Abstract
Lysosomal exonuclease phospholipase D (PLD) family PLD3 and PLD4 degrade single-stranded RNA or DNA and regulate TLR7 or TLR9 responses. Polymorphisms of these enzymes are associated with human diseases: PLD4 is associated with inflammatory diseases, and PLD3 is associated with neurodegenerative diseases. Here, we determine the structures of substrate-bound PLD3 and PLD4 by cryo-electron microscopy. Our structures reveal that PLD3 rebuilds a substrate-binding pocket, depending on the substrate, mainly via motion of the Phe335-containing loop. Furthermore, we captured the structure in a metastable state that appears during substrate rearrangement following product release. Together, our findings identify the residues that underlie the distinct activities of PLD3 and PLD4. This study provides a mechanistic basis for the exonuclease activity of PLD3 and PLD4 in single-stranded DNA degradation.
Similar content being viewed by others
Introduction
Nucleic acids (NAs) are fundamental molecules in living organisms that carry genetic information, yet exogenous NAs and their metabolites also play essential roles in host defense1,2. In innate immunity, pathogenic NAs are detected by NA sensors including Toll-like receptor (TLR) 3, 7-9 and 13 in the endosomal compartment3, as well as retinoic acid-inducible gene-I (RIG-I)-like receptors and cyclic GMP-AMP synthase (cGAS) in the cytoplasm4. These NA sensors trigger the NF-κB and IRF signaling to induce cytokine and type-I interferon production, promoting inflammatory and antiviral responses.
Nucleases restrict NA availability or produce activating ligands, playing indispensable roles in regulating innate immunity. Some sensors recognize double-stranded RNA (dsRNA), double-stranded DNA (dsDNA) or ribosomal RNA, while others recognize DNA/RNA degradation products, complicating the regulation mediated by nucleases. DNase II, an endonuclease for dsDNA with low sequence specificity, is required for generating TLR9 ligands5. However, DNase II deficiency leads to constitutive activation of cGAS-STING and AIM26, resulting in type I interferonopathy in human7 and embryonic lethality due to type I IFN- dependent anemia in mice8. RNase T2, which digests single-stranded RNA, regulates TLR7 and TLR8 but negatively regulates TLR39,10,11.
PLD3 and PLD4 are endolysosomal exonucleases that degrade single-stranded DNA (ssDNA) or single-stranded RNA (ssRNA)12,13. Genome-wide association studies have shown that PLD4 polymorphisms are associated with autoimmune diseases, including systemic lupus erythematosus14, systemic sclerosis15 and rheumatoid arthritis16 whereas PLD3 polymorphisms are associated with neurodegenerative diseases such as Alzheimer’s disease17, spinocerebellar ataxia18 and leukoencephalopathy syndrome19. Reflecting these distinct disease associations, PLD4 is expressed in limited tissues, mainly within immune cells, whereas PLD3 is ubiquitously expressed across various tissues. A recent study revealed that PLD3 degrades mitochondrial DNA (mtDNA) as a major substrate within lysosomes of neuronal cells20. In PLD3-deficient cells, mtDNA leakage into the cytosol is elevated, activating cGAS-STING-linked autophagy, suggesting a potential link between PLD3 dysfunction and APP metabolism20.
PLD3 and PLD4 negatively regulate TLR9 responses in immune cells12. They also contribute to the generation of TLR7 ligand in collaboration with RNase T221 or negatively regulate TLR13 responses13. PLD3 and PLD4 are specific for 5’-OH nucleotides, but not for 5’-phosphorylated nucleotides. Since RNase T2 and DNase II generate 3’-phosphorylated nucleotide and 5’-OH nucleotides, PLD3 and PLD4 can degrade the products generated by these nucleases. PLD3 and PLD4 belong to the phospholipase D (PLD) family, which contains the conserved HxK(x)4D/E (HKD/E) motif (Fig. 1a, Supplementary Fig. 1). Two HKD/E motifs, brought together either through homodimerization or internal duplication, form a catalytic cavity, in which His and Lys are involved in catalysis, while the Asp/Glu is located away from the catalytic center and contributes to stabilizing protein folding22,23. Despite the functional diversity within the PLD family, including lipid phospholipase, endonuclease, and exonuclease activities, members share a common catalytic mechanism. This mechanism proceeds via a ping-pong type reaction involving a covalent phosphohistidine intermediate24,25,26. One of the two conserved His in the HKD/E motif functions as a nucleophile to attack the substrate, resulting in the formation of a phosphohistidine intermediate. In the subsequent step, the other His serves as a general acid to activate a water molecule that attacks the phosphohistidine to cleave the phosphodiester bond and regenerate the free catalytic His residue.
