Abstract
In microbial communities, bacteria can inhibit or facilitate each other by altering their shared environment. Most studies of these interactions have focused on well-mixed environments, leaving spatial effects underexplored. Here, we show that in an antibiotic-treated community, bacterial spread depends on a facilitation mechanism that only emerges in spatial settings. The facilitating species enables the community’s range expansion but is then suppressed to a minority, making it a hidden initiator of the expansion. Focusing on two pathogens, immotile Klebsiella pneumoniae and motile Pseudomonas aeruginosa, we found that both tolerate a β-lactam antibiotic, with Pseudomonas being more resilient and dominating in well-mixed cultures. During range expansion, however, the antibiotic inhibits Pseudomonas’ ability to spread unless it is near Klebsiella—which creates a clear zone by degrading the antibiotic, at the expense of its own growth. As Pseudomonas spreads, it competitively suppresses Klebsiella. Our modeling and experimental analyses reveal that this facilitation operates at a millimeter scale. We also observed similar facilitation by a Bacillus species isolated from a hospital sink, in both pairwise and eight-member bacterial communities with its co-isolates. These findings suggest that spatially explicit experiments are essential to understand certain facilitation mechanisms and have implications for surface-associated microbial communities like biofilms and for polymicrobial infections involving drug-degrading immotile and drug-tolerant motile bacteria.
Similar content being viewed by others
Introduction
In nature, microbes typically exist in communities where they can inhibit or facilitate each other. The inherent complexity of natural microbial communities makes it challenging to uncover their underlying ecological mechanisms. To alleviate this challenge, simplified microbial communities with two to several members have been assembled to extract fundamental mechanistic insights, which can then be tested in increasingly complex communities1. Examples include predicting large community outcomes from pairwise interactions2, revealing limitations of pairwise information for larger communities3, demonstrating how interspecies interactions during chemotaxis and growth shape marine community assembly4, showing bioelectric signals mediate interspecies attractive interactions5, and illustrating how transient invaders can trigger state switching in a two-member community through pH modification6.
Bacteria often interact by altering their shared environment—for example, by breaking down complex resources into products utilizable by other microbes7,8,9,10,11,12,13, changing pH12,14 or modulating host immune responses15,16,17,18. Bacterial interactions also depend on context. For instance, in a synthetic mouse gut microbiota, Bacteroides caecimuris inhibited other members only when the polysaccharide inulin was the carbohydrate source12, acidifying the culture through inulin fermentation12. Likewise, Bacteroides uniformis can benefit or compete with butyrate-producing gut bacteria depending on the available carbohydrate sources11.
While previous studies on bacterial interactions in communities have focused predominantly on resource context and growth, spatial structure is a ubiquitous environmental context for microbes. Many microbes live in spatially structured communities, such as biofilms19,20,21. In such an environment, survival depends on both mobility and effectiveness in nutrient utilization22,23,24,25. The mobility can be intrinsic or through interactions with other community members. For example, Escherichia coli can be transported by Paenibacillus vortex26, and Pseudomonas aeruginosa variants hitchhike on shared biosurfactants27,28. Biosurfactant production enables collective spatial expansion (called swarming) but comes at a significant growth cost, and cheater variants that do not produce biosurfactants can destabilize this strategy during a prolonged spatial expansion of a population28. When invading a spatial environment, Pseudomonas’ biosurfactants can enable the displacement of competing Klebsiella, which previously colonized the environment29. These examples underscore the critical need for spatially explicit experiments to uncover microbial interactions.
Here, combining mathematical modeling and experiments, we reveal that in a microbial community treated with a β-lactam antibiotic, an immotile species facilitates the spatial expansion of a motile species when sufficiently close. The immotile species detoxifies the environment by degrading the antibiotic, which would otherwise suppress the dispersal—but not the growth—of the motile species. This enables the motile species to expand, and subsequently suppresses the growth of the immotile species through an apparent competition, reducing it to a minority and highlighting it as a hidden initiator of community range expansion.
We demonstrated this facilitation in two pairwise communities: 1) clinical isolates of β-lactamase (Bla)-producing Klebsiella pneumoniae (Klebsiella) as the immotile facilitator and Pseudomonas aeruginosa (Pseudomonas) as the motile species under cefotaxime (CTX) treatment, and 2) a nonpathogenic Bacillus paranthracis (Bp) with Bla genes (Supplementary Table 1) as the immotile facilitator and another, motile Pseudomonas strain (Pa22), co-isolated from the same hospital sink, under carbenicillin (CB) treatment. Finally, we demonstrated the applicability of Bp’s facilitation on Pa22 in a synthetic community with six additional isolates from the same sink sample.
Results
Klebsiella enabled Pseudomonas range expansion under β-lactam treatment at its own expense
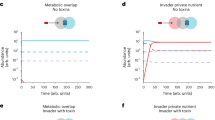
We tested the effect of a β-lactam antibiotic (CTX) treatment on the growth (i.e., biomass production) of Pseudomonas aeruginosa PA14 (Pseudomonas) and a Bla-producing clinical isolate of Klebsiella pneumoniae (Klebsiella)30 in well-mixed liquid cultures. Pseudomonas and Klebsiella are prevalent pathogens in many common infection types31,32 (Supplementary Fig. 1), can co-colonize patients33, and co-occur at human infection sites34 or urinary catheters35,36, suggesting that they have a strong niche overlap, making their community experiments clinically and ecologically relevant. We first grew Klebsiella and Pseudomonas in well-mixed pure cultures without or with CTX (2.5 μg ml−1) (Fig. 1a). Both species tolerated the CTX treatment, which reduced the biomass production of Klebsiella and Pseudomonas by 46% and 26%, respectively, when measured after 27 h (Fig. 1a, bottom). Klebsiella grew ~1.3-times faster than Pseudomonas without antibiotics (during the first 6 h) (Fig. 1a, top). With the CTX treatment, Klebsiella growth became detectable only after ~11 h of incubation (Fig. 1a, bottom), while Pseudomonas growth was virtually unaffected (Fig. 1a).
Kp: Klebsiella, Pa: Pseudomonas, CTX: Cefotaxime. Source data are provided as a Source Data file. a Pseudomonas tolerated CTX treatment better than Klebsiella. Liquid media were inoculated with Klebsiella (dark red) or Pseudomonas (cyan) at OD600 = ~2.1×10−5 (from dilutions of a cell suspension with OD600 > 0.05), without (top) or with CTX (2.5 μg ml−1, bottom). Curves and shades show average and standard deviation, respectively, from n = 2 biological replicates, each with three technical replicates. b Pseudomonas dominated Klebsiella under the CTX treatment in well-mixed co-cultures. Liquid media were inoculated with a 1:1 mixture of the two species at OD600 = ~2.1×10−5, incubated for 27 h, and then plated to determine relative abundances (Supplementary Fig. 4b). Symbols denote n = 3 independent replicates (solid/open markers show technical replicates); diamond indicates data from fluorescently labeled strains. Bars and error bars represent mean and standard deviation from independent experiments after averaging technical replicates. c Pseudomonas range expansion was suppressed by CTX and rescued by Klebsiella. 20 ml of swarming agar (0.55%) media were center inoculated with 1 μl of Klebsiella or Pseudomonas (OD600 = ~0.43). Colony images were captured at 24 h, processed, and analyzed for colony areas using a custom Python script (Methods; Supplementary Fig. 3). Images shown were equally adjusted for brightness/contrast. Dish diameter: 100 mm. See Supplementary Fig. 7 for raw images from n = 3 biological replicates. d The rescue of the coculture range expansion occurred at the expense of Klebsiella’s growth. In the same experiments as in (c), agar media were center inoculated with 1 μl of 1:1 mixture of the two species (OD600 = ~0.43) and imaged after 24 h. Then, whole colonies were harvested with 5 ml of saline, and relative abundances were estimated by plating (Supplementary Fig. 4b). Symbols indicate n = 3 independent experiments, with solid/open markers showing technical replicates. Bars and error bars show mean and standard deviation of independent experiments after averaging technical replicates. Images shown were equally adjusted for brightness/contrast.
The results above show that Klebsiella grew faster without antibiotic treatment, whereas Pseudomonas grew faster under the CTX treatment. When grown as a well-mixed pure culture, each species produced less biomass in medium conditioned by the other species than in unconditioned medium, demonstrating competition between the two species (Supplementary Fig. 2). If the two species primarily interact through competition in well-mixed cocultures, we expect the dominance of Klebsiella without the CTX treatment and the dominance of Pseudomonas with the CTX treatment. To this end, we tested the growth of mix cultures, each starting with equal fractions of Klebsiella and Pseudomonas, with or without CTX treatment. Indeed, without CTX treatment, Klebsiella reached a relative abundance of 75.0% after 27 h (Fig. 1b, top). With CTX (2.5 μg ml-1) treatment, Pseudomonas reached a relative abundance of 99.9% after 27 h (Fig. 1b, bottom).
