high, medium, and low groups, each of which contains four pyramidal cells (Cell #1-4) from four mice. Bar graph data are presented as mean ± SEM. ***, p < 0.001; **, p < 0.01. p < 0.0001 (o, Low vs Med); p = 0.0002 (o, Med vs High); p = 0.0051 (p). One-way ANOVA with Kruskal-Wallis multiple comparison test (o, p). Scale bars, 10 µm (g–I, n), 1 μm (b, f). AU indicates arbitrary units. Source data are provided as a Source Data file.
For more comprehensive mapping of functional AMPARs at excitatory synapses, we performed dual labeling of surface Halo-GluA2 (sHalo-GluA2) and intracellular PSD95-SNAP labeled with membrane-impermeable (100 nM FLAG2-JF646-HTL) and permeable (10 nM TMR-STL) ligands in single pyramidal cells of the somatosensory cortex layer 2/3 at P15 or P16 (Fig. 4e). We obtained z-stacks from four dually-labeled pyramidal cells derived from different animals (Supplementary Fig. 7f). PSD95-SNAP-labeled puncta were very often co-labeled for sHalo-GluA2 (Fig. 4f). Interestingly, the intensity of sHalo-GluA2 signals varied remarkably, with no detectable levels at a few PSD95-SNAP-labeled puncta (Fig. 4f, arrows). To quantify the amount of surface GluA2 at individual excitatory synapses, we semi-automatically registered 1004-1296 PSD95-SNAP-labled puncta as synaptic ROIs (Fig. 4g and Supplementary Fig. 7g). We found that the size of synaptic ROIs was tightly correlated with their integrated intensity for PSD95-SNAP over serial images (Supplementary Fig. 7h). Since the integrated intensity of fluorescently-labeled PSD95 is known to be proportional to the excitatory PSD size41, we concluded that the synaptic ROI size serves as a proxy of the excitatory PSD size. We next calculated and mapped the “signal density” of PSD95-SNAP and sHalo-GluA2 by normalizing their integrated intensity to the synaptic ROI size (Fig. 4h, i and Supplementary Fig. 7i, j). While the integrated intensity represents the total amount of PSD95-SNAP and sHalo-GluA2 at individual synapses, the signal density reflects the local concentration of these proteins. Notably, the signal density of sHalo-GluA2 was highly variable compared with that of PSD95-SNAP (CV: 0.53-0.63 for sHalo-GluA2 and 0.29–0.32 for PSD95-SNAP). When examining the relationship between the signal density and synaptic ROI size, we found a strong positive correlation for PSD95-SNAP, whereas sHalo-GluA2 density showed only a weak correlation (Fig. 4j, k and Supplementary Fig. 7k, l). Neither signal density measure correlated with the distance from the soma within the analyzed imaging volume (~ 60 µm from the soma) (Fig. 4l, m and Supplementary Fig. 7m, n), suggesting that both PSD95 and surface AMPARs preferentially accumulate at larger synapses regardless of their spatial location.
We then ranked synaptic ROIs based on their sHalo-GluA2 signal density, and grouped them into high (top 5%, presumably potentiated), medium (middle 90%), and low (bottom 5%, presumably silent) sHalo-GluA2 density groups40 (Fig. 4n). The average synaptic ROI size of the high sHalo-GluA2 density group was 1.3–2.4 (1.86 ± 0.23) times higher than the medium sHalo-GluA2 density group, while the low sHalo-GluA2 density group was 0.36–0.46 (0.43 ± 0.02) times lower. These findings suggest that potentiated synapses are relatively larger and silent synapses smaller in PSD size (Fig. 4o, p and Supplementary Fig. 7o). Thus, our platform enables single-cell multiplexed synaptome mapping of different protein subpopulations, providing insights into both whole-cell synaptic patterns within individual neurons and the spatial organization of presumably potentiated or silent synapses at the single-cell level.
Live imaging of temporally-distinct subpopulations of endogenous CaMKIIα
Live-cell pulse-chase labeling of HaloTag-fused proteins enables imaging of pre-existing and nascent subpopulations24. We first applied the pulse-chase labeling to endogenous Halo-CaMKIIα in hippocampal slice cultures, visualizing both the pre-existing and nascent (synthesized in 2 h) subpopulations of Halo-CaMKIIα within single pyramidal cells (Supplementary Fig. 8a). Pharmacological blockade of protein synthesis with anisomycin suppressed the nascent signals, validating the specificity for our pulse-chase labeling of the pre-existing and nascent subpopulations (Supplementary Fig. 8b, c). We then tested this application in mouse brains in vivo. We made a small hole on the skull to expose the somatosensory cortex, through which we performed in vivo pulse-chase labeling for the pre-existing and nascent subpopulations of Halo-CaMKIIα with JF549-HTL and JF646-HTL, respectively. The intervals between pulse and chase labeling were 2 or 72 h for distinct time windows of protein synthesis (Fig. 5a). The pulse-chase labeling with a 2 h-interval visualized both the pre-existing and nascent subpopulations with a comparable signal pattern across the cell (Fig. 5b, left and Supplementary Movie 1). In contrast, a 72 h-interval labeling provided stronger nascent and weaker pre-existing signals compared with a 2 h-interval labeling (Fig. 5b, right), demonstrating time-dependent turnover of endogenous CaMKIIα.
a Schematic of the in vivo pulse-chase labeling for pre-existing and nascent Halo-CaMKIIα in the neocortex. b 3D rendering of single pyramidal cells labeled for pre-existing (magenta) and nascent (green) Halo-CaMKIIα with a 2 (left) or 72 (right) h-interval between the pulse and chase labeling in the somatosensory cortex layer 2/3. Enlarged images of single dendritic branches are shown at the bottom. c Schematic of the in vivo pulse-chase labeling for pre-existing and nascent Halo-mEGFP-CaMKIIα in the neocortex. d In vivo two-photon microscopic images in the visual cortex of living mice at P35 showing signals for nascent (green) and total (cyan) pools of Halo-mEGFP-CaMKIIα in the presence (right) or absence (left) of 50 µM anisomycin. e Post-hoc confocal images with fixed control brains demonstrating comparable signal patterns for pre-existing (magenta) and nascent (green) Halo-mEGFP-CaMKIIα in the same area as observed by two-photon live imaging. Images in the middle column are obtained from the boxed area in the left panel. f A correlation between the fluorescent intensities for pre-existing and nascent Halo-mEGFP-CaMKIIα at individual dendritic spines in the presence or absence of anisomycin (n = 265 and 250 dendritic spines obtained from six pyramidal cells in four control and anisomycin-treated mice, respectively). Scale bars, 20 µm (b (top)), 2 μm (b (bottom), d, e). AU indicates arbitrary units. Source data are provided as a Source Data file.
Next, we tried to image both the total and pre-existing or nascent populations of CaMKIIα in living mice. For this aim, we tandemly fused HaloTag and mEGFP to endogenous CaMKIIα (Halo-mEGFP-CaMKIIα) in a sparse subset of neurons in the visual cortex (Fig. 5c). In this experiment, mEGFP fluorescence is a measure of the total Halo-mEGFP-CaMKIIα proteins, while HaloTag pulse or chase labeling signals indicate the pre-existing or nascent subpopulations 12. At P31-87, we pulse-labeled the pre-existing subpopulation with JF646-HTL, and chase-labeled the nascent subpopulation with TMRdirect-HTL after a 2 h interval (Fig. 5c and Supplementary Fig. 8f). In vivo two-photon time-lapse imaging revealed a ~ 30% reduction in the pre-existing signals at the same dendritic spines of pyramidal cells 2 h after the pulse-labeling, while total signal levels remained unchanged (Supplementary Fig. 8g–j). This indicates that the observed signal reduction is not due to photobleaching and confirms that this approach effectively quantifies the hourly degradation of CaMKIIα at individual spines in vivo. After the chase-labeling, we also observed the nascent signals at individual dendritic spines in living mice (Fig. 5d). Application of anisomycin blocked the nascent signals, demonstrating the specific labeling of the nascent Halo-mEGFP-CaMKIIα in vivo (Fig. 5d). To compare the pre-existing and nascent signals, we fixed the mice after the live imaging, and conducted confocal imaging for the pre-existing, nascent, and total populations. The pre-existing and nascent subpopulations perfectly overlapped with each other (Fig. 5e). As in the hippocampal slice culture (Supplementary Fig. 8d, e), we found a positive correlation in the signal intensity between the pre-existing and nascent subpopulations at individual dendritic spines (Fig. 5f). Again, we confirmed no nascent signals in the presence of anisomycin (Fig. 5e, f). Thus, in vivo pulse-chase labeling of HaloTag-fused endogenous proteins enables the detection of nascent protein subpopulations at individual synapses in living mouse brains.
Single-cell synaptome mapping of temporally-distinct subpopulations of endogenous PSD95 and βActin
Excitatory synapses capture postsynaptic scaffold proteins during late-phase (> 1 h) long-term potentiation that is dependent on protein synthesis42,43. Thus, a method to visualize the nascent subpopulation of a major postsynaptic scaffold, PSD95, at individual synapses within a single neuron would provide a valuable means for studying the mechanism of synaptic plasticity. To achieve this, we tagged PSD95 with HaloTag using SLENDR in single pyramidal cells of the somatosensory cortex, and conducted in vivo pulse-chase labeling for pre-existing and nascent PSD95-Halo with a 0.5, 1, or 2 h interval at P23-36 (Fig. 6a and Supplementary Fig. 9a). Since detecting nascent PSD95-Halo required an interval of at least 1 h (Supplementary Fig. 9b, c), we utilized a 2 h interval for the following analysis. We found fluorescent puncta co-labeled for pre-existing and nascent PSD95-Halo along dendritic branches in three pyramidal cells (Fig. 6b and Supplementary Fig. 9d). The distribution pattern of fluorescent signals was largely comparable between pre-existing and nascent PSD95-Halo, although a few puncta displayed a higher accumulation of nascent signals compared to pre-existing ones (Fig. 6b, arrows, and Supplementary Fig. 9d). We then spatially mapped the nascent-to-pre-existing ratio (NPR) of PSD95-Halo at 3239-4402 synaptic ROIs, which were registered with the pre-existing signal within a single pyramidal cell (Fig. 6c and Supplementary Fig. 9d). The synaptome plot revealed a relatively weak positive correlation between pre-existing and nascent signals at individual synapses, with a remarkably high NPR observed at a subset of synapses (R2 = 0.6708-0.7629; Fig. 6d and Supplementary Fig. 9e). This high NPR population preferred smaller synapses (Fig. 6e and Supplementary Fig. 9f), but not a specific distance from the soma (Fig. 6f and Supplementary Fig. 9g). Therefore, our in vivo pulse-chase labeling enables single-cell mapping of a minority of synapses that are enriched with nascent PSD95.
a Schematic of the in vivo pulse-chase labeling for pre-existing and nascent PSD95-Halo in the neocortex. b, g Representative images of a dendritic branch of the somatosensory cortex layer 2/3 pyramidal cells labeled for PSD95-Halo (b) and Halo-βActin (g) showing fluorescent intensities (top three panels) for pre-existing (magenta) and nascent (green) subpopulations, and heat maps of NPR (bottom). Arrows indicate synaptic ROIs (#2004, #2009, and #2031) with a high NPR. c, h Heat maps of NPR for PSD95-Halo (c) and Halo-βActin (h) at individual ROIs (n = 3239 and 3589) on a single pyramidal cell (gray). Boxed areas in c and h were enlarged in b and g, respectively. d–f Correlations between the fluorescent densities (d) for pre-existing and nascent PSD95-Halo or between the synaptic ROI size (e) or distance from the soma (f) and NPR of PSD95-Halo at individual synaptic ROIs. Solid spots in black indicate synaptic ROIs #2004, #2009, and #2031 shown in (b). i A correlation between the fluorescent densities for pre-existing and nascent Halo-βActin at individual synaptic ROIs. Scale bars, 10 µm (c, h), 2 µm (b, g). AU indicates arbitrary units. Source data are provided as a Source Data file.
