Abstract
Covalent inhibitors of oncoprotein KRAS have initial efficacy, but responses lack durability. Covalently modified oncoproteins are presented as MHC-restricted hapten-peptides (p*MHC) on the cancer cell surface, enabling combination of targeted therapy with immunotherapy to overcome drug resistance. Building on indirect evidence of KRASG12C-derived p*MHCs, we use immunopeptidomics to identify and directly quantify these synthetic neoantigens. To address challenges by their low copy number, we develop AETX-R114, a T cell engaging bispecific antibody with picomolar affinity for MHC-restricted sotorasib-modified KRASG12C peptides presented by three HLA-A3 supertype alleles. AETX-R114 dramatically increases the half-life and thereby the number of presented p*MHCs, enabling selective and potent killing of resistant cancer cells both in vitro and in vivo. To broaden the therapeutic potential of creating and targeting synthetic neoantigens, we further develop AETX-R302, which recognizes divarasib-modified KRASG12C peptides presented on alleles from the HLA-A2 and A3 supertypes. Cryo-EM structure determination reveals the molecular basis for breaking HLA supertype restriction. Collectively, our study illustrates how engineered antibodies can transform synthetic neoantigens into actionable cancer immunotherapy targets.
Similar content being viewed by others
Introduction
Tumor antigen-targeting immunotherapies in principle have superior safety and efficacy compared to systemic immunotherapy by immune checkpoint inhibitors, but identifying tumor-specific antigens remains a significant challenge. Unlike patient-specific neoantigens, which require individualized immunotherapeutic development1,2,3, commonly mutated oncoprotein-derived epitopes presented on class I major histocompatibility complex (MHC) molecules provide an opportunity to target cancer-specific antigens shared across patient populations4,5. However, their development has been limited by human leukocyte antigen (HLA) restriction and low pMHC presentation levels, resulting in a limited repertoire of well-validated neoantigen peptide-MHC (pMHC) targets6,7. Additionally, generating high-affinity, highly specific binders to pMHC targets using traditional TCR or TCR-mimicking (TCRm) antibodies remains challenging due to the subtle differences between many mutant and wild-type antigens8.
Mutated KRASG12X proteins are some of the most common tumor-specific, shared pMHC targets due to their high prevalence and well-characterized epitope landscape across high-frequency HLA alleles7,9,10,11. Despite their low surface density ( ~ 10 or fewer copies-per-cell), promising immunotherapies, primarily focused on KRASG12D and KRASG12V, are in development 9,10,12,13,14,15. Although KRASG12C mutations occur relatively frequently (9% in non-small cell lung cancer, 3% in colorectal cancer, and 1% in pancreatic cancer16), the development of KRASG12C-pMHC immunotherapies remains extremely limited. A key challenge arises from the cysteine residue, which is prone to heterogeneous post-translational modifications of the thiol group that result in multiple unknown isoforms, effectively diluting an already low-density target17. Mass spectrometry techniques to identify KRASG12C epitopes cleave thiol modifications to enable detection, preventing precise characterization of the cysteine’s modification state or states7,9. Thus, innovative strategies are required to fully exploit the therapeutic potential of KRASG12C neoepitopes.
Recently, we and others described a novel approach that addresses the limitations of targeting cysteine-containing neoantigens18,19. Synthetic neoantigens formed by proteasome-resistant modification of KRASG12C peptides with covalent inhibitors are loaded onto HLAs as so-called “haptenated peptides” or p* (peptides still carrying the covalent inhibitor) and presented as p*MHCs on the cancer cell surface as homogeneously modified epitopes. The distinct nature of these cancer-specific targets facilitates their high-specificity targeting by engineered antibodies and enables tumor-specific killing in vitro when antibodies are converted to bispecific T cell engagers (TCEs)18. This strategy has the potential to counteract disease progression driven by inherent or acquired resistance to covalent KRASG12C inhibitors, remaining effective in recurrent settings because the G12C mutation is typically retained in resistant cells20,21,22,23. Additionally, it leverages the immunomodulatory effects of covalent inhibitors such as increased HLA expression and T cell infiltration to enhance therapeutic impact24,25.
To date, only indirect evidence has supported the existence of such unique MHC antigens. We addressed this knowledge gap by characterizing KRASG12C-haptenated peptides using a custom mass spectrometry (MS) approach to identify and quantify p*MHCs on the cell surface. This novel insight guided the engineering of a TCE that, by design, increased the cell surface half-life and density of these endogenously unstable, ultra-low-density p*MHC neoantigens. This in turn enabled the first in vivo validation of a hapten-targeting, tumor-specific TCE. Furthermore, we demonstrate that engineering for high affinity does not preclude pan-HLA cross-reactivity within and even beyond a given HLA supertype. We describe the concerted design and development of two TCEs recognizing sotorasib- and divarasib-modified KRASG12C p*MHCs, presented on alleles covering 39% and 66% of the U.S. population, respectively.
Collectively, our study demonstrates that engineered bispecific antibodies targeting drug-modified KRASG12C neoantigens enhance MHC presentation and elicit potent, selective T cell-mediated cytotoxicity across HLA supertypes. Our study empowers and broadens the therapeutic utility of creating and targeting synthetic neoantigens and informs the combination of covalent inhibitors and TCEs to overcome drug resistance in KRASG12C-mutated cancers and other intracellular cancer specific targets amenable to covalent inhibition.
Results
Direct detection and characterization of KRASG12C haptenated MHC peptides
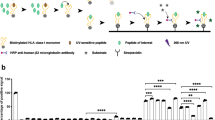
To directly assess p*MHC presentation, we developed a MS-based approach to detect and quantify two sotorasib (“soto”)-labeled KRASG12C hapten-peptides, soto-p7 and soto-p8, predicted to be presented by HLA-A*03:01 (A*03) and HLA-A*11:01 (A*11). This method employs a pair of synthetic, stable isotope-labeled internal standard peptides (SIL-IS, “heavy”) for each target. Double heavy isotope-labeled peptides guide the sensitive and selective acquisition of endogenous target spectra, and single heavy isotope-labeled peptides (hip) refolded into MHCs (hipMHCs) are used as embedded standards during peptide purification and processing for accurate copies-per-cell (CPC) quantification (Fig. 1a,b, Supplementary Fig. 1a)26,27,28. Using this protocol, we defined a limit-of-quantification (LOQ) of ~1 (p7) and ~10 (p8) CPC (Supplementary Fig. 1b).
a Sequences of KRASG12C epitopes. b Targeted MS workflow for identifying and quantifying haptenated MHC-I peptides. c Schematic of cell line engineering for generating HLA/KRASG12C overexpression A375-derived cell lines. d Mirror plot depicting the mass spectra in each sample for the double heavy-labeled synthetic (bottom, +14 m/z) and endogenous (top) soto-p7 peptides in cells treated for 48 h with DMSO (left) or 3 µM sotorasib (right). Annotated ions labeled in color depict identified b, y, a, or precursor ion fragments. Underlined amino acids are heavy isotope-labeled (V, K) or sotorasib-modified (C). e Estimated copies-per-cell of soto-p7 presented on A375-A11KC treated with 3 µM sotorasib between 20 h and 72 h. Data points represent n = 2 independent biological replicates. f Table of HLA-A3 supertype alleles summarizing the results of p7/soto-p7 targeted MS experiments, with HLA residues at the p*MHC/antibody binding interface identical to those of A*03:01 highlighted in green. A check mark = detected, “X” = undetected. g Estimated copies-per-cell of peptides presented on A375 A11KC cells with no treatment (p7/p8) or with 48 h of 3 µM sotorasib (soto-p7/8). Data points represent 3 technical replicates. h p/p*MHC complex half-life measured at 37 °C, calculated from 3 technical replicates per peptide. All error bars represent standard deviation (SD) from the mean.
Given the higher KRASG12C pMHC complex stability on A*11 than A*039, we engineered A375 cells (KRASWT, HLA-A*01:01/A*02:01) to overexpress KRASG12C and A*11 and confirmed that p7 and p8 were presented (“A375-A11KC,” Fig. 1c, Supplementary Fig. 2a)9. We next treated cells with sotorasib for 2–72 h and performed targeted immunopeptidomics. The mass spectra for endogenous soto-p7 and soto-p8 peptides matched those of heavy-labeled synthetic peptides, confirming their detection exclusively in sotorasib-treated cells. (Fig. 1d, Supplementary Fig. 2b). Although soto-p8 was detectable only at 48 h and below the LOQ, soto-p7 was detected as early as 2 h, peaking at 24–48 h, consistent with the reported ~24 h turnover rate of KRASG12C (Fig. 1e)29.The higher abundance of soto-p7 reflects the higher stability and epitope density of the non-haptenated KRASG12X-derived p7 peptides (10-mer) over p8 (9-mer), establishing soto-p7 as the primary hapten target9. Encouraged by our success in identifying sotorasib-labeled peptides, we treated A375-A11KC cells with adagrasib (“ada”) or divarasib (“diva”) for 48 h which revealed two novel hapten-peptides, ada-p7 and diva-p7. These results demonstrate the versatility of our approach with different KRASG12C covalent inhibitors (Supplementary Fig. 3).
MHC alleles are grouped into “supertypes” based on overlapping peptide binding properties, and A*11:01 belongs to the HLA-A3 supertype (hereafter, “A3 supertype”). To determine whether additional alleles of the A3 supertype can present the hapten-p7 peptide, we screened for those ( ≥ 1% U.S. population frequency) predicted to bind p7, hapten-p7 and/or favor lysine at the C-terminal anchor position, identifying seven alleles including A*03 (Supplementary Fig. 4a,b)30,31. The high sequence similarity amongst these alleles at the hapten-peptide/antibody binding interface, as seen in our previously reported soto-p7/A*03/A*11 cross-reactive clone R023, suggested the potential for broader antibody cross-reactivity across these alleles (Fig. 1f). Accordingly, we identified p7 presented on HLA-A*03:01, HLA-A*68:01 (A*68), and HLA-A*34:02, as well as soto-p7 presented on A*03 and A*68 using the A375 HLA/KRASG12C overexpression system (Supplementary Figs. 4c-e, 5a,b).
While these data provide the first direct evidence for p*MHC presentation in cells, the abundance of KRASG12C-derived p*MHCs was low, even in overexpression models. For instance, in A375-A11KC cells, haptenated p7 peptides were present at only 10 CPC, compared with 89 CPC of p7 in the same cell line (Fig. 1g). In endogenous cell systems, soto-p7 was either undetectable (HOP-62/A*03/A*11, NCI-H2030/A*11), or quantified below the 1 CPC LOQ in the NCI-H2122/A*03 model (Supplementary Fig. 5c) We measured complex stability of p7WT and soto-, ada-, and diva-p7 haptens at 37 °C and found all three drug haptens had a shorter complex half-life than p7WT, with A*03 showing the poorest overall stability (Fig. 1h). Whereas some post-translational modifications can enhance peptide stability and affinity, haptenization of p7 exhibits the opposite effect32. Reduced p*MHC stability, coupled with the dynamic nature of hapten presentation, renders these low-density hapten-p7 peptides challenging targets for pMHC-based immunotherapy. However, based on these findings, we hypothesized that binding of an antibody with slow dissociation from p*MHCs might enhance antigen stability on the cell surface and enable more effective TCE-mediated cell killing. To this end, we aimed to enhance R023 through affinity maturation while preserving binding specificity and cross-HLA recognition.
Affinity maturation of antibody R023
Cryo-EM-enabled structural characterization of R023 binding to sotorasib-conjugated p*MHCs revealed that R023 recognizes primarily the HLA and hapten moieties of its antigens, with mutational data confirming the central role of the hapten in antibody binding33. We used this information to design a tailored affinity maturation library for R023, aiming to decrease its dissociation rate from soto-p7/A*03 and A*11 while maintaining minimal competition with free sotorasib. R023 mainly interacts through its variable light (L) chain domain (VL) with the HLA, while its variable heavy (H) chain domain (VH) is mostly involved in hapten recognition 33. Thus, we focused the affinity maturation campaign on the VL of R023 to avoid enhancing binding solely to the hapten, which could lead to detrimental competition with free drug.