We then examined the two species and their interactions on an agar surface where Pseudomonas could exhibit swarming motility37. We tested the effect of the β-lactam antibiotic CTX (2.5 μg ml−1) on pure cultures of Pseudomonas or Klebsiella by inoculating them on agar and measuring their colonized areas and biomasses after 24 h of incubation (Fig. 1c; Supplementary Fig. 3, Supplementary Fig. 4a). The CTX treatment suppressed Pseudomonas growth by 28-fold when measured by colony areas (Fig. 1c; Supplementary Fig. 3), or 22-fold when measured by biomass (Supplementary Fig. 4a). This result is consistent with the studies that showed that Pseudomonas’ swarming is inhibited by antibiotic treatment38,39. In contrast, the CTX treatment did not significantly affect Klebsiella colony growth (Fig. 1c; Supplementary Fig. 3; Supplementary Fig. 4a). In both cases, Klebsiella colonies were smaller than those of Pseudomonas colonies (Fig. 1c; Supplementary Fig. 3).
We next measured the growth of a coculture starting with equal biomasses of the two species under the CTX treatment. The coculture colonized a 7.4-fold larger area and produced 6.2-fold greater biomass compared to Pseudomonas alone (Fig. 1c,d; Supplementary Fig. 3; Supplementary Fig. 4a). Without antibiotic treatment, Pseudomonas alone grew better than a mixture (Fig. 1c, d; Supplementary Fig. 3; Supplementary Fig. 4a), though the difference was statistically insignificant (A two-sample t-test assuming unequal variances, i.e., Welch’s t-test, was used to test the null hypothesis of no difference. The corresponding two-tailed P–values are 0.081 and 0.25 for colonized area and biomass, respectively). The collective range expansion by Klebsiella and Pseudomonas under the CTX treatment was also observed when different incubation temperatures were tested (30 °C and 40 °C instead of the 37 °C default, Supplementary Fig. 5).
When mixed colonies were harvested and the relative abundances of the two species were measured based on their colony forming unit (CFU) morphologies (Supplementary Fig. 4b), we found that Pseudomonas dominated in cocultures with (99.7%) or without (99.9%) CTX treatment (Fig. 1d). Distributing the biomass in the mixed colonies with respect to these relative abundances, we found that Klebsiella always suffered from the mixed inoculation, while Pseudomonas benefited from it under the CTX treatment (Supplementary Fig. 4a, compare the colored bars with and without a gray frame).
To facilitate the analysis of the colony composition in a spatially resolved manner, we used fluorescence proteins to label the two species (Pseudomonas-YFP and Klebsiella-mCherry). In a coculture consisting of these two labeled strains, we observed that Klebsiella was mostly confined at the seeding position, whereas the expanded colony front largely consisted of Pseudomonas (Supplementary Fig. 6).
Collectively, our results indicate that the CTX treatment suppressed Pseudomonas’ spatial expansion, which was rescued by Klebsiella in a co-culture (Fig. 1c,d; Supplementary Fig. 7). In turn, the activated expansion of Pseudomonas suppressed the growth of Klebsiella (Fig. 1d; Supplementary Fig. 4a), which could be explained by the exclusive spatial expansion ability of Pseudomonas (Fig. 1c) and its greater tolerance to CTX than Klebsiella (Fig. 1a).
The rescuing by Klebsiella was likely due to its ability to produce Bla, which degrades CTX30 (Supplementary Table 1). To test this notion, we collected supernatants from the Klebsiella pure cultures at ~6.3 h or media without cell inoculation as unconditioned control. After filter sterilizing the supernatants, we tested whether CTX-sensitive Escherichia coli could grow in them (Supplementary Fig. 8a, top). Following 21 h of incubation, E. coli exhibited detectable growth in the Klebsiella-supernatant but not in the unconditioned control, consistent with the CTX degradation by Klebsiella-produced Bla30 (Supplementary Fig. 8a, bottom; see Supplementary Fig. 8b for Klebsiella growth curves with the initial cell density of OD600 = ~5×10−4, which was used for this experiment).
CTX suppressed Pseudomonas’ spatial expansion in a dose-dependent manner
We next measured the suppression of Pseudomonas spatial expansion (in terms of colonized area) using varied initial doses of CTX (0 to 2.5 μg ml−1) (Supplementary Fig. 9). The suppression increased with the initial CTX dose (A0), approximately following a Hill inhibition function.
Pseudomonas growth was virtually unaffected by CTX (2.5 μg ml−1) treatment (Fig. 1a). Thus, the suppression in Pseudomonas’ spatial expansion (Fig. 1c; Supplementary Fig. 9) was likely due to the loss of motility and correlated with filamentous cell morphology (Supplementary Fig. 10), consistent with previous reports associating β-lactam-induced cell filamentation with reduced motility40,41. The extent of the filamentation of Pseudomonas cells was reduced when inoculated in a mixture with Klebsiella (Supplementary Fig. 10), which restored Pseudomonas’ spatial expansion (Fig. 1d). A similar filamentation of Pseudomonas was observed in well-mixed liquid cultures under CTX (2.5 μg ml−1) treatment, with or without Klebsiella (Supplementary Fig. 11). The spatial expansion of Pseudomonas was eventually stopped by the antibiotic concentration far (1.5–2 cm) away from the inoculation point. This is because Klebsiella did not move along with Pseudomonas (Supplementary Fig. 6) and, hence, could no longer reduce the antibiotics to an ineffective concentration. Likewise, actively swarming Pseudomonas still needed Klebsiella for swarming when inoculated on fresh antibiotic media at ~4times greater cell density than the original inoculum (Supplementary Fig. 12).
Theory reveals key determinants of the facilitation of spatial expansion
We conducted a theoretical analysis to gain insight into the Klebsiella-mediated collective range expansion during antibiotic treatment. We assumed that the Klebsiella population leads to an antibiotic sink and the growth media serves as a source for antibiotics (Fig. 2a, top)42. The terms sink and source were inspired by the infinite source and point sink reaction-diffusion framework we adapted below. We described the dependence of the effective spatial expansion ability (M) of Pseudomonas on the antibiotic concentration using the experimentally determined Hill function (Supplementary Fig. 9). Solving the system assuming cylindrical symmetry (see Supplementary Notes for details), we have:
where R > 0 is the boundary of the sink created by the Klebsiella population, S ≥ R is the domain boundary, \(d\in \left[R,\,S\right]\) is the distance of Pseudomonas from the sink boundary, and \({a}_{0}\in \left[0\,,\,\infty \right]\) is the initial antibiotic concentration normalized by its value for the half-maximum of Pseudomonas’ effective spatial expansion ability (\({a}_{0}=\frac{{A}_{0}}{{A}_{C}}\); see Supplementary Fig. 9 caption for AC and h). Here, the sink boundary R defines the radius of a circular clear zone within which the antibiotic concentration is an ineffective lower boundary value. For the latter, we considered zero for simplicity, though our theoretical framework can be applied to any arbitrary value. That is, each point on the clear zone boundary acts as a point sink for the antibiotics. Note that we expect the clear zone radius, R, to have two contributions: it scales with the physical size of the Klebsiella inoculum and has an additional term that increases nearly linearly with the characteristic diffusion length scale of Bla. This additional contribution is anticipated to depend only weakly on the Klebsiella seeding cell density (see Supplementary Notes, Supplementary Eq. (7) for details).
Source data are provided as a Source Data file. a Theoretical model: Top: Klebsiella–antibiotic interaction. Antibiotic concentration (\(a\)) forms a steady-state gradient, assuming a population of facilitator Klebsiella as an antibiotic sink, creating a clear zone with radius R, while growth media with a domain size of S acts as an infinite antibiotic source. \(a\) decreases near the sink, converging to an ineffective minimum (zero for simplicity; Supplementary Notes). Bottom: Klebsiella-antibiotic-Pseudomonas interactions. Equation (1) predicts that Pseudomonas’ effective spatial expansion ability (M) depends on its distance to sink boundary (d) and initial antibiotic concentration (\({a}_{0}\)). h = 6.642 (Supplementary Fig. 9). S/R = 10 was assumed. b Experiments corroborated the dependence of effective spatial expansion ability of Pseudomonas on its distance to Klebsiella. The experiment from Fig. 1d was repeated with 1 μl of Pseudomonas (left) and 1 μl of Klebsiella (right) inoculated side by side at varying distances (d, gold, in mm). Dishes were imaged at 23 h. Colonized areas were measured using a custom Python script and manually corrected with ImageJ if needed. Data were fitted to a Hill inhibition function (solid curve), yielding a Hill coefficient of 7.62 and a spatial scale of the facilitation of expansion (\({L}_{K}\): the distance at which colonized area is half-maximum) of 6.90 mm (\({R}^{2}\) = 0.990). Inoculum diameter: ~3 mm, dish diameter: 100 mm. Images shown were equally adjusted for brightness/contrast. Raw data from n = 7 independent replicates are in Supplementary Fig. 13. Supplementary Fig. 14 shows an experiment with fluorescently labeled strains. Half-maximum suppressing cefotaxime (CTX) dose: 1.29 μg ml−1 (Supplementary Fig. 9). c A coarse-grained kinetic model recapitulated the experimental range expansion patterns. Results from coarse-grained simulations at 48 h. Left: Black and white indicate absence and presence of bacteria, respectively. Right: Colonized area was calculated using a custom MATLAB script and plotted against d (gold, in mm). The data were fitted to a Hill inhibition function (solid curve), with a Hill coefficient of 92.4 and \({L}_{K}\) = 13.9 mm (\({R}^{2}\) = 0.999). Inoculum diameter: 10 mm. Supplementary Fig. 15 shows data at additional time points. Supplementary Table 2 lists model parameters and initial conditions.