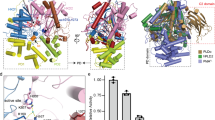
a, Domain diagram of human PLD3 and PLD4 with HKD/E motif sequence. PLD3 and PLD4 comprise the N-terminal transmembrane helix (TM, gray square and luminal HKD1 and HKE2 domains. HKD1 and HKE2, whose sequences are shown below, fold into a globular domain that serves as the exonuclease domain. Glycosylation sites are indicated by “Y”; sites where glycan chains are not resolved in the cryo-EM map are shown in light gray. Disulfide bonds are represented as orange lines. b, Overall cryo-EM map of hPLD3_poly(T). PLD3 forms a symmetric homodimer, with each protomer (purple and cyan) binding ssDNA (magenta). c, Cross section views of the catalytic cavity of hPLD3_poly(T) (left panel) and hPLD3_poly(A) (right panel). d, Detailed views of poly(T) recognition by hPLD3, including a 2D interaction diagram. Residues interacting with the ssDNA via hydrogen bonding (with heavy atom distances <3.9 Å) or π-π stacking interaction are shown in stick representation. Hydrogen bonds formed via side chains are shown in red dashed lines; those formed via main chains are shown in green dashed lines. π-π stacking interactions are indicated by ellipses. For comparison, residues that do not form hydrogen bonds in hPLD3_poly(T) but form them in hPLD3_poly(A) are shown in gray, with the distances indicated. e, as in d, detailed views of poly(A) recognition by hPLD3. f, Gel-based exonuclease assay of hPLD3 using 55-mer poly(A) or poly(T) ssDNA substrates. Each experiment was independently repeated three times with similar results.
Recently, the crystal structures of PLD3 and PLD4 have been reported27,28, revealing the overall architecture of its luminal domain and dimeric organization mediated by inter-helical interactions. The structure of PLD3 co-crystallized with oligonucleotide (ODN) represents the product-bound structure, as the ODN was degraded within the crystal27. PLD3 and PLD4 can function as a 5’-phosphatase for 5’-phosphorylated ssDNA (5’-Pi ssDNA), although their catalytic rate is more than 10,000-fold slower than that of exonucleases acting on 5’-OH ssDNA. The crystal structures of PLD3 and PLD4 co-crystallized with 5’-Pi ssDNA have also been reported28. In PLD3 structures, due to differences in crystallization times, distinct structures were obtained where the catalytic residue His was either phosphorylated via phosphate transfer from 5’-Pi ssDNA or dephosphorylated. In PLD4 structure, the catalytic residue His was phosphorylated, and the dephosphorylated ssDNA was trapped within the catalytic cavity. However, our understanding of substrate recognition, specificity, and exonuclease mechanism remains limited due to the insufficient electron density for the substrate or ssDNA product-bound form containing phosphohistidine intermediate in the previous works.
Here, we present the structures of PLD3 bound to different substrate ODNs by single particle cryo-EM analysis. The substrate adopts an L-shaped backbone structure at the 5’ end, accommodated within the narrow and deep catalytic cavity. Our structures reveal that PLD3 rebuilds its substrate-binding pocket depending on the binding nucleotides mainly by the motion of the Phe335-containing loop. Structural snapshots in the apo form, substrate-binding, product-binding and metastable states provide the structural basis for the conformational change in the process of degrading ssDNA by PLD3 and PLD4. We also identified the residues responsible the difference in exonuclease activity between PLD3 and PLD4, offering potential targets for drug design targeting PLD3 and PLD4.
Results
ssDNA binding to PLD3 is enhanced under acidic conditions
Human PLD3 (hPLD3) has been reported to undergo proteolytic cleavage after expression as a type-II transmembrane protein (Fig. 1a), yielding a luminal soluble protein in the lysosome29 that retains exonuclease activity comparable to that of the full-length protein30. The luminal domain of hPLD3 was purified from mammalian cells. We employed electrophoretic mobility shift assay (EMSA) to screen the conditions that form the stable complex of hPLD3 and its substrate prior to cryo-EM analysis. Unexpectedly, during this screening, we found that PLD3 H201A/H416A mutant (hPLD3_HA), in which the catalytic residues were mutated, nearly abolished the exonuclease activity, but a small residual activity remained (Supplementary Fig. 2a). This residual exonuclease activity was suppressed by the addition of sodium vanadate which serves as an inhibitor of phosphoryl transfer enzymes. We conducted EMSA assay with varying concentrations of hPLD3 (0, 2.5, 5.0, 10, and 20 μM) incubated with ssDNA. A band corresponding to ssDNA bound to hPLD3 was detected only at 20 μM at pH 6.0; however, at pH 5.5 or below, the band was observed at 10 μM (Supplementary Fig. 2b), indicating that acidic condition strengthens the interaction between hPLD3 and its substrate. This observation is consistent with previous reports that hPLD3 and hPLD4 require acidic environments for their exonuclease activity12.