We generated a combinatorial library by mutating complementarity-determining region (CDR) positions 29 - 32 of CDR-L1, 50 of CDR-L2, and 94 - 96 of CDR-L3, introducing at each explored position amino acid diversity informed by deep mutational scanning previously performed for R023 (Fig. 2a)33. Using yeast display34, we isolated clones that exhibited cross-reactive binding to soto-p7/A*03 and soto-p7/A*11 and slower dissociation from soto-p7/A*11 compared with the parental R023 clone (Fig. 2b). Several clones also showed decreased dissociation from soto-p7/A*03, with R114 exhibiting the strongest binding overall (Fig. 2b). As it also retained low free drug competition (Supplementary Figs. 6a), R114 was selected as the lead clone and paired with an anti-CD3 effector arm (hUCHT1-V9, anti-CD3ε) to generate a T-cell engager (TCE) in half-life extended, bispecific (scFv)2-scFc format (AETX-R114, Fig. 2c), for further biophysical and functional characterization35,36.
a Schematic representation of the structure guided affinity maturation of R023 to R114. The top panel shows a structural representation of the positions of R023 CDRs (shown as tubes) bound to soto-p7/A*11. The HLA moiety of the sotorasib p*MHC is shown in surface representation, while soto-p7 is represented as sticks (sotorasib) and tubes (p7). The side chains of R023 residues located at positions selected for the affinity maturation campaign are represented as spheres. The bottom panel shows CDR sequences of R023 and its descendant R114 clones, and a schematic representation of the affinity maturation process. R023 residues located at positions selected for the affinity maturation campaign are underlined, and mutations introduced in R114 are in bold. “Off-rate sorting” refers to Fluorescence-Activated Cell Sorting (FACS) of yeast libraries, performed upon staining aimed at the identification of binders with low dissociation from the target, as described below. b Binding of yeast-displayed scFv R023 and descendant matured clones to sotorasib p*MHC antigens in an “off-rate” setting. Target-bound yeast cells, after removal of unbound sotorasib p*MHCs, were incubated for 2 h with a 10-fold excess of a competitor molecule to prevent rebinding of dissociated target, and the signal from remaining bound molecules was measured. Different clones are indicated by colors (e.g., R023 is indicated by blue dots, and R114 is indicated by yellow dots). c Schematic representation of the TCE AETX-R114 in the (scFv)2-scFc format. d SPR sensorgrams of the interaction between AETX-TCEs and the indicated soto-p/MHC antigens. Biotinylated soto-p/MHCs were immobilized, and binding of soluble AETX-TCEs samples was measured using single-cycle kinetics. Kinetic values of fitted data are shown in the table below.
Surface plasmon resonance (SPR) kinetic measurements confirmed the higher affinities and slower dissociation rates of AETX-R114 compared with AETX-R023, with picomolar KD values at 37 °C for both soto-p7/A*03 (7 pM) and soto-p7/A*11 (160 pM). Importantly, AETX-R114 maintained its specificity and effector arm binding (Fig. 2d, Supplementary Fig. 6b,c). AETX-R114 also recognized soto-p7/A*68 with picomolar affinity (Supplementary Fig. 6d), further expanding the high-affinity cross-HLA recognition of the antibody.
TCE-mediated stabilization with AETX-R114 enhances p*MHC presentation
To test whether AETX-R114 could increase the half-life of p*MHCs, we incubated recombinant soto-p7 p*MHCs with AETX-R114 at 37 °C and measured complex stability. Indeed, AETX-R114 dramatically extended the half-life of soto-p7 on A3 supertype alleles with no effect on p7WT (Fig. 3a,b, Supplementary Fig. 7a,b). Half-life extension positively correlated with antibody affinity (Supplementary Fig. 7c,d), with AETX-R114 increasing soto-p7 half-life beyond 24 h on A*03 and A*11, reaching levels comparable to other tumor-associated antigens targeted by clinical TCE and TCR-T therapies (Fig. 3b)37,38,39. This effect was detected directly in NCI-H2122 cells, where incubation with sotorasib and AETX-R114 increased soto-p7 presentation from <1 CPC to ~14 CPC, a density shown to support effective TCE-mediated activity (Fig. 3c)37,40. Thus, consistent with our hypothesis, a high-affinity antibody with a slow dissociation rate, such as AETX-R114, can stabilize short half-life p*MHC targets to increase antigen surface density, overcoming the historic challenge of targeting an epitope with poor stability or low abundance (Fig. 3d,e).
a Proportion of intact p/p*MHC remaining after incubation at 37°C from 0 to 24 h. Complex half-life was determined using a one-phase decay, non-linear fit equation. Data show the mean of 3 technical replicates; error bars indicate SD. b Calculated p/p*MHC half-lives at 37°C. c Estimated copies-per-cell (CPC) of A*03 epitopes present on NCI-H2122 cells. Data points represent 3 technical replicates; error bars indicate SD. d Schematic of endogenous and (e) antibody-stabilized p*MHC presentation.
AETX-R114 directs T cell killing of sotorasib-treated tumor cells
We next assessed the ability of AETX-R114 to induce T cell-mediated cytotoxicity in sotorasib-resistant A*03/A*11 cancer cell lines (Supplementary Fig. 8a). AETX-R114 potently and selectively induced T cell-mediated killing in the presence of sotorasib, while the non-targeting isotype control (AETX-RSV) had no effect (Fig. 4a,b, Supplementary Table 2). Cytotoxicity was associated with activation of CD8+ T cells as well as IFNγ production, indicating that AETX-R114 induces a robust effector T cell response with demonstratable superiority to AETX-R023 (Supplementary Fig. 8b-d, Supplementary Fig. 9, Supplementary Table 2). AETX-R114 also elicited cytotoxicity and T cell activation using PBMCs as effector cells. Importantly, there was no evidence of fratricide observed against HLA-matched or unmatched donor cells, providing further evidence of TCE specificity (Supplementary Fig. 10).
a–d Incucyte analysis of AETX-R114-induced T cell cytotoxicity against cell lines endogenously expressing KRASG12C in the presence of sotorasib (soto) or vehicle (DMSO). Cell lines were plated in 2D (a–c) or as 3D spheroids (d) with a 10:1 E:T ratio of activated human T cells, and cytotoxicity was assessed at 48 h. Data points represent the mean of 3 technical replicates per condition; error bars indicate SD. The same color scheme applies in all panels. a T cell cytotoxicity from donor ID 110044729, representative of results obtained from 5 independent donors summarized in Supplementary Table 2. b Representative Incucyte images corresponding to the cytotoxicity assay shown in (a). c T cell cytotoxicity from NCI-H2122 HLA isogenic cell lines treated with 10 nM AETX-RSV or AETX-R114. A two-way ANOVA using Šidák’s multiple comparisons test was applied to sotorasib + AETX-RSV and sotorasib + AETX-R114 conditions across cell lines (ns not significant or P > 0.05). d T cell cytotoxicity from donor ID RG1200.
As additional validation of the cross-allele reactivity of AETX-R114, we generated an HLA-isogenic cell line panel by knocking out endogenous HLA-A*03 in NCI-H2122 cells and re-expressing HLA-A*03 (as a positive control), A*11, or A*68 (Supplementary Fig. 11). In a T cell killing assay, cytotoxicity was comparable to the parental cell line, confirming strong activity of AETX-R114 across A3 supertype alleles (Fig. 4c). We further investigated AETX-R114-specific activity in a 3D co-culture system, in which NCI-H2122eGFP cells were grown as spheroids with T cells. AETX-R114 showed dose-dependent cytotoxicity in the presence of sotorasib, while AETX-RSV had no effect (Fig. 4d).
These findings prompted us to test whether AETX-R114 could inhibit tumor growth in vivo. NCI-H2122 cells were grown subcutaneously as a co-mixed xenograft with human T cells in NOD SCID mice treated with sotorasib (Fig. 5a, Supplementary Fig. 12a-d). In a pilot study, AETX-RSV had no effect on tumor growth with or without sotorasib and therefore was used as a negative control (Supplementary Fig. 12). AETX-R114 significantly inhibited NCI-H2122 tumor growth and promoted survival with no body weight loss (Fig. 5b-e, Supplementary Fig. 13a). Flow cytometry analysis of the tumors 48 h after the final TCE dose revealed increased numbers of CD8+ and CD4+ T cells with AETX-R114 treatment, with a notable increase in the proportion of non-Treg CD4+ T helper cells (Fig. 5f,g, Supplementary Fig. 13b). Tumor-infiltrating T cells also exhibited increased expression of activation markers including CD25 and PD-1 (Fig. 5h,i, Supplementary Fig. 13c,d). Thus, AETX-R114 significantly inhibits sotorasib-resistant tumor growth in vivo through targeted T cell expansion and activation. This result was repeated across multiple donors (Supplementary Fig. 13e-g).
a Schematic of the in vivo study design. For a cell line-derived xenograft (CDX) mouse model, NCI-H2122 cells were implanted subcutaneously as a co-mixed xenograft with activated human T cells (donor ID RG1200, n = 10/group). Mice were dosed 30 mg/kg sotorasib p.o. daily, 1 × 106 T cells i.v. weekly, and 1 mg/kg AETX-RSV or 1 mg/kg AETX-R114 i.v. weekly. Tumors were harvested, weighed, and processed for flow cytometry analysis 48 h after the final TCE dose. b NCI-H2122 tumor growth with AETX-R114 vs AETX-RSV treatment. Data points represent the mean; error bars indicate standard error of the mean (SEM). Significance for time x treatment interaction assessed by mixed-effects model (Restricted Maximum Likelihood, REML), with time as a repeated factor and treatment as a fixed effect. c Kaplan-Meyer survival curve for AETX-RSV vs AETX-R114, significance determined by Mantel-Cox test. d Body weights normalized to starting weight, in which datapoints represent the mean and error bars indicate SEM (ns, not significant or P > 0.05 at any timepoint versus starting weight by mixed-effects model (REML) with Dunnett’s post hoc test for multiple comparisons). e Tumor weights measured in milligrams (mg); note that for two mice in the AETX-R114 group, no tumor was detected. f Numbers of tumor-infiltrating T cells normalized to tumor weight. g Proportion of each T cell subset in the tumors shown as mean minus SD; Treg are CD4+CD25+FOXP3+ cells. h and i Analysis of T cell activation markers including CD25 and PD-1 showing quantification for individual mice and representative flow cytometry plots. In e, f and h, i significance assessed by Student’s unpaired two-tailed t-test.
Expansion of the antibody sequence and structure space for p*MHC binder discovery
Structural characterization of previous p*MHC binders revealed distinct binding modes, differing notably from those of TCRs and TCRm antibodies33,41. Although synthetic antibody libraries have identified specific p*MHC binders18, restricting discovery to fixed scaffolds limits the mineable antibody sequence and structure space. For this reason, we built a large naïve human antibody library in Fab format for phage display selection, encompassing two sub-libraries, one with kappa L chain containing 4.6 × 1011 independent clones and the other with lambda L chain with 2.3 × 1011 independent clones42. We used this library to identify binders that overcome HLA supertype restriction by targeting divarasib-modified KRASG12C peptides presented on HLA-A*02 (diva-p5, KLVVVGAC[diva]GV), and A*03/A*11 (diva-p7). Combining Fab-phage display selection with scFv-yeast display sorting, we identified clone R302, which exhibited high affinity for all three targets (diva-p5/A*02, diva-p7/A*03, and diva-p7/A*11) while demonstrating no significant binding to the respective KRASWT pMHCs (p5WT, p7WT) and minimal free drug inhibition (Supplementary Fig. 14a,b). R302 binds divarasib p*MHCs with HLA-A*02, A*03 and A*11 alleles, potentially enabling a coverage of 66% of the U.S. population. Possible cross-reactivity to additional A3 and A2 supertype alleles would further increase the coverage. Upon conversion to a TCE in (scFv)2-scFc format, AETX-R302 exhibited single-digit nM affinity to all three divarasib p*MHCs and no binding to KRASWT pMHCs or sotorasib p*MHCs, thereby demonstrating cross-HLA high-affinity and specificity (Fig. 6a, Supplementary Fig. 14c,d).
a SPR sensorgrams of the interaction between AETX-R302 and the indicated divarasib p*MHC antigens. Biotinylated divarasib p*MHCs were immobilized, and binding of soluble AETX-R302 was measured using single-cycle kinetics. Kinetic values of fitted data are shown in the table below. b Incucyte-based cytotoxicity analysis of A375eGFP isogenic cell lines that were pulsed with diva-p5, diva-p7, p5WT, or p7WT peptides for 4 h prior to adding activated human T cells (10:1 E:T ratio) and AETX-R302 for 48 h. Plots show 3 technical replicates per condition; error bars indicate SD. The table shows EC50 values for fitted curves (n.d. = not determined). c Cartoon representation of the structures of Fab R302 (only the variable chains are shown) in complex with diva-p5/A*02 (left) and diva-p7/A*03 (right). HLAs are depicted in purple (A*02) and white (A*03), β2m in gray, haptens in yellow, conjugated peptides in green (p5) and dark blue (p7), and variable chains of R023 in light blue (VL) and blue (VH). The same color scheme, reported in the center of the figure, applies to the structural representations of the following panels. d Surfaces of diva-p5/A*02 (left) and diva-p7/A*03 (right) buried upon the binding of Fab R302 are colored in light blue. The hapten is represented by sticks. The bottom of the panel shows the sequence alignment of HLA-A*02, HLA-A*03 and HLA-A*11 (residues 1 - 180), performed with BLASTp. The only non-conserved residue among the alleles located in R302 epitope (residue 62, on HLA-A*03 only) is indicated on the aligned sequences (black triangle) and on the structures. e Alignment of R302_diva-p5/A*02 and R302_diva-p7/A*03 structures. The right side shows a cartoon representation of the aligned HLAs (residues 1 – 180), the hapten-peptide conjugates (divarasib is represented as sticks), and the R302 variable chains. The left side shows the hapten-peptide conjugates only, represented as sticks. Anchor residues of the two peptides are indicated with a *. Notably, V8 and V9 in diva-p5 and G10 and A11 in diva-p7 are all included in the R302 binding interface. They all share similar chemical properties (small aliphatic sidechains) and the same conformation.