For a constant d, M decreases from 1 and approaches 0 with increasing a0. For a given a0, M decreases with increasing d (Fig. 2a, bottom). For a sufficiently large a0, Pseudomonas’ effective spatial expansion ability is sharply activated when d < LK, where LK denotes the distance at which M is half-maximum. Thus, LK is a measure of the spatial scale of facilitation of spatial expansion. Critically, LK can be substantially larger than R, indicating a long-range interaction (Fig. 2a, bottom, LK = ~ 3.2 R for \({a}_{0}\) = 2).
Experiments corroborated the theoretical predictions
To test Eq. (1), we inoculated one Klebsiella population and one Pseudomonas population at varied distance (d) under CTX (2.5 μg ml−1) treatment. Then, we quantified the range expansion as the colonized areas at 23 h, observing a sharp change at a certain distance value (Fig. 2b), consistent with the prediction of the spatial scale of the facilitation of spatial expansion (LK) above. We demonstrated the robustness of the existence of \({L}_{K}\), repeating the same experiment, also with varied Klebsiella seeding cell density (Supplementary Fig. 13). The range expansion had a bias towards the Klebsiella population, which we confirmed by repeating this experiment with the fluorescently labeled strains (Supplementary Fig. 14).
A coarse-grained kinetic model recapitulated the experimental range expansion patterns
The spatial patterns of the range expansion (Fig. 2b) were not considered in our theoretical estimation of M and LK above. The spatial patterns are a function of not only the antibiotic gradient and the Pseudomonas’ spatial expansion ability—which the antibiotic gradient determines—but also the spatiotemporal dynamics of species and nutrient abundances. Thus, we next accounted for these factors and built a coarse-grained kinetic model, enabling a more constrained simulation of spatial expansion dynamics (Methods and Supplementary Table 2).
This model builds upon our previous models for Pseudomonas branching43 and Bla-mediated population dynamics in response to β-lactam treatment30 (see Methods). We simulated the range-expansion dynamics with varied seeding distance (d) between one Klebsiella population and one Pseudomonas population. Again, we quantified the total colonized area (at 48 h; see Supplementary Fig. 15 for different time points) and found a sharp onset of range expansion as d decreased below a critical value, consistent with the existence of LK (Fig. 2c). Simulations mimicking an intracellular degradation of antibiotics by an immotile species (zero dispersal for the antibiotic-degrading enzyme and the species that produces it) yielded similar results, suggesting that the antibiotic sink role could be fulfilled without released public goods (Supplementary Fig. 16).
The facilitation of spatial expansion occurred in a pairwise coculture of hospital sink co-isolates, Bacillus paranthracis and Pseudomonas aeruginosa
Colonization of hospital sink drains (e.g., p-traps) has been associated with pathogens such as K. pneumoniae44 and P. aeruginosa45. Moreover, biofilms formed in the p-traps of connected sinks and expanded upwards 2.5 cm day-1, possibly due to surface-associated bacterial motility46. If sufficiently close to the sink strainer, running sink water can lead to the spread of bacteria to the environment46, posing a health risk.
We next tested whether the facilitation of spatial expansion presented above could occur in communities of bacteria isolated from the same sink p-trap at Duke University Hospital. Two isolates, 192621 and 192622, were identified by 16S rDNA sequencing as Bacillus paranthracis and P. aeruginosa, respectively.
We first tested the effect of CB treatment on the growth of the two isolates. Specifically, we grew B. paranthracis 192621 (Bp) and P. aeruginosa 192622 (Pa22) in pure liquid cultures without or with CB (10 μg ml−1) (Fig. 3a). Both species tolerated the CB treatment, but Pa22 grew faster than Bp under both conditions. When grown as a well-mixed pure culture, each species produced less biomass in medium conditioned by the other species than in unconditioned medium, demonstrating competition between the two species (Supplementary Fig. 17). If the two species primarily interact through competition in well-mixed cocultures, we expect the dominance of Pa22 under both conditions. To this end, we tested the growth of mix cultures, each starting with equal fractions of Bp and Pa22, with or without CB treatment. Indeed, Pa22 dominated after 27 h for both cases, where Bp was undetectable (<0.1%) (Fig. 3b).
Bp: Bacillus paranthracis, Pa22: Pseudomonas aeruginosa 192622, CB: Carbenicillin. Source data are provided as a Source Data file. a Pa22 tolerated CB treatment better than Bp. Liquid media were inoculated with Bp (red) or Pa22 (light blue) at OD600 = ~2.1×10−5 (from dilutions of a cell suspension with OD600 > 0.05), without (top) or with CB (10 μg ml−1, bottom). Curves and shades show average and standard deviation, respectively, from n = 2 biological replicates, each with three technical replicates. b Pa22 dominated Bp in well-mixed cocultures. Liquid media were inoculated with a 1:1 mixture of the two species at OD600 = ~2.1×10−5, incubated for 27 h, and then plated to determine relative abundances (Supplementary Fig. 21b). Bars and error bars represent mean and standard deviation from n = 2 independent experiments (symbols) after averaging technical replicates (solid/open/lined markers). c Pa22’s range expansion was suppressed by CB treatment and rescued by Bp. 20 ml of swarming agar (0.55%) media were inoculated with 1 μl of Bp or Pa22 at OD600 = ~0.43 with or without CB (10 μg ml−1). Dishes were imaged at 24 h, processed, and analyzed for colony areas using a custom Python script (Methods). Images shown were equally adjusted for brightness/contrast. Dish diameter: 100 mm. Raw images from n = 2 independent replicates are in Supplementary Fig. 19. d The rescue of the coculture range expansion occurred at the expense of Bp’s growth. In the same experiments as in (c), media were center inoculated with 1 μl of 1:1 mixture of the two species at OD600 = ~0.43 (circles) or ~0.34 (triangles). The dishes were imaged after 24 h (see Supplementary Fig. 19 for raw images). Then, whole colonies were harvested with 5 ml of saline, and relative abundances were estimated by plating (Supplementary Fig. 21b). Bars and error bars show mean and standard deviation of n = 2 independent experiments (symbols) after averaging technical replicates (solid/open markers). Images shown were equally adjusted for brightness/contrast. Dish diameter: 100 mm.
When tested on agar, however, only a mixture of Bp and Pa22 exhibited spatial expansion after 24 h under treatment by CB (10 μg ml−1; ~2.4-times the concentration for the half-maximum suppression of the spatial expansion of Pa22 when alone, Supplementary Fig. 18) (Fig. 3c, d, Supplementary Fig. 19). The mixture’s spatial expansion under the CB treatment was maintained at 30 °C but not at 40 °C (Supplementary Fig. 20), indicating the context-dependence of mechanism. While mixing Pa22 with Bp promoted biomass production under the CB treatment by 4-fold, Bp itself became undetectable (<0.1%) in the final population (Fig. 3d; see Supplementary Fig. 21 for biomass data). Supernatants from CB-treated pure cultures of Bp allowed the growth of CB-sensitive E. coli (Supplementary Fig. 22), indicating the degradation of CB in the environment by Bp. These results suggest that Bp facilitated Pa22 only during community spatial expansion by degrading the antibiotics in the environment.
The principles we uncovered from the pairwise communities above (Figs. 1–3) are likely generalizable: When we used a variant of the CB-sensitive E. coli, which was engineered to express Bla conferring CB resistance47,48, it also rescued Pseudomonas swarming from CB inhibition (Supplementary Fig. 23).
The facilitation of spatial expansion occurred in a synthetic community of sink isolates
Pairwise communities above enabled us to investigate mechanistic underpinnings of the facilitation that enables collective range expansion under antibiotic treatment. To evaluate the generalizability of the mechanism, we next tested if Bp’s facilitation of Pa22’s spatial expansion could occur in the presence of other members of sink isolates. To this end, we assembled an eight-member community, which we termed SynkC, by introducing six additional isolates from the same sink sample from which we isolated both Bp and Pa22 (Table 1). SynkC consists of six distinct taxonomic groups based on 16S rDNA-based microbiome sequencing. Seven out of the eight SynkC members are genotypically and/or phenotypically distinct (Supplementary Fig. 24). In addition to Pa22, only one other SynkC member showed spatial expansion ability on its own (Supplementary Fig. 24a, Bacillus sp. ESY92). When tested alone, two SynkC members did not tolerate the CB treatment, and the others exhibited various degrees of tolerance (Supplementary Fig. 24b).