In contrast to PSD95, a cytoskeletal protein βActin is known to be rapidly accumulated at potentiated synapses during the early-phase long-term potentiation that is independent of protein synthesis42. For comparison with PSD95, we next labeled the pre-existing and nascent subpopulations of Halo-βActin, which was generated using SLENDR (Fig. 6g and Supplementary Fig. 9h–l). Dendritic spines labeled for Halo-βActin exhibited quite similar labeling patterns for pre-existing and nascent signals, resulting in a strong correlation at individual synapses compared with PSD95-Halo (R2 = 0.7954-0.8503; Fig. 6g–i and Supplementary Fig. 9i–l). The relatively more variation in NPR of PSD95 among synapses may be attributed to its longer retention time at dendritic spines compared with βActin, which has a retention time approximately 20 times shorter than that of PSD9544, while other mechanisms at each synapse to retain proteins may also affect the NPR.
Single-cell synaptome mapping of nascent-and-surface subpopulations of endogenous GluA2
Nascent AMPARs are known to be preferentially inserted into potentiated synapses upon learning5. However, it remains a challenge to image nascent-and-surface AMPARs in single neurons in the brain. We therefore applied our in vivo pulse-chase labeling to single-cell mapping of the nascent-and-surface subpopulations of endogenous GluA2 in the brain. Using SLENDR, we introduced Halo-GluA2 into single, layer 2/3 pyramidal cells in the somatosensory cortex. At P21-23, we pulse-labeled the pre-existing subpopulation of Halo-GluA2 with an excess amount of 1 µM membrane-permeable JF549-HTL through a cranial window. We fixed the brain 2 or 48 h after the pulse labeling, and chase-labeled the surface subpopulation of nascent Halo-GluA2 with 100 nM membrane-impermeable FLAG2-JF646-HTL (Fig. 7a). Confocal microscopy acquired z-stacks of 9 single HaloTag-labeled neurons from 50 or 100 µm-thick brain slices (Supplementary Fig. 10a). The labeled neurons exhibited many punctate signals for pre-existing Halo-GluA2 along dendritic branches with a significant decrease in the signal intensity between 2 and 48 h (Fig. 7b, c), suggesting time-dependent degradation of Halo-GluA2 at individual synapses12. Conversely, we detected bright signals for nascent-and-surface Halo-GluA2 on pre-existing Halo-GluA2-labeled puncta when protein synthesis was allowed for 48 h before fixation, but not 2 h (Fig. 7b, c). For single-cell synaptomic measurement of nascent-and-surface Halo-GluA2 signals in 48 h of the time window, we registered 1041–4025 pre-existing Halo-GluA2-labeled areas as synaptic ROIs on single pyramidal cells (3064 ± 368 ROIs form 9 cells) (Fig. 7d and Supplementary Fig. 10b–i). Single-cell mapping of nascent-and-surface Halo-GluA2 signals at individual synaptic ROIs revealed high signal intensity near the soma. Indeed, proximal ROIs (within 0-30 µm from soma) exhibited higher mean intensity for nascent-and-surface Halo-GluA2 than distal ROIs (within 50-80 µm from soma) in 8 out of 9 pyramidal cells analyzed (Fig. 7e–g and Supplementary Fig. 10b–i). Therefore, our in vivo pulse-chase labeling technique enables single-cell, large-scale, quantitative mapping of spatiotemporally distinct subpopulations of endogenous proteins at synapse resolution in the brain.
a Schematic of the in vivo pulse-chase labeling for pre-existing, and nascent-and-surface Halo-GluA2 in the neocortex. b Confocal images of dendritic branches of pyramidal cells labeled for pre-existing (magenta), and nascent-and-surface (green) Halo-GluA2 in the time windows of 2 (left) and 48 (right) h for protein synthesis in the somatosensory cortex layer 2/3. c Changes in the averaged density of pre-existing (top), and nascent-and-surface (bottom) Halo-GluA2 puncta on dendritic branches between the time windows of 2 (n = 7 neurons from 2 mice) and 48 (n = 9 neurons from 3 mice) h for protein synthesis. Data are presented as mean ± SEM. d Heat map of nascent-and-surface Halo-GluA2 density at 4025 synaptic ROIs in a single pyramidal cell. The color bar indicates % of the averaged intensity for nascent-and-surface Halo-GluA2 at all synaptic ROIs. Data are presented as mean ± SEM. e A correlation between the distance from the soma and nascent-and-surface Halo-GluA2 density at individual synaptic ROIs in the single pyramidal cell. Proximal (0–30 µm) and distal (50–80 µm) ROIs from the soma are highlighted in orange and blue. f Difference in nascent-and-surface Halo-GluA2 density between proximal (left) and distal (right) ROIs. Blank circles and lines represent means and medians with 25th and 75th percentiles, respectively. g Difference in the averaged density for nascent-and-surface Halo-GluA2 between proximal and distal ROIs in individual pyramidal cells (n = 9 neurons from 3 mice). ***, p < 0.001; **, p < 0.01; *, p < 0.05. p = 0.0311 (c, top); p = 0.0115 (c, bottom); p < 0.0001 (f); p = 0.0039 (g). Two-tailed Mann–Whitney U test (c,f) or two-tailed Wilcoxon matched-pairs signed rank test (g). Scale bars, 10 µm (d), 2 µm (b). AU indicates arbitrary units. Source data are provided as a Source Data file.
Discussion
Here we developed a simple and versatile platform for imaging not only total but also spatial (surface/intracellular), temporal (pre-existing/nascent), or temporal-and-spatial (nascent-and-surface) populations of endogenous proteins, such as synaptic and signaling proteins, in single cells in brain tissue. In combination with a semi-automatic analysis pipeline, we achieved single-cell synaptome mapping of various protein subpopulations at thousands of synapses in the brain, comprehensively visualizing the synapse diversity within a neuron of protein localization, trafficking and turnover.
There has been no simple method for quantitative imaging of spatially and temporally different subpopulations of endogenous proteins in brain tissue. Antibodies or chemical probes against the extracellular domain can label the surface subpopulation in living primary cultures or brain tissues45,46,47. However, they do not allow for single-cell, high-contrast imaging or label temporally-distinct subpopulations. pH-sensitive super-ecliptic pHluorin (SEP) or tandem fluorescent protein timers (tFTs) can help visualize subpopulations in living cells48,49. However, no single fluorescent protein tag is able to label both total and spatiotemporally distinct subpopulations of a tagged protein. Bio-orthogonal metabolic labeling methods can label nascent endogenous proteins50,51. However, these methods target nascent proteins in all cells in tissue and therefore are not suitable for single-cell, high-contrast imaging. The wide variety of chemical tag ligands that differ in colors and cell membrane permeability overcome these limitations, enabling labeling of different subpopulations of tagged endogenous proteins via single chemical tags in brain tissue.
SLENDR/vSLENDR, which is based on the HDR machinery following CRISPR-Cas9-mediated doubled-stranded breaks (DSBs), allows for single-cell endogenous protein labeling in the brain14,15. Other CRISPR-Cas9-mediated protein labeling techniques such as HITI, HiUGE, Orange, TKIT, or CRISPIE could also label endogenous proteins with chemical tags via the non-homologous end joining (NHEJ) machinery, possibly expanding our present protein labeling toolbox52,53,54,55,56. However, these NHEJ-based techniques rely on non-specific gene insertion into the DSB, unlike the precise gene insertion guided by the homologous sequence around the DSB in HDR. Thus, these NHEJ-based approaches may not be suitable for knock-in of multiple genes due to crosstalk between multiple knock-in loci. Sequential NHEJ-mediated knock-ins via a recombinase-driven switch of different gRNA expression could reduce this crosstalk, but it requires careful optimization of the recombination timing and knock-in vector amounts that differ from gene to gene57. In contrast, the HDR-based SLENDR/vSLENDR is compatible with multiplexed gene editing14,15. Multiplexed labeling of different proteins in the same cells demands high knock-in efficiency for each target. In this study, we achieved high knock-in efficiency of chemical tags by developing an improved version of SLENDR using an RNP complex (4.3-5.9% for CaMKIIα, βActin, and GluA2 genes). This enabled multiplexed labeling of three endogenous protein pairs of GluA2 and PSD95, GluN1, or CaMKIIα. Although this knock-in efficiency based on an RNP complex remains lower than the NHEJ-mediated knock-in efficiency54, this efficiency could likely be improved by performing IUE at earlier embryonic stages (e.g. E12-13 for the cortex), as demonstrated in the original SLENDR technique14.
We showed that the integrated cell-surface signals for GluA2 and GluN1 were both proportional to the spine-head volume at individual synapses. GluA2-containing AMPARs (GluA1/GluA2 or GluA2/GluA3 heterodimers) are a majority of AMPARs at excitatory synapses in pyramidal cells in the hippocampus or neocortex33,34. Therefore, our finding is consistent with the previous mapping of AMPAR- or NMDAR-mediated electrophysiological responses to focal stimulation of single dendritic spines in pyramidal cells35,36. Moreover, since electrophysiological responses can be attenuated during propagation from synapses to cell bodies, our method can provide a more precise measure of the surface AMPARs or NMDARs on the postsynaptic membrane at individual synapses. We should note that we did not distinguish the perisynaptic or extrasynaptic subpopulation of AMPARs or NMDARs from the synaptic subpopulation within dendritic spines, due to the limited spatial resolution of optical confocal imaging. Super-resolution microscopy, which we showed is compatible with our protein labeling platform, will be helpful in addressing this issue.
Single-cell synaptome mapping of protein localization requires specific, quantitative protein labeling as well as precise, high-throughput detection and measurement of signal intensity at thousands of synapses in each neuron. Consistent with the previous study22, we demonstrated specific, quantitative labeling of chemical tagged proteins across thick brain tissue using the small size (~ 1 kDa) of bright chemical tag ligands, which rapidly and evenly penetrated into deep tissue (Supplementary Fig. 3b-f). Our deep imaging of thick slices (> 100 µm in thickness) was assisted with a combination of a long working distance objective and additional tissue clearing to minimize axial signal attenuations (Supplementary Fig. 3e). In addition, we validated HaloTag signals for PSD95, GluN1, and GluA2 at excitatory synaptic sites using immunohistochemistry (Supplementary Fig. 2), confirming that these HaloTag-labeled puncta likely represent functional postsynaptic sites of excitatory synapses mostly formed on dendritic spines31,32. Based on the imaged signals, we determined synaptic ROIs with an iterative, semi-automatic approach (Supplementary Fig. 3g, h). Single-cell protein labeling easily discriminates true signals in labeled cells from neighboring noise signals, and is thus advantageous for this approach. These advantages enabled precise detection and measurement of signal intensity at individual synaptic ROIs. Indeed, using our approach, we mapped synaptic proteins such as GluA2, PSD95, or βActin at 1041-6311 synapses in single neurons in the brain. This large-scale mapping provides an unprecedentedly detailed characterization of the spatial distribution of endogenous synaptic proteins throughout an entire single neuron. Importantly, our single-cell synaptome analysis for sHalo-GluA2 and PSD95-SNAP showed a strong correlation between the integrated sHalo-GluA2 signals and synapse size (R2 = 0.7258 and 0.7532, Supplementary Fig. 7h). Since synapse size is known to correlate with spine-head volume (R2 = 0.774)41, this finding is fully consistent with our observed correlation between the integrated sHalo-GluA2 intensity and spine-head volume (R2 = 0.7207, Fig. 3d), as well as previous reports linking the AMPAR-mediated current to spine-head volume (R2 = 0.476-0.81)35. Thus, these consistent results across different experiments and methodologies strongly support the fidelity and reproducibility of our single-cell synaptome approach.
Our single-cell synaptome analysis showed no proximal-to-distal difference in the dendritic distribution of synapses rich in surface GluA2 (presumably potentiated synapses). On the other hand, GluA2 synthesized in 48 h was preferentially incorporated in the surface of dendritic spines located in proximal dendrites, while a few distal spines contained exceptionally abundant nascent-and-surface GluA2. It would be interesting to spatially map the overlapping population of active synapses, strong synapses and nascent AMPAR-fueled synapses in vivo, which has been a long-standing challenge in synaptic neuroscience. Our platform combined with Ca2+ imaging or active synapse tagging based on CaMPARI58 may allow for such mapping.