AETX-R302 mediated potent and selective T cell killing of A375 cells pulsed with divarasib-conjugated peptides across HLA-A*02, A*03, and A*11 alleles, confirming recognition of the hapten-peptide target across supertypes (Fig. 6b, Supplementary Fig. 14e). In drug-treated cells endogenously presenting the hapten peptide, cytotoxicity was observed only in HLA-A*11-expressing cells. This occurred despite AETX-R302 exhibiting comparable affinity across all 3 alleles, consistent with diva-p7 having the highest stability on A*11 (Fig. 1h, Supplementary Fig. 14f,g). These findings support the hypothesis that affinity maturation is necessary to enable effective targeting of less stable hapten-p*MHCs to fully leverage the potential of cross-HLA recognition. Therefore, at this stage R302 represents a biophysical proof-of-concept of cross-HLA supertype recognition.
Structural determinants for expanding high-affinity cross-reactivity of p*MHC binders across different HLA supertypes
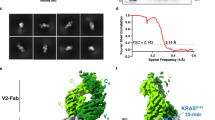
To understand how R302 achieves unprecedented single-digit nM binding affinity across HLA supertypes, we determined its cryo-EM structures in complex with diva-p5/A*02, and diva-p7/A*03. Overall nominal resolutions were 3.23 Å and 2.58 Å, respectively (Fig. 6c, Supplementary Fig. 15, 16). The structures reveal that although p5 and p7 have distinct sequences and present divarasib at different positions (the 8th and 6th, respectively), R302 binds both divarasib p*MHCs similarly, at an angle comparable to that of previously characterized hapten-peptide antibodies and distinct from that of TCRs or TCRm antibodies (Fig. 6c, Supplementary Fig. 17a)19,33,43. R302 recognizes an epitope that comprises parts of three different moieties of the p*MHCs: the HLA, the peptide, and the divarasib hapten (Fig. 6d, Supplementary Fig. 17b, Supplementary Table 3). The latter is partially buried in a shallow cleft formed between VL and VH of R302, leaving the methyl-pyrrolidine moiety of the drug solvent-exposed (Supplementary Fig. 18a).
The HLA moiety of the epitope is nearly identical in the two antibody-antigen complexes, and its widely shared portion is completely conserved in HLA-A*02, HLA-A*03, and HLA-A*11 (Fig. 6d, Supplementary Fig. 18b). Overlay of the two structures shows that the HLA and divarasib moieties are superimposable, with the main difference being the conformation of the peptides (Fig. 6e). As the p5 and p7 sequences are shifted by two positions, they present the cysteine-conjugated hapten from different locations within the peptide (Fig. 6e). Their conformational divergence arises from the 6th to the 9th positions (Fig. 6e), while anchor residues and peptide residues engaged in R302 binding share a similar conformation (Fig. 6e and Supplementary Fig. 18b). Overall, our structures reveal that R302 binds divarasib p*MHCs across HLA alleles belonging to the A2 and A3 supertypes by recognizing a shared epitope encompassing HLA residues, the hapten, and part of the peptide, while allowing the latter sufficient flexibility to compensate for frame shifting. Notably, the N-terminal sequence of CDR-H3 in R302 is “LY”, in place of the much more common “AR”, and it is a crucial determinant of the unique R302 paratope (Supplementary Fig. 18c). Because of its conservation in human antibodies, the “AR” sequence is frequently fixed in synthetic human antibody libraries44. The importance of having “LY” in this position validates our strategy of massively expanding the antibody sequence and structure space to identify unique p*MHC binders such as R302.
Discussion
Engineering highly specific, tumor-targeting immunotherapies is challenging due to the limited pool of shared neoantigens, their low pMHC surface density, and their high similarity to wild-type peptides. We have exploited the covalent modification of KRASG12C to generate synthetic neoepitopes as distinct tumor-specific targets and overcome the historic limitations of targeting cysteine-containing peptides. MS-based identification and quantification of these haptenated pMHCs revealed their short half-lives and ultra-low surface abundance, making them poor candidates for TCR or TCRm antibody targeting. However, by leveraging the hapten modification, we developed high affinity, slow off-rate antibodies that prolong p*MHC stability and increase presentation levels. The low-picomolar p*MHC affinity necessitates pairing with an intermediate-affinity anti-CD3 effector to promote immune synapse formation while preventing prolonged CD3 activation45,46. This approach ensures potent, on-target T cell-mediated killing in drug-resistant KRASG12C cancer models, while maintaining high specificity through the unique features of the haptenated peptide target.
KRASG12C inhibitor resistance is typically driven by mutations that sustain KRAS signaling or activate bypass pathways rather than secondary mutations of the cysteine residue itself. Consequently, our approach should remain effective in most patients20,21,22,23. Amplification of KRASG12C—a common resistance mechanism—would further increase synthetic neoepitope density, enhancing T cell-mediated killing through our strategy23,47,48.
Through affinity maturation of the sotorasib p*MHC-targeting antibody R023 we obtained R114, an antibody with single-digit pM affinity for A*03:01 and triple-digit pM affinities for A*11:01 and A*68:01, expanding targetability across three A3 supertype alleles. Comprehensive MS and stability analyses of soto-p7 guided R114 affinity optimization, prioritizing A*03 over A*11 due to the higher intrinsic stability and surface density of KRASG12C p*MHCs on A*11 and resulting in comparable potency across both high-frequency alleles. The enhanced binding of AETX-R114 subsequently enabled potent cross-allele T cell-mediated cytotoxicity in vitro and in vivo. Future in vivo studies using more physiologically relevant models, such as humanized or syngeneic systems, will be important to refine the evaluation of therapeutic efficacy and translational potential of AETX-R114. MS-based analysis identified hapten-peptides from three different KRASG12C inhibitors (sotorasib, adagrasib, and divarasib), demonstrating the feasibility of hapten presentation across different chemical structures. To assess the feasibility of extending our approach beyond sotorasib p*MHCs, we developed the divarasib p*MHC-targeting clone R302. Importantly, AETX-R302 expands the scope of p*MHC-targeting antibodies by recognizing with single-digit nanomolar affinities divarasib p*MHCs across two distinct HLA supertype families. Cryo-EM structural insights reveal that R302 achieves this unprecedented cross-reactivity by adopting the same binding mode for both A3 and A2 supertype targets, whereas previously reported cross-reactive binders, which showed significantly different affinities across HLA supertypes, engaged their targets with distinct binding modes33.
The ability of p*MHC-targeting antibodies to stabilize low-abundance or unstable tumor-specific antigens suggests a tractable approach for making previously intractable targets actionable. For instance, while KRASG12X mutant epitopes (e.g. KLVVVGAXGV) on HLA-A*02 have demonstrated immunogenic potential, they exhibit low affinity and poor stability, evading MS detection and rendering TCRm antibodies with nanomolar affinities ineffective in A02/KRASG12X mutant patients7,9,10,11,49,50,51. However, an antibody with low picomolar affinity and a slow off-rate may stabilize this epitope, enabling both MS detection and tumor-specific cell killing. Future efforts to affinity-mature R302 or develop alternative A02/KRASG12C binders will determine whether this approach can rescue previously disregarded epitopes, including non-haptenated mutation-containing epitopes. If so, then the repertoire of targetable MHC peptides could be expanded, increasing the number of patients who could benefit from “off-the-shelf” p/p*MHC-targeting immunotherapies.
While hapten modification uniquely enables high-affinity, cross-HLA recognition, the potential for off-target hapten modification and presentation of peptides on healthy tissue necessitates rigorous evaluation to avoid toxicity. Unlike conventional pMHC-targeting immunotherapies, standard cross-reactivity assays such as alanine scanning have limited suitability for p*MHCs, as antibody recognition is restricted to hapten-modified peptides. Thus, off-target reactivity of the antibody is dictated primarily by the specificity of the covalent modification. We addressed this issue by identifying peptide positions capable of presenting the hapten for antibody recognition and assessing whether off-target cysteine-containing sequences can bind relevant HLA alleles. Previously identified off-target cysteine sites—RTN4C1101 and HMOX2C282—are not predicted to bind A3 supertype alleles or to be recognized by AETX-R114 due to hapten positional constraints33, reducing the risk of off-target effects52,53. Nevertheless, comprehensive profiling of a small molecule’s off-target liabilities remains critical for ensuring clinical safety and guiding the selection of antibody clones to avoid unintended cross-reactivity.
While this approach has been explored for targets with existing cysteine-reactive covalent inhibitors, such as EGFR and BTK18, expanding its applicability will require novel chemistry to covalently modify proteins. Advances integrating chemoproteomics and immunopeptidomics may uncover new reactive cysteines presented on MHC molecules that are tumor-associated or tumor-specific54. Notably, covalent compounds may not need intrinsic function but could instead serve solely as handles for high-affinity, high-specificity antibody engineering. Optimized chemistry could stabilize rather than destabilize antigens, as observed with phosphorylated MHC peptides32,55. Beyond haptens, post-translationally modified HLA peptides—such as ubiquitination, SUMOylation, or phosphorylation modifications—may also present viable targets for this strategy55.
While prior studies have demonstrated enhanced antigen density through drug treatments to augment immunogenicity56,57,58,59,60, we establish an alternative approach—using the therapeutic itself. By leveraging covalent modifications on MHC peptides to generate tumor-specific neoepitopes, we establish a framework for developing high-affinity antibodies guided by detailed antigen characterization and molecular insights into binding and recognition. These engineered antibodies overcome challenges of low antigen density and poor stability, unlocking new opportunities for precision immunotherapy.
Methods
Cell lines, materials & plasmids
NCI-H2122 (CRL-5985), NCI-H2030 (CRL-5914), A375 (CRL-1619), and Raji (CCL-86) cells were purchased from the ATCC. HOP-62 cells were provided by the National Cancer Institute’s Division of Cancer Treatment and Diagnosis (NCI-DCTD) Tumor Repository. NCI-H2122, NCI-H2030, and HOP-62 cell lines and primary T cells were grown in R10 media comprising RPMI-1640 (Quality Biological, 10128), 10% FBS (Avantor, 76509-324), and 1% penicillin/streptomycin (Corning, 30-001-CI). A375 cells and derivatives were grown in complete DMEM (cDMEM) medium composed of DMEM (Corning, 45000-304), 10% FBS, and 1% penicillin/streptomycin. All cells were cultured at 37 °C with 5% CO2, and adherent cell lines were passaged using 0.05% Trypsin/0.53 mM EDTA (Corning, 25-051-CI). Lentiviral vectors and lentivirus were produced using VectorBuilder plasmid design and lentivirus preparation services. T cell engagers (TCEs) were prepared as 1 µM stocks in 1X Phosphate Buffered Saline (PBS) without calcium or magnesium (Corning, 21-040-CMR) and were stored at 4 °C for up to one month. IL-7 and IL-15 (Thermo Fisher, 200-07 and 200-15) were prepared at 10 µg/mL in sterile distilled water, diluted to 1 µg/mL in R10 medium, and stored at -80 °C as 1 mL aliquots. All cell lines were tested regularly and confirmed negative for mycoplasma contamination (Lonza, MycoAlert Mycoplasma Detection Kit, LT07-710).