Whole-genome sequencing of the SynkC members revealed that each member had at least one Bla gene. Bp had the most (17) Bla genes followed by Pa22 (7), while Bp was the only isolate carrying a class A Bla, according to Ambler’s classification scheme, based on amino acid sequence similarities of known Bla proteins (Supplementary Table 1). These sequencing results suggest a greater or a unique potential of Bp to degrade certain β-lactam antibiotics. Accordingly, in pairwise cultures, Bp was the only one that rescued the Pa22 spatial expansion from the CB treatment inhibition (Supplementary Fig. 25).
We ran agar experiments with SynkC with equal starting biomasses of all the eight members or by leaving out Bp (SynkC–Bp). Again, spatial expansion under the CB treatment occurred only when Bp was included (Fig. 4, images), though Bp was not detected in the final population of SynkC (Fig. 4, pie charts). In addition to promoting the spatial expansion and ~2.7-fold greater total biomass, the initial presence of Bp also increased community diversity under the CB treatment. Specifically, while only Pa22 was detected at the end of the SynkC–Bp experiment, both Pa22 and Enterobacter sp. were detected in the final SynkC community (Fig. 4, pie charts). This suggests that Enterobacter sp. should also be considered when analyzing the precise spatial distribution of bacteria. However, Enterobacter sp. cannot spread alone (Supplementary Fig. 24a), so any spatial expansion of Enterobacter sp. must depend on other species such as Pa22.
Bp: Bacillus paranthracis, Pa22: Pseudomonas aeruginosa 192622, CB: Carbenicillin. Source data are provided as a Source Data file. The agar (0.55%) surface was inoculated with 1 μl of a mixture containing equal biomasses of all the eight members (SynkC) or all but Bp (SynkC–Bp) at OD600 = ~0.34, with or without CB (10 μg ml−1). Images of the colonies were captured at 24 h. Then, whole colonies were harvested by flooding with ~5 ml of saline. The biomasses of the cell harvests were calculated by measuring the OD600 of each harvest suspension and multiplying it by the suspension volume. Unused inoculants and cell harvest suspensions were stored as frozen glycerol stocks and underwent 16S microbiome sequencing to determine the initial and final relative abundances of the community members (a 0.5% cut-off value applied). Each of the pie charts above shows six groups, with the Enterobacter sp. and Bacillus sp. groups representing the combined abundance of two isolates each, while all other groups represent a single isolate. For the final time point, relative areas of the pie charts represent the distribution of the respective biomasses measured. n = 2 independent experiments, each with technical replicates, yielded similar results. A representative set of data was shown here, with images equally processed for brightness/contrast. Dish diameter: 100 mm. See Supplementary Fig. 26 for the raw data from all replicates. Phenotypes of the individual members are in Supplementary Fig. 24.
Discussion
Our study provides a concrete demonstration that context—specifically spatial structure and chemical stress—can reveal a species’ facilitator role. By degrading β-lactam antibiotics, an immotile Bla-producing species (K. pneumoniae or B. paranthracis) creates a clear zone that restores the motility-based range expansion of a more resilient partner (P. aeruginosa), if sufficiently close. However, this facilitation is at the cost of the facilitators themselves—driving them to an extreme minority population, apparently through competitive exclusion by P. aeruginosa. Although the drug-degrading facilitator eventually becomes a minority due to competition, its transient facilitation is essential for spatial expansion and overall community success. This mechanism could contribute to the observed rapid biofilm formation in sink-associated microbial communities46.
Two contextual factors are critical for our system. One is spatial expansion, as mobility only matters in a spatially structured environment. This context-dependency is reminiscent of a recent example in which we show that the spatial expansion of Pseudomonas not only makes a cooperative trait essential27 but also allows non-cooperating variants (so-called cheaters) to leverage their growth advantage from the lack of cooperative trait expression, enabling them to outcompete the cooperating individuals28. The other contextual factor is antibiotic stress, which primarily suppresses mobility instead of the growth of Pseudomonas. Together, these factors shift the net effect of the drug-degrading bacteria from competitive to facilitative. This shift aligns with the stress gradient hypothesis, which suggests that positive interactions dominate negative interactions in more stressful environments49,50,51,52.
The principles uncovered here are likely generalizable. Other species or consortia could similarly modify their environment—through pH shifts12, enzyme secretion53,54, nutrient release/break-down11,53,54, or other chemical changes—to unlock traits like motility55,56, biofilm growth57,58,59, or nutrient utilization (i.e., necrotrophy53) relevant only in spatially structured habitats. Such facilitation may be common in natural and engineered microbial communities but remain untapped due to a lack of spatially explicit experimental inquiry.
Clinically, consistent with the notion of private versus public benefits of Bla producers in bacterial communities60, our work suggests that certain Bla producers may play a crucial, even if transient, role in facilitating the spread of more virulent pathogens, such as Pseudomonas, even if they remain undetected in standard assays. Current microbiome analyses often exclude low-abundance taxa61, potentially missing these transient facilitators and underestimating their influence on infection dynamics.
In conclusion, recognizing the context-dependent and transient facilitative interactions enriches our understanding of microbial ecology, offers insights for controlling infections, and opens new avenues for studying hidden facilitators in complex microbial ecosystems.
Methods
Bacterial growth media and procedures
In every experiment, bacteria were first streaked on a lysogeny broth (LB) agar plate from their frozen glycerol stock, and a single colony was incubated overnight in liquid LB media at 37 °C and 225 r.p.m. The overnight cultures were then centrifuged at 1150 g for 5 min, and the resulting cell pellets were resuspended in phosphate-buffered saline (PBS) supplemented with 8 g l-1 of casamino acids before using in the experiment.
For all experiments, PBS (2.4 g l−1 Na2HPO4 anhydrous, 3 g l−1 KH2PO4 anhydrous, 0.5 g l−1 NaCl, 1 mM MgSO4, and 0.1 mM CaCl2) supplemented with casamino acids (8 g l−1, unless otherwise noted) was used. Agar (final concentration of 0.55%) was used when applicable.
The media was prepared following a recipe adapted from Xavier et al.62: To make 1 liter of 5× phosphate buffer stock solution, 12 g Na2HPO4 anhydrous, 15 g KH2PO4 anhydrous, and 2.5 g NaCl were dissolved in deionized water and sterilized by autoclaving. The casamino acids stock solutions were made at 20% (w/v) by microwaving 40 g casamino acids (Gibco™ Bacto™ 223120) in deionized water, sterilized by filtering (0.22 μm) and stored at 4 °C. The main solutions of agar were made at 1.25% (w/v) on the day of the experiment dissolving granulated agar (BD Difco™ 214530) in deionized water and sterilizing by autoclaving. Then, appropriate amounts were immediately used to prepare the experimental media.
In the experiments and overnight cultures with the Pseudomonas-YFP and Klebsiella-mCherry variants, gentamicin (15 μg ml−1) was added for the maintenance of the plasmids. Anhydrotetracycline (aTc; 100 ng ml−1) was also added for inducing the mCherry expression. All antibiotic solutions used in the study were made fresh on the day of the experiment. The cultures with aTc were protected from light.
All OD600 measurements were taken using 200 μl samples and a plate reader (Tecan Infinite 200 with its i-control software 3.9.1.0).
Well-mixed liquid cultures
We grew 2–3 ml cells in 16 ml culture tubes at 37 °C and 225 r.p.m.
Growth curve experiments
We measured the growth curves of bacteria using a plate reader (Tecan Infinite 200 with its i-control software 3.9.1.0). In each well of a 96-well plate, we added 195 μl liquid media and 5 μl cell inoculum culture (prepared by centrifuging overnight-grown cells at 1150 g for 5 min and then resuspending in the liquid medium). To prevent evaporation, we used Nunc Edge multi-well plates (Thermo Scientific) with built-in water reservoirs. In addition to the built-in water reservoirs, we also reserved all the edge wells for water only. The cells were then incubated in the plate reader at 37 °C, and OD600 measurements were taken at 10 min intervals after the plate being shaken rigorously. For technical replicates of a particular condition, cells from the same inoculum were inoculated in different wells of a 96-well plate and measured. Background signals measured from media containing no cells were subtracted from the data. Custom MATLAB scripts were used for data analysis, during which a four time point moving average was applied to the raw OD600 time series data.
Agar media experiments
20 ml growth media was solidified in 100-mm petri dishes. Overnight-grown cells were centrifuged at 1150 g for 5 min and then resuspended in the liquid medium to prepare a cell inoculum culture. The seeding droplet volume from cell inoculum cultures was 1 μl. The seeded plates were incubated at 37 °C, unless otherwise noted, agar side up.
