Abstract
Cutaneous tuberculosis (CTB) is an infectious disease highly associated with extracellular matrix remodeling and granuloma-driven fibrosis. Fibroblasts play crucial roles in this fibrotic process, but their specific roles in Mycobacterium tuberculosis (Mtb) skin infections remain unclear due to the lack of proper in vitro models. Here, we demonstrate that skin organoids (SKOs) derived from human induced pluripotent stem cells can model CTB infected by Mtb. Single-cell RNA analyses reveal an increase in fibroblasts, upregulation of genes involved in collagen synthesis, and enhanced collagen degradation induced by MMP2 and MMP14 in Mtb-infected SKOs. This is accompanied by the destruction of nerve cells and adipocytes. Importantly, the onset of fibrosis in Mtb-infected SKOs is dependent on the activation of the PI3K-AKT signaling pathway and transcription factor AP1 in fibroblasts. Pharmacological inhibition of PI3K-AKT and AP1 alleviates fibrosis and collagen deposition. Our findings have uncovered distinct alterations in cell populations during Mtb-induced skin fibrosis, highlighting the crucial roles of PI3K-AKT and AP1. The study demonstrates the utility of SKOs for investigating CTB pathogenesis and evaluating potential antifibrotic treatments.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is an ancient disease over 5000 years and remains prevalent in many developing countries1. It can also pose a potential global health threat due to the ever-increasing international migration. Caused primarily by Mycobacterium tuberculosis (Mtb), TB is a chronic granulomatous infection that can affect any organ in the body. While pulmonary TB is the predominant clinical type, extrapulmonary TB have been increasing, accounting for 8 to 24% of all TB cases2. Among them, cutaneous tuberculosis (CTB) comprises about 1.5–3% of these cases3,4,5. In recent years, there has been a resurgence of CTB in regions with high rates of HIV infection and multi-drug-resistant pulmonary TB1,6,7. Therefore, this poses a public health concern and requires further investigation for better diagnosis and management.
CTB more frequently affects women, especially young adults, with lesions mainly located on the face, neck, and abdomen8. However, due to Mtb’s slow replication rate9 and low reproductive efficiency in the skin8, skin infections often present with concealed symptoms, leading to delayed diagnosis and treatment10. Furthermore, CTB exhibits diverse clinical features related to infection routes, including exogenous and endogenous transmission of Mtb. The exogenous mechanism involves direct inoculation into the skin, while the endogenous mechanism is secondary to a previous infection with transmission through contiguous, lymphatic or hematogenous dissemination7,11,12. Notably, CTB often induces extracellular matrix (ECM) remodeling, leading to fibrosis and granulomas13,14. In this process, fibroblasts play a crucial role as the primary structural cells involved in fibrosis and collagen production. Yet, due to the lack of suitable in vitro models, CTB remains a complicated and largely unexplored disease in terms of its pathogenesis, clinical manifestations, and diagnosis.
To better understand the pathophysiology of CTB and develop more effective treatments, biologically relevant experimental models that can accurately replicate skin features are essential. Since Mtb is exclusively a pathogen to humans, no animal model can fully replicate all the pathological features observed in human disease. Advanced human organotypic skin models derived from human induced pluripotent stem cells (hiPSCs) can reproduce the key physical and physiological characteristics of human skin and possess many crucial accessory organs, including hair follicles, sebaceous glands, nerves, fat, and melanocytes15,16,17. These unique features make skin organoids (SKOs) a valuable tool for investigating skin-related diseases, microbial infections, and drug development18,19,20. Recently, hiPSCs-derived SKOs have been used in studies focusing on host-microbe interactions21,22,23.
In this study, we established SKOs using the method described by Koehler16 to investigate the pathology of CTB. To simulate the dissemination route of Mtb through the circulatory or lymphatic systems within the body, our in vitro model of CTB employed dermal infection, rather than direct epidermal infection. Comprehensive and multidimensional analyses of SKOs with and without Mtb infection revealed significant changes in the pathological structure and cell types of Mtb-infected SKOs. The increase in fibroblast proportion, along with the decrease in nerve cell and adipocyte populations, offers important insights for the treatment of CTB. Notably, the role of the PI3K-AKT-AP1 pathway in promoting fibrosis and collagen deposition, as well as the effective pharmacological inhibition in SKOs, provide new ideas for the development of novel treatment strategies for CTB-related fibrosis. The results demonstrate that hiPSCs-derived SKOs can serve as a pathological model for evaluating and advancing new therapeutic strategies for CTB.
Results
The hair-bearing SKOs harbor adipocytes and express diverse collagen subtypes
To establish an in vitro model of CTB, SKOs derived from hiPSCs were prepared following a previously reported16 protocol (Fig. 1A). After 130 days of cultivation, we successfully replicated the SKOs described in the literature (Supplementary Fig. 1A, B). The SKOs histologically exhibited structures resembling human skin tissue, characterized by a mature stratified epidermis (with a Loricrin-positive granular layer and KRT5-positive basal layer, Supplementary Fig. 1C, D), a Vimentin- and P75-positive dermis, and complex hair follicles with CD71-positive inner root sheath, CD82-positive cuticle, and SOX2-expressing dermal papilla cells (Supplementary Fig. 1E–G). Various skin cell types including neural cells were also present in the SKOs (Supplementary Fig. 1H–J). In alignment with native human skin tissue, the SKOs exhibited abundant collagen types, including I, III, and IV, with type I collagen being the most prominent (Fig. 1B), though SCD1-positive adipocytes and sebaceous glands were found in the dermal layer of the SKOs (Fig. 1C). These comprehensive analyses demonstrate that our SKOs faithfully recapitulate the key structural and functional characteristics of native human skin. The robust reproducibility of SKOs containing hair follicles, adipocytes, neural components, and proper ECM composition makes them a reliable and physiologically relevant in vitro model system for cutaneous biology research and dermatopathological investigations.
A Schematic representation of the experimental workflow of skin organoids (SKOs) culture, Mycobacterium tuberculosis (Mtb) infection and scRNA-seq analysis. Created in BioRender. Xiang, M. (2025) https://BioRender.com/dko3bm8. B Representative immunohistochemical staining with antibodies against collagen I, III and IV in SKOs. This was repeated three times with similar results. Scale bar = 500 µm or 50 µm, as indicated in the image. C Representative immunofluorescence staining with antibodies against the adipocyte marker SCD1 in SKOs. This was repeated three times with similar results. Scale bar = 50 or100 µm, as indicated in the image.
Infection and confirmation of SKOs as an in vitro model of CTB
To establish an in vitro model of CTB, the mature SKOs (after induction for 125 days) were co-cultured with logarithmic-phase Mtb bacteria for 0, 3, 9, 15 days. Then the SKOs were collected at different time points to assess their infection status. Intracellular Mtb infection was confirmed on day 9 post infection, with purple-red rod-shaped bacteria detected at the edge of the SKOs through acid-fast staining (Fig. 2A, B). On day 15 post-infection, Mtb had aggregated at the edge of the SKOs but did not invade deeper structures, suggesting that the SKOs possessed a functional skin barrier. The infected SKOs exhibited pathological changes resembling CTB, manifested by damage to the epidermal structure on days 9 and 15 post-Mtb infection (Fig. 2C, D). Furthermore, skin stratification and integrity were assessed by immunofluorescence staining. Uninfected SKOs exhibited well-defined epidermal stratification without Vimentin+ dermal fibroblasts present in the epidermis. Upon Mtb infection, the epidermal basal layer (CK5 + ) was significantly disrupted, accompanied by thickening of the granular layer (Loricrin + ) at the damaged sites, and the migration of dermal fibroblasts (Vimentin + ) into the epidermal compartment (Fig. 2E).