Structure determination of PLD3 and PLD4 bound to ssDNA
We successfully determined the structures of hPLD3_HA bound to several different substrates, poly(T) ODN (hPLD3_poly(T)) and poly(A) ODN (hPLD3_poly(A)) at 2.9 Å resolution in addition to hPLD3 WT apo-form (hPLD3_apo) structure at 2.9 Å resolution (Supplementary Fig. 3-5, Supplementary Table 1). We also determined the cryo-EM structures of hPLD4 WT apo-form (hPLD4_apo) and in the presence of poly(T) substrate at 3.3 Å and 3.1 Å resolution, respectively (Supplementary Table 1, Supplementary Figs. 6, 7). Sodium vanadate was added at a low concentration (0.1 mM sodium vanadate) to the sample of hPLD3 HA bound to substrate. However, vanadate was not observed in the cryo-EM map, suggesting that it is unlikely to bind to hPLD3 under the cryo-EM conditions.
PLD3 forms a homodimer through two α-helices, and each protomer consists of HKD1 and HKE2, which adopt the α/β hydrolase fold. The conserved His and Lys residues of each HKD/E motif (His201 and Lys203 in HKD1 and His416 and Lys418 in HKE2) are positioned facing each other to form the catalytic site. In contrast, the Asp/Glu (Asp208 in HKD1 and Glu423 in HKE2) stabilizes hPLD3 structure by forming hydrogen bonds with the main chain amides (Supplementary Fig. 8a). The overall structure of hPLD3_apo is similar to the recently reported crystal structure of hPLD327 (Supplementary Fig. 8b).
The protomer structure of hPLD4 is also similar to that of hPLD3, and both proteins possess essentially the same dimer interface (Supplementary Fig. 9a). In sharp contrast to PLD3, PLD4 forms a tetramer in our structure, adopting a face-to-face configuration with a twist, mediated by the additional interactions between two dimers (A-B dimer and C-D dimer) (Supplementary Fig. 9). The tetramer formation of PLD4 is also supported by size exclusion chromatography, in which PLD4 eluted faster than PLD3 (Supplementary Fig. 10), consistent with a larger molecular assembly. The cryo-EM samples were adjusted to approximately 1 mg/mL (~18 μM), at which concentration hPLD3 was predominantly observed as a dimer, whereas hPLD4 was mainly observed as a tetramer in the cryo-EM analysis. The two PLD4 dimers contact each other through two interfaces with a total area of ~900 Å2. One interface involves the hydrogen bonds between Gln290 and the main chain of His293, and π-π stacking interactions between Phe289 and His285 or His285 and His293. The other interface is essentially the same, but the interacting pairs in each protomer are inverted (Supplementary Fig. 9b). Either the A–B dimer or the C–D dimer is flipped across the two-fold axis. This leads to a misalignment with the opposite dimer (C–D or A–B, respectively), indicating that strict two-fold symmetry is absent between the two dimers (Supplementary Fig. 9c).
It has been reported that hPLD3 can be N-glycosylated at five sites based on mass spectrometry analysis27. In previously determined crystal structures, only two N-linked glycans (linked to Asn236 and Asn284 in hPLD3) were modeled for both human and mouse PLD327,28. In contrast, four N-linked glycans, corresponding to all sites except for Asn97, were observed in our cryo-EM structure (Fig. 1a). For PLD4, no experimental data are available to identify specific glycosylation sites. In the crystal structure of human PLD4, no glycans were modeled28, whereas in our cryo-EM structure, six N-linked glycans were observed (Fig. 1a).
L-shaped substrate captured in the catalytic cavity of PLD3
We determined the structures of hPLD3_poly(T) and hPLD3_poly(A). In both structures, the substrates are captured within a narrow and deep catalytic cavity surrounded by a negatively charged surface (Supplementary Fig. 11). The substrates enter the cavity almost perpendicularly, rather than extending along the protein surface (Fig. 1b). A prominent feature of the binding mode is a sharp bend in the backbone between the 5′-end and the second nucleotide, producing an L-shaped conformation in which the sugar-phosphate backbone is directed inward and the base moiety of the 5′-end nucleotide is oriented outward (Fig. 1c). This L-shaped configuration positions the 5′-end nucleotide near the cavity exit, facilitating the release of the 3′-phosphorylated mononucleotide, the product of PLD3 cleavage. It is widely accepted that ssDNA exhibits considerable flexibility. Therefore, the observed L-shaped conformation of the ssDNA substrate is a consequence of structural constraints imposed by the binding pocket. Interestingly, our structures also revealed a substrate-dependent conformational change in both PLD3 and the bound substrate, which will be discussed in a later section.