Small molecules
For in vitro and in vitro studies, sotorasib (Selleck Chemical, S8830) divarasib (MedChemExpress, HY145928) and adagrasib (MedChemExpress, HY130149) were reconstituted in DMSO (MilliporeSigma, D2650), aliquoted, and stored at -20 °C until use. Puromycin (Mirus, MIR5940) was used at 1 µg/mL and blasticidin (InvivoGen, Ant-bl-1) was used at 10 µg/mL for selection of transduced cells based on dose response studies. For in vivo studies, sotorasib was purchased from MedChemExpress (HY114277) and prepared in 1% (v/v) Tween 80 (MilliporeSigma, P8074) + 2% (w/v) hydroxypropyl methylcellulose (MilliporeSigma, 423203) in sterile water.
Generation of NCI-H2122A*03-KO
NCI-H2122 cells were transfected with a vector containing Cas9-GFP and an HLA-A*03-targeting single-guide RNA (sgRNA). The vector was generated using a cloning plasmid ordered from VectorBuilder which contains a restriction site allowing for insertion of an sgRNA. The plasmid was linearized using BbsI-HF (NEB, R3539S). The sgRNA sequence (CATCCTGGATACTCACGACG) was designed using Alt-RTM CRISPR HDR Design Tool by IDT. Primers to synthesize the sgRNA (forward: 5’-CACCGCATCCTGGATACTCACGACG-3’ and reverse: 5’-AAACCGTCGTGAGTATCCAGGATGC-3’) were generated by IDT and annealed using T4 polynucleotide kinase (NEB, M0201S). The sgRNA was cloned into the linearized vector using T4 DNA ligase (NEB, M0202S). Cells were transfected using FuGene 4 K Transfection Reagent (Promega, E5911). After 48 h, Cas9+ cells were sorted based on their GFP expression, and single cell clones were expanded. Editing of the A*03 locus was validated by NGS amplicon sequencing using Illumina technology (performed by Azenta). Knockout of the HLA-A*03 allele was validated by flow cytometry with an anti-human HLA-A3 antibody (1:200 PE-HLA-A*03; clone GAP-A3, BD Pharmingen, 566605) and immunopeptidomics.
Generation of engineered cell lines
Cells were plated at 5 × 105 cells/well in 6-well plates in appropriate media. After 24 h, media was exchanged for fresh media with 10% FBS (no antibiotics) containing 5–10 µg/mL polybrene (MilliporeSigma, TR-1003) and lentivirus at an MOI of 1–10. After overnight incubation, media was exchanged for fresh media supplemented with 1 µg/mL puromycin and/or 10 µg/mL blasticidin to select for transduced cells. Cells were allowed to grow for 1–2 days before passaging into larger flasks with fresh selection marker. After 5–7 days in culture, viability, HLA expression, and/or eGFP expression was validated by flow cytometry using a BD FACSymphony A3 (FACSDiva Software v9.3.1). HLA expression was further validated by immunopeptidomics. KRASG12C overexpression was confirmed by western blotting. To ensure that there were no changes in the growth of engineered cell lines, 2 × 103 eGFP-expressing cells were plated in the presence of vehicle (DMSO) or 300 nM sotorasib in 96-well flat bottom plates. Plates were imaged every 6 h for 120 h using an Incucyte S3 (Sartorius, v2024A) at 20x magnification with phase and green (300 ms) channels and four images/well. eGFP+ cell confluence was quantified using an area-based mask and by averaging all four images.
Western blot
NCI-H2122 cells (5-10×106) were harvested and resuspended in lysis buffer comprising 1x RIPA Buffer (Thermo Fisher, 89900), 1x Halt Protease Inhibitor (Thermo Fisher, 78429), Benzonase (MilliporeSigma, E1014), and water. Samples were rotated at 4 °C for 10 min to lyse. Cell debris was cleared by centrifugation, and protein in the soluble fraction was quantified using the Pierce BCA Assay Kit (Thermo Fisher, 23227), following the manufacturer’s instructions. Samples were resolved on Invitrogen NuPage 4–12% Bis-Tris Protein Gels (Invitrogen, NP0329) and transferred to nitrocellulose membranes using the iBlot 2 Drying system (Thermo Fisher). Membranes were probed with Anti-Vinculin Recombinant Rabbit Monoclonal Antibody (Thermo Fisher, clone 42H89L44, 700062, 1:1000) and Anti-PanRas Mouse Monoclonal Antibody (MilliporeSigma, clone Ab-3, OP40, 1:1000,) overnight at 4 °C. Blots were washed with 1x TBS-T (Thermo Fisher) and incubated with the secondary antibodies Goat anti-Rabbit IgG (H + L) HRP Conjugate (Bio-Rad, #706515, 1:10,000) or Goat anti-Mouse IgG (H + L) HRP Conjugate (Bio-Rad, 1706516, 1:10,000) for 1 h at room temperature. Chemiluminescence detection was performed with the Pierce ECL Western Blotting Substrate (Thermo Fisher, 32209) and the iBright FL1500 imager (Invitrogen).
Preparation of activated human T cells
Peripheral blood mononuclear cells (PBMCs, STEMCELL Technologies, 70025.2) were thawed, transferred to 20 mL R10 media in T75 flasks, and incubated overnight to allow adherent cells to attach. The following day, suspension cells were counted and mixed with CD3/CD28 Dynabeads (Thermo Fisher, 11131D) using 25 µL beads/1 × 106 cells. Cells and beads were plated at a cell concentration of 1 × 106 cells/mL in R10 medium with 10 ng/mL each of IL-7 and IL-15 and incubated for 3 days before magnetic separation of T cells from the beads. T cells were replated and expanded to maintain a concentration of 1 × 106 cells/mL in fresh R10 medium with 10 ng/mL each of IL-7 and IL-15. After removing from the beads, T cells were expanded for at least 10 days before use in assays to ensure downregulation of activation markers. Beyond 10 days of expansion, T cell aliquots were frozen down in 90% FBS with 10% DMSO as aliquots of 40 − 80 × 106 cells/vial. After thawing in R10 medium with 10 ng/mL each of IL-7 and IL-15, frozen T cells were allowed to recover at least 2 days prior to use in assays.
Validation of sotorasib resistance
To assess sotorasib sensitivity, 2 − 3 × 103 NCI-H2122, NCI-H2030, or HOP-62 cells were seeded in 96-well, clear, flat bottom plates in R10 media. After 24 h, media were replaced with fresh media containing a titration of sotorasib. After 72 h, viability was assessed using CellTiterGlo (Promega, G9241). Luminescence was measured using a Spark plate reader (Tecan, SPARKCONTROL v3.2). Data were graphed and fitted using GraphPad Prism version 10.5.0.
Incucyte-based cytotoxicity analysis
eGFP-expressing cell lines were plated 1×104 cells/well in clear, flat bottom 96-well plates with 300 nM sotorasib, 400 nM divarasib or vehicle (0.03% DMSO). After 18–24 h, media were aspirated and exchanged for fresh media with 300 nM sotorasib, 400 nM divarasib or DMSO, human T cells at a 10:1 E:T ratio or human PBMCs at a 5:1 E:T ratio, and TCEs. After 48–72 h, plates were imaged using an Incucyte S3 (Sartorius) at 20x magnification with phase and green (300 ms) channels and four images/well. eGFP+ cell confluence was quantified using an area-based mask and by averaging all four images.
For peptide pulse experiments, A375eGFP cell lines were plated 1 × 104 cells/well in clear, flat bottom 96-well plates. After 18–24 h, media were aspirated and cells were treated with WT KRAS or divarasib-labeled p7 or p5 KRASG12C peptides prepared in cDMEM media. After 4 h peptide incubation, activated human T cells at a 10:1 E:T ratio and TCEs were added. After 48 h, plates were imaged and eGFP+ cell confluence was quantified as above.
For spheroid analysis, 1 × 104 NCI-H2122eGFP cells with 10 × 104 human T cells (10:1 E:T ratio) were plated in clear, round bottom 96-well plates with 300 nM sotorasib. After 18–24 h, fresh media was added containing 300 nM sotorasib and TCEs. After 48 h, plates were imaged using an Incucyte S3 at 4x magnification with phase and green (300 ms) channels and 1 image/well. The spheroid analysis software package (Sartorius) was used to quantify eGFP mean intensity within the spheroid boundary. All 2D and 3D cytotoxicity data was normalized to 0 nM TCE and transformed using the function Y = 100 – Y to produce cytotoxicity curves. Data were graphed and fitted using GraphPad Prism.
Activation flow cytometry
T cells or PBMCs from cytotoxicity assays were transferred to round bottom 96-well plates for staining at 48–72 h after plating. Cells were stained with a surface stain cocktail that included 1:50 BB515-CD3 (clone SK7, BD, 564560), 1:50 RB780-CD8 (clone RPA-T8, BD, 568684), 1:50 PerCP-Cy5.5-CD4 (clone SK3/Leu3a, BD, 566923), 1:100 PE-CD25 (clone M-A251, BioLegend, 985802), 1:100 APC-CD69 (clone FN50, BioLegend, 985206), and 1:2000 eFluor 780 viability dye (Invitrogen, 65-0865-14) for 30 min at 4 °C. After washing, cells were resuspended in FACS buffer [1X PBS, 2% FBS, 1 mM EDTA (Invitrogen, 15575020), 0.01% sodium azide (MilliporeSigma, S2002)] and analyzed using an iQue 3 HTS cytometer (Sartorius, Forecyt Standard Edition 9.0 R1). Flow cytometry data were analyzed using FlowJo version 10.10.0.
ELISA
Supernatants were collected from cytotoxicity assays at 48 h after plating. IFNy was quantified using the LEGENDplex Human CD8/NK Panel (13-plex) V02 (BioLegend, 741187). Standards were reconstituted and prepared as per the manufacturer’s instructions to generate an 8-point standard curve for each analyte. Supernatants were diluted by 2-4-fold with BioLegend assay buffer before shaking incubation together with standards in a V-bottom plate with analyte-specific capture beads for 2 h. Samples and standards were tested in technical triplicates and duplicates, respectively. Samples and standards were washed and incubated for 1 h with biotinylated detection antibody followed by addition of streptavidin-phycoerythrin for 30 min. After washing, samples and standards were analyzed on an iQue3 HTS cytometer. FCS files were uploaded to LEGENDPlex’s Data Analysis Software Suite, which enables cytokine quantification based on size and corresponding fluorescence and quantifies each cytokine based on the standard curve for each cytokine.
In vivo studies
CDX prophylactic mouse model
NCI-H2122 cells (1 × 106) were mixed with 10 × 106 expanded human T cells (STEMCELL Technologies, Donor ID RG1200, RG3564, or CE0009519) in 50% Matrigel/PBS and implanted subcutaneously in the hind flank of 8-week-old female NOD SCID mice (The Jackson Laboratory, strain 001303) on study day 0. Subsequently, 1 × 106 human T cells were prepared in PBS and administered intravenously (i.v.) via the tail vein weekly starting on day 0. Also on day 0, daily oral dosing with sotorasib (30 mg/kg) was initiated. TCEs or vehicle (PBS) (4–10 mice per group) were prepared in sterile PBS and administered i.v. starting on day 1. Body weights and tumor volumes (0.5 × height × width2, manually measured using digital calipers) were measured twice weekly following implantation. Donor characteristics are listed in Supplementary Table 1. The in vivo studies were carried out in a biosafety level 2 (BSL-2), specific pathogen-free (SPF) vivarium. Experimental and control mice were bred at The Jackson Laboratory and co-housed in the vivarium. Mice were monitored daily and euthanized via CO₂ inhalation before reaching the IACUC-approved maximum tumor volume of 2000 mm³, experiencing weight loss exceeding 15%, or meeting other humane endpoint criteria. Matching the XX genotype of the xenografted NCI-H2122 cancer cell line, only female mice were used.