A,B Detection of Mtb infection in SKOs using acid-fast staining (A) and qPCR analysis (B) at different time points. Arrows indicated acid-fast Mtb. Scale bar = 400 or 50 µm, as indicated in the image, n = 3 biological replicates and error bars represent mean ± SEM. C Histological evaluation of the structural changes in SKOs at different time points following Mtb infection using H&E staining, Scale bar = 100 µm. This was repeated three times with similar results. D Immunofluorescence staining images of epidermal marker KRT5 in SKOs following Mtb infection at different time points, Scale bar = 100 µm. This was repeated three times with similar results. E Representative immunofluorescence staining with antibodies against epithelial layer markers KRT5 (basal layer), Loricrin (granular layer) and dermal layer marker Vimentin in SKOs pre- and post-infection, Scale bar = 100 µm. This was repeated three times with similar results. F Identifying the different cell types present in both uninfected SKOs (termed as normal) and Mtb-infected SKOs (termed as case) groups. Each cell type is labeled with a unique color. G The expression levels of marker genes within each cell types. H Comparison of UMAP visualization for single-cell data between the normal group and the case group. I Stacked bar chart of the proportion of all cell types in each sample between the normal group and the case group. Source data are provided as a Source Data file.
To further characterize the cellular changes, single-cell RNA sequencing (scRNA-seq) analysis of the Mtb-infected SKOs was conducted. After quality control, a CTB-SKOs scRNA-seq atlas was established, comprising 126,918 cells, with 40,193 cells in the Mtb-infected SKOs group (termed as case group in the figure), and 86,725 cells in the uninfected SKOs group (termed as normal group in the figure). In CellMarker 2.0 and Cell Taxonomy database, 12 cell types were identified in the normal SKOs based on the specific markers: fibroblast (COL1A1, COL1A2, DCN), basal cell (KRT5, KRT14), melanocyte cell (PMEL, DCT, MLANA), glial cell (SOX2, TTYH1), endothelial cell (CLDN5), spinous cell (KRT10), chondrocyte (ACAN, COL2A1), schwann (SOX10, S100B, CDH19), adipocyte (APOE, SCD), neuron (TUBB3, BEX1), mitotic cell (HMGB2, MKI67, TOP2A), smooth muscle cell (TPM2, DES) (Fig. 2F–H and Supplementary Fig. 2A). First, we analyzed our scRNA-seq data along with the reported data from SKOs cultured after 133-day24 (Supplementary Fig. 1B, C). In comparison, in addition to the adipocytes only detected in our SKOs, there was an increase in the number of endothelial cells and nerve-related cells in our normal SKOs and the nerve-related cells including neurons, schwann cells, and glial cells, were more mature in our normal SKOs (Supplementary Fig. 1D, E). Upon Mtb infection, the proportions of several cell lineages were altered, with an increased proportion of fibroblasts and chondrocytes, and a decreased proportion of adipocytes, neurons and glial cells (Fig. 2I, and Supplementary Fig. 2F).
Mtb infection induces ECM remodeling and skin fibrosis
Differential gene expression analysis of individual cell subpopulations revealed elevated expression of various collagen-related genes in Mtb-infected SKOs in most of the cell subpopulations (Fig. 3A). Among them, the type I collagen gene COL1A2 was significantly upregulated in majority of cell types (9/12, p < 0.05), except in schwann cells, neurons and glial cells. Significantly elevated expression of COL3A1 was observed in fibroblast, basal cell, spinous cell, mitotic cell and smooth muscle cell subpopulations (p < 0.05). Moreover, COL4A1 expression was markedly elevated in schwann cells and adipocytes (p < 0.05), while COL4A2 showed specific upregulation in adipocyte subpopulations (p < 0.05) (Fig. 3A). However, both in normal and case group, most genes associated with the Wnt pathway, tight junctions complexes, TGF-β pathway and ECM showed low expression levels (Supplementary Fig. 3). Only CTNNB1, SFRP1, TIMP1, DCN and FN1 were relatively highly expressed. Compared with collagen-related genes, these ECM-associated genes were negligibly affected by Mtb infection.
A Violin plot of expression levels of MMPs and collagen family genes in all cell types. B The enrichment pathways of the upregulated genes identified through the differential analysis between the normal group and the case group. Enrichment significance was assessed by hypergeometric test, with p values adjusted using the Benjamini-Hochberg FDR method. C Sirius red staining and Masson’s trichrome staining images (left) and quantitative analysis (right) of collagen content in the SKOs pre- and post-Mtb infection, Scale bar = 500 µm, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. D qPCR analysis of mRNA expression levels of type I, III and IV collagen genes in SKOs pre- and post-Mtb infection, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. E, F Immunohistochemistry results show the expression levels (E) and quantitative analysis (F) of type I, III and IV collagen proteins in the SKOs pre- and post-Mtb infection, Scale bar = 500 µm, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. G qPCR analysis of MMP14 and MMP2 mRNA expression levels in SKOs pre- and post-infection, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. H ELISA analysis of MMP14 and MMP2 levels in SKOs pre- and post-infection, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. I Representative immunofluorescence staining with antibodies against MMP14 and MMP2 in the SKOs pre- and post-Mtb infection, Scale bar = 500 µm. This was repeated three times with similar results. Source data are provided as a Source Data file.
The induced expression of collagen genes suggests that Mtb infection drives ECM remodeling in SKOs, which may contribute to the development of skin fibrosis. This notion is further supported by the activation of ECM-related pathways in Mtb-infected SKOs (Fig. 3B). Histological analysis using Sirius red and Masson’s trichrome staining confirmed an overall increase in total collagen content in Mtb-infected SKOs compared to uninfected controls (Fig. 3C). qPCR analysis corroborated the upregulation of various collagen-related genes upon Mtb infection (Fig. 3D). Immunohistochemical analysis revealed that the contents of collagen types I, III and IV were significantly increased in Mtb-infected SKOs (Fig. 3E, F). scRNA-seq analysis revealed alterations in the expression of matrix metalloproteinases (MMPs) in Mtb-infected SKOs. MMPs are crucial for ECM degradation and tissue remodeling. Specifically, MMP2 expression was upregulated in fibroblasts, basal cells, and adipocytes, whereas MMP14 expression was increased in fibroblasts (Fig. 3A). These findings were further validated by qPCR, ELISA and immunofluorescence staining analyses (Fig. 3G–I). Collectively, these results demonstrate that Mtb infection in SKOs triggered the upregulation of collagen expression. Subsequently, MMP2 and MMP14 promoted ECM remodeling by degrading collagen.
Impact of Mtb infection on other cell populations in SKOs
After Mtb infection, there was an increase in fibroblast proportion, a decrease in nerve cells and adipocytes within the SKOs cell population, along with the occurrence of fibroblast differentiation into chondrocyte. In Mtb-infected SKOs, the proportion of neuron and glial cells were reduced, while the subpopulations of neuroendocrine cells and merkel progenitor cells were significantly increased among the subtypes of neuron cells (neurecto-epithelial cell, neuron stem cell, serotonergic neuron, neuroendocrine cell, ASCL1-neuroendocrine cell and merkel progenitor cell) (Fig. 4A–C and Supplementary Figs. 2F, 4A). A pseudo-chronological analysis of the overall neuronal cell subpopulation indicates that neural cells with high differentiation potential were the majority, while a smaller percentage was consisted of more mature serotonergic neurons and neurecto-epithelial cells (Fig. 4D). This suggests that Mtb infection may lead to the destruction of nerve cells and impede the effective differentiation of neural stem cells into functional neurons.
A Representative immunofluorescence staining with antibodies against neuronal cells marker TUJ1 in the SKOs pre- and post-Mtb infection, Scale bar = 500 µm. This was repeated three times with similar results. B Identify each subpopulation of neuron cells in both the normal group and the case group. C Stacked bar chart of the proportion of each subpopulation of neuron cells in normal and case groups. D The pseudo-temporal analysis reveals distinct differentiation trajectories of various subpopulations of neuron cells across normal and case groups. E Enrichment analysis reveals the activation of pathways associated with neurodegeneration. F Representative immunofluorescence staining with antibodies against PSMD7 and TUJ1 in the SKOs pre- and post-Mtb infection, Scale bar = 200 µm. This was repeated three times with similar results. G Representative immunofluorescence staining with antibodies against adipocytes marker SCD1 in the SKOs pre- and post-Mtb infection, Scale bar=500 µm. This was repeated three times with similar results. H GO enrichment pathway analysis of differentially expressed genes in adipocytes. Bubble size indicates the number of enriched genes, with color changes reflecting p values adjust significance levels. Enrichment significance was assessed by hypergeometric test, with p values adjusted using the Benjamini-Hochberg FDR method.