There are a few things to note about our platform. The fusion of chemical tags to endogenous proteins may affect their native structures or functions. To avoid such potential disturbance, we inserted chemical tags at the same amino acid position as the previous fluorescent protein tagging59,60,61. If chemical tags are targeted to uncharacterized amino acid positions, it is necessary to test the function or localization pattern of the tagged proteins in a careful manner. Machine learning-based algorithms that predict the three-dimensional protein structure could be helpful in addressing this issue62. Another thing to note is that labeling of the cell-surface and intracellular subpopulations should be carefully applied to living tissues. Since transmembrane proteins are dynamically trafficked between the surface and intracellular subpopulations, the time lag between membrane-impermeable and -permeable ligand application may affect the specificity of labeling of each subpopulation. pH-sensitive fluorophore ligands could overcome this problem63, because the difference in pH levels between the surface and intracellular subpopulations allows for their separable imaging with pH-sensitive probes.
Synapses are diverse in their function and molecular composition, playing a crucial role in shaping the complexity and functionality of neural circuits. Many techniques, including electrophysiological and biochemical approaches, rely on “population-averaging” and do not account for individual and spatial differences amongst synapses. Most of the imaging techniques focus on only a small population of synapses within single neurons, displaying a quite partial representation of synapse diversity. To gain a more comprehensive understanding of synapse diversity in the brain, researchers employ brain-wide synaptome analysis11,12,13. However, this method conflates variation between cells with synapse variation within a single neuron. Our single-cell synaptome mapping of protein subpopulations visualizes the spatial organization of synapse diversity in protein localization, trafficking and turnover, providing a more accurate and informative representation of the synaptic landscape in single neurons. Furthermore, our platform enables the detection and characterization of minority but influential synapses that might be overshadowed in conventional bulk analysis, as we spatially mapped silent and potentiated synapses using our platform. Therefore, our platform allows scientists to delve into the complexities of each single neuron at the synapse level, enabling the study of physiology, development or pathology of distinct neurons in a variety of experimental paradigms. For example, in the context of memory formation64,65,66,67, our platform may be able to spatially map the synaptic tagging and capture of memory-related proteins68 in the whole engram cell structure. This technical advance, thus has the power to provide new insights into neuronal organization and computations in learning and memory.
Methods
Animals
Animal care and experimental protocols were approved by the Animal Experiment Committee of Niigata University and were carried out under the Guidelines for the Care and Use of Laboratory Animals of Niigata University (protocol number: SA01663, SA01664). To obtain Cas9 line (RBRC11579), CAG-floxed STOP Cas9 line was first created using almost exactly the same method as CAG-floxed STOP tdTomato reporter line (MGI:6192640) shown in the previous study69. Briefly, the targeting vector was designed to insert a CAG promoter, floxed Neo-STOP cassette and hspCas9 gene into intron 1 of the ROSA26 locus, and it was introduced into RENKA70, a C57BL/6N-derived ES cell line, by electroporation. After selecting G418-resistant clones, genome recombination was confirmed by Southern blotting. Male chimera mice were generated by injection of recombinant ES cells into eight-cell stage embryos from ICR mice, and then heterozygous F1 mice were obtained by natural mating with C57BL/6 N female mice. To remove the Neo-STOP cassette, heterozygous mice were crossed with Actb-iCre line71 which ubiquitously expresses Cre recombinase, generating Cas9 mice. Slc:ICR mice were purchased from SLC Japan. All mice used in experiments were maintained under a 12 hr light/dark cycle. The day on which the vaginal plug was detected was designated as embryonic day 0 (E0). The first 24 h after birth was referred to as postnatal day 0 (P0). Fetal mice of Slc:ICR female at E14 or 15 were used for in utero electroporation experiments. P1-P5 pups of heterozygous Cas9 mice were used for cortical AAV injection. P5–P6 pups of Slc:ICR and heterozygous Cas9 mice were used for organotypic slice cultures. Both male and female mice were used and randomly assigned to experimental groups, because this study primarily focuses on method development.
Organotypic slice cultures
Organotypic brain slice cultures were prepared as described previously72. In brief, hippocampal or cortical slices of 325 μm thickness were prepared with a McIlwain tissue chopper (The Mickle Laboratory Engineering) from both male and female mice at P5–6 or P0-2, respectively. The slices were plated on hydrophilic PTFE membranes (Millicell, Millipore) and maintained in a culture medium containing MEM (Sigma-Aldrich), 20% heat-inactivated horse serum (GIBCO), 1 mM glutamax (GIBCO), 1 mM CaCl2, 2 mM MgSO4, 12.9 mM D-Glucose, 5.2 mM NaHCO3, 30 mM HEPES, 0.075% Ascorbic acid, 1 µg/ml Insulin at 37 °C in a 5% CO2 humidified atmosphere. The culture medium was exchanged with fresh medium every other day.
DNA constructs
For SLENDR/vSLENDR, we designed plasmid vectors carrying HDR donor template DNAs. The ~2 kb homology sequence of target genes flanking the insertion site for the HaloTag or SNAP-tag sequence was synthesized and subcloned into the pUC57 or pUCAmp vector (Genewiz or Azenta). The synthesized HaloTag sequence with a short linker (ctcgagccaaccactgaggatctgtactttcagagcgataacgat)73 was incorporated into the pUC57 or pUCAmp vector carrying the homology sequence, resulting in pUC-HDR vectors for the HaloTag. The synthesized SNAP-tag sequence18 was incorporated into pUC-HDR vectors in place of the HaloTag sequence. For the plasmid-based HaloTag knock-in, target-specific gRNA sequences were subcloned into the pX330 plasmid vector encoding the Cas9 protein and gRNA backbone (Addgene plasmid # 42230). For the CRISPR-Cas9-mediated knockout of SynGAP1 gene, a DNA sequence containing three distinct SynGAP1-specific gRNAs (tcagcacgttatctactcgg, tgcctgtcggcaaggtcacg, and ctgcacatagcatccggtaa), each of which was downstream of the U6 promoter, was synthesized and subcloned into the pCAG-Cas9 plasmid vector. The Cas9 sequence in this vector was C-terminally linked to the TagBFP via a P2A cleavage sequence, resulting in pCAG-Cas9-2A-TagBFP-SynGAP1KO. To generate AAV-HDR for Halo-CaMKIIα or Halo-mEGFP-CaMKIIα, we engineered pAAV-HDR-mEGFP-CaMKIIα (Addgene plasmid #104589)15. The HaloTag sequence was substituted for or inserted upstream of mEGFP, resulting in pAAV-HDR-Halo-CaMKIIα or pAAV-HDR-Halo-mEGFP-CaMKIIα, respectively. All obtained constructs were verified by DNA sequencing.
In utero electroporation
In utero electroporation (IUE) was performed as previously described in the original SLENDR publication14. In brief, mice were deeply anesthetized by intraperitoneal injection of medetomidine (0.75 mg/kg of body weight; Zenoaq), midazolam (4.0 mg/kg of body weight; Sandoz), and butorphanol (5.0 mg/kg of body weight; Meiji Seika Pharma)74. To relax the myometrium, ritodorin hydrochloride (0.7-1.4 μg/g of body weight; Sigma-Aldrich) was applied to the exposed uterine horns. Cas9-gRNA RNPs were obtained by incubating a crRNA/tracrRNA hybrid with Streptococcus pyogenes (Sp) Cas9 protein (Integrated DNA Technologies) for 5 min at room temperature, and then mixed with plasmid vectors for DNA donor template and fluorescent proteins as IUE markers. The final concentration of a crRNA/tracrRNA, Cas9 protein, DNA donor template plasmids, and fluorescent protein plasmids was 20 μM, 0.33 μg/μl, 2 μg/μl, and 1 μg/μl, respectively. The CRISPR-Cas9-mediated knockout of SynGAP1 gene was obtained by supplementing RNP/plasmid solution with the pCAG-Cas9-2A-TagBFP-SynGAPKO vector (1 μg/μl). For the plasmid-based knock-in, the target-specific pX330 vectors (1 μg/μl) were included in a mixture of plasmids, instead of the RNP complex. For multiplexed HaloTag and SNAP-tag labeling, half of the amount of RNPs for each target or 1 µg/µl of target-specific pX330 vectors was used. RNP/plasmid solution was mixed with Fast Green (0.1 mg/ml, Sigma-Aldrich), and 1-2 μl of the solution was injected into the lateral ventricle with a microdispenser (Drummond). Electroporation was performed at E14 or E15. Electric pulses (45 V for 50 ms, 4 times with 950 ms intervals) were delivered with forceps-shaped electrodes (CUY650P5; Nepa Gene) connected to an electroporator (NEPA21, Nepa Gene). The position and angle of the electrode were set as described previously14. For the recovery from anesthesia, mice were intraperitoneally injected with atipamezole (0.75 mg/kg of body weight; Zenoaq).
AAV vector production and viral transduction in vitro and in vivo
All custom AAV vectors were produced by Vigene Biosciences. AAV1-HDR-Halo-CaMKIIα and AAV9-HDR-Halo-mEGFP-CaMKIIα were pseudotyped with serotype 1 and 9, and titrated as 1.5×1014 and 7.0×1013 GC/ml, respectively. For viral transduction into hippocampal organotypic slice cultures, AAV1-HDR-Halo-CaMKIIα was directly added onto the cultures at DIV5–8. The viral amount applied to each slice was 1.5×1011 GC. Cultures transduced with AAV were analyzed at DIV14-17. For viral transduction into neonatal brains in vivo, CAG-Cas9 mice at P0-2 were cryoanesthetized for 2-3 min before injection, and gently held by hand. A mixture of AAV9-HDR-Halo-mEGFP-CaMKIIα (7.0×1013 GC/ml) and Fast Green (0.1 mg/ml, Sigma-Aldrich) was injected into the unilateral cerebral cortex by a hand-held microdispenser (Drummond). To obtain a large injected area, the total volume of 2-4 μl viral solution was administrated for multiple injection sites 1-2 mm separate from each other. The injection result was confirmed by coloring the entire cortex with Fast Green. After injection, the pups were placed on a heat blanket for several minutes and then returned to their mother.
Fixation and tissue preparation
Hippocampal or cortical organotypic slice cultures were immersed in the fixative of 4% paraformaldehyde in 0.1 M phosphate buffer (4% PFA/PB, pH 7.4) for 1 h. Mice were anesthetized with medetomidine (0.75 mg/kg of body weight; Zenoaq), midazolam (4.0 mg/kg of body weight; Sandoz), and butorphanol (5.0 mg/kg of body weight; Meiji Seika Pharma) as described above, and then fixed by transcardial perfusion with 4% PFA/PB or 9% glyoxal/8% acetic acid adjusted to pH 4-5 (2.5-3 ml/g of body weight) for 10 min75. Glyoxal-fixed brains were further postfixed overnight. The brain was removed from the skull and rinsed with PBS. Coronal sections (50-300 μm in thickness) were prepared using a vibratome (VT1200S, Leica) before or after chemical tag labeling. For immunohistochemistry, brain slices were permeabilized for 30 min with 0.1% Triton X-100 in PBS (PBST), blocked for 30 min with 10% normal goat serum, and incubated for 1 h or overnight with the following primary antibodies: mouse anti-Gephyrin (1:1000, Synaptic Systems #147021), rabbit anti-GFP (1 µg/ml, Nittobo76), rabbit anti-GluA2 (1 µg/ml, Nittobo77), rabbit anti-Shank2 (1 µg/ml, Nittobo78), rabbit anti-VGluT1 (1 µg/ml, Nittobo79), or rabbit anti-VIAAT (1 µg/ml, Nittobo80). Finally, they were fluorescently labeled for 1 or 2 h with Alexa Fluor Plus 405- or 647-conjugated species-specific secondary antibodies (1 μg/ml, Invitrogen). PBST was used as a washing and dilution buffer. All the procedures were done at room temperature.
Development of a membrane-impermeable FLAG2-JF646-HaloTag ligand (FLAG2-JF646-HTL)
General synthetic organic chemistry methods
The reference numbers for all chemical compounds utilized in this investigation are presented in Supplementary Fig. 5. The HaloTag ligand building block was acquired commercially (Promega). This building block is designated HaloTag(O2)-NH2 in our chemical nomenclature. This compound is assigned the number 5 in our numbering system. DYKDDDDK-Cysteine peptide was also acquired from a commercial source (LifeTein). The complete amino acid sequence of this peptide is DYKDDDDKC. This peptide is also known by the name “FLAG-Cysteine” peptide. This peptide is designated as compound 10 in our numbering system. Other commercial reagents were obtained from various chemical suppliers. These reagents were the best quality available and used exactly as received from the suppliers without any additional purification steps.