Tumor processing and flow cytometry analysis
On study day 31, all animals were euthanized by CO2 inhalation, and the tumors were excised and weighed. A portion (50–150 mg) of excised tumors was used for flow cytometry, and the remainder was stored at -80 °C. Flow cytometry samples were digested by using the Tumor Dissociation System (Miltenyi, 130-095-9292) and then stained with a surface stain cocktail including 1:60 Zombie Aqua viability stain (BioLegend, 423102), 1:10 APC-TCRα/β (clone IP26, BioLegend, 306718), 1:200 APC-Cy7-CD4 (clone RPA-T4, BioLegend, 300518), 1:200 BV650-CD8 (clone T8, BioLegend, 301042), 1:40 BV711-CD69 (BioLegend, 310944), 1:60 BV421-CD25 (clone M-A251, BioLegend, 356114), 1:20 PerCP-eFluor710-HLA-DR (clone L243, Invitrogen, 46-9952-42), 1:60 PE-Dazzle594-ICOS (clone C398.4 A, BioLegend, 313532), and 1:100 Fc block (clone S17011E, BioLegend, 156604). After washing, samples were fixed with the Foxp3 Transcription Factor Staining Buffer Set (eBioscience, 00-5523-00) prior to staining with 1:20 PE-FOXP3 (clone 259D, BioLegend, 320208). Stained tumor samples were analyzed using a Thermo Fisher Attune NxT instrument.
CDX dose-finding and optimization
NCI-H2122 cells (1 × 106) were implanted as a co-mixed xenograft with 2 × 106 activated human T cells (STEMCELL Technologies, Donor ID 1110044729) in 8-week-old female NOD SCID mice. When tumors reached ~200 mm3 (D-1), daily oral dosing with 30 mg/kg sotorasib was initiated and a single dose of 1 × 106 T cells was administered intravenously. The next day (D0), AETX-RSV or AETX-R114 (each at 1 mg/kg) was dosed intravenously.
For pharmacokinetic (PK) dose-finding analysis, retroorbital bleeds were performed on mice at 24 h, 48 h, 72 h, 96 h, 168 h, and 336 h (n = 3 per timepoint) after treatment for analysis of plasma, and tumors were harvested from mice at 24 h, 72 h, 168 h (n = 3 per timepoint), and 336 h (n = 6) for tumor PK analysis. Blood was centrifuged to yield plasma and tumors were homogenized. White high bind plates (Greiner, 655094) were coated with streptavidin prepared in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, 7.7 mM NaH3, pH 9.6) and blocked with TBS, 0.05% (v/v) Tween-20, 5% (w/v) BSA, pH 7.4. Biotinylated human CD3δ/ε heterodimer (Acro Biosystems, CDD-H82W6) were added to coated plates. Duplicate samples of diluted plasma or diluted tumor homogenate were transferred to coated plates in parallel with a dose response of standard (AETX-R114) and incubated. Plates were washed, 1:10,000 HRP-Goat anti-Human IgG detection antibody (Jackson ImmunoResearch, 109-035-088) was added, and chemiluminescence was detected by using the SuperSignal ELISA Pico reagent (Thermo Fisher, 37070).
For pharmacodynamic (PD) analysis, tumors (n = 3 per timepoint) were obtained at 6 h, 24 h, 48 h, 72 h, and 168 h after treatment. Tumors were processed and analyzed as described under “Tumor processing and flow cytometry analysis”.
Protein engineering: p/p*MHC production
Production of β2m
Human recombinant Beta-2 microglobulin (β2m) containing a 6x Histidine-tag was expressed in Escherichia coli. Bacteria were cultivated overnight in Super Broth medium (Quality Biological, NC1115169) supplemented with 100 µg/mL carbenicillin (Teknova, C2135). When OD600 reached 0.5, cultures were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Thermo Fisher, AM9464). Following induction, the bacteria were harvested by centrifugation at 4500 g for 15 min using 500-mL conical tubes. Pellets were lysed in B-PER Complete Lysis Buffer (Thermo Fisher, 89822), followed by sonication. The insoluble fraction, containing β2m, was collected and solubilized overnight in 8 M urea and 20 mM Tris-HCl (pH 8.0). Solubilized β2m was loaded onto a 5-mL HisTrap HP immobilized metal affinity chromatography (IMAC) column (Cytiva, 17-5248-02) installed on a NGC Discover (Bio-Rad, 7880009) operated using ChromLab software (Bio-Rad, v6.1.29.0). A refolding gradient was applied in-column by gradually transitioning from the 8 M urea solubilization buffer to a native buffer consisting of 50 mM Tris-HCl, 250 mM NaCl, pH 8.0. This gradient was executed over 60 column volumes, and β2m was subsequently eluted using the native buffer supplemented with 500 mM imidazole.
The eluate was buffer-exchanged into the native buffer to remove imidazole, and the 6x Histidine-tag was cleaved by adding NEB TEV Protease (NEB, P8112S) at 1000 units of protease per 2 mg of tagged β2m and rotating overnight at 4 °C. Then, the mixture was centrifuged at 2000 g for 2 min to remove insoluble debris. A 5-mL sample of the reaction, containing approximately 15 mg of β2m, was loaded onto a HiLoad 16/600 Superdex 75 pg column (Cytiva, 28989333) installed on a Bio-Rad NGC Discover 10 Chromatography System and operated at a flow rate of 1 mL/min with 1x PBS as the running buffer. The tagless β2m was effectively separated from residual TEV protease and uncleaved His-tagged β2m.
Production of HLAs
HLAs containing a 6x Histidine-tag and an AviTag for biotinylation were expressed in E. coli. Bacteria were cultivated for 4 h in Super Broth medium supplemented with 50 µg/mL carbenicillin and 25 µg/mL chloramphenicol (MilliporeSigma, R4408), and at OD600 of 0.5, cultures were induced by adding 1 mM IPTG. Chloramphenicol and 50 µM biotin were added to co-express BirA, which catalyzed biotinylation of the HLAs. Following induction, bacteria were harvested by centrifugation at 4,500 g for 15 min in 500-mL conical tubes, and pellets were lysed in Thermo Fisher B-PER Complete Lysis Buffer and sonicated. The insoluble fraction, containing HLAs, was collected and solubilized overnight in a buffer containing 8 M urea and 20 mM Tris-HCl (pH 8.0). Solubilized HLAs were loaded onto a 5-mL HisTrap HP IMAC column installed on a Bio-Rad NGC Discover 10 Chromatography System operated using ChromLab software. The column was equilibrated with the 8 M urea solubilization buffer, and HLAs were eluted in the same buffer supplemented with 500 mM imidazole.
Peptide synthesis
All synthetic peptides were custom synthesized from Biosynth ( > 95% peptide purity, >99% isotopic purity for SIL amino acids). SIL valine: 13C6, SIL lysine: 13C6, 15N2.
Hapten-peptides conjugation and purification
Sotorasib, adagrasib, and divarasib were dissolved in N,N-dimethylformamide (DMF, MilliporeSigma, 270547) to a concentration of 40 mM. KRASG12C peptides were dissolved in water to 4 mM and conjugated to drugs at a molar ratio of 1:2 peptide:drug overnight at room temperature, protected from light. The final reaction composition was 45% DMF (v/v) in water, 4 mM drug, 2 mM KRASG12C peptide, and 50 mM Tris-HCl (pH 8.0).
Conjugated peptide-haptens were purified by reverse phase HPLC using a Zorbax Eclipse Plus C18, 95 A, 4.6 × 250 mm, 5-µm column (Agilent, 959990-902). The mobile phase comprised (buffer A) water + 0.1% (v/v) trifluoroacetic acid (TFA) and (buffer B) acetonitrile + 0.5% (v/v) water + 0.1% (v/v) TFA. A gradient of 10–95% buffer B over 20 min was used to separate the unconjugated KRASG12C peptide, the conjugated peptide-hapten, and the free drug. The isolated peptide-hapten was dried by vacuum centrifugation and re-dissolved in water.
p/p*MHC refolding and purification
p/p*MHCs were assembled in 50-mL tubes by first adding β2m (3 µM) and peptide-hapten (30 µM) to PBS. Following brief mixing, HLA (6 µM) was introduced, and the mixture was gently rotated overnight at 4 °C, protected from light. Tubes were then centrifuged at 4500 g for 10 min. The protein mixture was concentrated 10-fold by using Amicon Ultra protein concentrators (MilliporeSigma, UFC8010, 10 kDa MWCO). Concentrated samples were centrifuged at 2000 g for 2 min to remove insoluble aggregates, and a 1-mL aliquot was loaded onto a Superdex 75 Increase 10/300 GL column (Cytiva, 29148721) installed on a Bio-Rad NGC Discover 10 Chromatography System operated using ChromLab software. Size exclusion chromatography (SEC) was performed to separate the refolded p*MHC complex from soluble aggregates, excess β2m, and unbound peptide-hapten. Following purification, eluted p/p*MHC complexes were concentrated to approximately 10 µM.
UV-mediated peptide exchange for hipMHCs
UV-mediated peptide exchange was performed using single SIL-labeled synthetic peptides and recombinant, biotinylated Flex-T monomers (BioLegend, 280005 & 280007)26. Stable complex concentrations after peptide exchange were measured using the Flex-T HLA Class I ELISA assay (BioLegend) according to the manufacturer’s instructions (BioLegend, 280301, 421701, 280302, 280303, 423501).
Antigen characterization
p/p*MHCs stability analysis
Stability of p*MHCs in vitro was analyzed by multiplex bead binding assay61, as follows: Biotinylated p*MHCs were immobilized at 5% nominal saturation to beads (Dynabeads M-270 Streptavidin, Thermo Fisher, 65305) labeled with different concentrations of DyLight 650 (Thermo Fisher, 84547) to allow pooling of multiple samples. Bead pools were resuspended in BSS (500 mM Tris-HCl pH 7.5, 1.5 M NaCl, 50 g/L BSA) and incubated in absence or presence of 10 nM TCEs at 37 °C for varying amounts of time, as reported in the figures (typically 24, 8, 4, 2, 1, 0.5, 0.25 h). After incubation, samples were placed on ice, washed with BSST (BSS + 0.05% Tween 20), and residual folded p/p*MHCs were detected with PE anti-human β2m antibody (clone 2M2, 1:500, BioLegend, 316305). Flow cytometry data were collected using a BD FACSymphony A3 or an IntelliCyt iQue3 flow cytometer (Sartorius). The signal intensity of PE was normalized to the values in the absence of incubation (100% folded p/p*MHC) and in the absence of antigen (0% folded p/p*MHC), for each sample. Half-life values were obtained by fitting the data with a one phase decay equation built in Prism.
Peptide MHC binding prediction
Peptide binding predictions were performed by using NetMHCpan 4.131. Reported values correspond to EL (eluted ligand) binding scores, where threshold scores of 0.5% and 2% identify predicted strong and weak binders, respectively.
HLA allele frequency
HLA allele frequency was estimated across subpopulations from the U.S. National Marrow Donor Program62, accessed via the Allele Frequency Net Database63. Phenotypic frequency (PF) was derived from allele frequency (AF) by assuming Hardy-Weinberg equilibrium. Overall frequency for all races/ethnicities was population-weight corrected by using Census 2020 data64.
Immunopeptidomics
HLA peptide isolation and purification
At the time of harvest, tissue culture cells were washed with 1X PBS, removed from plates by using 0.05% Trypsin-EDTA, recovered by centrifugation at 1000 g for 3 min, washed once with 1X PBS, and snap frozen in liquid N2. Cell pellets were lysed for 10 min in 600 µL of lysis buffer [20 mM Tris-HCl pH 8.0, 100 mM NaCl, 6 mM MgCl2, 1 mM EDTA, 1.5% (v/v) Triton X-100 (MilliporeSigma, T9284), 60 mM octyl B-D-glucopyranoside (MilliporeSigma, O8001, 1 mM phenylmethanesulfonyl fluoride solution (MilliporeSigma, 93482), and 1X Halt Protease Inhibitor] and 1 µL Pierce Universal Nuclease (Thermo Fisher, 88702) per 5 × 107 cells. Lysates were cleared by centrifugation for 5 min at 10,000 g and 4 °C. For quantitative analyses, 1 fmol of p7/soto-p7 and 5 fmol of p8/soto-p8 were spiked into cell lysates prior to immunoprecipitation.
HLA complexes were immunoprecipitated by incubating 40 µL per 5 × 107 cells of GammaBind Plus Sepharose Beads (Cytiva, 17088601), pre-charged for 3 h, with 100 µg per 5 × 107 cells of anti-human MHC Class I (HLA-A, HLA-B, HLA-C) antibody (clone W6/32, BioXCell, BE0079) and rotating overnight at 4 °C. HLA-I complexes were eluted from the beads by using an adapted, high-throughput, plate-based approach7,9. The following wash and elution sequence (1 mL) was performed using 96-well filter plates with a 25-µm polyethylene frit (Agilent, 204495): 1 × methanol, 1 x acetonitrile, 1 × 1% (v/v) formic acid, 1 × 10 mM Tris-HCl pH 8.0, add lysate and beads, 2 x Tris HCl pH 8.065,66. Peptides were eluted twice in 200 µL of 10% (v/v) formic acid for 5 min into a 96-deep well plate. Peptides were isolated from antibody, HLA, and β2m using a passivated 10-kDa molecular weight cutoff filter (Cytiva, OD010C34) by centrifugation26.