Enrichment analysis of differentially expressed genes (DEGs) in each neuron cell subpopulation indicated the suppression of small molecule metabolism, as the genes of FABP5, PTGDS, PGK1, and TTR being collectively downregulated (Supplementary Fig. 4B, C). This was further supported by immunofluorescence staining showing a significant decrease in PTGDS expression in Mtb-infected SKOs (Supplementary Fig. 4D). This may lead to disorders in energy and lipid metabolism, and oxidative damage, ultimately impairing neuronal metabolism and survival. Furthermore, there was a significant activation of neurodegenerative pathways, along with the dysregulation of PSMD and NDUF family genes (Fig. 4E). Immunofluorescence analysis confirmed the upregulation of PSMD7 in Mtb-infected SKOs (Fig. 4F), suggesting the involvement of proteasome-mitochondria-oxidative stress mechanisms in neurodegeneration. Moreover, the reduced proportion of adipocytes (Fig. 4G), was accompanied by the activation of multiple necroptosis pathways leading to necrotic cell death (Fig. 4H). Our findings indicate that Mtb infection leads to a reduction in neuronal and adipocyte cell populations through the activation of necroptotic pathways, suggesting a potential mechanism for the impairment of neurological and metabolic functions associated with CTB.
Analysis of the key signaling molecules involved in fibroblast differentiation revealed significantly elevated expression of LOX/LOXL2 in Mtb-infected fibroblasts and other cell subpopulations (Supplementary Fig. 4E). Immunofluorescence staining confirmed the upregulation of LOX/LOXL2 in Mtb-infected SKOs (Supplementary Fig. 4F). The finding suggest that Mtb infection induced overexpression of LOX/LOXL2, which may enhance α-SMA expression and drive fibroblast-to-myofibroblast differentiation, contributing to skin fibrosis. Unexpectedly, chondrocytes showed a notable rise in Mtb-infected SKOs (Supplementary Fig. 2F), with the upregulation of COL2A1 expression confirmed by qPCR and immunofluorescence analyses (Supplementary Fig. 5A-C). Cell trajectory analysis suggested a differentiation pathway from fibroblasts to chondrocytes, where chondrocytes exhibited a more advanced differentiation state (Supplementary Fig. 5D, E). Although the implications of fibroblast-chondrocyte differentiation are unknown, it may represent an aberrant differentiation event specific to the SKOs model rather than genuine pathological differentiation of CTB.
Mtb infection induces skin fibrosis through activation of the PI3K-AKT signaling pathway
To understand the mechanisms leading to fibrosis, we next conducted a comprehensive analysis on the differentiation of fibroblasts after Mtb infection. Analysis of cell populations show an increasing trend in the proportion of fibroblast after Mtb infection (Supplementary Fig. 2F). Fibroblasts were further categorized into eight subtypes, including fibroblast, EGR1+ fibroblast, VEGFA+ fibroblast, reticular fibroblast, papillary fibroblast, FN1+ fibroblast, myofibroblast and APOE+ fibroblast (Fig. 5A, B). Analysis of the cell proportions by pseudo-chronological analysis revealed an increase in the proportion of the EGR1+ fibroblast subpopulation in the Mtb-infected SKOs, indicating enhanced cell differentiation (Fig. 5C). Differential enrichment analysis detected the activation of multiple ECM remodeling-related signaling pathways, with a particular emphasis on the PI3K-AKT pathway in a subset of fibroblasts (Fig. 5D, E). Subsequent analysis of genes enriched in the PI3K pathway among DEGs in the fibroblasts identified multiple upregulated genes associated with fibrosis (Fig. 5F–H).
A Distribution of fibroblasts in the overall data. B The expression levels of specific markers for further classifying fibroblasts into distinct subpopulations. C Identification of distinct fibroblast subpopulations. Stacked bar chart of the proportion of each fibroblast subpopulations in normal and case groups. D Pseudo-temporal analysis reveals the differentiation trajectories of different subpopulations of fibroblasts in normal and case groups. E Fibroblasts, chondrocyte and schwann cells exhibit upregulation of genes associated with collagen-related pathways in the case group. Enrichment significance was assessed by hypergeometric test, with p values adjusted using the Benjamini-Hochberg FDR method. F Upregulation of PI3K pathway-related genes in fibroblasts of the case group. The color intensity represents the magnitude of gene expression upregulation. G qPCR analysis of mRNA expression levels of PI3K-AKT pathway-related genes in SKOs pre- and post-Mtb infection, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. H Immunofluorescence staining of proteins associated with the PI3K-AKT pathway pre- and post-Mtb infection, Scale bar = 100 or 500 µm, as indicated in the image. Source data are provided as a Source Data file.
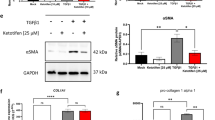
The PI3K-AKT pathway has been implicated in the pathogenesis of fibrotic diseases in various organs, including the lung25, kidney26, liver27, intestine28 and skin29. To determine whether targeting the PI3K-AKT signaling pathway could ameliorate the activation of fibroblasts and collagen gene expression in a Mtb-infected SKO model, we utilized the PI3K inhibitor LY294002 to suppress PI3K-AKT pathway. Immunoblotting analysis revealed that phosphorylated AKT (p-AKT) protein expression was upregulated in Mtb-infected SKOs (Fig. 6A, B), indicating the activation of the PI3K-AKT pathway. In contrast, treatment with 10 μM LY294002 attenuated the expression of p-AKT protein and recovered close to normal levels (Fig. 6A, B). Subsequent qPCR analysis demonstrated that the mRNA expression of myofibroblast markers α-SMA and Fibronectin (FN1) was upregulated in Mtb-infected SKOs and downregulated following LY294002 treatment (Fig. 6C). Immunofluorescence analysis further confirmed these findings, showing that the protein levels of α-SMA and FN1 were increased following Mtb-infection and partially recovered upon LY294002 treatment (Fig. 6D, E). Furthermore, treatment with LY294002 attenuated the collagen production induced by Mtb infection (Fig. 6F, G).
A, B Western blot analysis of phosphorylated AKT expression levels in different treatment groups (A), and according quantitative analysis (B), n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. C qPCR analysis of mRNA expression levels of α-SMA and FN1 in different treatment groups, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. D, E Immunofluorescence staining images (D) and quantitative analysis (E) of α-SMA and FN1 expression in different treatment groups. Scale bar = 250 µm, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. F, G Masson’s trichrome staining images (F) and quantitative analysis (G) of collagen content in different treatment groups. Scale bar = 100 µm, Mtb+LY294002: Mtb infection with LY294002 treatment group, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. Source data are provided as a Source Data file.
To further validate the critical role of the PI3K-AKT pathway in Mtb infection-induced fibrosis, we also used Wortmannin, another PI3K-AKT inhibitor, to treat Mtb-infected SKOs. qPCR analysis confirmed that inhibiting PI3K-AKT pathway significantly attenuated skin fibrosis (Supplementary Fig. 6A). These results suggest that targeting the PI3K-AKT signaling axis may be a promising therapeutic approach to mitigate skin fibrosis in the context of CTB.
Mtb infection activates AP1 transcriptional activity
AP1 (activating protein-1) is a collective term referring to dimeric transcription factors composed of JUN, FOS, ATF or MAF subunits that bind to a common AP1-binding site on DNA, and regulates the transcription of target genes30. Accumulating evidence has indicated that the aberrant activation of AP1 is associated with the development of various skin diseases and organ fibrosis31,32,33. As PI3K-AKT signaling pathway has been demonstrated to serve as a mediator of AP1 activation34, and both of these pathways have been shown to play crucial roles in the progression of fibrotic diseases35,36, we continued to examine AP1 expression of the SKOs. scRNA-seq results showed that the expression levels of multiple AP1 subunits were upregulated in the fibroblasts from Mtb-infected SKOs (Fig. 7A, B). qPCR analysis of the AP1 transcription factor family subunits revealed that the FOS subunits c-FOS, FOSL1 and FOSL2, the JUN subunits c-JUN and JUND, the ATF subunits ATF3 and ATF4, as well as the MAF subunits MAFA and MAFF, were all upregulated to varying degrees in Mtb-infected SKOs (Fig. 7C), indicating a comprehensive upregulation of AP1 transcription factors under Mtb infection.