Solvents used for chemical reactions were anhydrous grade. These solvents were packaged in Sure/Seal bottles (Aldrich). Sometimes these bottles were refilled with anhydrous argon gas. Argon gas is abbreviated as Ar(g) where the (g) indicates the gaseous state. Sometimes the bottles were used once and then discarded rather than refilling them with Ar(g) to ensure the use of pristine anhydrous solvent. Reactions were conducted in glass vessels. These glass vessels contained Teflon-coated magnetic stir bars to allow stirring by a magnetic stirring hotplate. The Teflon coating prevents the stir bars from reacting with sensitive compounds. Two types of glass vessels were used. The first type was round-bottomed flasks. These flasks were sealed with rubber septa. The second type was septum-sealed crimp-top microwave reaction vials (Biotage). All reactions were protected from light exposure. Light protection was achieved using aluminum foil (Reynolds). Light protection is necessary because many organic compounds can undergo unwanted photochemical reactions when exposed to light. All reactions were also conducted under an inert atmosphere. This inert atmosphere consisted of anhydrous Ar(g). The Ar(g) was introduced into the reaction vessels using a needle through the septum. Reaction mixtures were heated when elevated temperatures were required. Heating was accomplished using aluminum blocks (IKA Works). These aluminum blocks were specially designed to fit on top of stirring hotplates (IKA Works) and conduct heat. Temperatures during reactions were monitored continuously. Temperature monitoring was achieved using an electronic contact thermometer. The temperature was controlled by adjusting settings on the stirring hotplate controls. These controls allow for precise temperature regulation during chemical transformations.
Reaction progress was monitored using two analytical methods. The first method was thin layer chromatography. Thin layer chromatography is commonly abbreviated as TLC. TLC was performed on precoated glass plates. These plates contained silica gel 60 F254 as the stationary phase. The F254 designation indicates that the silica contains a fluorescent indicator. The silica gel layer had a thickness of 250 μm. Compounds separated on TLC plates were visualized using two different techniques. The first visualization technique was UV illumination. Under UV light, many organic compounds appear as dark spots against the fluorescent background. The second visualization technique involved chemical staining. Two different staining reagents were employed. The first staining reagent was ceric ammonium molybdate. This reagent reacts with many organic functional groups to produce colored spots. The second staining reagent was potassium permanganate (KMnO4). This reagent is particularly useful for detecting alkenes and other oxidizable functional groups. The second monitoring method was tandem liquid chromatography–mass spectrometry. This analytical technique is commonly abbreviated as LC–MS. The specific model of LC–MS system used was the LCMS 2020 (Shimadzu). This system was equipped with a specialized chromatography column. The column was a Kinetex model (Phenomenex). The column had specific dimensions: 30 mm in length and 2.1 mm in internal diameter. The column was packed with C18 stationary phase particles. These particles had a diameter of 2.6 μm and a pore size of 100 Å. Sample injection volumes for LC–MS analysis ranged from 1 to 10 μL depending on sample concentration. The mobile phase consisted of a linear gradient. This gradient ran from 5% to 98% CH3CN in H2O containing 0.1% by volume formic acid (HCO2H). The formic acid assists with ionization in the mass spectrometer. Each analytical run lasted exactly 6 minutes. The flow rate was maintained at 1 mL/min throughout each analysis. The mass spectrometer used electrospray ionization. Electrospray ionization is abbreviated as ESI. The ionization was operated in positive ion mode, meaning that positively charged ions were detected.
Reaction products were purified using two different chromatographic purification methods. The selection of purification method depended on the scale of the reaction and the properties of the target compound. The first purification method was SiO2 gel chromatography. This was performed using an Isolera automated purification system (Biotage). The system utilized prepacked Sfär SiO2 gel columns (Biotage). These columns come prepacked with a high-quality silica gel stationary phase. The second purification method was preparative high-pressure liquid chromatography. This technique is commonly abbreviated as preparative HPLC. Preparative HPLC allows for purification of larger quantities of material compared to analytical HPLC. The specific model used for preparative HPLC was the Agilent 1200 series (Agilent) and was equipped with a preparative-scale chromatography column (Gemini–NX model, Phenomenex). The column had dimensions appropriate for preparative separations: 150 mm in length and 30 mm in internal diameter. The large internal diameter allows for injection of larger sample volumes. The column was packed with C18 stationary phase particles. These particles had a diameter of 10 μm and a pore size of 110 Å. The flow rate for preparative separations was 42 mL/min. This high flow rate is necessary to maintain reasonable separation times when using large-diameter columns. Solvent gradient conditions were optimized individually for each compound being purified using mobile phases consisting of CH3CN and H2O containing 0.1% by volume trifluoroacetic acid (TFA) to improve peak shape and provide consistent ionization conditions.
Analytical high-pressure liquid chromatography analyses were performed to assess compound purity and identity. These analyses used two different HPLC systems depending on whether mass spectrometric detection was required. The first analytical system combined liquid chromatography with mass spectrometry. This system was an Agilent 1200 series LC system coupled to a mass spectrometer. The chromatography column used was a Gemini–NX column (Phenomenex). This column had analytical-scale dimensions: 150 mm in length and 4.6 mm in internal diameter. The smaller internal diameter is appropriate for analytical separations with smaller sample volumes. The column was packed with C18 stationary phase particles. The stationary phase particles were 5 μm in diameter and had a 110 Å pore size. These specifications match the preparative column except for the particle size and column dimensions. The flow rate for analytical separations was 1 mL/min. The second analytical system was a standalone HPLC instrument without mass spectrometric detection (UFLC system, Shimadzu). UFLC stands for ultra-fast liquid chromatography. The “ultra fast” designation refers to the instrument’s capability to perform separations at higher pressures, which allows for faster analysis times. The higher-pressure capability was not used. The column specifications for this system were identical to those used in the LC–MS system. Briefly, the column was a Gemini–NX from Phenomenex with dimensions of 150 × 4.6 mm and 5 μm C18 particles with 110 Å pore size. The flow rate was maintained at 1 mL/min and used linear gradients of the mobile phases CH3CN and H2O containing 0.1% TFA. Both analytical HPLC systems were equipped with diode array detectors. Diode array detectors are commonly abbreviated as DAD. These detectors can simultaneously monitor multiple wavelengths of UV-visible light. This capability allows for compound identification based on UV-visible absorption spectra in addition to retention time information. Retention times, abbreviated as “tR”, are reported in minutes and represent the time elapsed from sample injection to the appearance of the compound’s peak maximum at the detector.
High-resolution mass spectrometry data were obtained by the High Resolution Mass Spectrometry Facility at the University of Iowa. High-resolution mass spectrometry is abbreviated as HRMS. This technique provides accurate mass measurements that can confirm molecular formulas of synthesized compounds. The mass spectrometer used ESI was and operated in positive ion mode.
Nuclear magnetic resonance spectroscopy was performed to characterize the structure of synthesized compounds. Nuclear magnetic resonance is commonly abbreviated as NMR. NMR spectra were recorded using a specialized high-field NMR spectrometer (Avance 400 MHz spectrometer, Bruker). The 400 MHz designation refers to the frequency at which protons resonate in the magnetic field of this instrument. The recorded NMR spectra were processed using specialized computer software (MestReNova, Mestrelab Research). Deuterated solvents were used for all NMR spectroscopy experiments. Deuterated solvents are versions of common solvents where hydrogen atoms have been replaced with deuterium atoms. The use of deuterated solvents is necessary because they do not produce signals in 1H NMR spectra, allowing for clear observation of sample signals. These deuterated solvents were used as purchased from chemical suppliers. No additional purification steps were performed on these solvents because the suppliers provide them at sufficient purity for NMR applications.
Chemical shift values for both 1H NMR and 13C NMR spectra were referenced to established standards. Two different reference standards were used depending on the solvent. The first reference standard was tetramethylsilane. Tetramethylsilane is commonly abbreviated as TMS. TMS is an organosilicon compound with the chemical formula Si(CH3)4. TMS is used as a reference because it produces a single, sharp peak that does not overlap with most organic compound signals. The second reference method used residual solvent peaks. Residual solvent peaks arise from small amounts of protonated solvent present in deuterated solvents. These peaks have well-established chemical shift values that can serve as internal references. 19F spectra were referenced to CFCl3, which also provides a single sharp peak.
Data reporting for 1H NMR spectra followed standardized conventions used throughout the scientific literature. The data reporting format included several specific pieces of information for each observed signal. The first piece of information was the chemical shift value, which is commonly designated by the symbol “δ”. Chemical shift values are reported in parts per million, which is abbreviated as ppm. Parts per million is a dimensionless unit that represents the frequency difference between the sample signal and the reference signal divided by the spectrometer frequency. The second piece of information was the multiplicity of each signal. Signal multiplicity describes the splitting pattern caused by coupling between neighboring nuclei. Different multiplicities are designated using standardized abbreviations. A singlet, which appears as a single peak, is abbreviated as “s”. A doublet, which appears as two peaks of equal intensity, is abbreviated as “d”. A triplet, which appears as three peaks with intensity ratio 1:2:1, is abbreviated as “t”. A quartet, which appears as four peaks with intensity ratio 1:3:3:1, is abbreviated as “q”. A pentet, which appears as five peaks, is abbreviated as “p”. A doublet of doublets, which appears as four peaks of equal intensity, is abbreviated as “dd”. Signals that cannot be clearly analyzed are designated as multiplets and abbreviated as “m”. The third piece of information reported for proton NMR signals was the coupling constant. Coupling constants describe the magnitude of signal splitting caused by neighboring nuclei. Coupling constants are measured in Hz. The fourth and final piece of information was the integration value. Integration values represent the relative number of protons contributing to each signal. These values are determined by measuring the area under each signal in the 1H NMR spectrum. Data reporting for 13C NMR spectra used a different format than 1H NMR data. The primary information reported for 13C NMR signals was the chemical shift value measured in ppm. Additional structural information was provided regarding the hydrogen multiplicity of each carbon atom. This multiplicity information indicates the number of hydrogen atoms directly attached to each carbon. Quaternary carbon atoms, which have no attached hydrogens, are designated as “C”. Tertiary carbon atoms, which have one attached hydrogen, are designated as “CH”. Secondary carbon atoms, which have two attached hydrogens, are designated as “CH2”. Primary carbon atoms, which have three attached hydrogens, are designated as “CH3”. This hydrogen multiplicity information was obtained using a specialized NMR technique called DEPT spectroscopy. DEPT is an abbreviation for “distortionless enhancement by polarization transfer.” DEPT is a pulse sequence technique that provides information about carbon multiplicity by selectively enhancing signals from carbons with different numbers of attached hydrogens. This technique allows for easy identification of carbon types within molecular structures. Data reporting for 19F NMR spectra used a similar format to 13C NMR data. The primary information reported for 19F NMR signals was the chemical shift value measured in ppm.
Experimentals and characterization for all new compounds
3,3"-bismethoxycarbonyl-6-tert-butoxycarbonyl-JF646 (3)
An oven-dried microwave reaction vial was charged with triflate 120 (581 mg, 786 µmol, 1 equiv), Pd2dba3 (72 mg, 79 µmol, 0.1 equiv), XPhos (112 mg, 236 µmol, 0.3 equiv), methyl azetidine-3-carboxylate hydrochloride (2, 358 mg, 2.36 mmol, 3 equiv), and Cs2CO3 (1.2 g, 3.77 mmol, 4.8 equiv). The vial was sealed and backfilled with Ar(g) (3×), after which degassed anhydrous dioxane (8 mL) was added. After stirring this reaction mixture at 100 °C for 4 h, the reaction mixture was cooled and filtered through a pad of celite, washing with CH2Cl2. The filtrate was adsorbed on celite, concentrated under reduced pressure, and purified by SiO2 gel chromatography (100 g SiO2 column, 0–40% EtOAc/hexanes, linear gradient) to provide 3 as a foamy light-yellow solid (509 mg, 97%). 1H NMR (CDCl3, 400 MHz) δ 8.12 (dd, J = 8.0, 1.3 Hz, 1H), 7.96 (d, J = 8.0 Hz, 1H), 7.81 (s, 1H), 6.88 (d, J = 8.7 Hz, 2H), 6.70 (d, J = 2.6 Hz, 2H), 6.33 (dd, J = 8.7, 2.6 Hz, 2H), 4.11 (t, J = 7.9 Hz, 4H), 4.07 – 4.03 (m, 4H), 3.73 (s, 6H), 3.60 – 3.53 (m, 2H), 1.55 (s, 9H), 0.66 (s, 3H), 0.59 (s, 3H). 13C NMR (CDCl3, 101 MHz δ 173.11 (C), 170.16 (C), 164.27 (C), 155.21 (C), 150.14 (C), 137.23 (C), 136.04 (C), 133.17 (C), 129.94 (CH), 128.82 (C), 127.56 (CH), 125.65 (CH), 124.95 (CH), 115.91 (CH), 112.91 (CH), 91.33 (C), 82.32 (C), 54.46 (CH2), 54.44 (CH2), 52.28 (CH3), 33.49 (CH), 28.12 (CH3), 0.04 (CH3), −0.73 (CH3). HRMS (ESI) calculated for C37H41N2O8Si [M + H]+ = 669.2627, found 669.2623.