PLD3 adjusts the catalytic cavity size to recognize substrates with distinct conformations
In the cryo-EM map of hPLD3_poly(T), four nucleotides at the 5’ end (dT1-dT4) are clearly resolved and modeled (Supplementary Fig. 12a). The first three nucleotides are accommodated in the catalytic cavity of PLD3, while the fourth nucleotide lies on the cavity (Fig. 1c). The subsequent nucleotides are not well resolved due to the dynamic nature in the PLD3-bound state. The 5’-end dT1 is captured at the innermost region of the cavity, and its 3’-phosphate group faces inward, forming interactions with Lys203, Asn218, Lys418 and Asn432, while the base group faces outward, engaging in π-π stacking with Tyr411 (Fig. 1d). The H201A and H416A mutants also orient their side chains toward the phosphate group of dT1, which is suitable for hydrolyzing 5’ nucleotide of the substrate. Additionally, the 5’-hydroxyl group interacts with the phenolic group of Tyr126 and the main chain carbonyl group of Tyr411. This moiety is confined within a narrow internal space, indicating that 5’-phosphorylation of substrates would cause the steric clash with PLD3 (Fig. 1c, d), thereby preventing accommodation. This structural observation aligns with previous study13 and current findings showing that PLD3 lacks exonuclease activity against 5’-phosphorylated ODN (Supplementary Fig. 13a). The 3’-phosphate group of dT2 interacts with the side chains of Asn328, Trp367 and Tyr437, while the 3’-phosphate group of the third nucleotide interacts with the side chains of His369 and Arg488 (Fig. 1d). The base moiety of dT2, that of dT3 and Phe335 are stacked up in this order through a π-π stacking interaction. dT4 is largely exposed from the catalytic cavity of PLD3 and forms an off-plane interaction with the aromatic ring of Phe335.
In the cryo-EM map of hPLD3_poly(A), five nucleotides at the 5’ end (dA1-dA5) are observed (Supplementary Fig. 12b). The 5’-end dA1 interacts with PLD3 in essentially the same way as dT1 in the poly(T) substrate, and dA2 also adopts the same position as that in the poly(T) substrate (Fig. 1c, e). Strikingly, the base moiety of dA3 is oriented in the opposite direction relative to dA2. As a result, the base moiety of dA2 directly forms an aromatic stacking interaction with Phe335, along with additional polar interactions with Thr441 (Fig. 1c, e). The flipped dA3 forms an aromatic stacking interaction with His369. The following nucleotides, dA4 and dA5, do not interact directly with PLD3. In addition to the distinct conformations of the substrates, exonuclease assay revealed that PLD3 exhibits higher exonuclease activity toward poly(T) than poly(A), suggesting that PLD3 has a sequence preference in its exonuclease activity (Fig. 1f).
The PLD3 structures of the apo form and the substrate-bound forms are well superimposed, suggesting that no large conformational changes occur upon substrate binding, except for the loop containing Phe335. This loop forms a π-π stacking interaction with dT3 in the poly(T) substrate or dA2 in poly(A) substrate. When the poly(A) binds to PLD3, Phe335 retains the same orientation as in the apo-form, allowing dA2 to be snugly accommodated within a confined space (Fig. 1e),formed by Tyr437 and Thr441 at the base, and flanked by Phe125 and Val199 on one side and Phe335 on the other (Supplementary Fig. 14). In contrast, when poly(T) binds PLD3, the benzene ring of Phe335 flips outward to create space for dT3, enabling the formation of a π-π stacking interaction network involving dT2, dT3 and Phe335 (Supplementary Fig. 14). If poly(A) were to adopt the same arrangement as poly(T), steric clashes would occur between dA2 and dA3 due to the larger purine base, indicating that PLD3 can recognize purine and pyrimidine base in a different manner by the motion of Phe335.
Recognition of the 5’ three to four nucleotides by PLD3 is important for the regulation of TLR9 signaling
To validate the functional importance of residues that interact with the substrate, we conducted an end-labeled fluorescence-quenched oligonucleotide (EFQO) assay to monitor the exonuclease activity, using purified PLD3 and an ODN labeled with 5’-FAM and 3’-quencher. ODN1668, which is one of type B CpG ODN serving as a TLR9 ligand, was used in the EFQO assay. We mutated the residues involved in the catalytic reaction (His201/His416, and Glu229 which is predicted to accept proton from His416 to initiate nucleophile attack by His416), in phosphate group interactions (Lys203, Asn328, Trp367, His369, Lys418 and Tyr437) and in π-π stacking interactions (Phe335 and Tyr411). All of the above mutants reduced activity, supporting the importance of phosphate-mediated and π-π stacking interactions in substrate recognition (Fig. 2a, b). The EFQO assay revealed that the HA mutant retained only 0.2% of the specific activity of the wild type, indicating that only a minimal level of activity remains.
a, EFQO assay. PLD3 WT or PLD3 mutant were incubated with 5’FAM-ssDNA-BHQ1 (FASMAC) with the sequence TCCATGACGTTCCTGATGCT (ODN1668), and the released fluorescence was recorded over time. b, Activity measured by the EFQO assay. Compared with WT activity (100%), mutant activities were categorized as follows less than 10% (green), 10% to less than 40% (yellow), and more than 40% (cyan). Bar plots represent mean ± s.e.m of the linear regression at each time point (n = 11, 1–11 min). c, TLR9 response of PLD3 WT or mutants. Cells overexpressing PLD3 WT, each mutant, or control (−) were incubated with CpG ODN (200 nM), and TLR9 response was assessed as activation of an NF-κB reporter. Graphs show mean ± SE of n = 3 independent experiments. Bar plots represent mean ± s.e.m. P values were determined by one-way ANOVA with Turkey test (*P < 0.05, ****P < 0.0001).