A The transcriptional profile of fibroblasts contains multiple transcription factors associated with AP1. B A comparison of standardized transcription factor activity levels of AP1 related transcription factors between the normal and case group. C qPCR analysis of mRNA expression levels of AP1 related transcription factors in the SKOs pre- and post-Mtb infection, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. D, E Western blot analysis of phosphorylated JUN and FOS expression levels in the SKOs pre- and post-Mtb infection (D), and corresponding quantitative analysis (E), n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. (F) qPCR analysis of mRNA expression levels of JUN and FOS in different treatment groups, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. G qPCR analysis of mRNA expression levels of α-SMA, COL1A1 and FN1 in different treatment groups, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. H, I Immunofluorescence staining images (H) and quantitative analysis (I) of α-SMA and FN1 expression in different treatment groups. Scale bar = 250 µm, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. J, K Masson’s trichrome staining images (J) and quantitative analysis (K) of collagen content in different treatment groups. Mtb+T5224: Mtb infection with T5224 treatment group. Scale bar = 100 µm, n = 3 biological replicates and error bars represent mean ± SEM. Statistical differences according to an unpaired two-tailed t-test. Source data are provided as a Source Data file.
To investigate the potential of targeting AP1 transcription factors as a therapeutic strategy for Mtb-induced skin fibrosis, we utilized a selective AP1 inhibitor (T5224) that is known to suppress the DNA-binding activity of FOS and JUN subunits37. Western blot analysis confirmed that the phosphorylation levels of JUN and FOS were upregulated in Mtb-infected SKOs (Fig. 7D, E). Correspondingly, the mRNA levels of the JUN and FOS were downregulated following T5224 treatment (Fig. 7F). Furthermore, qPCR analysis revealed that T5224 treatment reversed the Mtb-induced upregulation of fibrosis-related genes, including α-SMA, FN1 and COL1A1 (Fig. 7G). Immunofluorescence staining showed that the fluorescence intensities of α-SMA and FN1 were significantly reduced in the T5224 treated Mtb-infected SKOs, compared to the non-treated Mtb-infected SKOs, approaching the levels of uninfected SKOs (Fig. 7H, I). Masson’s trichrome staining confirmed that the T5224 treated SKOs showed similar pathological manifestations to the control group without Mtb infection, with reduced collagen deposition (Fig. 7J, K). Furthermore, treatment of Mtb-infected SKOs with SR11302, another AP1 inhibitor, significantly reduced skin fibrosis, as confirmed by qPCR analysis (Supplementary Fig. 6B). Our findings demonstrate that targeted inhibition of AP1 transcription factors attenuated Mtb-induced skin fibrosis.
The results presented here indicate that inhibiting the PI3K-AKT-AP1 pathway helps alleviate the fibrosis caused by Mtb infection, suggesting that this molecular pathway could represent a potential therapeutic target for the management of CTB.
Discussion
In clinical practice, chronic CTB (such as lupus vulgaris and tuberculosis cutis verrucosa) is often accompanied by fibrosis and epidermal rupture following prolonged inflammation, manifesting as abnormal deposition of collagen in the dermis and scar formation13,14. To facilitate the investigation on CTB pathology, we established an in vitro model using hiPSCs-derived SKOs. Characterization of the pathological profiles show shared tissue repair/fibrosis mechanisms in both clinical and our SKOs models. Though the accumulation of fibrotic structures acts as an isolation to resist the invading pathogens in granulomas38, excessive fibrosis within these structures may impede the effective diffusion of antibiotics into bacterial loci within the granulomas39, potentially impacting host organ function by leaving scar tissue after regression40,41. To obtain a comprehensive picture of Mtb invasion and its significant pathological changes, we applied in-deepth analyses by comparing SKOs with and without Mtb infection, including qPCR, scRNA-seq, acid-fast staining and immunofluorescence staining. The study on SKOs enabled us to unveil several novel mechanisms involved in the fibrotic process of CTB, which were not described in the studies on clinical samples.
Following Mtb infection, there was a rise in the proportion of fibroblasts and a decline in nerve cells and adipocytes within the SKOs cell population. Fibroblasts are highly dynamic cells that are responsible for producing and remodeling ECM contributing to tissue repair and fibrosis41,42. Opposite to the fibroblast, nerve cells and adipocytes were reduced in SKOs after Mtb infection. Since nerve cells are one of the targets for treatment of Mtb43,44, the decrease may be linked to the activation of the neuronal apoptosis pathway. Further study is needed to understand how Mtb induces neuronal apoptosis and its implications on CTB patients. The reduction of adipocytes in SKOs following Mtb infection aligns with previous reports that shown an acute loss of fat cells in mice post aerosol Mtb infection45. Adipose tissue pathophysiology is crucial in consumption45 and may thus influence the activation and reactivation of Mtb infection. Changes in these cell populations may modulate CTB pathology during Mtb infection, thus, our findings offer new perspectives on the infection mechanism of CTB. It is also worthwhile to explore this pathological phenomenon of Mtb infection in other organs such as the lungs and central nervous system.
Mtb infection in SKOs caused ECM remodeling and increased fibroblast populations, leading to collagen deposition resembling fibrosis observed in granuloma formation. This process may be a result of elevated levels of MMP2 and MMP14 in fibroblast and myofibroblast populations induced by Mtb infection. MMPs are a family of enzymes that play a crucial role in ECM remodeling and collagen degradation, which are tightly regulated in various physiological and pathological processes46,47,48,49. Previous studies have linked MMP promoter polymorphisms to TB pathogenesis50. Specifically, MMP1, MMP3 and MMP9 have been identified as key regulatory factors in TB pathology51. However, our data did not indicate significant changes in their expression after Mtb infection, possibly because these MMPs are mainly produced by neutrophils and macrophages52,53, which SKOs lack. MMP2 and MMP14 were upregulated in early TB lung granulomas of rhesus macaques54. They can degrade various ECM components such as collagen types IV, V, VII, and X, elastin and fibronectin53. MMP14 is also known to activate pro-MMP13 and pro-MMP2 and directly degrade collagen types I, II, and III55,56. Consistent with this, Mtb-infected SKOs showed varying degrees of deposition of type I, III and IV collagen. Mtb infection induces upregulation of MMP and disrupts the balance with tissue inhibitor of metalloproteinase (TIMP), leading to altered ECM deposition and granulomatous pathological outcomes57. Consequently, MMP2 and MMP14 are likely major contributors to the ECM remodeling in Mtb-infected SKOs, leading to collagen deposition and influencing pathological outcomes.
The most interesting finding is the upregulation of the genes associated with the PI3K-AKT signaling pathway and AP1 related transcription factors in Mtb-infected SKOs. Previous studies have demonstrated that the interaction between the PI3K-AKT pathway and TGF-β promotes the formation of fibrosis58. Activation of PI3K-AKT can also induce fibrosis by regulating its downstream targets like mTOR59. The transcription factor AP1 is closely linked to the PI3K-AKT pathway and plays a role in cell proliferation60. Activation of the PI3K-AKT pathway can increase AP1 DNA binding activity, contributing to liver fibrosis36. Notably, AP1 can be activated by the TLR-JNK-AP1 axis and regulate fibrosis-related proteins33. Despite lacking evidence of upstream signal activation, which may be related to the timing of infection, our findings are in line with these studies, indicating that activation of the PI3K-AKT signaling pathway and AP1 activity may be a common mechanism for fibrotic pathogenesis. Future work should monitor the activation mode and expression patten of the key molecules in these pathways during Mtb infection, potentially by manipulating PI3K-AKT, AP1 or TGF-β signals to understand their role in CTB skin fibrosis. This may help developing a topical ointment targeting AP1 or PI3K-AKT signaling pathways to minimize systemic side effects and alleviate skin damage from long-term anti-TB treatment.
Hair-bearing SKOs have been demonstrated as valuable experimental models for investigating host-microbe interactions with pathogens like mpox virus and enterovirus 71, known to cause HFMD23,61. These models helped to advance dermatology research, for understanding of infection mechanisms, immune responses and drug reactions. In this study, we demonstrated the feasibility of hiPSCs-derived SKOs to model Mtb infection. Though the model allowed us to discover differential host defense responses elicited by Mtb infection, it has limitations by lacking sweat glands, blood vessels and immune cells15,62, which are important players during the progression of infectious disease.