For both data dependent acquisition (DDA) and p7/p8-targeted methods, filtered peptides were reduced and alkylated to enable cysteine detection by mass spectrometry (MS), and all peptides were desalted, frozen, and dried by vacuum centrifugation as detailed9. Peptides were resuspended in 3% (v/v) acetonitrile / 5% (v/v) formic acid prior to MS analysis.
MS data acquisition & analysis
MS data were acquired by using an Orbitrap Ascend Tribrid MS (Thermo Scientific, v4.0.4091), Vanquish Neo Ultra-High Performance Liquid Chromatography system (Thermo Scientific, v2.1.1.51769), and Aurora Ultimate XT 25 cm × 75 ID 1.7 um C18 analytical chromatography columns (IonOpticks, AUR4-25075C18-XT) operated at 55 °C. The specified injection volume of peptides was directly loaded onto the Aurora Ultimate XT chromatography column. All MS files, analysis type, cell number used, and associated figures, can be found in Supplementary Data 1.
DDA
Standard MS parameters were as follows: spray voltage, 1.9 kV; no sheath or auxiliary gas flow; heated capillary temperature, 300 °C. Peptides were eluted over a 120-min gradient ranging from 3–44% buffer B (acetonitrile + 0.1% (v/v) formic acid), at a rate of 200 nL/min. Buffer A is 0.1% formic acid.
Full scan mass spectra (350-1,400 m/z, 60,000 resolution) were detected in the Orbitrap analyzer with an automatic gain control (AGC) target of 4e5, automatic maximum injection time (auto max IT). Precursor ions were filtered with a minimum intensity threshold of 2.5e4, precursor fit error of 50%, precursor fit window of 1.1, and dynamic exclusion time of 60 s after a fragmentation event. For each full scan, MS2 spectra were collected within a 3-s cycle time. Ions were isolated in the quadrupole (0.7 m/z isolation window) with an AGC target of 5e4, auto max IT, and fragmented by higher-energy collisional dissociation (HCD) with a normalized collision energy (nCE) of 25% at a 45,000 resolution. Charge states 2, 3, and 4 were fragmented with priority, and +1 ions were fragmented with secondary priority and filtered for a mass range of 800-1,400 m/z.
DDA mass spectra were analyzed with Proteome Discoverer (version 3.0.1.27) and searched by using Sequest HT + INFERYS rescoring against the human SwissProt database (version 2023-06-28) plus a custom KRASG12C database with dynamic methionine oxidation and static cysteine carbamidomethyl modifications. MS/MS spectra were matched with a precursor mass tolerance of 10 ppm on 20 mmu for fragment ions. Identified peptides were filtered for search engine rank = 1, percolator q-value ≤ 0.01.
Targeted MS analysis
Prior to analysis, 100 fmol of each double SIL-synthetic peptide for the acquisition target(s) were spiked into each sample before injection. Standard MS parameters for SureQuant acquisition were as follows: spray voltage: 1.9 kV, no sheath or auxiliary gas flow, heated capillary temperature: 300 °C. The custom SureQuant acquisition template available in Thermo Scientific Orbitrap Ascend 4.0 was used for this method. More detailed instructions for building an MHC, isotopologue-based SureQuant acquisition method have been described27,67.
Briefly, full scan mass spectra (350-1,700 m/z, 120,000 resolution) were detected in the Orbitrap analyzer with an AGC target of 1e6, auto max IT). The targeted mass filter was used to identify double stable isotope-labeled (SIL)-synthetic peptide precursor ions matching the defined m/z values (+/- 5 ppm). Precursors exceeding a defined intensity threshold were isolated (isolation window 0.4 m/z) and fragmented (nCE: 18% for soto-p7/p8, 28% for ada-p7, 28% for p7/p8, 20% for diva-p7) with a scan range of 150-1,700 m/z, 22,500 resolution, 43 second max IT, AGC target of 5e5. A product ion trigger filter next performed pseudo-spectral matching, triggering an MS2 event of the endogenous target peptide and (optionally) the single SIL-synthetic peptide (hipMHC) if n ≥ 5 product ions were detected from the defined list (+/- 10 ppm). If triggered, MS2 scans were initiated at the defined mass offsets to capture the endogenous and hipMHC peptides with the same isolation window, AGC target, and collision energy as the double SIL-synthetic peptide MS2 scan, but with a resolution of 120,000 and max IT of 350 ms. LC gradients were customized for each target to optimize data acquisition and sensitivity.
All targeted MS data were analyzed by using Skyline (version 23.1.0.455)68. Mirror plots of endogenous and SIL-synthetic peptides to confirm spectral matches were generated by using the Interactive Peptide Spectral Annotator tool69. The top 200 most abundant ions for each spectra were extracted from Thermo Scientific FreeStyle Data Visualization software (version 1.8.63.0) and plotted with a fragment tolerance of +/- 5 ppm, matching threshold of 0.25%. Integrated peak areas of the top 6 more abundant product ions (b, y, a) for the endogenous and single SIL-synthetic internal standard were exported from Skyline and summed. The endogenous peptide concentration was estimated by taking the ratio of endogenous to single SIL-synthetic internal standard (hipMHC peptide) and multiplying by the concentration of hipMHC. The number of molecules was divided by the number of cells to estimate copies-per-cell (CPC). The linear intensity response of the single-point embedded calibrant was validated between the lower and upper limits of defined quantitation (see Supplementary Fig. 1b).
Antibody discovery, production, and characterization
Affinity maturation of R023
Affinity maturation of R023 was performed by yeast display sorting of a structure-guided
combinatorial library, as schematized in Fig. 2a. In the first two rounds, the library was sorted to isolate yeast clones binding to both soto-p7/A*03 and soto-p7/A*11. In the third round, “off-rate” sorting was performed to isolate clones dissociating more slowly from soto-p7/A*11 than from the parental clone R02333. Briefly, yeast cells from the second round of sorting and displaying parental scFv R023, were stained with 10 nM biotinylated soto-p7/A*11 for 30 min, followed by the removal of unbound antigen and subsequent incubation with 100 nM non-biotinylated competitor (non-biotinylated soto-p7/A*03) for 2 h at room temperature. The binding signal from the parental scFv R023 was used as reference, and clones showing binding signal higher than the parental were recovered after sorting on an S3e FACS (Bio-Rad). Single clones derived from the third sorting round were isolated and re-analyzed in the same “off-rate” setting for binding to both soto-p7/A*11 and soto-p7/A*03 (Fig. 2b).
TCE production
AETX-R114 and AETX-RSV were produced and purified by WuXi Biologics using CHO cells. Harvested supernatant were purified using Protein A affinity chromatography and polished using SEC. The purified proteins were exchanged to 10 mM HEPES, 150 mM NaCl, 3 mM EDTA (pH 7.4). Both purified AETX-R114 and AETX-RSV TCEs were >99% pure and monodiperse, as evaluated by SEC-HPLC. Endotoxin contamination of the purified TCEs was assessed by using EndoSafe LAL Cartridges (Charles River, PTS-2005). Both TCEs had endotoxin levels lower than 0.1 EU/mg. TCE AETX-R302 was expressed in Expi293F cells (Thermo Fisher, A14527) and purified using a Protein A HP column (Cytiva, 17040201) and subsequently loaded onto a Superdex 200 column (Cytiva, 28989336) for polishing.
Surface plasmon resonance
The affinities of TCEs for all cognate antigens (p/p*MHCs) exploited in this study were determined by using a Biacore 1 K+ instrument (Cytiva, Insight Control v5.0.18.22102) loaded with a Series S high-affinity streptavidin (SA) sensor chip (Cytiva, BR100531) and 6 flow cells. The sensor chip was prepared by a 10-min regeneration step with a solution composed of 50 mM NaOH and 1 M NaCl followed by three 30-s regeneration steps with 10 mM glycine-HCl (pH 2.0). The running buffer was 1xHBS-EP+ (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20). Biotinylated p/p*MHCs were diluted in 1xHBS-EP+ running buffer to a concentration of 5 nM and captured in each individual flow cell at a flow rate of 10 µL/min. Flow cell 1 was not captured with any biotinylated antigen and was used for reference subtraction. For kinetic (kon and koff) and thermodynamic (Kd) measurements, single-cycle kinetics were performed at 25 °C or 37 °C with 1xHBS-EP+ as running buffer. The TCEs were diluted in 1xHBS-EP+ running buffer, and injected into all 6 flow cells as analytes at 37 °C at a flow rate of 30 µL/min with concentrations of 0.3125 nM, 0.625 nM, 1.25 nM, 2.5 nM, and 5 nM for AETX-R114 binding to soto-p7/A*03/11, 1.25 nM, 2.5 nM, 5 nM, 10 nM, and 20 nM for AETX-R114 binding to soto-p7/A*68 and AETX-R023 binding to soto-p7/A*03, and 5 nM, 10 nM, 20 nM, 40 nM, and 80 nM for AETX-R023 binding to soto-p7/A*11. Association was for 2 min, and dissociation was monitored for 10 min or 20 min. For measuring the affinities of AETX-R302 TCE binding to diva-p5/A*02, diva-p7/A*03/11 and soto-p7/A*03 at 25 °C, TCEs were diluted in 1xHBS-EP+ running buffer and injected into all 6 flow cells at a flow rate of 30 µL/min with concentrations of 1 nM, 3 nM, 9 nM, 27 nM and 81 nM. For single-cycle kinetics of AETX-R114 and AETX-R023 to CD3, biotinylated CD3δ/ε heterodimer (Acro, CDD-H82W6) was captured on Series S high-affinity streptavidin (SA) sensor chip. AETX-R114 and AETX-R023 were diluted in 1xHBS-EP+ running buffer and injected as analytes at 37 °C and a flow rate of 30 µL/min with concentrations of 1.25 nM, 2.5 nM, 5 nM, 10 nM, and 20 nM. Sensorgram data were double referenced (subtraction of running buffer alone and flow cell 1) and fit to a 1:1 binding kinetics fitting model in Biacore evaluation software (Biacore Insight Evaluation Software Version 5.0.18.22102) to yield association and dissociation rate constants kon and koff, respectively. Kd was calculated as koff/kon.
For one-cycle binding measurements of AETX-R114 to biotinylated ligands HLA-A*3 (Kactus, MHC-HM44RB) or HLA-A*11 (Kactus, MHC-HM41RB) loaded with soto-p7, p7WT or G12W mutated peptide p7W12, biotinylated ligands were captured on a Series S high-affinity streptavidin (SA) sensor chip, and AETX-R114, diluted in 1xHBS-EP+ to a final concentration of 80 nM, was injected into all 6 flow cells at 25 °C at a flow rate of 30 µL/min. For one-cycle binding measurement of AETX-R302 to ligands diva-p5/A*02, diva-p7/A*11, p5WT/A*02, p7WT/A*03 and p7WT/A*11, biotinylated ligands were captured on a Series S high-affinity streptavidin (SA) sensor chip, and AETX-R302, diluted in 1xHBS-EP+ to a final concentration of 100 nM, was injected into all 6 flow cells at 25 °C and a flow rate of 30 µL/min.
AEthena library generation
A human naïve antibody phage display library in Fab format (AEthena library) was constructed as described70. Briefly, total RNA from bone marrow mononuclear cells (BMMC) or PMBC of 15 healthy donors (STEMCELL Technologies, 70001 & 70025) was extracted using TRI reagent (Molecular Research Center, TR118) and reverse transcribed. The resultant cDNAs were used as templates for PCR amplification of the V domains of antibody light (VL: Vκ or Vλ) and heavy (VH) chains. The same amount of amplified Vκ, Vλ, and VH from each donor was mixed and used as templates for assembly with the corresponding light chain constant domain (CL: Cκ or Cλ) to create VL-CL-VH - DNA fragments with randomly paired VL and VH. The assembled DNA cassette was digested with SfiI and ligated upstream of CH1-Δp3 into the SfiI-digested phagemid using T4 DNA ligase, yielding phagemids encoding Fabs linked to the N-terminus of a fragment of protein III (ΔpIII: amino acids 230–406) of filamentous phage M1342. The ligated phagemid library was eletroporated into E. coli strain SS320 (Biosearch Technologies, 60512). The kappa and lambda sub libraries of the AEthena library comprised 4.9 × 1011 and 2.3 × 1011 independent Fab clones, respectively.