To further examine the functional importance in cellulo, BaF cells were stimulated with CpG-B and NFκB activation was evaluated. Overexpression of wild type PLD3 (PLD3 WT) attenuated TLR9-dependent NFκB activation. In contrast, the response was restored to near-control level when PLD3 mutants K203A, E229A, H369A or K418A were overexpressed. PLD3 H201A/H416A, F335A or W367A mutants showed weaker attenuation of the response than PLD3 WT, whereas N328A, Y411A or Y437F mutants attenuated NFκB activation to a similar extent as PLD3 WT (Fig. 2c, Supplementary Fig. 15). These results further support the importance of PLD3 exonuclease activity in the regulation of TLR9 signaling.
Structural comparison identified residues that contribute to the difference in exonuclease activity between PLD3 and PLD4
Our detailed analysis confirmed that PLD3 exhibits higher exonuclease activity than PLD4 (Supplementary Fig. 13b), consistent with the previous report28. The structures of PLD3 bound substrates allow us to re-examine the difference in activity between PLD3 and PLD4. Consistent with the sequence similarity, the PLD3_apo and PLD4_apo structures are superimposed well, with a root-mean-square deviation (r.m.s.d.) of 0.76 Å, although several loop regions exhibit structural differences (Fig. 3a). Similarly, PLD3_poly(T) is well aligned with PLD3_apo (r.m.s.d. of 0.74 Å), and each protomer of PLD4, whether in the apo form or in the presence of poly(T), also aligns well, with an r.m.s.d. ranging from 0.3 to 0.5 Å, suggesting that PLD3 and PLD4 show no significant conformational change upon substrate binding. Poly(T) substrate fits into PLD4 with only a little steric clash, strongly suggesting that PLD4 can recognize the substrate in a manner similar to PLD3 (Fig. 3b). Structure-based sequence alignment revealed that several substrate-interacting residues in PLD3 are not conserved in PLD4. Asn328, His369 and Arg488 in hPLD3 (Supplementary Fig. 1), which interact with the 3’-phosphates of the substrate, correspond to Glu341, Asn382 and Val503 in hPLD4, respectively. To test the functional consequences of these differences, these residues are converted or vice versa (Fig. 3b). E341N mutation in PLD4 may eliminate the electrostatic repulsion with 3’-phosphate of the second nucleotide. Similarly, V503R mutation in hPLD4 may enhance interaction with 3’-phosphate of the third nucleotide. In our enzymatic assay, E341N mutant of hPLD4 exhibited higher activity than WT and E341N/V503R exhibited markedly increased activity (Fig. 3c). These mutants retained activity even at neutral pH (pH 7.0). Conversely, hPLD3 N328E mutant, which is supposed to cause the electrostatic repulsion with 3’-phosphate of the second nucleotide, exhibited reduced activity compared to WT (Fig. 3c). These results suggest that these sequence discrepancies contribute to the difference in exonuclease activity between PLD3 and PLD4.
a, Superposition of the structures of hPLD4_apo (pink) and hPLD3_poly(T) (purple) to hPLD3_ apo (light green). Loops of hPLD3 and hPLD4 that do not align well are highlighted in cyan (hPLD3_poly(T), light green (hPLD3_apo) and magenta (hPLD4_apo), respectively. “h3” and “h4” denote hPLD3 and hPLD4, respectively. b, Close-up view of the catalytic cavity in hPLD3_poly(T) are shown with a stick model. Most of the residues shown in a interact with the phosphate group of dT2 or dT3. c, Gel-based exonuclease assay using substrate-I with PLD4 or PLD3 mutant at different pH conditions. Each experiment was independently repeated three times with similar results.
Structure of PLD4 in the presence of poly(T) substrate revealed dynamic nature of substrate
In the C1 symmetry cryo-EM map of the hPLD4 tetramer in the presence of poly(T) substrate, poly(T) molecules are observed in both catalytic sites of one dimer, while densities corresponding to poly(T) are ambiguous in the other dimer. Interestingly, the poly(T) densities differ between the two PLD4 protomers. We were able to model two unconnected 3’-phospho dT nucleotides in one protomer and connected three dT nucleotides in the other protomer (Supplementary Fig. 16). Since PLD4 digests ssDNA to generate deoxynucleotide 3’-phosphate, unconnected two 3’-phospho dT nucleotides are assigned as digestion products, including the newly formed 5’ end of the poly(T) substrate, suggesting that this structure represents the product bound form (referred to as “product-bound hPLD4”). The three connected dT nucleotides are assigned as the 5’ portion of poly(T) substrate, indicating that the other protomer represents the substrate-bound form (referred to as “substrate-bound hPLD4”).