Another limitation of the SKOs is the unintended, off-target differentiation of chondrocytes, which has been observed previously16,19. Although human dermal fibroblasts exhibited the ability to differentiate into osteoblasts and chondrocytes in vitro63,64,65, however, there is no corresponding clinical reports of increased chondrocytes in human Mtb-infected skin lesions. Thus, the observed chondrocyte differentiation in this study may potentially reflect an abnormal differentiation event specific to the SKO model rather than a genuine pathological process. This finding should be validated through comparative studies in authentic CTB pathological models or biopsy samples when becoming available. With the limitations, further refinements of the existing SKOs culture system are necessary to improve its complexity and biological relevance, such as integration of macrophage cells or/and induction of vascularization24,66.
One technical limitation in this study is quantification of infected bacteria, for which we used the qPCR detection of 16s rRNA and acid staining instead of the gold standard method of Colony-Forming Unit (CFU) enumeration67,68. Although the 16s rRNA and acid staining are frequently used for quantification of live bacteria for diagnosis and monitoring of TB69,70,71,72, these methods provide indirect evidence of the ongoing growth and survival of Mtb within the organoids.
In summary, this study reports a novel model for investigating CTB in vitro. Most importantly, it provides the significant information regarding to the pathological mechanisms of CTB. The identification of the crucial role of the PI3K-AKT signaling pathway and AP1 transcription factors in initiating fibrotic responses and corresponding pharmacological inhibition experiments provide valuable information for development of pharmacological treatments to effectively attenuate fibrosis and collagen accumulation in CTB. Finally, the potential relationship between the reduction in the overall proportion of functional nerve cells and adipocytes, particularly those in a terminally differentiated state, and the degree of CTB disease severity warrants further investigation.
Methods
Human pluripotent stem cell lines and culture
Culture experiments were performed with two human induced pluripotent stem cell lines (RC01001-A and RC01004), purchased from Shownin Biotechnologies. Cells were cultured on 6-well plates coated with Matrigel (354277, Corning) and maintained in mTeSR1 medium (85850, StemCell Technologies) supplemented with 100 μg/mL Normocin (ant-nr-05, Invivogen). The medium was replenished every other day or daily depending on cell confluency. Cells were passaged at around 80% confluency (generally every 4 to 5 days). The passaging was performed in tiny clusters with ReLeSR (100-0483, StemCell Technologies), and in mTeSR1 medium containing 10 μM Y27632 (04-0012-02, Stemgent).
Generation of SKOs from hiPSCs
Hair-bearing SKOs were generated following the previously established protocol15. A comprehensive step-by-step differentiation method is detailed in the Nature Protocol publication titled “Generation and characterization of hair-bearing skin organoids from human pluripotent stem cells.”
Briefly, cells were dissociated into single-cell suspensions using Accutase Cell Dissociation Reagent (A1110501, Gibco). Subsequently, 3500 cells resuspended in 100 μL of Essential 8 Flex medium (A2858501, Gibco) supplemented with 20 μM Y27632 were dispensed into each well of a U-bottom ultra-low attachment 96-well plate (7007, Corning). The plate was centrifuged (200 × g, 6 min) and incubated for 24 h. After the initial incubation (day 1), 100 μL of fresh E8 medium was added to each well to dilute the Y27632. On day 0 of differentiation, cells were cultured in E6 medium (A1516401, Gibco) supplemented with: 2% Matrigel (v/v, 354230, Corning), 10 μM SB431542 (04-0010-05, Stemgent), 4 ng/mL basic FGF (100-18B, PeproTech), 5 ng/mL BMP4 (120-05, PeproTech). This medium formulation is referred to as “E6SFB” in the protocol. On day 0, all cell aggregates were transferred to a new 96-well U-bottom plate in 100 μL of E6SFB medium. On day 3, the medium was supplemented with 200 ng/mL LDN (a BMP inhibitor, 04-0074, Stemgent) and 50 μg/mL bFGF. On day 6, 75 μL of fresh E6 medium was added to each well. On day 9, half of the medium was replaced with fresh E6 medium. On day 12, all aggregates were individually transferred to 24-well low-attachment plates (3473, Corning) containing 500 μL of organoid maturation medium (OMM) supplemented with 1% Matrigel. The OMM consists of: 1:1 mixture of Advanced DMEM/F-12 (12634010, Gibco) and Neurobasal Medium (21103049, Gibco), 1× GlutaMax (35050061, Gibco), 0.5× B-27 minus vitamin A (12587010, Gibco), 0.5× N-2 (17502048, Gibco), 0.1 mM 2-mercaptoethanol (21985023, Gibco) and 100 μg/mL Normocin. Plates were then placed on an orbital shaker (65 × rpm) in a 37 °C, 5% CO₂ incubator. On day 15, half of the spent medium was replaced with fresh OMM containing 1% Matrigel. From day 18 to day 45, half-medium changes were performed every 3 days using OMM. For experiments extending beyond day 45, medium changes were adjusted to every other day. Additionally, depending on organoid size (e.g., larger aggregates) and medium consumption (indicated by yellowing), the total medium volume was increased to 1 mL per well to ensure adequate nutrient supply.
Bacterial strains and culture
In this study, Mycobacterium tuberculosis (Mtb) H37Rv strain (ATCC35743, JZ101702) was cultured on Middle Brook 7H11 Agar Base (LA7240, Solarbio) supplemented with 10% Middlebrook OADC Growth Supplement (LA9560, Solarbio) and 0.5% Glycerol at 37 °C. When the bacteria reached mid-exponential phase, bacterial clones were harvested in saline. The Mtb suspension was then dispersed, counted using a Bacterial ultrasonic dispersion counter (TB Healthcare) and adjusted to 1MCF.
SKOs and Mtb co-culture
SKOs were cultured in antibiotic-free OMM medium for three days prior to the experiment. Then, the SKOs were transferred to a new 24-well plate with pre-cut wide pipette tips, and fresh antibiotic-free OMM medium was added. To achieve Mtb infection, 33 μL of 1MCF Mtb (1 × 107/mL) was added to each well for 24 h (Each organoid contained 3–5 million cells, and the multiplicity of infection (MOI) was approximately 2-3:1), then washed the SKOs three times with PBS to remove extracellular bacteria, continued culturing the SKOs at 37 °C (5% CO2) for several days according to the needs of study. During Mtb infection, 90% of the medium was replaced every 2-3 days to ensure a constant density of Mtb in the medium.
In all figures, the control groups are labeled as “Uninfection” unless otherwise specified. The “Infection” group represents SKOs exposed to Mtb and cultured for 9 days. The “Uninfection” group represent SKOs that are cultured in parallel without Mtb exposure.
Sections preparation
SKOs were fixed in 4% formaldehyde overnight at 4 °C and then processed through a dehydration gradient. After the tissues were embedded in paraffin, they were cut into 4 μm thick sections for hematoxylin and eosin (H&E) staining, Acid-Fast staining, Immunohistochemistry, immunofluorescence analysis, Sirius Red staining and Masson’s trichrome staining.
Histological analysis
Four micrometer paraffin sections were deparaffinized using xylene, followed by rehydration with graded ethanol, and subsequent staining with hematoxylin and eosin staining solution (G1120, Solarbio) according to the manufacturer’s instructions.
Acid-Fast staining
The sections were stained with Ziehl-Neelsen staining dye at room temperature for 4 h, followed by a 5 min wash with slow-running water. Subsequently, they were decolorized with a destaining solution for several seconds, and counterstained with hematoxylin solution for 5 min. After another round of washing and decolorization, the sections were sealed with neutral gum.
Immunohistochemistry
For immunohistochemical analysis, the sections underwent deparaffinization, rehydration, and heat-induced antigen retrieval in a microwave to boil for at least 15 min using an antigen retrieval buffer. Endogenous catalase was then neutralized with 0.3% H2O2 for 30 min after cooling. Following this, sections were blocked with 10% goat serum in PBS for 1 h and then incubated with the primary antibodies at 4 °C overnight. The sections were then washed in PBS. and subsequently incubated with HRP-conjugated secondary antibodies for 1 h. Next, sections were washed three times again in PBS and incubated with DAB working solution for 5 min. After counterstaining with hematoxylin, the sections were mounted with glycerol gelatin (Sigma-Aldrich, Cat No. 1002946952). The ImageJ (Fiji, version 1.8.0, National Institute of Health) and GraphPad Prism 9 (version 9.5.1, GraphPad Software) is used to process data from cell images and histograms. The primary antibodies are listed in Supplementary Table 1.