3,3"-bismethoxycarbonyl-6-carboxy-JF646 (4)
Compound 3 (49 mg, 73 µmol, 1 equiv) was dissolved in CH2Cl2 (4 mL). TFA (0.7 mL) was added and the resulting blue solution was stirred at ambient temperature for 8 h. The reaction mixture was concentrated under reduced pressure and azeotroped with toluene (2×4 mL) to yield 4 as a blue solid (53 mg, 99%). This material was used in the subsequent synthetic step without any additional purification. 1H NMR (400 MHz, CDCl3) δ 8.22 (dd, J = 8.1, 1.4 Hz, 1H), 8.07 (d, J = 8.0 Hz, 1H), 7.94 (d, J = 1.3 Hz, 1H), 6.84 (d, J = 8.8 Hz, 2H), 6.79 (d, J = 2.6 Hz, 2H), 6.37 (dd, J = 8.9, 2.6 Hz, 2H), 4.25 (t, J = 8.4 Hz, 4H), 4.18 – 4.14 (m, 4H), 3.75 (s, 6H), 3.67 – 3.60 (m, 2H), 0.62 (s, 3H), 0.55 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 172.81 (C), 169.35 (C), 169.21 (C), 150.07 (C), 139.22 (C), 134.55 (C), 133.26 (C), 131.04 (C), 130.74 (CH), 130.46 (CH), 127.33 (CH), 127.24 (CH), 117.62 (CH), 113.66 (CH), 54.90 (CH2), 54.87 (CH2), 52.58 (CH3), 33.29 (CH), 0.01 (CH3), −1.37 (CH3). Analytical HPLC: tR = 12.4 min, 99.0% purity (10–95% CH3CN/H2O linear gradient over 20 min with constant 0.1% v/v TFA, 1 mL/min flow rate, detection at 650 nm). An analytical sample was prepared by HPLC purification to confirm the identity. 1H NMR (CD3CN, 400 MHz) δ 8.17 (dd, J = 8.0, 1.4 Hz, 1H), 8.02 (dd, J = 8.0, 0.8 Hz, 1H), 7.77 (dd, J = 1.3, 0.7 Hz, 1H), 6.82 (d, J = 2.6 Hz, 2H), 6.79 (d, J = 8.7 Hz, 2H), 6.35 (dd, J = 8.8, 2.7 Hz, 2H), 4.13 (t, J = 8.3 Hz, 4H), 4.04 – 4.02 (m, 4H), 3.69 (s, 6H), 3.60 – 3.52 (m, 2H), 0.62 (s, 3H), 0.53 (s, 3H). HRMS (ESI) calculated for C33H33N2O8Si [M + H]+ = 613.2001, found 613.2009.
3, 3"-bismethoxycarbonyl-JF646-HTL (6)
Compound 4 (53 mg, 73 µmol, 1 equiv) was dissolved in DMF (2 mL). To this solution was added DIEA (128 µL, 734 µmol, 10 equiv) and N,N,N′,N′-Tetramethyl-O-(N-succinimidyl)uronium tetrafluoroborate (TSTU; 27.6 mg, 92 µmol, 1.25 equiv). The reaction mixture was stirred for 10 min at ambient temperature, after which HaloTag(O2)-NH2 (5, 28.6 mg, 110 µmol, 1.5 equiv) was added. After stirring the reaction mixture for 16 h at ambient temperature, DMF was removed under reduced pressure. Purification by SiO2 gel chromatography (25 g SiO2 column, 0–100% EtOAc/hexanes, linear gradient) provided 6 as a light green solid (40.3 mg, 67.1% over two steps). 1H NMR (CDCl3, 400 MHz) δ 7.96 (dd, J = 8.0, 0.7 Hz, 1H), 7.89 (dd, J = 8.0, 1.4 Hz, 1H), 7.68 (t, J = 1.0 Hz, 1H), 6.85 (d, J = 5.4 Hz, 1H), 6.77 (d, J = 8.7 Hz, 2H), 6.67 (d, J = 2.7 Hz, 2H), 6.28 (dd, J = 8.7, 2.7 Hz, 2H), 4.10 (dd, J = 8.6, 7.3 Hz, 4H), 4.06 – 4.02 (m, 4H), 3.73 (s, 6H), 3.65 – 3.59 (m, 6H), 3.58 – 3.52 (m, 4H), 3.49 (t, J = 6.7 Hz, 2H), 3.38 (t, J = 6.7 Hz, 2H), 1.75 – 1.68 (m, 2H), 1.49 (q, J = 7.1 Hz, 2H), 1.42 – 1.34 (m, 2H), 1.33 – 1.27 (m, 2H), 0.63 (s, 3H), 0.56 (s, 3H). 13C NMR (CDCl3, 101 MHz) δ 173.13 (C), 169.88 (C), 166.21 (C), 155.03 (C), 150.28 (C), 139.97 (C), 136.78 (C), 133.16 (C), 128.85 (C), 128.00 (CH), 127.60 (CH), 126.08 (CH), 123.52 (CH), 116.11 (CH), 112.88 (CH), 71.33 (CH2), 70.35 (CH2), 70.11 (CH2), 69.62 (CH2), 54.58 (CH2), 54.56 (CH2), 52.35 (CH3), 45.11 (CH2), 40.11 (CH2), 33.59 (CH), 32.59 (CH2), 29.47 (CH2), 26.74 (CH2), 25.46 (CH2), 0.32 (CH3), −1.12 (CH3). HRMS (ESI) calculated for C43H53ClN3O9Si [M + H]+ = 818.3234, found 818.3234.
3, 3"-carboxy-JF646-HTL (7)
Compound 6 (40.3 mg, 49 µmol, 1 equiv) was dissolved in a mixture of THF:CH3OH (1:1 v/v; 4 mL). An aqueous solution of NaOH (1 M, 0.6 mL) was added and the resulting solution and stirred for 5 h. The reaction mixture was diluted with H2O (5 mL) and the acidified to pH ~1 using an aqueous solution of HCl (1 M). The mixture was extracted with EtOAc (4 × 2 mL) and the combined organics were concentrated under reduced pressure. This crude product was purified by preparative HPLC purification using a 5–95% CH3CN/H2O linear gradient with constant 0.1% v/v TFA. Product-containing fractions were combined and lyophilized to obtain 7 as a blue solid (TFA salt, 32.2 mg, 72.3%). 1H NMR (CD3CN, 400 MHz) δ 8.08 (d, J = 8.1 Hz, 1H), 7.99 (dd, J = 8.1, 1.6 Hz, 1H), 7.59 (d, J = 1.5 Hz, 1H), 7.28 (t, J = 5.6 Hz, 1H), 6.86 (d, J = 2.7 Hz, 2H), 6.82 (d, J = 9.0 Hz, 2H), 6.32 (dd, J = 9.0, 2.7 Hz, 2H), 4.25 (t, J = 8.8 Hz, 4H), 4.15 (t, J = 7.3 Hz, 4H), 3.62 – 3.45 (m, 12H), 3.33 (t, J = 6.5 Hz, 2H), 1.73 – 1.65 (m, 2H), 1.46 – 1.39 (m, 2H), 1.38 – 1.30 (m, 2H), 1.29 – 1.21 (m, 2H), 0.60 (s, 3H), 0.52 (s, 3H). 13C NMR (CD3CN, 101 MHz) δ 174.26 (C), 169.23 (C), 166.80 (C), 152.27 (C), 140.76, 132.68, 132.13, 130.35, 128.94 (CH), 128.42, 125.68, 113.53 (CH), 71.59 (CH2), 70.91 (CH2), 70.75 (CH2), 69.89 (CH2), 55.32 (CH2), 46.21 (CH2), 40.69 (CH2), 33.84 (CH), 33.31 (CH2), 30.21 (CH2), 27.34 (CH2), 26.11 (CH2), −0.16 (CH3), −1.41 (CH3). HRMS (ESI) calculated for C41H49N3O9ClSi [M + H]+ = 790.2921, found 790.2925.
Maleimide2-JF646-HTL (8)
Compound 7 (11.7 mg, 12.9 μmol, 1 equiv) was dissolved in DMF (2 mL). To this solution was added N,N-diisopropylethylamine (DIEA; 22.6 μL, 129 μmol, 10 equiv), 1-(2-aminoethyl)−1H-pyrrole-2,5-dione (TFA salt, 13.1 mg, 51.7 μmol, 4 equiv), and 1-[bis(dimethylamino)methylene]−1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU, 12.8 mg, 33.6 μmol, 2.6 equiv). The reaction mixture was stirred for 2.5 h at ambient temperature. The solvent was removed under reduced pressure, and the product was purified by preparative HPLC purification using a 30–80% CH3CN/H2O linear gradient over 18 mins with a constant 0.1% v/v TFA. Product-containing fractions were combined and lyophilized to obtain 8 as a blue solid (TFA salt, 9.6 mg, 65%). 1H NMR (CD3OD, 400 MHz) δ 8.73 (t, J = 5.4 Hz, 1H), 8.26 – 8.13 (s, 2H), 8.08 (d, J = 7.9 Hz, 1H), 7.69 (t, J = 1.1 Hz, 1H), 6.90 – 6.81 (m, 3.5H), 6.80 – 6.78 (m, 3.5H), 6.38 (d, J = 2.6 Hz, 2H), 6.36 (d, J = 2.6 Hz, 2H), 4.37 – 3.95 (m, 8H), 3.65 – 3.54 (m, 12H), 3.43 – 3.37 (m, 4H), 3.43 – 3.37 (m, 6H), 1.70 (p, J = 6.8 Hz, 2H), 1.49 (p, J = 6.8 Hz, 2H), 1.43 – 1.30 (m, 4H), 0.62 (s, 3H), 0.55 (s, 3H). 19F NMR (CD3OD, 376 MHz) δ − 77.15. Analytical HPLC: tR = 11.8 min, >99.0% purity (10–95% CH3CN/H2O linear gradient over 20 min with constant 0.1% v/v TFA, 1 mL/min flow rate, detection at 650 nm). HRMS (ESI) calculated for C53H61N7O11ClSi [M + H]+ = 1034.3881, found 1034.3891.
FLAG2-JF646-HTL (11)
Compound 9 (9.6 mg, 8.4 μmol, 1 equiv) was dissolved in 3:1 v/v PBS:CH3CN (8 mL). To this solution was added FLAG-Cysteine (10, 42.6 mg, 38.2 μmol, 4.5 equiv), and the reaction mixture was stirred overnight at ambient temperature. The product was purified by preparative HPLC purification using a 5–40% CH3CN/H2O linear gradient over 15 min with constant 0.1% v/v TFA. Product-containing fractions were combined and lyophilized to obtain 11 as a blue solid (TFA salt, 22.0 mg, 77.8%). Analytical HPLC: tR = 8.3 min, >99.0% purity (10–95% CH3CN/H2O linear gradient over 20 min with constant 0.1% v/v TFA, 1 mL/min flow rate, detection at 650 nm). HRMS (ESI) calculated for C141H1954ClN29O53S2Si [M + 5H]5+ = 654.0472, found 654.0463. Precommercial samples of FLAG2-JF646-HTL can be requested by contacting L.D. Lavis.