In product-bound hPLD4, one 3’-phospho dT forms salt bridges with His214, Lys216, His428 and Lys430 in the conserved HKD/E motif through its 3’ phosphate group and its base moiety is sandwiched between Phe423 and Leu183 via a π-π stacking interaction (Fig. 4a). These interactions are similar to those observed for the 5’-end dT1 of the poly(T) substrate bound to PLD3, suggesting that this nucleotide corresponds to the product dT 3-phosphate. The newly formed 5’ end of the substrate, generated by PLD4 digestion, is located adjacent to the product and its 5’ hydroxyl group faces the 5’ hydroxyl group of the product dT 3-phosphate. The base moiety is sandwiched between Val212 and Phe348, while the 3’-phosphate group does not participate in interactions with PLD4 (Fig. 4a). The nucleotides trailing the newly formed 5’ end dT1 were not modeled due to ambiguous density, which may reflect a conformational flexibility (Supplementary Fig. 16a).
a, The product recognition by hPLD4 (left panel) and the corresponding 2D interaction diagram. Residues interacting with nucleotides via hydrogen bonds (with heavy atom distances ≤ 4.0 Å) or π-π stacking interactions are shown in stick representation. Hydrogen bonds are indicated with dashed lines. b, Substrate recognition by hPLD4 in the metastable state. Hydrogen bonds via side chains are shown as red dashed lines. The π-π stacking interaction is indicated with an ellipse.
In substrate-bound hPLD4, dT1 is sandwiched between Phe423 and Leu183 in a manner similar to product-bound hPLD4. Importantly, in sharp contrast to product-bound hPLD4 and hPLD3_poly(T)/hPLD3_poly(A), the dT1 3’ phosphate group is positioned away from the catalytic residues. His214 forms a salt bridge with the dT1 3’- phosphate group; however, the side chains of Lys216, His428 or Lys430 are more than 5.5 Å away from the 3’-phosphate group of dT1. Instead, phenolic group of Tyr449 forms bifurcated hydrogen bonds with the 3’ phosphate of dT1, contributing to substrate binding. dT2 is sandwiched between Val212 and Phe348 and dT3 stacks on Phe348 (Fig. 4b). These observations strongly suggest that the observed substrate-bound hPLD4 represents a metastable state rather than a substrate-cleaving state, in which the substrate is properly positioned for catalysis. We speculate that this state represents a transient intermediate during substrate rearrangement following product release, reflecting the repositioning of the substrate after initial digestion by PLD4.
Reconstructing catalytic dynamics from structural snapshots
Structural comparisons were performed among hPLD3_apo, hPLD3_poly(T), hPLD3_poly(A), mouse PLD3 phospho-histidine intermediate (PDB ID; 8V06) form (mPLD3_intermediate), mouse PLD3 phosphate-bound form (PDB ID; 8V07) (phosphate-bound mPLD3), and human PLD3 product-bound form (PDB ID; 8Q1X) (product-bound hPLD3) to gain further insights into the catalytic mechanism. The conformations of the catalytic residues, as well as those interacting with the substrate, are well conserved between human and mouse, and across various catalytic states. The catalytic residue His416 in human PLD3 (corresponding to His414 in mouse) exhibits a rotamer shift only in the mPLD3_intermediate and the product-bound hPLD3, moving toward His201 in human (His199 in mouse) (Fig. 5a). Tyr411 in human (Tyr409 in mouse) which stacks with the base of the first nucleotide, exhibits structural variability. Phe335 in human (Phe333 in mouse), which stacks with the base of the second or third nucleotide, undergoes conformational changes. The other residues involved in the substrate binding show no significant changes. The first nucleotide of the bound substrate (poly(T) or poly(A)) in the PLD3_poly(T)/poly(A) occupies the same site as that in the product-bound hPLD3, and their phosphate groups are well aligned with that of the mPLD3_intermediate. Taken together, the structural changes during the enzymatic cycle appear to be minor except for the Phe335-containing loop as described above.
a, Structural comparisons among hPLD3_apo (light green), hPLD3_poly(T) (purple), hPLD3_poly(A) (yellow), mPLD3_intermediate (PDB 8V06) (pink), phosphate-bound_mPLD3, (PDB 8V07) (orange) and product-bound hPLD3 (PDB 8QIX) (cyan) around the catalytic site. Residues interacting with the substrate are aligned across these structures. “h” and “m” denote human and mouse, respectively. b, Structural comparison of hPLD3 in the substrate-cleaving state (hPLD3_poly(T), shown in gray) and hPLD4 in either the substrate-cleaved state (product-bound hPLD4, shown in green; left panel) or the substrate-moving state (substrate-bound hPLD4, shown in magenta). c, Conformational changes associated with the proposed exonuclease mechanism. The details are provided in the main text. d, A proposed scheme of PLD3 and PLD4 exonuclease action.