Immunofluorescence analysis
For immunostaining, the sections were deparaffinized and rehydrated, and heat-induced antigen retrieval by boiling in a microwave for at least 15 min. Subsequently, the sections were blocked in 10% normal goat serum for 1 h at room temperature. Primary antibodies were then added at the indicated dilutions in 10% normal goat serum and incubated overnight at 4 °C. Following three washes with PBS, secondary antibodies diluted 1:500 in 3% normal goat serum were added for 2 h at room temperature protected from light. After repeating the previous washing steps, DAPI staining (#4083, Cell Signaling) was added in DPBS for 15 min at room temperature in the dark. Finally, samples were mounted with fluorescence mounting medium (Dako, S3023) and stored at 4 °C until imaging using an inverted confocal laser-scanning microscope (Nikon). The ImageJ (Fiji, version 1.8.0, National Institute of Health) and GraphPad Prism 9 (version 9.5.1, GraphPad Software) is used to process data from cell images and histograms. The primary antibodies are listed in Supplementary Table 1.
Sirius Red staining
After deparaffinization, the paraffin sections were stained with Weigert’s iron hematoxylin solution for 8 min, followed by rinsing with running water. Subsequently, the sections were stained with Sirius Red staining solution for 20 min, rinsing in deionized water. Finally, rapid dehydration was rapidly dehydrated in gradient concentrations of ethanol and xylene and sealed with resin. The ImageJ (Fiji, version 1.8.0, National Institute of Health) and GraphPad Prism 9 (version 9.5.1, GraphPad Software) is used to process data from cell images and histograms.
Masson’s trichrome staining
After deparaffinization, the sections were soaked in potassium dichromate overnight, then rinse with tap water. Then, the sections were stained with Weigert’s iron hematoxylin solution for 3 min, followed by rinsing with running water. The samples were differentiated with 1% hydrochloric acid in alcohol for 10–15 s, followed by staining with Masson’s bluing solution, rinsing in deionized water. Subsequently, the sections were stained with Ponceau-acid fuchsin solution for 5 min, differentiated in phosphomolybdic acid, and then stained with aniline blue solution for several minutes. After rinsing with deionized water, the sections are placed in acetic acid working solution for 1 min. Finally, rapid dehydration was rapidly dehydrated in gradient concentrations of ethanol and xylene and sealed with resin. The ImageJ (Fiji, version 1.8.0, National Institute of Health) and GraphPad Prism 9 (version 9.5.1, GraphPad Software) is used to process data from cell images and histograms.
Real-time quantitative polymerase chain reaction (qPCR)
Bacterial load within SKOs was assessed using absolute quantification of 16S rRNA. Initially, 1 mL of 1MCF H37Rv suspension was centrifuged at 13,400 × g for 10 min, followed by RNA extraction with TRIzol reagent (284910, Life Technologies). The extracted RNA was reverse transcribed into cDNA using a TAKARA reverse transcription kit. Subsequently, the cDNA was serially diluted tenfold (ranging from 300 to 300,000,000 copies) and equal volumes of cDNA templates were used for qPCR. A standard curve was established according to CT values and corresponding bacterial copy numbers. SKOs were washed thrice with PBS at specified time intervals, then homogenized with 1 mm diameter grinding beads under low-temperature conditions. Following RNA extraction using TRIzol reagent, reverse transcription and qPCR reactions were carried out, and the obtained CT values were substituted into the linear equation of the above standard curve to obtain the corresponding bacterial copy number.
To detect mRNA changes of DEGs in SKOs, qPCR was performed using TB Green Premix Ex Taq™ (RR420A, Takara) according to the manufacturer’s instructions. The resulting CT values were normalized to the mRNA expression level of β-Actin, and differential expressions between the groups were calculated by 2-△△CT. The primer sequences are shown in the Supplementary Table 2.
Western blotting analysis
After removing the organoid medium, the SKOs were washed with PBS, followed by the addition of RIPA Lysis Buffer containing 1% phosphatase inhibitor and protease inhibitor, to extract proteins using a low-temperature tissue grinder. Protein samples (40 μg) were subjected to 10% SDS-PAGE polyacrylamide electrophoresis and transferred to PVDF membranes. The membranes were then blocked with 10% skim milk at room temperature for 2 h. To detect the activation of the PI3K-AKT signaling pathway and AP1 transcription factor, PVDF membranes were probed with primary antibodies overnight at 4 °C. Following incubation with a specific HRP-conjugated secondary antibodies for 1 h, the immunoblot signal was visualized using the BioRad ChemiDOC XRSP Detection System (BioRad), and the intensity of the western blot bands was quantitatively analyzed using Image J (Fiji, version 1.8.0, National Institute of Health) and GraphPad Prism 9 (version 9.5.1, GraphPad Software). Primary antibodies used are listed in Supplementary Table 1.
PI3K-AKT and AP1 signal inhibition test
Before infection, SKOs were divided into three groups: Control (no treatment), Mtb group (infected with Mtb), and Mtb+inhibitor group (infected with Mtb while treated with 10 μM T5224 (HY-12270, MedChemExpress) or 10 μM SR11302 (2476, TOCRIS) or 10 μM LY294002 (HY-10108, MedChemExpress) or 1 μM Wortmannin (HY-10197, MedChemExpress)). The SKOs underwent simultaneous infection with Mtb and inhibitor treatment, with subsequent medium changes and inhibitor supplementation every 2-3 days. The SKOs were harvested on the 9th day.
SKOs dissociation for scRNA-seq
The SKOs were washed three times with PBS using wide-mouth pipette tips, minced as finely as possible and digested for 30 min in Advanced DMEM/F12 supplemented with 1 mg/mL type I collagenase (17018029, Gibco). Following collagen lysis, the digestion was terminated with RPMI1640, and the mixture was centrifuged at 300 × g for 5 min at 4 °C. The pellet was resuspended in 2–3 mL of TrypLE express (12604013, Gibco) and incubated at 37 °C for 10 min. Subsequently, 0.4% bovine serum albumin (BSA)/PBS solution was added to stop the dissociation reaction, centrifuged for 5 min at 300 × g at 4 °C and washed again with 0.4% BSA/PBS. The cell suspension was pipetted repeatedly and passed through a 70 μm cell sieve twice to obtain the single-cell suspension. Cell viability and live cell count were calculated to ensure that the final cell concentration and viability were applicable for sequencing with trypan blue staining.
scRNA-seq library preparation and sequencing
The cell suspension was used immediately for scRNA-seq library preparation with a target recovery of 10,000 cells and loaded to a 10× Genomics Chromium Controller for Gel beads in emulsion generation and barcoding using the Chromium Single Cell 3′ Reagent Kit v.3.1 Dual Index (10× Genomics) according to the manufacturer’s instructions. scRNA-seq 3’ gene expression libraries were generated using the 10× Genomics Chromium single cell system and sequenced using the Illumina NovaSeq 6000 sequencing system.
scRNA-seq data processing
We used the Cell Ranger Software version 6.0.2 to align raw reads to human transcripts (GRCh38) and to generate feature-barcode matrices. The R package Doublefinder version 2.0.3 was utilized to identify and remove potential double cells. Analysis of scRNA-seq data was conducted using the R package Seurat version 4.3.0 and associated tools. Cells with fewer than 300 expressed genes and less than 500 counts were filtered out to ensure data quality. Additionally, cells with mitochondrial gene expression exceeding 20% and erythrocyte gene expression exceeding 3% were excluded from further analysis.
Identification of DEGs
We ran principal component analysis (PCA) after normalizing our single-cell data set and identifying variable genes using the R package Seurat. R package Harmony was utilized for sample integration and removal of batch effects. Cell classification and clustering were carried out using the FindNeighbors and FindClusters functions from Seurat. The UMAP algorithm was used for dimensionality reduction. Specific expression marker genes for each cluster were identified using the FindAllMarkers function from Seurat with |log2Foldchange | ≥0.5, cell percentage ≥ 0.25, and p value ≤ 0.05. Cell types were determined by comparing expression patterns with cell type-specific markers from the CellMarker2.0 and Cell Taxonomy databases. Cluster annotation was done accordingly.