HaloTag labeling with heterologous cell cultures
HEK293T cells (#HCL4517, Thermo Fisher Scientific) were transfected with plasmid vectors encoding HaloTag and mEGFP during the logarithmic growth phase. Two days after transfection, HEK293T cells were fixed with 4% paraformaldehyde/0.1 M phosphate buffer for 10 min, and then labeled with either cell membrane-permeable JF646-HTL (100 nM) or impermeable FLAG2-JF646-HTL (100 nM)/PBS in the presence or absence of 0.1% TritonX-100 to permeabilize the cell membrane for 30 min. For live cell labeling (Supplementary Fig. 1a, b), HEK293T cells were labeled with 100 nM JF646-HTL in the culture medium for 30 min before fixation.
HaloTag-fused protein labeling for fixed brain imaging
Single labeling of total populations of HaloTag-fused proteins
Fixed slice cultures and brain sections were incubated with membrane-permeable HTLs (50 nM JF549-HTL or 100 nM JF646-HTL) for 30-60 min. In Supplementary Fig. 5i, 100 nM HTLs (FLAG2-JF646-HTL) achieved 70-80% of their maximum signal intensity. This concentration of HTLs would provide an optimal balance between imaging quality and cost-effectiveness. HTLs were diluted with PBS, and all the labeling procedures were done at room temperature throughout this study.
Dual labeling of the surface and intracellular subpopulations of HaloTag-fused proteins
Prior to preparing brain slices, fixed whole brains were sequentially incubated with a membrane-impermeable HTL (1 µM FLAG2-JF646-HTL) for 30 min for the surface labeling, and membrane-permeable HTL (50 nM JF549-HTL) for 30 min for the intracellular labeling. To confirm complete surface labeling of HaloTag-fused proteins with FLAG2-JF646-HTL, fixed whole brains were sequentially labeled with two different membrane-impermeable HTLs (0.1-2 µM FLAG2-JF646-HTL for 30 min and 1 µM AF488-HTL for 30 min).
Dual labeling of the pre-existing and nascent subpopulations of HaloTag-fused proteins in vitro
Hippocampal slice cultures obtained from Cas9 mice were transduced with AAV1-HDR-Halo-CaMKIIα at DIV2, pulse-labeled for the pre-existing subpopulation with 1 µM JF646-HTL for 30 min at DIV14-17. The slices were thoroughly washed with culture media for 5 min three times to completely remove JF646-HTL from hippocampal slices, and further incubated for 2 h to obtain de novo synthesis of HaloTag-fused proteins. The slices were then fixed with 4% PFA/PB for 1 h, and chase-labeled for the nascent subpopulation with 50 nM JF549-HTL for 1 h. To validate nascent signals, a protein synthesis inhibitor, anisomycin (50 µM, Sigma-Aldrich), was applied to culture media after the pulse labeling.
Dual labeling of the pre-existing and nascent or nascent-and-surface subpopulations of HaloTag-fused proteins in vivo
Under anesthesia with a mixture of medetomidine, midazolam, and butorphanol as described above, mice were subjected to a craniotomy over the primary somatosensory cortex at P21-23 for Halo-CaMKIIα or P38-44 for PSD95-Halo and Halo-βActin, as described previously81. After removing the dura mater, a cranial window was treated with an excess amount of JF549-HTL (1 µM) for 30 min to pulse-label pre-existing subpopulation, washed thoroughly with PBS to remove unbound ligands, and then covered with PEO-CYTOP nanosheets82 for 0.5, 1, 2 or 48 h to protect the brain surface. Subsequently, mice were fixed with 4% PFA/PB as described above. Fixed whole brains were chase-labeled for the nascent or nascent-and-surface subpopulation with the membrane-permeable JF646-HTL (100 nM) or -impermeable FLAG2-JF646-HTL (100 nM), respectively, for 1 h. The depth-dependent difference in the HaloTag signals could be caused by multiple factors: a low concentration of HaloTag ligands, high expression levels of HaloTag-fused proteins, or long distance from the cortical surface beneath the cranial window. In this study, no significant difference in the mean intensity for pulse signals of PSD95-Halo was confirmed between the upper quarter (near the brain surface: 671.8 ± 65.4 AU, 6 images) and the lower quarter (150 µm deeper: 674.8 ± 81.0 AU, 6 images) of the confocal images (p = 0.48, t-test), suggesting that 1 µM JF549-HTL exhibits high penetration capability to effectively label pre-existing HaloTag-labeled proteins within the layer 2/3 of the neocortex beneath the cranial window.
Multiplexed HaloTag- and SNAP-tag-fused protein labeling for fixed brain imaging
Halo-GluA2 and SNAP-CaMKIIα
Fixed cortical slice cultures were sequentially incubated with a membrane-impermeable HTL (100 nM FLAG2-JF646-HTL) to label the surface subpopulation of Halo-GluA2 and a membrane-permeable SNAP-tag ligand (10 nM TMR-Star SNAP-tag ligand (TMR-STL)) to label the total population of SNAP-CaMKIIα. The labeling time was 30 min for each. Throughout this study, STLs were diluted with TBS (pH 8.0) which provided more efficient SNAP-tag labeling than dilution with PBS.
Halo-GluN1 and SNAP-GluA2
Brain slices fixed at P27 or P35 were sequentially labeled with membrane-permeable HTL and STL (100 nM JF646-HTL and 10 nM TMR-STL) for 30 min each.
Halo-GluA2 and PSD95-SNAP
Whole brains fixed at P15 or P16 were treated with a membrane-impermeable HTL (100 nM FLAG2-JF646-HTL) for 30 min to label the surface pool of Halo-GluA2. Because of an insufficient penetration of a membrane-permeable TMR-STL caused by its non-selective absorption to the meninges, the labeled brains were sliced with a vibratome (VT1200S, Leica). Subsequently, the brain slices were incubated with 10 nM TMR-STL for 30 min to label the total population of PSD95-SNAP.
Fixed brain imaging
Labeled slices were mounted with Fluoromount-G (Southern Biotech), unless otherwise noted. For whole-neuron imaging, thick slices (300 µm in thickness) were cleared according to the SeeDB2G protocol29. Briefly, slices were successively treated with 2% saponin/PBS for 12 h, 33% Omnipaque/2% saponin/PBS for 6 h, 50% Omnipaque/2% saponin/PBS for 6 h, and 90% Omnipaque/2% saponin/PBS for 12 h, and then mounted with 100% Omnipaque. For expansion microscopy by the CUBIC-X protocol83, brain slices (50 µm in thickness) was sequentially treated with ScaleCUBIC-1 reagent (5 wt% Quadrol, 25 wt% urea, 15 wt% TritonX-100) at 37 °C for 2 h and 20% imidazole at room temperature for 4 h, which allowed for an approximately twice linear expansion of brain slices. Brain slices were imaged using confocal laser-scanning microscopes (FV1200 or FV3000, Olympus) equipped with 20x dry (UPLXAPO20X; numerical aperture = 0.8, working distance = 0.6 mm, Olympus), 30x silicone-immersion (UPLSAPO30XS; numerical aperture = 1.05, working distance = 0.8 mm, Olympus), and 60x silicone-immersion (UPLSAPO60XS2; numerical aperture = 1.3, working distance = 0.3 mm, Olympus) objectives and 405, 488, 559, and 633 laser lines (Coherent) or a spinning disc confocal microscope (BC43, Andor) equipped with a 100x oil-immersion objective (CFI Plan Apochromat Lambda D 100X Oil, Nikon). For quantitative analysis, single or z-stack images were acquired as described in Supplementary Data 3.
Super-resolution imaging
Super-resolution images were acquired using an inverted microscope (Zeiss ELYRA S.1) equipped with a Plan-Apochromat 63×/1.4 NA oil immersion lens. Structured Illumination Microscopy (SIM) images were captured using an Andor iXon 885 EMCCD camera and processed with ZEN 2011 software (Zeiss). TetraSpeck fluorescent beads (0.1 µm, Thermo Fisher Scientific) were used as alignment standards. Excitation was performed with 488 nm and 561 nm lasers at 5% power, with an exposure time of 100 msec. Structured illumination patterns (5 phases × 3 rotations) were generated using gratings with periods of 28.0 µm (488 nm) and 34.0 µm (561 nm). Reconstructed SIM images were processed using software-recommended parameters, with a final pixel size of 40 nm in the xy-plane and 110 nm along the z-axis.
Live-cell HaloTag-fused protein imaging in slice cultures
Cortical slice cultures carrying Halo-ERK2 and CRTC1-Halo at DIV14-15 were incubated in culture media containing 2 nM TMRdirect-HTL and 1 µM tetrodotoxin (TTX, Abcam) for 24 h. The slices were then transferred to artificial cerebrospinal fluid (ACSF) consisting of 119 mM NaCl, 2.5 mM KCl, 0.5 mM CaCl2, 5 MgCl2 mM, 26 mM NaHCO3, 1 mM NaH2PO4, and 11 mM glucose, supplemented with 1 µM TTX and gassed with 5% CO2 / 95% O2. Neurons labeled with mEGFP and TMRdirect-HTL were imaged using a two-photon galvo-galvo scanning microscope (TCS SP5 MP, Leica) equipped with a 20x water immersion objective (HCX PL APO; numerical aperture = 1.0, working distance = 2.0 mm, Leica). mEGFP and TMRdirect-HTL were excited at 840 nm with a Ti:sapphire laser (Coherent). Green and red fluorescent signals from mEGFP and TMRdirect-HTL were separated by a beam splitter (560 nm) and band pass filter (525/50 nm for the green channel, 585/40 nm for the red channel) before the HyD (GaAsP) detectors. To stimulate neuronal activity in brain slices, the slices were perfused with ACSF containing 50 µM bicuculline and 100 µM 4-aminopyridine. The acquisition of images (speed: 0.2 Hz, zoom: 20x, size: 512×512 pixels, line average: 4x) was started 2 min before the stimulation and repeated every 2 min. The laser power at the slices was kept under 10 mW throughout imaging to minimize phototoxicity and photobleaching.
Live-cell nascent HaloTag-fused protein imaging in living mice
The neocortex of Cas9 mice at P0 was injected with AAV-HDR-Halo-mEGFP-CaMKIIα. Under anesthesia with a mixture of medetomidine, midazolam, and butorphanol as described above, a craniotomy was performed over the primary visual cortex at P35. After removing the dura mater, a cranial window was treated with JF646-HTL (1 µM) for 30 min to label pre-existing CaMKIIα, and washed with PBS to remove unbound ligands. Two hours later, the cranial window was subject to TMRdirect-HTL (100 nM) for 30 min to label nascent CaMKIIα, and then sealed with plastic wrap or dual layered cover glasses. Dual color imaging for JF646-HTL (pre-existing CaMKIIα) or TMRdirect-HTL (nascent CaMKIIα) and mEGFP (total CaMKIIα) was performed with a two-photon resonant-galvo scanning microscope (Bergamo II, Thorlabs) equipped with a 25x water immersion objective (XLPLN25XWMP2; numerical aperture = 1.05, working distance = 2.0 mm, Olympus) and a tunable femtosecond laser (InSight X3, Spectra Physics) at the excitation laser wavelength of 840 nm. Green, red, and far-red fluorescent signals were collected using band pass filter (525/50 nm for the green channel, 607/70 nm for the red channel) or high pass filter (647 nm for far-red channel). The acquisition of dendritic images from the superficial layer of the primary visual cortex was done with a zoom factor of 17.1x and an image size of 512×512 pixels. After two-photon imaging, mice were fixed by transcardial perfusion with 4% PFA/PB (2.5-3 ml/g of body weight). Brain sections (50 µm in thickness) were prepared with a vibratome (VT1200S, Leica) and imaged with a confocal microscope (FV3000, Olympus). For confocal microscopy, JF646-HTL (pre-existing CaMKIIα) was detected together with TMRdirect-HTL (nascent CaMKIIα) and mEGFP (total CaMKIIα) from the same brain region as observed by two-photon microscopy. To validate the specificity of nascent signals, anisomycin (50 µM) was added to PBS on the cranial window following the pre-existing CaMKIIα labeling with a JF646-HTL.