CTX/CB environmental degradation assays
The protocol used was adapted with minimal changes from our previous study30: 3 ml liquid media with/without the facilitator bacteria (Klebsiella or Bp) inoculated (initial cell density of OD600 = ~5×10−4) and with/without the antibiotic (2.5 μg ml−1 CTX or 10 μg ml−1 CB) addition were incubated at 37 °C and 225 r.p.m. At ~6.3 h, 1 ml samples from the media were removed, in which clavulanic acid (5 μg ml−1 final concentration) was then added to prevent any further Bla activity. Next, supernatants were prepared by filtration of these 1 ml samples using 0.22 μm cellulose acetate filters (VWR). Finally, the supernatants were inoculated with E. coli MC4100Z1 at initial cell density of OD600 = ~5×10−4, at which the E. coli is sensitive to the antibiotic treatments here. Then, OD600 measurements were taken after 21 h incubation at 37 °C at 225 r.p.m.
Spent media assays
Similar to the CTX/CB environmental degradation assays above, 3 ml liquid culture media were incubated for ~6.3 h at 37 °C and 225 r.p.m. with no inoculant (for an unconditioned control media) or with Klebsiella or Pseudomonas (or with Bp or Pa22) at initial cell densities of OD600 = ~5×10−4. Then, supernatants were collected by filtration using 0.22 μm cellulose acetate filters (VWR). Within each pairwise system, the filtered supernatants of each species were cross-inoculated with the other species (OD600 = ~5×10−4). Subsequently, growth curve measurements were performed in a plate reader at 37 °C, as described above.
Relative abundance measurements based on colony forming unit (CFU) morphology
As shown in Supplementary Fig. 4b and Supplementary Fig. 21b, CFUs were grown on LB agar (1.5%) for 20 h at 37 °C to determine the relative abundance of the constituent species in pairwise mixed cultures. To do so, cell harvests were serially diluted in sterile saline (11.6 g l−1 NaCl) solution down to ~100 CFUs per 100 μl spread on a 100-mm LB agar plate.
Bacterial strains
P. aeruginosa PA14 (Pseudomonas), a clinical K. pneumoniae isolate D-005 from Deverick J. Anderson’s laboratory (Klebsiella), E. coli MC4100Z1, E. coli MC4100Z1 with a plasmid that encodes an engineered cytoplasmic β-lactamase (E. coli Bla+)47, and the hospital sink p-trap isolates described below (Table 1) were used. For some experiments variant strains that expressed spectrally distinct fluorescent proteins from a plasmid, Pseudomonas-YFP28 and Klebsiella-mCherry (this study, see Methods and Supplementary Table 3), were used.
Construction of the Klebsiella-mCherry strain
Strain and transformation
Zymo Mix & Go Transformation kit (Zymo Research) was used to make K. pneumoniae D-005 chemically competent with one modification: the overnight culture of D-005 was re-grown in super optimal broth (SOB) supplemented by 0.7 mM (final concentration) ethylenediaminetetraacetic acid (EDTA)63.
Plasmid construction
ptetmCherryGMR was constructed from the parent plasmids ptetmCherry64, pUCP30T-eCFP plasmid65, and pJ1996_v266. The ptetmCherry plasmid without the chloramphenicol resistance gene, the gentamicin resistance gene, and a DNA fragment containing two transcription terminators were PCR amplified from the ptetmCherry, pUCP30T-eCFP, and pJ1996_v2 plasmids, respectively, by DNA primers with overhangs. The resulting linear DNA fragments were digested by DpnI and subsequently column purified with DNA Clean & Concentor-5 (Zymo Research). Then, they were ligated together with NEBuilder HiFi DNA Assembly Master Mix (NEB #E2621) to yield the final plasmid. The final plasmid was transformed into Top10F’ to propagate, miniprepared with Zymo Plasmid Miniprep-Classic kit (Zymo Research), and sequenced by Sanger sequencing (Genewiz/Azenta Life Sciences) to validate. All the DNA primers were purchased from Integrated DNA Technologies and listed in Supplementary Table 3.
SynkC: Isolation of bacteria from the sink p-traps and their taxonomic identification
The members of SynkC (Table 1) were isolated by propagations of a sample collected from a sink p-trap in an intensive care unit in a recently constructed bedtower at the Duke University Hospital. Sterile tubing was fed down into the p-trap and a 50 ml syringe was used to agitate the fluid to collect biofilm portions and then withdraw a sample67. 50 μl of p-trap water was used to inoculate 950 μl of tryptic soy broth in a 2 ml deep well plate (Thermo Scientific) and grown for 16 h at 37 °C with shaking at 700 r.p.m. The resulting culture was glycerol stocked (25% glycerol) and frozen. In parallel, it was serially diluted to 10−6 in phosphate buffered saline and 10 μl of dilutions from 10−3 to 10−6 were plated on tryptic soy agar (TSA) with and without 100 μg ml−1 carbenicillin. The plates were incubated at 37 °C for 10 h and at room temperature for 66 h.
Pa22 (ESY55) and Bp (ESY57) were isolated via restreaking different parts of the resulting colonies on fresh agar plates and incubating for 18 h at 37 °C. A new set of overnight cultures were then made by inoculating 2 ml of tryptic soy broth with individual colonies on the restreaked plates and incubating for 16 h at 37 °C. These cultures were then glycerol stocked as the eventual Pa22 and Bp. Subsequent colonies from these stocks were sent for 16S rDNA sequencing at Azenta Life Sciences for species identification.
ESY66 and ESY73 were isolated as follows: First, the glycerol stock of the original overnight culture described above was inoculated in LB and then grown for ~8 h at 37 °C with shaking at 225 r.p.m. The resulting culture was glycerol stocked (25% final concentration of glycerol) and frozen. This stock is called the ‘master stock’ hereafter. The master stock was diluted ~104-fold in 11.6 g l−1 saline, and 100 μl of the dilution was plated on TSA plates. The plates were incubated at 37 °C for ~20 h. Individual colonies were restreaked on LB agar plates, and the plates were incubated at 37 °C for ~20 h, again. Then, overnight cultures were grown by inoculating 2 ml of LB with individual colonies from the restreaked plates and incubating for ~18 h at 37 °C with shaking at 225 r.p.m. These cultures were then glycerol (25%) stocked as the eventual isolates used in this study. Individual colonies from these stocks were sent for 16S rDNA sequencing at Azenta Life Sciences for species identification.
ESY80, ESY83, ESY92, and ESY93 were isolated as follows: First, ~1 μl from the master stock was inoculated on swarming agar media and incubated for ~24 h at 37 °C. The final colony was suspended in 11.6 g l−1 saline and then glycerol (25%) stocked and frozen. This new frozen stock was diluted ~105-fold in 11.6 g l−1 saline, and 100 μl of the dilution was plated on TSA plates. The plates were incubated at 37 °C for ~20 h. Individual colonies were restreaked on LB agar plates, and the plates were incubated at 37 °C for ~20 h, again. Then, overnight cultures were grown by inoculating 2 ml of LB with individual colonies from the restreaked plates and incubating for ~18 h at 37 °C with shaking at 225 r.p.m. These cultures were then glycerol (25%) stocked as the eventual isolates used in this study. Individual colonies from these stocks were sent for 16S rDNA sequencing at Azenta Life Sciences for species identification.
16S microbiome sequencing
16S V3/V4 region sequencing (20,000 reads) service of SeqCenter, LLC was used. Aliquots from cell harvests were supplemented by glycerol (25% final concentration) before being frozen at −80 °C. The frozen aliquots were shipped overnight to the company’s facility on dry ice. The following protocols were provided by SeqCenter, LLC along with the corresponding sequencing results.
DNA extraction
All standard DNA extractions at SeqCenter follow the ZymoBIOMICS™ DNA Miniprep Kit. 20−200 μl from the samples submitted were transferred into 550 μl of lysis solution. Cells suspended in lysis solution were transferred into the ZR BashingBead™ Lysis Tubes and mechanically lysed using the MP FastPrep-24™ lysis system with 1 min of lysis at maximum speed and 5 min of rest for 5 cycles. Samples were then centrifuged at 10,000 g for 1 min. 400 μl of supernatant was transferred from the ZR BashingBead™ Lysis Tube to a Zymo-Spin™ III-F Filter and centrifuged at 8000 g for 1 min. 1200 μl of ZymoBIOMICS™ DNA Binding Buffer was added to the effluent and mixed via pipetting. 800 μl of this solution was transferred to a Zymo-Spin™ IICR Column and centrifuged at 10,000 g for 1 min. This step was repeated until all material was loaded onto the Zymo-Spin™ IICR Column. DNA bound to the Zymo-Spin™ IICR Column was washed 3 times with 400 μl and 700 μl of ZymoBIOMICS™ DNA Wash Buffer 1 and then 200 μl of ZymoBIOMICS™ DNA Wash Buffer 2 with a 1-min spin down at 10,000 g for each, respectively. Washed DNA was eluted using 75 μl of ZymoBIOMICS™ DNase/RNase Free Water following a 5-min incubation at room temperature and a 1-min spin down at 10,000 g. The Zymo-Spin™ III-HRC Filter was prepared using 600 μl of the ZymoBIOMICS™ HRC Prep Solution and a centrifugation at 8000 g for 3 min. Eluted DNA was then purified by running the effluent through the prepared Zymo-Spin™ III-HRC Filter. Final DNA concentrations were determined via Qubit.