The Findmarker function was utilized to identify DEGs between normal and case SKOs with p value < 0.05 and |log2FC > 0.25 |. To specify enriched biological processes in DEGs for each cell type under each condition, the R package clusterProfiler version 4.6.2 was employed alongside the org.Hs.eg.db version 3.16.0 annotation package. Enrichment analysis was conducted on the differential genes (p value < 0.05), and the R package GseaVis version 0.0.8 was utilized for visualizing the GSEA enrichment analysis results.
Network analysis
We utilized a combination of bioinformatics tools to analyze the differentiation trajectories within the entire cell atlas. The R package Monocle3 version 1.3.4 was employed to construct the differentiation trajectory model and assess the evolution and differentiation status of different cell types. Furthermore, the R package Monocle version 2.26.0 was utilized to analyze the quasi-temporal relationships within the neuron cell subpopulation, while the Python package CytoTRACE version 0.3.3 was utilized to evaluate the differentiation status and potential of Fibroblast cell subpopulations. Additionally, the python package pySCENIC version 0.12.1 was utilized to calculate transcription factor activity and construct regulatory networks at the single-cell level for a comprehensive understanding of transcriptional regulation mechanisms.
Statistics and Reproducibility
All experiments were repeated three times with similar results. Data are all indicated as mean ± SD. An unpaired two-tailed t-test was performed for statistical analyses unless otherwise specified. P < 0.05 was considered as significant for all analyses, and the p values were directly indicated in the figure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The processed scRNA-seq data generated in this study have been deposited in the Genome Sequence Archive for Human database under accession code HRA011953) (https://ngdc.cncb.ac.cn/search/specific?db=hra&q=HRA011953). The human skin organoid data of day 133 used in this study are available in the Gene Expression Omnibus database under accession code GSE231607. Source data are provided with this paper.
Code availability
The analysis code is available on Zenodo under the following link https://doi.org/10.5281/zenodo.17118354).
References
Santos, J. B. et al. Cutaneous tuberculosis: epidemiologic, etiopathogenic and clinical aspects - part I. Bras. Dermatol. 89, 219–228 (2014).
2021, G.T.R. World Health Organization. https://www.who.int/teams/global-tuberculosis-programme/tb-reports (WHO, 2021).
Kumar, B. & Muralidhar, S. Cutaneous tuberculosis: a twenty-year prospective study. Int. J. Tuberc. Lung Dis. 3, 494–500 (1999).
Varshney, A. & Goyal, T. Incidence of various clinico-morphological variants of cutaneous tuberculosis and HIV concurrence: a study from the Indian subcontinent. Ann. Saudi Med. 31, 134–139 (2011).
van Zyl, L., du Plessis, J. & Viljoen, J. Cutaneous tuberculosis overview and current treatment regimens. Tuberculosis 95, 629–638 (2015).
Kwan, C. K. & Ernst, J. D. HIV and tuberculosis: a deadly human syndemic. Clin. Microbiol. Rev. 24, 351–376 (2011).
Barbagallo, J., Tager, P., Ingleton, R., Hirsch, R. J. & Weinberg, J. M. Cutaneous tuberculosis: diagnosis and treatment. Am. J. Clin. Dermatol. 3, 319–328 (2002).
Hernández Solis, A. et al. Skin biopsy: a pillar in the identification of cutaneous Mycobacterium tuberculosis infection. J. Infect. Dev. Ctries 6, 626–631 (2012).
Franco-Paredes, C. et al. Cutaneous mycobacterial infections. Clin. Microbiol. Rev. 32, e00069-18 (2018).
Hamada, M., Urabe, K., Moroi, Y., Miyazaki, M. & Furue, M. Epidemiology of cutaneous tuberculosis in Japan: a retrospective study from 1906 to 2002. Int. J. Dermatol. 43, 727–731 (2004).
Chen, Q., Chen, W. & Hao, F. Cutaneous tuberculosis: a great imitator. Clin. Dermatol. 37, 192–199 (2019).
Karas, L., Lu, C. Y., Agrawal, P. B. & Asgari, M. M. The impact of the orphan drug act on food and drug administration-approved therapies for rare skin diseases and skin-related cancers. J. Am. Acad. Dermatol. 81, 867–877 (2019).
Terziroli Beretta-Piccoli, B., Mainetti, C., Peeters, M. A. & Laffitte, E. Cutaneous granulomatosis: a comprehensive review. Clin. Rev. Allergy Immunol. 54, 131–146 (2018).
Moos, P. J. et al. Single cell analysis of peripheral TB-associated granulomatous lymphadenitis. bioRxiv: the preprint server for biology (2024).
Lee, J. & Koehler, K. R. Skin organoids: a new human model for developmental and translational research. Exp. Dermatol 30, 613–620 (2021).
Lee, J. et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582, 399–404 (2020).
Lee, J. et al. Generation and characterization of hair-bearing skin organoids from human pluripotent stem cells. Nat. Protoc. 17, 1266–1305 (2022).
Sandoval, A. G. W., Gim, K. Y., Huang, J. T. & Koehler, K. R. Applications of human pluripotent stem cell-derived skin organoids in dermatology. J. Investig. Dermatol. 143, 1872–1876 (2023).
Ramovs, V. et al. Characterization of the epidermal-dermal junction in hiPSC-derived skin organoids. Stem Cell Rep. 17, 1279–1288 (2022).
Kim, M. J. et al. Modeling of solar UV-induced photodamage on the hair follicles in human skin organoids. J. Tissue Eng. 15, 20417314241248753 (2024).
Ma, J. et al. Establishment of human pluripotent stem cell-derived skin organoids enabled pathophysiological model of SARS-CoV-2 infection. Adv. Sci. 9, e2104192 (2022).
Jung, S. Y. et al. Wnt-activating human skin organoid model of atopic dermatitis induced by Staphylococcus aureus and its protective effects by Cutibacterium acnes. iScience 25, 105150 (2022).
Li, P. et al. Mpox virus infection and drug treatment modelled in human skin organoids. Nat. Microbiol 8, 2067–2079 (2023).
Gopee, N. H. et al. A prenatal skin atlas reveals immune regulation of human skin morphogenesis. Nature 635, 679–689 (2024).
Margaria, J. P., Moretta, L., Alves-Filho, J. C. & Hirsch, E. PI3K signaling in mechanisms and treatments of pulmonary fibrosis following sepsis and acute lung injury. Biomedicines 10, 756 (2022).
Kattla, J. J., Carew, R. M., Heljic, M., Godson, C. & Brazil, D. P. Protein kinase B/Akt activity is involved in renal TGF-beta1-driven epithelial-mesenchymal transition in vitro and in vivo. Am. J. Physiol. Ren. Physiol. 295, F215–F225 (2008).
Xiu, A. Y., Ding, Q., Li, Z. & Zhang, C. Q. Doxazosin attenuates liver fibrosis by inhibiting autophagy in hepatic stellate cells via activation of the PI3K/Akt/mTOR signaling pathway. Drug Des., Dev. Ther. 15, 3643–3659 (2021).
Rahmani, F. et al. Rigosertib potently protects against colitis-associated intestinal fibrosis and inflammation by regulating PI3K/AKT and NF-kappaB signaling pathways. Life Sci. 249, 117470 (2020).
Huang, Y. et al. Enhancement of zyxin promotes skin fibrosis by regulating FAK/PI3K/AKT and TGF-beta signaling pathways via integrins. Int J. Biol. Sci. 19, 2394–2408 (2023).
Karin, M., Liu, Z. & Zandi, E. AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 (1997).
Teofoli, P. et al. Expression of Bcl-2, p53, c-jun and c-fos protooncogenes in keloids and hypertrophic scars. J. Dermatol. Sci. 22, 31–37 (1999).
Pi, Z. et al. ATRA ameliorates fibrosis by suppressing the pro-fibrotic molecule Fra2/AP-1 in systemic sclerosis. Int. Immunopharmacol. 121, 110420 (2023).
Lee, H. S. et al. Mycobacterium tuberculosis induces connective tissue growth factor expression through the TLR2-JNK-AP-1 pathway in human lung fibroblasts. FASEB J. 33, 12554–12564 (2019).
Li, J. et al. PI-3K and Akt are mediators of AP-1 induction by 5-MCDE in mouse epidermal Cl41 cells. J. Cell Biol. 165, 77–86 (2004).