Quantitative analysis of individual cell bodies
Confocal images were analyzed with the ImageJ software unless otherwise noted in the following quantification. To measure HaloTag knock-in efficiency, the number of mEGFP-positive cells with fluorescence intensity above a predetermined threshold was counted across entire coronal sections. HDR-mediated knock-in occurs selectively in dividing neuronal progenitors, but not in non-dividing mature neurons14. Each cell division halves the copy number of mEGFP-encoding plasmids introduced into neuronal progenitors via IUE, leading to a reduction or disappearance of mEGFP fluorescence in individual HDR-mediated knock-in neurons. However, this reduction can be limited to a small subset of cells transfected by IUE, thereby resulting in a minimal effect on the estimation of the HaloTag knock-in efficiency. For measuring the fluorescent signals in individual neurons, the background noise was subtracted from each raw fluorescent image. Regions of interest (ROIs) were semi-automatically or manually registered for cell bodies labeled for mEGFP (Supplementary Fig. 3) or pre-existing Halo-CaMKIIα (Supplementary Fig. 8), respectively. The semi-automatic detection of ROIs used “Analyze Particles” function based on a binary mask generated by thresholding images with “moments”. Mean fluorescent intensities for each channel were measured from individual ROIs. In Supplementary Fig. 3, the mean fluorescent intensity for HaloTag or immunohistochemical signals was further normalized to mEGFP signals in the same cells. The data were obtained from two or three brain slices from two or three mice and pooled together.
Quantitative analysis of individual synapses in dendritic segments
A series of ROIs for each spine head protruding from a given dendritic branch were manually registered across multiple z-stack sections. After the background subtraction, the integrated fluorescent intensity for each channel was measured from all the ROIs corresponding to each spine head. mEGFP signals were used for the estimation of spine-head volume. For the data presentation and statistical analysis, the integrated fluorescent intensity at individual spine-heads was normalized to the maximum intensity in individual mice. All the data were obtained from three fragments of tertiary apical dendrites of pyramidal cells in three mice and pooled together.
Single-cell synaptome analysis based on a simple image thresholding
For simple quantification for single-cell synaptome analysis in Figs. 3, 4, 6, 7, Supplementary Fig. 6, 7, 9 and 10, a z-stack containing a single labeled neuron was divided into multiple substacks composed of 5 serial images. Since some synaptic puncta were distributed across adjacent images, they could be split between two adjacent substacks. To avoid analyzing these split synaptic puncta as distinct synapses, single images were skipped between two adjacent substacks based on the stereological quantification method84. Averaged images of individual substacks were subjected to background subtraction, and manually thresholded for a binary mask to distinguish synaptic puncta from noise signals. In each averaged substack, individual synaptic puncta were then registered as synaptic ROIs with “Analyze Particles” function based on the binary masks. Tiny dot-like noise signals with a size of <5 square pixels (<0.024 µm2) were omitted for this registration across the images. The above image processing was assisted with the customized ImageJ Macro. If the synaptic ROIs were distant from arrays of synaptic puncta (>3 µm), or overlapped with each other due to low imaging resolution, they were proofread manually. The signal density (equal to the mean intensity) for each channel at individual synaptic ROIs was measured from each averaged substack. The size, coordinate, and distance from the soma were also obtained from individual synaptic ROIs. Based on these values, the heat map for the signal density or integrated signal intensity for synaptic proteins and ROI size at individual synaptic ROIs was generated with a custom-written MATLAB code. Heat maps presented in Figures were processed to dilate colored ROIs via the maximum filter with the radius value of 2.5 or 5 pixels by a Photoshop software. In Fig. 4 and Supplementary Fig. 7, dendritic traces of labeled neurons were generated with a “Simple Neurite Tracer” plugin. Data were obtained from 2 (Fig. 6 and Supplementary Fig. 9), 3 (Fig. 7 and Supplementary Fig. 10), 4 (Fig. 4 and Supplementary Fig. 7), or 12 (Fig. 3 and Supplementary Fig. 6) mice. The corresponding ImageJ Macro and Matlab codes are publicly deposited.
Whole-neuron synaptome analysis with a deep learning-based algorithm
Deep learning-based detection of individual Halo-GluA2-labeled puncta in a whole single neuron
In Fig. 2 and Supplementary Fig. 4, z-stacks of images containing whole labeled neurons (1600 × 1600 or 2048 × 2048 pixels, 0.5 µm of z-interval, 512, 465, or 495 slices) was subjected to the deconvolution process85, and then analyzed with a deep learning-assisted method for a robust and accurate detection of Halo-GluA2-labeled puncta in a whole single neuron. Deep neural networks required the annotation of Halo-GluA2-labeled puncta in the input z-stack to learn both the foreground (Halo-GluA2-labeled puncta) and the background areas. For this, we adopted an interactive approach in which a user was able to iteratively correct the output from the AI. This allowed the AI to re-train the deep neural model on the basis of the user’s correction, thereby enabling more accurate detection of Halo-GluA2-labeled puncta. The details are as follows:
-
1.
Initial annotation: Given a part of the analyzing z-stack (36 out of 512 slices) that contained several arrays of Halo-GluA2-labeled puncta, an expert annotated the position of a subset of Halo-GluA2-labeled puncta (imperfect annotation). The position of bright noise puncta, which were distant from arrays of Halo-GluA2-labeled puncta, were annotated as negative samples (Supplementary Fig. 3g, (1)). The number of puncta annotated initially was 172 and 214 for Halo-GluA2-labeled and noise puncta, respectively.
-
2.
Punctum extraction: We used a position heat map, which had achieved good performance for cell detection86. To effectively capture the image features, we applied a partial maximum intensity projection (MIP) technique. This transformed a 3D volume into a series of 2D images, each of which was a local MIP generated from several z-slices. Given a set of the local MIP images and annotated points, U-net87 was trained to predict a heat map so that a centroid of Halo-GluA2-labeled puncta could fit a peak with a Gaussian distribution (Supplementary Fig. 3g, (2)). To train the detection network from incomplete annotation, we used a masked loss function, which gave the loss only around a given position of the annotated Halo-GluA2-labeled puncta88. This masked loss was capable of training the detection network using partially annotated positions of Halo-GluA2-labeled puncta. Then, the trained detection network predicted the position heat map for the entire z-stack, and the local peak of the heat map was detected as candidate positions for Halo-GluA2-labeled puncta.
-
3.
Noise removal: Candidate positions potentially included noises as well as Halo-GluA2-labeled puncta. To remove noises from the candidate positions, we used an SVM classifier89. Notably, Halo-GluA2-labeled puncta were apposed with close proximity to each other. In contrast, noise signals were sparsely distributed, compared to Halo-GluA2-labeled puncta. Therefore, we used the distance from the candidate positions to their neighbor Halo-GluA2-labeled puncta as discriminative features for classification. More precisely, the distances from a given candidate position to the ten nearest neighbors (a vector ∈R^(10×1)) were measured for initially annotated Halo-GluA2-labeled puncta (positive samples) and noises (negative samples) as the feature for the SVM. We trained the SVM on the basis of the features to determine if the candidate was a Halo-GluA2-labeled punctum or noise (Supplementary Fig. 3g, (3)).
-
4.
Correction: If the prediction included noises or false negatives (miss detection) due to the small size of training data, an expert (user) provided the additional annotations for the position of noises or false negatives (Supplementary Fig. 3g, (4)). Then, the detection network and SVM were re-trained using the user’s correction information, providing the improved result for the user. When the result still contained noises, a user iteratively added more annotations. This process allowed for a significant reduction of the total annotation cost. In addition, the re-trained model also can be applied to annotation-free data.
Deep learning-based reconstruction of dendritic branches
We took an interactive approach again to accurately reconstruct dendritic branches of Halo-GluA2-labeled neurons on the basis of the network of Halo-GluA2-labeled puncta as follows:
-
1.
Soma annotation: Since several Halo-GluA2-labeled somata could be included in a 3D volume, their positions were annotated as seeds (Supplementary Fig. 3h, (1)).
-
2.
Tree generation: We made a tree that originated in the annotated soma by connecting neighbor Halo-GluA2-labeled puncta. In the tree, nodes and edges corresponded to single Halo-GluA2-labeled puncta and connections between their nearest neighbors, respectively. Since multiple neurons were included in the analyzed volume, dendritic branches from different cells could be overlapped or connected with each other, causing an inaccurate reconstruction of dendritic branches. To avoid this problem, a tree was generated based on the following rule: If a given Halo-GluA2-labeled punctum had a shorter path length along the tree to the soma of Neuron X than any other soma, it belonged to Neuron X (Supplementary Fig. 3h, (2)).
-
3.
Correction: Our system discriminated between different trees with different colors (e.g., blue, red, green). Since a set of Halo-GluA2-labeled puncta (partial tree) were unconnected to any main trees due to miss detection or large distance, a user provided additional annotations by painting the regions of the unconnected partial tree in specified colors (Supplementary Fig. 3h, (3)). Finally, dendritic branches that belonged to a single Halo-GluA2-labeled neuron were reconstructed on the basis of the corrected tree.
-
4.
Segmentation and measurement: For detailed analysis, reconstructed branches were separated into segments between two bifurcation points by a breadth-first search from the root (soma) (Supplementary Fig. 3h, (4)). Halo-GluA2-labeled puncta were assigned to a parent segment. For each segment, we obtain the length, the number, mean intensity, and intensity variance of Halo-GluA2-labeled puncta, and the distance from the soma.
Statistics and reproducibility
All experiments were independently repeated at least three times with biological replicates. Data from all animals used in the experiments were included in the statistical analysis and data plots. Quantitative data are presented as mean ± SEM. The details of sample number (n), including exact values, are indicated in the main text and/or figure legends. Two-tailed Mann-Whitney test, two-tailed Wilcoxon matched-pairs signed rank test, one-way ANOVA with the Friedman test, one-way ANOVA with the Kruskal-Wallis test, or the Kolmogorov–Smirnov test was used when two or more independent samples were compared. Statistical analyses were conducted using Prism 10 software (GraphPad Software). Differences between data sets were judged to be significant at p < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Microscopic images presented or analyzed in this paper are publicly available at figshare (https://doi.org/10.6084/m9.figshare.29887013). The data sources are listed in Supplementary Data 3. Source data are provided with this paper.
Code availability
All original code used in this paper is deposited and publicly available at Zenodo (https://doi.org/10.5281/zenodo.17396593).
References
Delva, E. & Kowalczyk, A. P. Regulation of Cadherin Trafficking. Traffic 10, 259–267 (2009).
Lau, C. G. & Zukin, R. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat. Rev. Neurosci. 8, 413–426 (2007).
Niessen, C. M., Leckband, D. & Yap, A. S. Tissue Organization by Cadherin Adhesion Molecules: Dynamic Molecular and Cellular Mechanisms of Morphogenetic Regulation. Physiological Rev. 91, 691–731 (2011).
Diering, G. H. & Huganir, R. L. The AMPA Receptor Code of Synaptic Plasticity. Neuron 100, 314–329 (2018).
Matsuo, N., Reijmers, L. & Mayford, M. Spine-Type-Specific Recruitment of Newly Synthesized AMPA Receptors with Learning. Science 319, 1104–1107 (2008).
Hafner, A.-S., Donlin-Asp, P. G., Leitch, B., Herzog, E. & Schuman, E. M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364, eaau3644 (2019).
Monday, H. R., Kharod, S. C., Yoon, Y. J., Singer, R. H. & Castillo, P. E. Presynaptic FMRP and local protein synthesis support structural and functional plasticity of glutamatergic axon terminals. Neuron 110, 2588–2606.e2586 (2022).
Iascone, D. M. et al. Whole-Neuron Synaptic Mapping Reveals Spatially Precise Excitatory/Inhibitory Balance Limiting Dendritic and Somatic Spiking. Neuron 106, 566–578.e568 (2020).
Micheva, K. D., Busse, B., Weiler, N. C., O’Rourke, N. & Smith, S. J. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68, 639–653 (2010).
Reshetniak, S. et al. A comparative analysis of the mobility of 45 proteins in the synaptic bouton. EMBO J. 39, e104596 (2020).
Zhu, F. et al. Architecture of the Mouse Brain Synaptome. Neuron 99, 781–799.e710 (2018).
Bulovaite, E. et al. A brain atlas of synapse protein lifetime across the mouse lifespan. Neuron 110, 4057–4073.e4058 (2022).
Cizeron, M. et al. A brainwide atlas of synapses across the mouse life span. Science 369, 270–275 (2020).
Mikuni, T., Nishiyama, J., Sun, Y., Kamasawa, N. & Yasuda, R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell 165, 1803–1817 (2016).
Nishiyama, J., Mikuni, T. & Yasuda, R. Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron 96, 755–768.e755 (2017).