Sequencing
Samples were prepared using Zymo Research’s Quick-16S kit with phased primers targeting the V3/V4 regions of the 16S gene. The specific primer sequences are listed in Table 2. Following clean up and normalization, samples were sequenced on a P1 600cyc NextSeq2000 Flowcell to generate 2×301 bp paired end (PE) reads. Quality control and adapter trimming was performed with bcl-convert (v4.2.4).
Analysis
Sequences were imported to Qiime268 for analysis. Primer sequences were removed using Qiime2’s cutadapt69 plugin using the degenerate primer queries listed in Table 3.
Sequences were then denoised using Qiime2’s dada2 plugin70. Denoised sequences were assigned operational taxonomic units (OTUs) using the Silva 138 99% OTUs full-length sequence database and the VSEARCH71 utility within Qiime2’s feature-classifier plugin. OTUs were then collapsed to their lowest taxonomic units, and their counts were converted to reflect their relative frequency within a sample.
Note that, for SynkC, neither the SeqCenter microbiome sequencing nor the Azenta Life Sciences 16S sequencing-based species identification pipelines could distinguish the members within the Enterobacter sp. and Bacillus sp. taxonomic groups. The identities of Pa22, Bp, Citrobacter murliniae, and Enterobacter wuhouensis were better resolved to species level from the best fits of the Azenta Life Sciences pipeline, so we used those identities. We confirmed that these four isolates did not fall into either the Enterobacter sp. or the Bacillus sp. taxonomic groups in the SeqCenter pipeline, which was used to generate the relative abundance data in Fig. 4 and Supplementary Fig. 26.
Whole-genome sequencing
Whole-genome sequencing service of SeqCenter, LLC was used. The strains were streaked on LB agar plates from their frozen stocks. After overnight incubation at 37 °C, the plates were shipped overnight to the company’s facility. The following protocols were provided by SeqCenter, LLC along with the corresponding sequencing results.
DNA extraction
DNA extractions were performed by SeqCenter, LLC following the ZymoBIOMICS™ DNA Miniprep Kit: A loopful of cells (~50–100 mg) were aseptically scraped from the submitted agar plates and resuspended in 750 μl of lysis solution.
Cells suspended in lysis solution were transferred into the ZR BashingBead™ Lysis Tubes and mechanically lysed using the MP FastPrep-24™ lysis system with 1 min of lysis at maximum speed and 3 min of rest for 2 cycles. Samples were then centrifuged at 10,000 g for 1 min. 400 μl of supernatant was transferred from the ZR BashingBead™ Lysis Tube to a Zymo-Spin™ III-F Filter and centrifuged at 8000 g for 1 min. 1200 μl of ZymoBIOMICS™ DNA Binding Buffer was added to the effluent and mixed via pipetting. 800 μl of this solution was transferred to a Zymo-Spin™ IICR Column and centrifuged at 10,000 g for 1 min. This step was repeated until all material was loaded onto the Zymo-Spin™ IICR Column.
DNA bound to the Zymo-Spin™ IICR Column was washed 3 times with 400 μl and 700 μl of ZymoBIOMICS™ DNA Wash Buffer 1 and then 200 μl of ZymoBIOMICS™ DNA Wash Buffer 2 with a 1-min spin down at 10,000 g for each, respectively. Washed DNA was eluted using 75 μl of ZymoBIOMICS™ DNase/RNase Free Water following a 5-min incubation at room temperature and a 1-min spin down at 10,000 g. The Zymo-Spin™ III-HRC Filter was prepared using 600 μl of the ZymoBIOMICS™ HRC Prep Solution and a centrifugation at 8000 g for 3 min. Eluted DNA was then purified by running the effluent through the prepared Zymo-Spin™ III-HRC Filter. Final DNA concentrations were determined via Qubit.
Illumina sequencing
Illumina sequencing libraries were prepared using the tagmentation-based and PCR-based Illumina DNA Prep kit and custom Integrated DNA Technologies 10 bp unique dual indices with a target insert size of 280 bp. No additional DNA fragmentation or size selection steps were performed. Illumina sequencing was performed on an Illumina NovaSeq X Plus sequencer in one or more multiplexed shared-flow-cell runs, producing 2×151 bp paired-end reads. Demultiplexing, quality control and adapter trimming was performed with bcl-convert (a proprietary Illumina software for the conversion of bcl files to basecalls, v4.2.4).
Illumina short-read assembly and annotation
Short read assembly was performed with Unicycler (0.5.0, default parameters)72. Assembly statistics were recorded with QUAST (5.2.0, default parameters)73. Samples were annotated with Bakta (1.8.1, default parameters; db version 5.0)74.
Imaging
Bacterial colonies on agar plates were imaged with a UVP Colony Doc-It Imaging Station (using its original software 7.0.3) with epi white light, using the same brightness and contrast settings for all images. The same z-plane focus was applied to all images within the same independent experiment.
Microscopic imaging was performed using a Keyence BZ-X710 microscope and Keyence BZX Software Suite 1.3.1.1. To image a whole swarming colony, its entire plate was scanned and imaged, and the resulting images were stitched using the built-in algorithm of the software.
Image analysis
A custom Python script was used for processing and analyzing the colonized area for bacterial colony images. This script firsts finds and removes the plate boundaries. Then, the bacterial colony (or colonies when applicable) is automatically segmented using a user-defined threshold intensity value (The script can also generate a segmented image on white background for display purposes). When applicable, the script calculated the seeding distance between different species as the Euclidean distance between two positions, defined by the user based on the distinct appearance of the inoculum zones. For each case, the segmentation results were manually validated and corrected using Image J manual tools when necessary.
Custom MATLAB scripts were used for the coarse-grained kinetic model simulations and the corresponding analyses.
Coarse-grained kinetic model
Pseudomonas cell density time change is given by Eq. (2):
where \(P\) is Pseudomonas cell density (cell density unit: cu), \({\alpha }_{P}\) is the growth rate constant of Pseudomonas (h−1), \(\Omega\) indicates the entire colony domain, and ∫Ω indicates an integral over the entire colony domain. \({G}_{P}\) is the Pseudomonas growth function as defined by Eq. (3):
where N is the local nutrient (casamino acids) concentration (g l−1), KN is the local nutrient concentration for half-maximum growth rate (g l−1), \({C}_{m}\) is the local cell density for the half-maximum suppression of the growth rate (cu), P is the local Pseudomonas cell density (cu), E is the local Klebsiella cell density (cu), and KC is the local carrying capacity (cu).
Pseudomonas branch extension is governed by Eq. (4):
where Li is the length of branch i (mm), \(\gamma\) is Pseudomonas spatial expansion efficiency (mm h−1 cu−1), \(W\) is the branch width previously measured from experimental data (mm)43, A is the local cefotaxime antibiotic concentration (mol l−1), \({A}_{C}\) is the cefotaxime concentration for half-maximum suppression of Pseudomonas effective spatial expansion ability (mol l−1), h is the steepness of the suppression of Pseudomonas effective spatial expansion ability by cefotaxime, \({\Omega }_{i}\) indicates the domain of branch i, and \({\int }_{{\Omega }_{i}}\,\) indicates an integral over branch \(i\) domain.
Klebsiella cell density time change is given by Eq. (5):
where \({D}_{E}\) is the diffusive dispersal rate of Klebsiella (mm2 h−1), and \({G}_{E}\) and \({L}_{E}\) are Klebsiella growth and lysis functions, respectively, as defined by Eqs. (6),(7):
where \({\mu }_{E}\) is the growth rate constant of Klebsiella in proportion to that of Pseudomonas.
where \({\phi }_{E}\) is the proportionality constant between the growth and lysis rates of Klebsiella, \({A}_{E}\) is the cefotaxime concentration for half-maximum Klebsiella lysis rate (mol l−1), and \({h}_{2}\) is the steepness of the onset of Klebsiella lysis by cefotaxime.
Nutrient concentration time change is defined by Eq. (8):
where \({\beta }_{N}\) and \({\beta }_{{EN}}\) are the rate constants for nutrient consumption by Pseudomonas and Klebsiella, respectively, cell density growth (g l−1 h−1 cu−1 for both). Note that \({\beta }_{{EN}}\) must be constrained tightly as Pseudomonas would otherwise avoid Klebsiella-colonized territory due to nutrient depletion. DN is the diffusivity of nutrients (mm2 h−1).