Wygrecka, M. et al. TGF-beta1 induces tissue factor expression in human lung fibroblasts in a PI3K/JNK/Akt-dependent and AP-1-dependent manner. Am. J. Respir. Cell Mol. Biol. 47, 614–627 (2012).
Wang, J. et al. Kupffer cells mediate leptin-induced liver fibrosis. Gastroenterology 137, 713–723 (2009).
Avouac, J. et al. Inhibition of activator protein 1 signaling abrogates transforming growth factor beta-mediated activation of fibroblasts and prevents experimental fibrosis. Arthritis Rheum. 64, 1642–1652 (2012).
Pagán, A. J. & Ramakrishnan, L. The Formation and Function of Granulomas. Annu. Rev. Immunol. 36, 639–665 (2018).
Hawn, T. R., Matheson, A. I., Maley, S. N. & Vandal, O. Host-directed therapeutics for tuberculosis: can we harness the host? Microbiol. Mol. Biol. Rev. MMBR 77, 608–627 (2013).
Mornex, J. F., Leroux, C., Greenland, T. & Ecochard, D. From granuloma to fibrosis in interstitial lung diseases: molecular and cellular interactions. Eur. Respir. J. 7, 779–785 (1994).
Talbott, H. E., Mascharak, S., Griffin, M., Wan, D. C. & Longaker, M. T. Wound healing, fibroblast heterogeneity, and fibrosis. Cell Stem Cell 29, 1161–1180 (2022).
Lynch, M. D. & Watt, F. M. Fibroblast heterogeneity: implications for human disease. J. Clin. Investig. 128, 26–35 (2018).
Randall, P. J. et al. Neurons are host cells for Mycobacterium tuberculosis. Infect. Immun. 82, 1880–1890 (2014).
Ruhl, C. R. et al. Mycobacterium tuberculosis sulfolipid-1 activates nociceptive neurons and induces cough. Cell 181, 293–305 (2020).
Ayyappan, J. P. et al. Adipose tissue regulates pulmonary pathology during TB infection. mBio 10, e02771-18 (2019).
Dobaczewski, M., Gonzalez-Quesada, C. & Frangogiannis, N. G. The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J. Mol. Cell. Cardiol. 48, 504–511 (2010).
Xu, Y. et al. Matrix metalloproteinase inhibitors enhance the efficacy of frontline drugs against Mycobacterium tuberculosis. PLoS Pathogens 14, e1006974 (2018).
Taylor, J. L. et al. Role for matrix metalloproteinase 9 in granuloma formation during pulmonary Mycobacterium tuberculosis infection. Infect. Immun. 74, 6135–6144 (2006).
Khokha, R., Murthy, A. & Weiss, A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat. Rev. Immunol. 13, 649–665 (2013).
Elkington, P. T., Ugarte-Gil, C. A. & Friedland, J. S. Matrix metalloproteinases in tuberculosis. Eur. Respir. J. 38, 456–464 (2011).
Cabrera, S. et al. Overexpression of MMP9 in macrophages attenuates pulmonary fibrosis induced by bleomycin. Int. J. Biochem. Cell Biol. 39, 2324–2338 (2007).
Santos, M. P. et al. Doxycycline aggravates granulomatous inflammation and lung microstructural remodeling induced by Schistosoma mansoni infection. Int. Immunopharmacol. 94, 107462 (2021).
Cabral-Pacheco, G. A. et al. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int. J. Mol. Sci. 21, 9739 (2020).
Mehra, S. et al. Transcriptional reprogramming in nonhuman primate (rhesus macaque) tuberculosis granulomas. PLoS ONE 5, e12266 (2010).
Knauper, V. et al. Cellular activation of proMMP-13 by MT1-MMP depends on the C-terminal domain of MMP-13. FEBS Lett. 532, 127–130 (2002).
Leeman, M. F., Curran, S. & Murray, G. I. The structure, regulation, and function of human matrix metalloproteinase-13. Crit. Rev. Biochem. Mol. Biol. 37, 149–166 (2002).
Marquis, J. F. et al. Fibrotic response as a distinguishing feature of resistance and susceptibility to pulmonary infection with Mycobacterium tuberculosis in mice. Infect. Immun. 76, 78–88 (2008).
Sun, Y., Zhang, Y. & Chi, P. Pirfenidone suppresses TGF‑β1‑induced human intestinal fibroblasts activities by regulating proliferation and apoptosis via the inhibition of the Smad and PI3K/AKT signaling pathway. Mol. Med. Rep. 18, 3907–3913 (2018).
Conte, E. et al. PI3K p110γ overexpression in idiopathic pulmonary fibrosis lung tissue and fibroblast cells: in vitro effects of its inhibition. Lab. Investig. J. Tech. Methods Pathol. 93, 566–576 (2013).
Dang, S. et al. MicroRNA‑218 inhibits the malignant phenotypes of glioma by modulating the TNC/AKT/AP‑1/TGFβ1 feedback signaling loop. Int. J. Mol. Med. 48, 205 (2021).
Li, J. et al. A skin organoid-based infection platform identifies an inhibitor specific for HFMD. Nat. Commun. 16, 2513 (2025).
Sun, H., Zhang, Y. X. & Li, Y. M. Generation of skin organoids: potential opportunities and challenges. Front Cell Dev. Biol. 9, 709824 (2021).
Sommar, P. et al. Engineering three-dimensional cartilage- and bone-like tissues using human dermal fibroblasts and macroporous gelatine microcarriers. J. Plast. Reconstr. Aesthet. Surg. 63, 1036–1046 (2010).
Tuan, R. S. Skin and bones (and cartilage): the dermal fibroblast connection. Nat. Rev. Rheumatol. 5, 471–472 (2009).
Outani, H. et al. Direct induction of chondrogenic cells from human dermal fibroblast culture by defined factors. PLoS ONE 8, e77365 (2013).
Rimal, R. et al. Vascularized 3D human skin models in the forefront of dermatological research. Adv. Health. Mater. 13, e2303351 (2024).
Thurnheer, T. & Belibasakis, G. N. Integration of non-oral bacteria into in vitro oral biofilms. Virulence 6, 258–264 (2015).
Inoyama, D. et al. A Preclinical Candidate Targeting Mycobacterium tuberculosis KasA. Cell Chem. Biol. 27, 560–570 e510 (2020).
Zemb, O. et al. Absolute quantitation of microbes using 16S rRNA gene metabarcoding: a rapid normalization of relative abundances by quantitative PCR targeting a 16S rRNA gene spike-in standard. Microbiologyopen 9, e977 (2020).
Li, R. et al. Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep. 7, 5752 (2017).
Vilcheze, C. & Kremer, L. Acid-fast positive and acid-fast negative mycobacterium tuberculosis: the Koch paradox. Microbiol. Spectr. 5, 2003–2015 (2017).
Lamanna, C. The nature of the acid-fast stain. J. Bacteriol. 52, 99–103 (1946).
Acknowledgements
We are grateful to Professor Jin-Ping Li at Uppsala University for the critical reading of the manuscript and providing constructive comments. This work was supported by the National Natural Science Foundation of China (No. 62422318 to H.C.), and National Key Research and Development Program of China (No.2024YFA1307700 to X.B. and No. 2023YFF0725500 to H.C.), and the State Key Laboratory of Medical Proteomics (SKLP-K202407 to H.C.).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.T., X.B., X.Q., and H.C.; Data curation: L.Y., Y.L., P.Z., and L.F.; Investigation: L.Y., Y.L., P.Z., and L.F.; Methodology: L.Y., Y.L., P.Z. and L.F.; Resource: X.Q.; Software: L.F., Y.L. and M.X.; Formal analysis: J.W., J.Z., L.G., C.W.; Validation: L.Y., P.Z., Z.Y., Q.S., and C.L.; Visualization: L.Y., Y.L., P.Z., and L.F.; Supervision: Y.T., X.B., X.Q., and H.C.; Funding acquisition: Y.T. and H.C.; Writing-original: L.Y., P.Z., and L.F; Writing-review & editing: Y.T. The manuscript was read and approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chang-Hwa Song, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yue, L., Liang, Y., Zhong, P. et al. Human pluripotent stem cell-derived skin organoids enabled pathophysiological model of Mycobacterium tuberculosis infection. Nat Commun 16, 10831 (2025). https://doi.org/10.1038/s41467-025-65848-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65848-z