Watanabe, M. et al. Selective scarcity of NMDA receptor channel subunits in the stratum lucidum (mossy fibre-recipient layer) of the mouse hippocampal CA3 subfield. Eur. J. Neurosci. 10, 478–487 (1998).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008).
Keppler, A. et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21, 86–89 (2003).
Gautier, A. et al. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 (2008).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Erdmann, R. S. et al. Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags. Cell Chem. Biol. 26, 584–592.e586 (2019).
Kohl, J. et al. Ultrafast tissue staining with chemical tags. Proc. Natl Acad. Sci. USA 111, E3805–E3814 (2014).
Poc, P. et al. Interrogating surface versus intracellular transmembrane receptor populations using cell-impermeable SNAP-tag substrates. Chem. Sci. 11, 7871–7883 (2020).
Wang, H.Y., Lin, Y.-P., Mitchell, C.K., Ram, S. & O’Brien, J. Two-color fluorescent analysis of connexin 36 turnover: relationship to functional plasticity. J. Cell Sci. jcs.162586. https://doi.org/10.1242/jcs.162586 (2015).
Presman, D. M. et al. Quantifying transcription factor binding dynamics at the single-molecule level in live cells. Methods 123, 76–88 (2017).
Ramakrishna, S. et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24, 1020–1027 (2014).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24, 1012–1019 (2014).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3, e04766 (2014).
Ke, M. T. et al. Super-Resolution Mapping of Neuronal Circuitry With an Index-Optimized Clearing Agent. Cell Rep. 14, 2718–2732 (2016).
Sheng, M. & Hoogenraad, C. C. The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu Rev. Biochem 76, 823–847 (2007).
Nusser, Z. et al. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron 21, 545–559 (1998).
Racca, C., Stephenson, F. A., Streit, P., Roberts, J. D. B. & Somogyi, P. NMDA Receptor Content of Synapses in Stratum Radiatum of the Hippocampal CA1 Area. J. Neurosci. 20, 2512–2522 (2000).
Lu, W. et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron 62, 254–268 (2009).
Wenthold, R. J., Petralia, R. S., Blahos, J. II & Niedzielski, A. S. Evidence for multiple AMPA receptor complexes in hippocampal CA1/CA2 neurons. J. Neurosci. 16, 1982–1989 (1996).
Matsuzaki, M. et al. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4, 1086–1092 (2001).
Noguchi, J., Matsuzaki, M., Ellis-Davies, G. C. R. & Kasai, H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron 46, 609–622 (2005).
Gamache, T. R., Araki, Y. & Huganir, R. L. Twenty years of SynGAP research: from synapses to cognition. J. Neurosci. 40, 1596–1605 (2020).
Llamosas, N. et al. Syngap1 regulates experience-dependent cortical ensemble plasticity by promoting in vivo excitatory synapse strengthening. Proc. Natl Acad. Sci. USA 118 https://doi.org/10.1073/pnas.2100579118 (2021).
Hoelzel, C. A. & Zhang, X. Visualizing and Manipulating Biological Processes by Using HaloTag and SNAP-Tag Technologies. Chembiochem 21, 1935–1946 (2020).
Kerchner, G. A. & Nicoll, R. A. Silent synapses and the emergence of a postsynaptic mechanism for LTP. Nat. Rev. Neurosci. 9, 813–825 (2008).
Cane, M., Maco, B., Knott, G. & Holtmaat, A. The relationship between PSD-95 clustering and spine stability in vivo. J. Neurosci. 34, 2075–2086 (2014).
Bosch, M. et al. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82, 444–459 (2014).
Meyer, D., Bonhoeffer, T. & Scheuss, V. Balance and stability of synaptic structures during synaptic plasticity. Neuron 82, 430–443 (2014).
Gray, N. W., Weimer, R. M., Bureau, I. & Svoboda, K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 4, e370 (2006).
Mammen, A. L., Huganir, R. L. & O’Brien, R. J. Redistribution and Stabilization of Cell Surface Glutamate Receptors during Synapse Formation. J. Neurosci. 17, 7351–7358 (1997).
Ojima, K. et al. Ligand-directed two-step labeling to quantify neuronal glutamate receptor trafficking. Nat. Commun. 12, 831 (2021).
Wakayama, S. et al. Chemical labelling for visualizing native AMPA receptors in live neurons. Nat. Commun. 8, 14850 (2017).
Khmelinskii, A. et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 30, 708–714 (2012).
Sankaranarayanan, S., De Angelis, D., Rothman, J. E. & Ryan, T. A. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 79, 2199–2208 (2000).
tom Dieck, S. et al. Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015).
Iwasaki, S. & Ingolia, N. T. The Growing Toolbox for Protein Synthesis Studies. Trends Biochemical Sci. 42, 612–624 (2017).
Fang, H., Bygrave, A.M., Roth, R.H., Johnson, R.C. & Huganir, R.L. An optimized CRISPR/Cas9 approach for precise genome editing in neurons. Elife 10 https://doi.org/10.7554/eLife.65202 (2021).
Gao, Y. et al. Plug-and-Play Protein Modification Using Homology-Independent Universal Genome Engineering. Neuron 103, 583–597.e588 (2019).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Willems, J. et al. ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18, e3000665 (2020).
Zhong, H. et al. High-fidelity, efficient, and reversible labeling of endogenous proteins using CRISPR-based designer exon insertion. eLife 10, e64911 (2021).
Droogers, W.J., Willems, J., MacGillavry, H.D. & Jong, A.P.H.D. Duplex Labeling and Manipulation of Neuronal Proteins Using Sequential CRISPR/Cas9 Gene Editing. eNeuro 9. https://doi.org/10.1523/ENEURO.0056-22.2022 (2022).
Perez-Alvarez, A. et al. Freeze-frame imaging of synaptic activity using SynTagMA. Nat. Commun. 11, 2464 (2020).
Barria, A. & Malinow, R. Subunit-specific NMDA receptor trafficking to synapses. Neuron 35, 345–353 (2002).
Shi, S., Hayashi, Y., Esteban, J. A. & Malinow, R. Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105, 331–343 (2001).
Shi, S. H. et al. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Sci. (N. Y., N. Y.) 284, 1811–1816 (1999).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Martineau, M. et al. Semisynthetic fluorescent pH sensors for imaging exocytosis and endocytosis. Nat. Commun. 8, 1412 (2017).
Guenthner, C. J., Miyamichi, K., Yang, H. H., Heller, H. C. & Luo, L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784 (2013).
Kawashima, T. et al. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods 10, 889–895 (2013).
Hayashi-Takagi, A. et al. Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature 525, 333–338 (2015).
Choi, J. H. et al. Interregional synaptic maps among engram cells underlie memory formation. Science 360, 430–435 (2018).
Redondo, R. L. & Morris, R. G. Making memories last: the synaptic tagging and capture hypothesis. Nat. Rev. Neurosci. 12, 17–30 (2011).
Nakayama, H. et al. Microglia permit climbing fiber elimination by promoting GABAergic inhibition in the developing cerebellum. Nat. Commun. 9, 2830 (2018).
Mishina, M. & Sakimura, K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res 58, 105–112 (2007).
Zhou, L. et al. Deletion of exons encoding carboxypeptidase domain of Nna1 results in Purkinje cell degeneration (pcd) phenotype. J. Neurochem 147, 557–572 (2018).
Stoppini, L., Buchs, P. A. & Muller, D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods 37, 173–182 (1991).
Ohana, R. F. et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 68, 110–120 (2009).
Kawai, S., Takagi, Y., Kaneko, S. & Kurosawa, T. Effect of Three Types of Mixed Anesthetic Agents Alternate to Ketamine in Mice. Exp. Anim. 60, 481–487 (2011).
Konno, K., Yamasaki, M., Miyazaki, T. & Watanabe, M. Glyoxal fixation: An approach to solve immunohistochemical problem in neuroscience research. Sci. Adv. 9, eadf7084 (2023).
Takasaki, C. et al. Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur. J. Neurosci. 32, 1326–1336 (2010).
Shimuta, M. et al. Postsynaptic modulation of AMPA receptor-mediated synaptic responses and LTP by the type 3 ryanodine receptor. Mol. Cell Neurosci. 17, 921–930 (2001).
Matsuda, K. et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328, 363–368 (2010).
Miyazaki, T., Fukaya, M., Shimizu, H. & Watanabe, M. Subtype switching of vesicular glutamate transporters at parallel fibre-Purkinje cell synapses in developing mouse cerebellum. Eur. J. Neurosci. 17, 2563–2572 (2003).
Miura, E. et al. Expression and distribution of JNK/SAPK-associated scaffold protein JSAP1 in developing and adult mouse brain. J. Neurochem 97, 1431–1446 (2006).
Holtmaat, A. et al. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 4, 1128–1144 (2009).
Takahashi, T. et al. PEO-CYTOP Fluoropolymer Nanosheets as a Novel Open-Skull Window for Imaging of the Living Mouse Brain. iScience 23, 101579 (2020).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Fiala, J. C. & Harris, K. M. Extending unbiased stereology of brain ultrastructure to three-dimensional volumes. J. Am. Med Inf. Assoc. 8, 1–16 (2001).
Van Kempen, G. M. P., Van Vliet, L. J., Verveer, P. J. & Van Der Voort, H. T. M. A quantitative comparison of image restoration methods for confocal microscopy. J. Microsc. 185, 354–365 (1997).
Hayashida, J., Nishimura, K. & Bise, R. Consistent Cell Tracking in Multi-frames with Spatio-Temporal Context by Object-Level Warping Loss. 3-8 Jan. 1759–1768, (2022).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. held in Cham, 2015//. N. Navab, J. Hornegger, W.M. Wells, and A.F. Frangi, eds. (Springer International Publishing), 234–241, (2015).
Fujii, K., Suehiro, D., Nishimura, K. & Bise, R. Cell Detection from Imperfect Annotation by Pseudo Label Selection Using P-classification. held in Cham, 2021//. M. de Bruijne, P.C. Cattin, S. Cotin, N. Padoy, S. Speidel, Y. Zheng, and C. Essert, eds. (Springer International Publishing), 425–434, (2021).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
Acknowledgements
The authors thank L. Colgan for critical reading of the manuscript; the T.M. lab members for discussion; and Y. Nakagami for technical assistance. This work was supported by Grants-in-Aid for Scientific Research (JP20K21461, JP20H03349, JP20H05918, JP23K18160, and JP24K02130 to M.U.; and JP19H05206, JP19K22467, JP20H05055, JP20H03337, JP21H00188, JP21F21113, JP22K19355, JP22H05491, JP22K21353, JP23H04672, JP23H02574, JP23K27265, JP24H01229, JP24K22000, and JP25H02490 to T.M.), Japan Agency for Medical Research and Development (AMED) (JP25wm0625221 to M.U.; JP19dm0207080, JP21wm0525014, and JP24wm0625117 to T.M.), Japan Science and Technology Agency (JST) (JPMJFR231M to M.U. and JPMJPR16F9 to T.M.), Human Frontier Science Program (CDA00043/2019-C to T.M.), Kowa Life Science Foundation (M.U.), Narishige Neuroscience Research Foundation (M.U.), Uehara Memorial Foundation (T.M.), TORAY Science Foundation (T.M.), Takeda Science Foundation (T.M.), Brain Science Foundation (T.M.), and the Howard Hughes Medical Institute (HHMI) (P.K. and L.D.L.).
Author information
Authors and Affiliations
Contributions
Conceptualization, M.U. and T.M.; Methodology, M.U. and T.M.; Investigation, M.U., R.I., P.K., X.L., M.Isogai, C.H., M.N., and M.Igarashi; Formal analysis, M.U., R.I., K.F., K.Shiku., R.B., and T.M.; Software, M.U., K.F., K.Shiku., R.B., and T.M.; Resources, P.K., M.A., Y.O., K.Sakimura. and L.D.L.; Writing – original draft, M.U., R.B., L.D.L., and T.M.; Writing – review & editing, T.M.; Funding acquisition, M.U. and T.M.; Supervision, T.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Hyung-Bae Kwon and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Uchigashima, M., Iguchi, R., Fujii, K. et al. Single-cell synaptome mapping of endogenous protein subpopulations in mammalian brain. Nat Commun 16, 9705 (2025). https://doi.org/10.1038/s41467-025-65813-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65813-w