Antibody discovery: R302
Antibody R302 was identified through 4 rounds of phage display selection of the AEthena library, followed by additional rounds of yeast display sorting, as follows. Briefly, the phage display library was incubated with 100 nM biotinylated diva-p7/A*03 and 100 nM diva-p7/A*11 bound to streptavidin magnetic beads (Promega, Z5482) at room temperature for 2 h. The magnetic beads were then captured by using a magnetic separation stand (Promega, Z5342), washed 4 times with washing buffer, resuspended in PBS, and used to infect E. coli strain ER2738 (Biosearch Technologies, 60522) for subsequent phage propagation. From rounds 2 to 4, the selection was divided into 2 arms to recover diva-p7/A*03 and diva-p7/A*11 binders, respectively. In both selections, the phage pool was pre-cleared with 100 nM biotinylated p7WT/A*03 and 100 nM p7WT/A*11 bound to streptavidin magnetic beads at room temperature for 2 h and then subjected to positive selection with 100 nM diva-p7/A*03 or 100 nM diva-p7/A*11. After 4 rounds of selection, enrichment of phage binders for diva-p7/A*03/A*11 over p7WT/A*03/A*11 was observed in both the kappa and lambda sub libraries. The 4th round phage output pool was cloned into the yeast display vector pYD1 (Creative Biogene, OVT2893) in scFv format and subjected to two rounds of positive sorting on a Melody FACS (BD, FACSChorus 3.0). For the first round, yeast cells were sorted in the presence of 100 nM of divarasib p*MHC complexes (diva-p5/A*02, diva-p7/A*03 and diva-p7/A*11). This procedure was repeated in the second round in the presence of 1 µM free divarasib. Single clones isolated from the second round were analyzed on an IntelliCyt iQue3 flow cytometer (Sartorius), and clone R302, isolated from the diva-p5/A*02 sort, was chosen for further characterization (Supplementary Fig. 12a).
The antibodies described in this study are proprietary to Aethon Therapeutics and are not publicly available. Reasonable requests for access to these materials for non-commercial academic research purposes may be considered by the corresponding authors, subject to a material transfer agreement.
Structural characterization of R302-p*MHC complexes
Fab expression
Fab R302 was expressed and purified as reported33. Briefly, genes encoding antibody R302 were cloned into a vector for the expression of Fab with a rigidified elbow sequence71. Fab R302 was produced in E. coli strain 55244 (ATCC), grown overnight at 30 °C in Super Broth medium, and purified using a 5-mL HiTrap Protein G affinity column (Cytiva, 17040501). Purity was confirmed by SDS-PAGE.
Purification of R302-p*MHC complexes
Fab:antigen complexes were formed by mixing purified divarasib p*MHCs with Fab R302 at a molar ratio of 1:1.2 and incubating on ice for 30 min. Complexes were purified by SEC on a Superdex 75 Increase 10/300 GL column equilibrated in PBS. Fractions containing the complexes, as confirmed by SDS-PAGE, were concentrated to >2.5 g/L and stored in PBS at −80 °C until use.
Cryo-EM sample preparation
Samples were prepared for single particle cryo-EM as reported33. Briefly, fluorinated octyl maltoside (Anatrace, O310F) was added to a final concentration of 0.7 mM to cryo-EM samples (3 µL for each sample; R302_diva-p5/A*02 at 2.6 g/L and diva-p7/A*03 at 4.2 g/L), right before their application to the grids (Quantifoil 0.6/1 300 mesh grids, coated in-house with gold foil72) and subsequent freezing. The grids were preventively glow-discharged for 30 s in the PELCO easiGlow Glow Discharge Cleaning System (Ted Pella), immediately before sample application. All grids were frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) set as follows: chamber temperature 4 °C, chamber humidity 100%, no waiting time, blot time 4 s, blot force 5.
Cryo-EM data collection and processing
Cryo-EM datasets were collected at the New York University (NYU) Cryo-EM Laboratory using Leginon 3.616,73. All data acquisitions were performed on a Titan Krios (Thermo Fisher Scientific) with a K3 camera (Gatan) at a magnification of 105,000×, corresponding to a super-resolution pixel size of 0.4125 Å (or a nominal pixel size of 0.825 Å after binning by 2). Zero-loss images were obtained by using an energy filter slit width of 20 eV. For R302_diva-p5/A*02, movies were collected at a dose rate of 26.1e−/Å2/s with a total exposure of 1.8 s, for an accumulated dose of 47.0 e−/Å2. For R302_diva-p7/A*03, movies were collected at a dose rate of 26.4 e−/Å2/s with a total exposure of 1.8 s, for an accumulated dose of 47.5 e−/Å2. Intermediate frames were recorded every 0.04 s for a total of 40 frames per micrograph. All images were collected at a nominal defocus range of 0.5 to 2.7 μm. Ice thickness was determined as described74. On-the-fly processing was performed using MotionCor2 v 1.6.475 and CTFFIND4 v 4.1.1476 under control of Appion77. A total of 6,339 images were collected for sample R302_diva-p5/A*02, and 4,371 images were collected for sample R302_diva-p7/A*03.
Further data processing was performed by importing the micrographs into cryo-SPARC v4.6.278 and following a common strategy for both samples, as schematized in Supplementary Fig. 15 and specified hereafter. Using Blob Picker, an initial set of particles was identified on a subset of micrographs (500 to 1000) with a contrast transfer function (CTF) fit of better than 6.0 Å. Particles were then extracted (384 pixels box size binned to 128 pixels) and subjected to multiple subsequent steps of 2D classification and selection for a first round of particle cleaning. The resulting particle subsets were used as templates to pick a new set of particles with Template Picker, this time from the whole set of micrographs, followed by particle extraction (384 pixels box size binned to 128 pixels) and multiple subsequent steps of 2D classification and selection. The new subset of particles was used to generate four 3D maps (classes), performing a heterogeneous ab initio reconstruction. The resulting 3D maps and the associated particles obtained were further processed by multiple steps of heterogeneous refinement, using decoy volumes to further clean the particles subsets. The particles obtained were re-extracted without binning (384 pixels box size). Then, after a step each of global and local CFT refinement, and the removal of particle duplicates, they were used for a step of nonuniform refinement79. Finally, local refinement was performed by using a mask that enclosed the p*MHC and R302 variable chains (made using UCSF Chimera 1.18 software80), resulting in the final map. For R302_diva-p5/A*02, final local refinement was performed with 65,281 particles, giving a map with an overall resolution of 3.23 Å. For R302_diva-p7/A*03, final local refinement was performed with 166,767 particles, giving a map with an overall resolution of 2.58 Å. These two maps were used to build structure models using Coot 0.9.881. As a starting model, we used PDBID 8VRA82. The final structures were validated using the PDB deposition tool, obtaining the statistics summarized in Supplementary Table 4. Cryo-EM maps and structures were visualized and imaged using UCSF Chimera X 1.1083. Interaction interfaces were then analyzed using PDBePISA 1.48 (https://www.ebi.ac.uk/pdbe/prot_int/pistart.html)84, and geometric analysis of the docking angles of antibody-p*MHCs complexes was performed using the web server TCR3d85.
Gating schemes
Exemplary gating schemes for all flow cytometry-based experiments are depicted in Supplementary Fig. 19.
Statistics & reproducibility
For in vivo and in vivo studies, no statistical method was used to predetermine sample size. For in vitro immunological studies, 4–6 different human T-cell donors were used to demonstrate validity and reproducibility. No mice were excluded from any analyses. For in vitro studies, all biological and technical replicates were included except for Fig. 3a, Supplementary Fig. 14f,and Supplementary Fig. 7b-c, where outliers were excluded. Outliers were defined as data points that were outside 1.5*IQR or were Incucyte images (Supplementary Fig. 14f) which contained an imaging error. These exclusion criteria were not pre-established. Regarding randomization, for all in vivo studies involving tumor-bearing mice, subcutaneous xenograft size was measured prior to assigning mice to treatment groups, ensuring each group had a comparable average tumor size. The investigators were not blinded to allocation during experiments and outcome assessment.
Ethical statement
Animal protocols were approved by the Institutional Animal Care and Use Committees of ATP R&D (IACUC, ATP-105) and Mispro (IACUC, 2024-AET-01) and were performed according to the NIH Guide for the Care and Use of Laboratory Animals.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry data files have been deposited in MassIVE repository under the dataset identifier MSV000097193. Supplementary Data 1 contains a file map of MS files. The cryo-EM maps and structure data have been deposited in the Electron Microscopy Database and PDB (EMD-4936186 (https://www.emdataresource.org/EMD-49361), EMD-4936287 (https://www.emdataresource.org/EMD-49362), PDBID 9NFB88 (https://www.wwpdb.org/pdb?id=pdb_00009nfb), and PDBID 9NFC89 (https://www.wwpdb.org/pdb?id=pdb_00009nfc). The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Tran, E. et al. Cancer immunotherapy based on mutation-specific CD4 + T cells in a patient with epithelial cancer. Science (1979) 344, 641–645 (2014).
Borgers, J. S. W. et al. Personalized, autologous neoantigen-specific T cell therapy in metastatic melanoma: a phase 1 trial. Nat. Med 31, 1–13 (2025).
Yarchoan, M. et al. Personalized neoantigen vaccine and pembrolizumab in advanced hepatocellular carcinoma: a phase 1/2 trial. Nat. Med 30, 1044–1053 (2024).
Xie, N. et al. Neoantigens: promising targets for cancer therapy. Signal Transduct. Target Ther. 8, 1–38 (2023).
Katsikis, P. D., Ishii, K. J. & Schliehe, C. Challenges in developing personalized neoantigen cancer vaccines. Nat. Rev. Immunol. 24, 213–227 (2023).
Ragone, C., Cavalluzzo, B., Mauriello, A., Tagliamonte, M. & Buonaguro, L. Lack of shared neoantigens in prevalent mutations in cancer. J. Transl. Med 22, 1–17 (2024).
Gurung, H. R. et al. Systematic discovery of neoepitope–HLA pairs for neoantigens shared among patients and tumor types. Nat. Biotechnol. 42, 1107–1117 (2023).
Chang, A. Y. et al. Opportunities and challenges for TCR mimic antibodies in cancer therapy. Expert Opin. Biol. Ther. 16, 979 (2016).
Choi, J. et al. Systematic discovery and validation of T cell targets directed against oncogenic KRAS mutations. Cell Rep. Methods 1, 100084 (2021).
Douglass, J. et al. Bispecific antibodies targeting mutant RAS neoantigens. Sci. Immunol. 6, eabd5515 (2021).
Bear, A. S. et al. Biochemical and functional characterization of mutant KRAS epitopes validates this oncoprotein for immunological targeting. Nat. Commun. 12, 4365 (2021).
Zhang, M. et al. Identification and affinity enhancement of T-cell receptor targeting a KRASG12V cancer neoantigen. Commun. Biol. 7, 1–13 (2024).
Poole, A. et al. Therapeutic high affinity T cell receptor targeting a KRASG12D cancer neoantigen. Nat. Commun. 13, 1–15 (2022).
Tran, E. et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016).
Benton, A. et al. Mutant KRAS peptide targeted CAR-T cells engineered for cancer therapy. Cancer Cell 43, 1365–1376 (2025).
Salem, M. E. et al. Landscape of KRASG12C, associated genomic alterations, and interrelation with immuno-oncology biomarkers in kras-mutated cancers. JCO Precis Oncol. 6, e2100245 (2022).
Li, X., Gluth, A., Zhang, T. & Qian, W. J. Thiol redox. Proteom.: Charact. Thiol-based Post-Transl. Modif. Proteom. 23, e2200194 (2023).
Hattori, T. et al. Creating MHC-restricted neoantigens with covalent inhibitors that can be targeted by immune therapy. Cancer Discov. 13, 132–145 (2023).
Zhang, Z. et al. A covalent inhibitor of K-Ras(G12C) induces MHC class I presentation of haptenated peptide neoepitopes targetable by immunotherapy. Cancer Cell 40, 1060–1069 (2022).