Antibiotic concentration time change is given by Eq. (9) as follows:
where B is the local concentration of extracellular Bla (mol l−1), \({\kappa }_{B}\) is the rate constant for cefotaxime antibiotic degradation by Bla (l mol−1 h−1), and \({D}_{A}\) is the diffusivity of antibiotics (mm2 h−1).
Extracellular Bla concentration time change is given by Eq. (10):
where \({B}_{{{\rm{in}}}}\) is the intracellular concentration of Bla (mol cell−1; applies to Klebsiella only), \({d}_{B}\) is the spontaneous degradation rate constant for Bla (h−1), and \({D}_{B}\) is the diffusivity of Bla (mm2 h−1).
We have formulated the above mathematical model building upon our previous models for Pseudomonas branching pattern formation43 and Bla-producing bacteria population dynamics under β-lactam treatment30:
As we lack a complete mechanistic understanding of how Pseudomonas colonies develop branches, our model defines the colony morphology formation based on a set of explicit rules43: (1) A colony initiates from a circular inoculum. (2) Branches emerge with a defined branch width W and branch density D (number of branch tips per unit area), both measured from experimental data. (3) Each branch i extends at an extension rate \({\rm{d}}{L}_{i}/{{\rm{d}}t}\), which is proportional to cell growth within the branch. (4) Each branch extends following the local nutrient gradient (towards where the nutrient concentration increases most steeply) and bifurcates when the local cell density reaches the threshold D.
At each timestep, we compute cell growth, nutrient consumption, antibiotics degradation, etc., and determine the extension rate from cell growth using the equations described above. For simplicity, we assume that the cell density redistributes and becomes uniform within the colony at each timestep.
Based on this framework, we incorporated antibiotic inhibition into the branch extension to account for the effect of antibiotics on colony expansion. Also, from our previous model for Bla-producing bacteria population dynamics under β-lactam treatment30, we incorporated Klebsiella growth and lysis due to antibiotics, Bla production by lysing Klebsiella, and antibiotic degradation by Bla.
We used the parameter values from the two original papers30,43, with some minor modifications, when possible. The diffusivities of the antibiotic and Bla were chosen based on their molecular weights relative to nutrients (i.e., casamino acids), as suggested by Einstein’s relation. The parameter values for the cefotaxime concentration for the half-maximum suppression of Pseudomonas’ effective spatial expansion ability (\({A}_{C}\)) and the sharpness of this suppression (h) were used from the Hill-fits to our experimental data above (see Supplementary Fig. 9a caption).
We assumed no spontaneous degradation of the antibiotic or nutrient recycling after lysis. Finally, we added another parameter to account for our experimental observation above that Pseudomonas spatial expansion is effectively biased towards Klebsiella (Fig. 2b and Supplementary Fig. 14). This parameter essentially determines the relative dominance of Pseudomonas’ range expansion on nutrient gradients versus antibiotic gradients depending on their global profiles: Pseudomonas would move towards decreasing antibiotic concentration (i.e., the Klebsiella neighborhood), except when d = 0, if the spatial global average of antibiotic concentration is greater than \({A}_{C}\) and if the global minimum of nutrient concentration is greater than a factor (\({A}_{g}\) = 0.7 as the default value) of the nutrient concentration for half maximum specific growth rate (\({K}_{N}\)). For \(d=0\), this factor was applied as \({A}_{g}\) = 1016 to represent an infinitely large value.
The partial differential equations were numerically solved using a custom MATLAB implementation of the alternating direction implicit method by applying no-flux boundary conditions. The numerical simulations were run in a 1001-by-1001-pixel spatial domain, where each pixel dimension was equivalent to a ~0.106 mm spatial length based on the model parameters used. Each population was started in a circular zone equivalent of 5 mm radius. The simulation timestep used was equivalent to 0.02 h, again, based on the model parameters used.
See Supplementary Table 2 for the values of the model parameters as well as the initial conditions used.
Statistics and reproducibility
No statistical method was used to predetermine sample size. Cultures were randomly allocated to treatment groups (antibiotic or control) and to incubator positions. Wells within multi-well plates were not randomized; however, all plates were handled consistently and incubated under uniform conditions. Investigators were not blinded to allocation during experiments and outcome assessment. For microbiome sequencing data presentation, a cut-off threshold of 0.5% relative abundance was applied to remove taxa absent from the SynkC design that appeared in the results, likely reflecting noise or minor contamination.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
16S rDNA sequencing results supporting Table 1 and whole-genome sequencing results supporting Supplementary Table 1 have been deposited in NCBI database (accession numbers: PX457886-PX457893 and BioProject ID: PRJNA1345047, respectively). Source data are provided with this paper.
Code availability
Codes are available at https://github.com/youlab/FacilitatedExpansion (https://doi.org/10.5281/zenodo.17343543).
References
Großkopf, T. & Soyer, O. S. Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77 (2014).
Friedman, J., Higgins, L. M. & Gore, J. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 1, 0109 (2017).
Chang, C.-Y., Bajić, D., Vila, J. C. C., Estrela, S. & Sanchez, A. Emergent coexistence in multispecies microbial communities. Science 381, 343–348 (2023).
Clerc, E. E. et al. Chemotaxis, growth, and inter-species interactions shape early bacterial community assembly. ISME J. 19, wraf101 (2025).
Humphries, J. et al. Species-independent attraction to biofilms through electrical signaling. Cell 168, 200–209.e212 (2017).
Amor, D. R., Ratzke, C. & Gore, J. Transient invaders can induce shifts between alternative stable states of microbial communities. Sci. Adv. 6, eaay8676 (2020).
Ze, X., Duncan, S. H., Louis, P. & Flint, H. J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543 (2012).
Centanni, M. et al. Bifidobacterium pseudolongum in the ceca of rats fed hi-maize starch has characteristics of a keystone species in bifidobacterial blooms. Appl. Environ. Microbiol. 84, e00547–00518 (2018).
Cartmell, A. et al. A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat. Microbiol. 3, 1314–1326 (2018).
Chng, K. R. et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat. Ecol. Evol. 4, 1256–1267 (2020).
Feng, J. et al. Polysaccharide utilization loci in Bacteroides determine population fitness and community-level interactions. Cell Host Microbe 30, 200–215.e212 (2022).
Weiss, A. S. et al. Nutritional and host environments determine community ecology and keystone species in a synthetic gut bacterial community. Nat. Commun. 14, 4780 (2023).
Flynn, J. M., Niccum, D., Dunitz, J. M. & Hunter, R. C. Evidence and role for bacterial mucin degradation in cystic fibrosis airway disease. PLoS Pathog. 12, e1005846 (2016).
Sadiq, F. A. et al. Dynamic social interactions and keystone species shape the diversity and stability of mixed-species biofilms—an example from dairy isolates. ISME Commun. 3, 118 (2023).
Kropp, C. et al. The keystone commensal bacterium Christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep. 11, 11494 (2021).
Hajishengallis, G., Darveau, R. P. & Curtis, M. A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725 (2012).
Hajishengallis, G. & Lamont, R. J. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24, 477–489 (2016).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).
Fenchel, T. Microbial behavior in a heterogeneous world. Science 296, 1068–1071 (2002).
Costerton, J. W., Geesey, G. G. & Cheng, K. J. How bacteria stick. Sci. Am. 238, 86–95 (1978).
Or, D., Smets, B. F., Wraith, J. M., Dechesne, A. & Friedman, S. P. Physical constraints affecting bacterial habitats and activity in unsaturated porous media – a review. Adv. Water Resour. 30, 1505–1527 (2007).
Le, S. et al. Bacteriophage protein Dap1 regulates evasion of antiphage immunity and Pseudomonas aeruginosa virulence impacting phage therapy in mice. Nat. Microbiol. 9, 1828–1841 (2024).
Mitchell, J. G. & Kogure, K. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 55, 3–16 (2006).
Tsao, Y.-F. et al. Phage morons play an important role in Pseudomonas aeruginosa phenotypes. J. Bacteriol. 200, https://doi.org/10.1128/jb.00189-00118 (2018).
Shah, M. et al. A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa. Mol. Cell 81, 571–583.e576 (2021).
Finkelshtein, A., Roth, D., Jacob, E. B. & Ingham, C. J. Bacterial swarms recruit cargo bacteria to pave the way in toxic environments. MBio 6, https://doi.org/10.1128/mbio.00074-00015 (2015).
Caiazza, N. C., Shanks, R. M. Q. & O’Toole, G. A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187, 7351–7361 (2005).
Luo, N. et al. The collapse of cooperation during range expansion of Pseudomonas aeruginosa. Nat. Microbiol. 9, 1220–1230 (2024).
Pradhan, D. et al. Toroidal displacement of Klebsiella pneumoniae by Pseudomonas aeruginosa is a unique mechanism to avoid competition for iron. MBio 16, e01149–01125 (2025).