We also examined structural differences among substrates bound to PLD3 and PLD4. The structural comparison between hPLD3_poly(T) and product-bound hPLD4 can reflect conformational changes associated with substrate cleavage. The 3’-phosphate group of the product remains in approximately the same position as in hPLD3_poly(T), whereas the 5’-OH group of the product is shifted by 8.1 Å due to ribose-ring flipping after cleavage (Fig. 5b left). Accompanying this conformational change, the phosphate group of the newly formed 5’-end nucleotide is displaced outward (~6.9 Å) to avoid steric clashes with the 5’-OH group of the product (Fig. 5b left). In addition, the 5’-OH of the newly formed nucleotide is displaced outward (~4.9 Å) and forms a hydrogen bond with the 5’-OH group of the product. In contrast, the structural comparison between the substrate-bound hPLD4 and hPLD3_poly(T) reveals conformational changes associated with the transition toward a substrate-cleaving state. The 3’-phosphate of dT1 or dT2 in the PLD4 complex is displaced by 4.8 or 7.9 Å, respectively, relative to the complex with PLD3, whereas the base moiety of dT1 or dT2 remains almost properly aligned due to π-π stacking interaction with Phe423 or Phe348 (Fig. 5b right). These observations suggest that the base moiety may serve as a structural scaffold to guide the phosphate into its proper position. In the case of dT3, the base moiety and the phosphate group exhibit substantial displacement (4.9 and 10.6 Å, respectively). In summary, unlike PLD3/PLD4, which remains largely unchanged in overall structure, the substrate undergoes significant conformational changes across different states of the enzymatic reaction.
Discussion
We determined the complex structure with the substrate prior to cleavage and revealed that the binding pocket changes depending on the substrate. By successfully capturing other states as well, we were able to propose an integrative model of the enzymatic cycle in which PLD3 and PLD4 actively function. During the cycle, the protein undergoes minimal conformational changes except in the Phe335-containing loop, and after cleavage, the bound substrate is repositioned into a conformation suitable for further cleavage through coordinated movements of the product and the remaining substrate. We can provide insights into the structural details relevant to substrate recognition and catalysis. Most nucleases in the PLD family (such as Nuc25, BfiI31 and TDP132,33) possess a large catalytic cavity, allowing oligonucleotides to bind by sliding along their protein surface (Supplementary Fig. 11). In contrast, PLD3 and PLD4 feature a narrow and deep catalytic cavity into which the oligonucleotide enters almost vertically. The substrate bends into an L-shape at the phosphodiester bond between 5’ end and the second nucleotide, a conformation that appears important for product release. This substrate-capturing mechanism by PLD3 and PLD4 is distinctive among known nucleases.
Interpolation of PLD3 and PLD4 structures with a poly(T) substrate suggests a putative trajectory for the exonuclease mechanism (Fig. 5c, d and supplementary movie). First, the substrate enters the catalytic cavity and adopts an L-shaped folded structure for proper cleavage by PLD3 and PLD4 (substrate-cleaving state). The substrate is cleaved through a phosphohistidine intermediate (substrate-cleaving intermediate), resulting in the production of a deoxynucleotide 3’-phosphate and a newly formed 5’ end nucleotide of the substrate (substrate-cleaved state). The 3’-phosphate group of the product remains in approximately the same position, whereas the 5’-OH group of the product is shifted due to ribose-ring flipping after cleavage (Fig. 5c). After cleavage, the product is released outside, after the 3’-phosphate group of the newly formed 5’ end (corresponding to the phosphate group between dT2 and dT3 before cleavage) is pushed out to solvent probably facilitated by a steric clash with the 5’-OH group of product (Fig. 5c left). Subsequently, the substrate shifts to a position suitable for the next cleavage step. The following nucleotides are disordered in the cryo-EM map, likely due to their conformational flexibility, which may facilitate substrate repositioning. In a recent report on the crystal structure of PLD3 in the presence of substrate analogue phosphorothioate ODN, the analogue was degraded into the product. While the product deoxynucleotide 3’-phosphate was observed, cleaved portion of the substrate was disordered27, further supporting the notion of the conformational flexibility following substrate cleavage. After product release, the substrate moves to a proper position for the next cleavage through moving-state. Our snapshots of PLD4 suggest that substrate moving undergoes through a meta-stable state (substrate-moving state), which can guide suitable arrangement of subsequent cleavage (substrate-cleaving state).