Batrash, F., Kutmah, M. & Zhang, J. The current landscape of using direct inhibitors to target KRASG12C-mutated NSCLC. Exp. Hematol. Oncol. 12, 1–19 (2023).
Liu, J., Kang, R. & Tang, D. The KRAS-G12C inhibitor: activity and resistance. Cancer Gene Ther. 29, 875–878 (2021).
Isermann, T., Sers, C., Der, C. J. & Papke, B. KRAS inhibitors: resistance drivers and combinatorial strategies. Trends Cancer 11, 91–116 (2024).
Awad, M. M. et al. Acquired resistance to KRAS G12C inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Hamarsheh, S., Groß, O., Brummer, T. & Zeiser, R. Immune modulatory effects of oncogenic KRAS in cancer. Nat. Commun. 11, 1–11 (2020).
Kemp, S. B. et al. Efficacy of a Small-Molecule Inhibitor of KrasG12D in Immunocompetent Models of Pancreatic Cancer. Cancer Discov. 13, 298–311 (2023).
Stopfer, L. E., Mesfin, J. M., Joughin, B. A., Lauffenburger, D. A. & White, F. M. Multiplexed relative and absolute quantitative immunopeptidomics reveals MHC I repertoire alterations induced by CDK4/6 inhibition. Nat. Commun. 11, 1–14 (2020).
Stopfer, L. E. et al. Absolute quantification of tumor antigens using embedded mhc-i isotopologue calibrants. Proc. Natl. Acad. Sci. USA 118, e2111173118 (2021).
Gallien, S., Kim, S. Y. & Domon, B. Large-scale targeted proteomics using internal standard triggered-parallel reaction monitoring (IS-PRM). Mol. Cell. Proteom. 14, 1630–1644 (2015).
Canon, J. et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575, 217–223 (2019).
Sidney, J., Peters, B., Frahm, N., Brander, C. & Sette, A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9, 1–15 (2008).
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48, W449–W454 (2020).
Mohammed, F. et al. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for presentation of transformed self. Nat. Immunol. 9, 1236 (2008).
Maso, L. et al. Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes. Proc. Natl. Acad. Sci. USA 121, e2319029121 (2024).
Boder, E. T. & Wittrup, K. D. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 14, 55–62 (1998).
Rodrigues, M. L., Shalaby, M. R., Werther, W., Presta, L. & Carter, P. Engineering a humanized bispecific F(ab’)2 fragment for improved binding to T cells. Int J. Cancer Suppl. 7, 45–50 (1992).
Giffin, M. J. et al. AMG 757, a half-life extended, DLL3-targeted bispecific T-cell engager, shows high potency and sensitivity in preclinical models of small-cell lung cancer. Clin. Cancer Res. 27, 1526–1537 (2021).
Middleton, M. R. et al. Tebentafusp, a TCR/Anti-CD3 bispecific fusion protein targeting gp100, potently activated antitumor immune responses in patients with metastatic melanoma. Clin. Cancer Res. 26, 5869–5878 (2020).
Rohaan, M. W. et al. MART-1 TCR gene-modified peripheral blood T cells for the treatment of metastatic melanoma: a phase I/IIa clinical trial. Immuno-Oncol. Technol. 15, 100089 (2022).
Sailer, N. et al. T-cells expressing a highly potent PRAME-specific T-cell receptor in combination with a chimeric PD1-41BB Co-stimulatory receptor show a favorable preclinical safety profile and strong anti-tumor reactivity. Cancers (Basel) 14, 1998 (2022).
Liddy, N. et al. Monoclonal TCR-redirected tumor cell killing. Nat. Med 18, 980–987 (2012).
Pandey, A. et al. Therapeutic targeting and structural characterization of a sotorasib-modified KRAS G12C–MHC I complex demonstrate the antitumor efficacy of hapten-based strategies. Cancer Res 85, 329–341 (2025).
Rader, C. Generation and selection of phage display antibody libraries in fab format. Cold Spring Harb. Protoc. https://doi.org/10.1101/PDB.TOP107764 (2024).
Maso, L. et al. Generation of actionable, cancer-specific neoantigens from KRAS(G12C) with adagrasib. Proc. Natl. Acad. Sci. USA. 122, e2509012122 (2025).
Miller, K. R. et al. T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS One 7, e43746 (2012).
Robertson, I. B. et al. Tuning the potency and selectivity of ImmTAC molecules by affinity modulation. Clin. Exp. Immunol. 215, 105 (2023).
Tokatlian, T. et al. Chimeric antigen receptors directed at mutant KRAS exhibit an inverse relationship between functional potency and neoantigen selectivity. Cancer Res. Commun. 2, 58 (2022).
Yaeger, R. et al. Molecular characterization of acquired resistance to KRASG12C-EGFR inhibition in colorectal cancer. Cancer Discov. 13, 41–55 (2023).
Zhao, Y. et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 599, 679–683 (2021).
Baleeiro, R. B. et al. Optimized anchor-modified peptides targeting mutated RAS are promising candidates for immunotherapy. Front. Immunol. 13, 902709 (2022).
Dao, T. et al. Impact of tumor heterogeneity and microenvironment in identifying neoantigens in a patient with ovarian cancer. Cancer Immunol. Immunother. 70, 1189 (2020).
Scheinberg, D. A., Dao, T. & Liu, C. Monoclonal antigen-binding proteins to intracellular oncogene products. U.S. Patent 11, 405 (2022).
Wang, Y. et al. Global profiling of AMG510 modified proteins identified tumor suppressor KEAP1 as an off-target. iScience 26, 106080 (2023).
Patricelli, M. P. et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 6, 316–329 (2016).
Zhang, C. et al. A platform for mapping reactive cysteines within the immunopeptidome. Nat. Commun. 15, 1–15 (2024).
Kacen, A. et al. Post-translational modifications reshape the antigenic landscape of the MHC I immunopeptidome in tumors. Nat. Biotechnol. 41, 239–251 (2022).
Reits, E. A. et al. Radiation modulates the peptide repertoire, enhances MHC class I expression and induces successful antitumor immunotherapy. J. Exp. Med 203, 1259–1271 (2006).
Liu, W. M., Fowler, D. W., Smith, P. & Dalgleish, A. G. Pre-treatment with chemotherapy can enhance the antigenicity and immunogenicity of tumours by promoting adaptive immune responses. Br. J. Cancer 102, 115–123 (2010).
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Brea, E. J. et al. Kinase regulation of human MHC class i molecule expression on cancer cells. Cancer Immunol. Res 4, 936–947 (2016).
Stopfer, L. E. et al. MEK inhibition enhances presentation of targetable MHC-I tumor antigens in mutant melanomas. Proc. Natl. Acad. Sci. USA 119, e2208900119 (2022).
Hattori, T. et al. Multiplex bead binding assays using off-the-shelf components and common flow cytometers. J Immunol Methods 490, 112952 (2021).
Gragert, L., Madbouly, A., Freeman, J. & Maiers, M. Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol. 74, 1313–1320 (2013).
Gonzalez-Galarza, F. F. et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 48, D783–D788 (2020).
US Census Bureau. 2020 Census: Redistricting File (Public Law 94-171) Dataset. https://www.census.gov/data/datasets/2020/dec/2020-census-redistricting-summary-file-dataset.html (2021).
Abelin, J. G. et al. Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction. Immunity 51, 766–779 (2019).
Chong, C. et al. High-throughput and sensitive immunopeptidomics platform reveals profound interferon γ-mediated remodeling of the human leukocyte antigen (HLA) ligandome. Mol. Cell. Proteom. 17, 533–548 (2018).
Leddy, O. et al. Validation and quantification of peptide antigens presented on MHCs using SureQuant. Nat. Protoc. 20, 1196–1222 (2025).
MacLean, B. et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Brademan, D. R., Riley, N. M., Kwiecien, N. W. & Coon, J. J. Interactive peptide spectral annotator: a versatile web-based tool for proteomic applications. Mol. Cell Proteom. 18, S193–S201 (2019).
Peng, H. & Rader, C. Generation of antibody libraries for phage display: human fab format. Cold Spring Harb Protoc https://doi.org/10.1101/PDB.PROT108597 (2024).
Bailey, L. J. et al. Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination. J. Mol. Biol. 430, 337–347 (2018).
Russo, C. J. & Passmore, L. A. Ultrastable gold substrates: Properties of a support for high-resolution electron cryomicroscopy of biological specimens. J. Struct. Biol. 193, 33–44 (2016).
Cheng, A. et al. Leginon: New features and applications. Protein Sci. 30, 136–150 (2021).
Rice, W. J. et al. Routine determination of ice thickness for cryo-EM grids. J. Struct. Biol. 204, 38–44 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017 14:4 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Lander, G. C. et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Pettersen, E. F. et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Maso, L., Bang, I. & Koide, S. Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate. Protein Data Bank (2024).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Pierce, B. G. & Weng, Z. A flexible docking approach for prediction of T cell receptor-peptide-MHC complexes. Protein Sci. 22, 35–46 (2013).
Maso, L. EMD-49361, Structure of the cross-HLA supertype antibody R302 bound to a class I MHC presenting a divarasib-modified KRAS-G12C peptide on HLA-A*02. Electron Microscopy Database https://www.ebi.ac.uk/emdb/EMD-49361 (2025).
Maso, L. EMD-49362, Structure of the cross-HLA supertype antibody R302 bound to a class I MHC presenting a divarasib-modified KRAS-G12C peptide on HLA-A*03. Electron Microscopy Database https://www.ebi.ac.uk/emdb/EMD-49362 (2025).
Maso, L. 9NFC, Structure of the cross-HLA supertype antibody R302 bound to a class I MHC presenting a divarasib-modified KRAS-G12C peptide on HLA-A*03. Protein Data Bank https://www.rcsb.org/structure/9NFBC (2025).
Maso, L. 9NFB, Structure of the cross-HLA supertype antibody R302 bound to a class I MHC presenting a divarasib-modified KRAS-G12C peptide on HLA-A*02. Protein Data Bank https://www.rcsb.org/structure/9NFB (2025).
Acknowledgements
We thank Nicole Barucci, Lori Lopresti-Morrow, and George Mourgkos from ATP R&D for supporting the in vivo studies, Elizabeth Elliot for her operational support at Aethon Therapeutics and assistance with manuscript submission, and William Rice, Bing Wang, and Huihui Kuang from the NYU Langone Health’s Cryo–Electron Microscopy Laboratory (RRID: SCR_019202).
Author information
Authors and Affiliations
Contributions
L.M., S.A.M., B.G.N., S.K., R.C., P.D.S.J., C.R., and L.E.S. conceived and designed strategies and experiments to demonstrate that T.C.E.s targeting drug-modified KRASG12C neoantigens enhance M.H.C. presentation and elicit potent, selective T cell-mediated cytotoxicity across HLA supertypes. S.A.M., D.N.M., S.S., M.S. and L.E.S. performed cell line engineering, validation, and maintenance. L.E.S. and D.M. performed immunopeptidomics. L.M., S.A.R.-A., and E.R. performed p/p*MHC, Fab, and TCE expression, purification, and validation. S.T.T. advised on hapten-peptide conjugation, purification, and characterization. A.P., T.H., A.K., and W.L. performed phage display library generations and selections. L.M. and A.P. performed yeast display library generations and screenings. B.V. advised on antibody sequence and structure analyses. W.L. performed surface plasmon resonance assays. L.M. performed cryo-EM sample preparation, data collection, and data processing. S.A.M., M.S. and D.N.M. performed in vitro cytotoxicity experiments. S.A.M. and T.A. performed PDX mouse model in vivo and ex vivo studies. L.M. and S.S. performed p/p*MHC stability studies. L.M., S.A.M., C.R., and L.E.S. analyzed all data and wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
L.M., S.A.M., S.A.R.-A., A.P., D.N.M., M.S., S.S., T.A., B.V., W.L., S.T.T., P.D.S.J., R.C., C.R., and L.E.S. are or were previously employees of Aethon Therapeutics and hold equity. BGN and SK hold equity in Aethon Therapeutics. BGN’s spouse holds equity in Revolution Medicines and owned equity in Mirati Therapeutics and Amgen during the course of this work. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Johannes Huppa, Christian Klein and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Maso, L., Mosure, S.A., Rodriguez-Aponte, S.A. et al. Engineered antibodies that stabilize drug-modified KRASG12C neoantigens enable selective and potent cross-HLA immunotherapy. Nat Commun 16, 11264 (2025). https://doi.org/10.1038/s41467-025-66132-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66132-w