Meredith, H. R. et al. Applying ecological resistance and resilience to dissect bacterial antibiotic responses. Sci. Adv. 4, eaau1873 (2018).
Magill, S. S. et al. Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370, 1198–1208 (2014).
Rennie, R. P., Jones, R. N. & Mutnick, A. H. Occurrence and antimicrobial susceptibility patterns of pathogens isolated from skin and soft tissue infections: report from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 2000). Diagnostic Microbiol. Infect. Dis. 45, 287–293 (2003).
Marchaim, D. et al. “Swimming in resistance”: co-colonization with carbapenem-resistant Enterobacteriaceae and Acinetobacter baumannii or Pseudomonas aeruginosa. Am. J. Infect. Control 40, 830–835 (2012).
Palavecino, E., Ramirez, K., Greene, S. R. & Kilic, A. Co-existence of VIM-2-producing Pseudomonas aeruginosa and KPC-2 and OXA-232-co-producing Klebsiella pneumoniae in the United States. Ann. Lab Med. 40, 267–269 (2020).
Frank, D. N., Wilson, S. S., St. Amand, A. L. & Pace, N. R. Culture-independent microbiological analysis of foley urinary catheter biofilms. PLoS ONE 4, e7811 (2009).
Noorian, P. et al. A model, mixed-species urinary catheter biofilm derived from spinal cord injury patients. BioRxiv https://doi.org/10.1101/2024.11.04.621968 (2024).
Rashid, M. H. & Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. 97, 4885–4890 (2000).
Arslan, E., Coşkun, M. K., Çobanoğlu, Ş, Aslan, M. H. & Yazıcı, A. Effects of four antibiotics on Pseudomonas aeruginosa motility, biofilm formation, and biofilm-specific antibiotic resistance genes expression. Diagnostic Microbiol. Infect. Dis. 106, 115931 (2023).
Kumar, L., Brenner, N., Brice, J., Klein-Seetharaman, J. & Sarkar, S. K. Cephalosporins interfere with quorum sensing and improve the ability of Caenorhabditis elegans to survive Pseudomonas aeruginosa infection. Front. Microbiol. 12, 115931 (2021).
Liu, Y., Lehnert, T. & Gijs, M. A. M. Effect of inoculum size and antibiotics on bacterial traveling bands in a thin microchannel defined by optical adhesive. Microsyst. Nanoeng. 7, 86 (2021).
Oliveira, N. M. et al. Suicidal chemotaxis in bacteria. Nat. Commun. 13, 7608 (2022).
Berg, H. C. Random Walks in Biology REV—Revised (Princeton University Press, 1993).
Luo, N., Wang, S., Lu, J., Ouyang, X. & You, L. Collective colony growth is optimized by branching pattern formation in Pseudomonas aeruginosa. Mol. Syst. Biol. 17, e10089 (2021).
Starlander, G. & Melhus, Å. Minor outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J. Hospital Infect. 82, 122–124 (2012).
Hota, S. et al. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect. Control Hospital Epidemiol. 30, 25–33 (2009).
Kotay, S., Chai, W., Guilford, W., Barry, K. & Mathers, A. J. Spread from the sink to the patient: In situ study using green fluorescent protein (GFP)-expressing Escherichia coli to model bacterial dispersion from hand-washing sink-trap reservoirs. Appl. Environ. Microbiol. 83, e03327–03316 (2017).
Tanouchi, Y., Pai, A., Buchler, N. E. & You, L. Programming stress-induced altruistic death in engineered bacteria. Mol. Syst. Biol. 8, 626 (2012).
Şimşek, E. et al. A ‘rich-get-richer’ mechanism drives patchy dynamics and resistance evolution in antibiotic-treated bacteria. Mol. Syst. Biol. 20, 880–897 (2024).
Bertness, M. D. & Callaway, R. Positive interactions in communities. Trends Ecol. Evol. (Amst.) 9, 191–193 (1994).
Bronstein, J. L. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. (Amst.) 9, 214–217 (1994).
Hoek, T. A. et al. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 14, e1002540 (2016).
Fetzer, I. et al. The extent of functional redundancy changes as species’ roles shift in different environments. Proc. Natl Acad. Sci. 112, 14888–14893 (2015).
Thiery, S. & Kaimer, C. The predation strategy of Myxococcus xanthus. Front. Microbiol. 11, 2 (2020).
Elyasi Far, B., Ahmadi, Y., Yari Khosroshahi, A. & Dilmaghani, A. Microbial alpha-amylase production: progress, challenges and perspectives. Adv. Pharm. Bull. 10, 350–358 (2020).
Jin, F., Conrad, J. C., Gibiansky, M. L. & Wong, G. C. L. Bacteria use type-IV pili to slingshot on surfaces. Proc. Natl Acad. Sci. 108, 12617–12622 (2011).
Mattick, J. S. Type IV pili and twitching motility. Annu Rev. Microbiol 56, 289–314 (2002).
Fortune, G. T., Oliveira, N. M. & Goldstein, R. E. Biofilm growth under elastic confinement. Phys. Rev. Lett. 128, 178102 (2022).
Seminara, A. et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc. Natl Acad. Sci. 109, 1116–1121 (2012).
Zhang, Q. et al. Morphogenesis and cell ordering in confined bacterial biofilms. Proc. Natl Acad. Sci. 118, e2107107118 (2021).
Ma, H. R., Xu, H. Z., Kim, K., Anderson, D. J. & You, L. Private benefit of β-lactamase dictates selection dynamics of combination antibiotic treatment. Nat. Commun. 15, 8337 (2024).
de Cena, J. A., Zhang, J., Deng, D., Damé-Teixeira, N. & Do, T. Low-abundant microorganisms: the human microbiome’s dark matter, a scoping review. Front. Cell. Infect. Microbiol. 11, 689197 (2021).
Xavier, J. B., Kim, W. & Foster, K. R. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79, 166–179 (2011).
Fournet-Fayard, S., Joly, B. & Forestier, C. Transformation of wild type Klebsiella pneumoniae with plasmid DNA by electroporation. J. Microbiol. Methods 24, 49–54 (1995).
Payne, S. et al. Temporal control of self-organized pattern formation without morphogen gradients in bacteria. Mol. Syst. Biol. 9, 697 (2013).
Barbier, M. & Damron, F. H. Rainbow vectors for broad-range bacterial fluorescence labeling. PLoS ONE 11, e0146827 (2016).
Santos-Moreno, J., Tasiudi, E., Stelling, J. & Schaerli, Y. Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 11, 2746 (2020).
Warren, B. G. et al. Identification of carbapenem-resistant organism (CRO) contamination of in-room sinks in intensive care units in a new hospital bed tower. Infect. Control Hospital Epidemiol. 45, 302–309 (2024).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics 7, 000685 (2021).
Acknowledgements
The authors thank Lawrence A. David, Joanna B. Goldberg, David K. Karig, Jeffrey Letourneau, Jia Lu, Alice S. Prince, and David S. Weiss for helpful feedback. We thank Yasa Baig, Minsu Kim, Philip N. Rather, and Stuart A. West for their critical reading of an earlier version of the manuscript. This work was partially supported by grants from National Institutes of Health (LY: R01GM098642), Office of Naval Research (LY: N00014-20-1-2121), Defense Advanced Research Projects Agency (L.Y.: 2501-203-2015861), National Science Foundation (L.Y. and C.T.L.: MCB-1937259), and by the Engineering Research Centers Program of the National Science Foundation under NSF Cooperative Agreement No. EEC-2133504. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.Ş.: Conceived the study, designed, and conducted the research. Wrote the manuscript. Jointly supervised the research with L.Y. C.A.V.: Assisted with experiments in Fig. 1 and Supplementary Fig. 10. Equally contributed with K.S. K.S.: Assisted with experiments in Supplementary Fig. 13. Equally contributed with C.A.V. Z.Z. Assisted with experiments in Supplementary Fig. 8a and Supplementary Fig. 22a. Wrote the cylindrically symmetric antibiotic gradient solution. N.L.: Performed the experiment in Supplementary Fig. 23. D.L.: Constructed the fluorescent protein-labeled variant of Klebsiella (Klebsiella-mCherry). Assisted in the experiments that led to Supplementary Fig. 10. H.R.M.: Assisted in the isolation of the hospital sink bacteria. D.J.A.: Obtained and provided the p-trap water samples which led to the isolation of the hospital sink bacteria. C.T.L.: Assisted with data interpretation, writing the manuscript, and articulating the broader ecological context. Acquired funding. L.Y.: Conceived the study and assisted with research design and data interpretation. Wrote the manuscript. Jointly supervised the research with E.Ş. Acquired funding. All authors contributed to the reviewing and editing of manuscript drafts and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Ákos T Kovács, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Şimşek, E., Villalobos, C.A., Sahu, K. et al. Spatial proximity dictates bacterial competition and expansion in microbial communities. Nat Commun 16, 10885 (2025). https://doi.org/10.1038/s41467-025-65892-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65892-9