A recent report28, along with our findings, demonstrates that PLD3 exhibits higher exonuclease activity than PLD4. Our structure-based mutational analysis revealed that the differences in residues interacting with the 3’-phosphate of the substrate contribute to the distinct activities of PLD3 and PLD4. PLD4 contains an extra hydrophobic loop (residues 179-188) absent in PLD3 (Fig. 3a, b), which may influence substrate selectivity28. Their distinct exonuclease activities and substrate specificities may reflect distinct physiological roles since PLD4 polymorphisms are associated with autoimmune diseases14,15,16, whereas PLD3 polymorphisms are associated with neurodegenerative diseases17,18,19. Therefore, the distinctive features of their substrate-binding pocket may serve as valuable targets for developing selective inhibitors or activators. In the PLD4 tetramer, the N-terminal regions of the two dimers (A-B dimer and C-D dimer) extending from the transmembrane domain are oriented toward each other (Supplementary Fig. 9). It appears unlikely that full-length PLD4 adopts this tetrameric arrangement on the membrane, although approximately 40 residues between the TM region and the luminal domain are missing in the cryo-EM structure. The crystal structure of hPLD4 has, in fact, been reported to form a homodimer28. The functional relevance of the tetrameric assembly remains unclear.
The substrate sequence preference of PLD3 exonuclease activity has been reported, but previous studies focused mainly on a 5’ end nucleotide30. Our cryo-EM structural analysis revealed that PLD3 and PLD4 recognize the first three to four nucleotides at the 5’ end of substrates, suggesting that the substrate specificities extend to more complex combinations of multiple nucleotides at the 5’ end. The structures of PLD3 bound to different substrates showed that each substrate adopts a distinct conformation in the catalytic cavity of PLD3 and interacts with PLD3 through different interactions, suggesting that PLD3 and probably PLD4 flexibly remodel their catalytic pockets depending on the substrate sequence. Further research is needed to clarify the sequence preferences of PLD3 and PLD4 in more detail.
Recently the crystal structure of hPLD4 in the complex with the inhibitory ligand 5’-Pi ssDNA (5’-Pi-TTTTT-3’) was reported, in which His428 is phosphorylated via phosphate transfer from the ligand, and the dephosphorylated ssDNA is trapped in the catalytic cavity28. Comparison with our PLD3 and PLD4 structures has revealed structural differences between the crystal structure and the various catalytic states captured in our study (Supplementary Fig. 17). Compared with the structure of hPLD3_ poly(T), the phosphorylated His428 in the crystal structure obstructs the proper positioning of the 3’-phosphate of 5’ end nucleotide, likely disrupting the interaction between PLD4 and 5’-OH ssDNA. Thus, the crystal structure in the presence of 5’-Pi ssDNA likely represents an inhibitory state, consistent with the inhibitory effect of 5’-Pi ssDNA on exonuclease activity in vitro28. In this inhibitory state, the base moiety of dT1 is sandwiched by Leu183 and Phe423, as observed in our structure in the substrate-moving state. In sharp contrast to our structures, dT2 flips outward and dT3 engages in a π-π stacking interaction with Phe348 in the inhibitory state. These structural differences underscore the conformational flexibility of ssDNA-bound PLD3 or PLD4 and highlight the role of π-π stacking interaction through the base moiety as a scaffold to guide phosphate to the correct position. Overall, this study provides structural snapshots to expand and deepen our understanding of the ssDNA degradation mechanism mediated by PLD3 and PLD4.
Methods
Protein expression and purification
The cDNAs of human PLD3 (hPLD3) and human PLD4 (hPLD4) were provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. The DNA sequence corresponding to the luminal domain (residues 61-490 for hPLD3 or residues 91-506 for hPLD4) were amplified by the polymerase chain reaction (PCR) and cloned into the pEZT-BM vector. These proteins were fused with an N-terminal secretion signal sequence derived from rabbit IgG heavy chain and a C-terminal HRV3C protease cleavage site and protein A tag. Site-directed mutations were introduced by PCR-based mutagenesis. All plasmids were verified by DNA sequencing. Baculoviruses were generated in Spodoptera frugiperda Sf9 cells using the Bac-to-Bac system (Invitrogen). For protein expression, Expi293F cells (Invitrogen) were cultured in Expi293 expression medium at 37 °C under 8% CO2 in a CO2 incubator (PHC). When the cell density reached 3.0 − 6.0 ×106 cells per mL, cells were diluted to 3.0 ×106 cells per mL and P4 virus was added at a final concentration of 7% (v/v). Cells were cultured at 30 °C for 5 days in the presence of 10 mM sodium butyrate to enhance protein expression.
The cell culture supernatant was obtained by centrifugation at 7,000 g for 10 min. The collected supernatant was applied to IgG Sepharose 6 Fast Flow (Cytiva), which specifically binds to protein A fusion conjugates. The mixture was gently stirred at 4 °C for an hour. The resin was washed with buffer A (20 mM Tris-HCl pH 7.0 containing 150 mM NaCl) and subsequently, proteins were eluted using buffer B (100 mM glycine pH 3.5, 150 mM NaCl). The eluted proteins were instantly neutralized using a pH adjustment buffer (400 mM Tris-HCl pH7.0, 150 mM NaCl). After cleavage of protein A tag by HRV3C protease, the eluted proteins were further purified by gel filtration chromatography (Superdex 200 increase, Cytiva) in buffer A. The peak fractions were concentrated using Amicon Ultra centrifugal filter (Merk Millipore), frozen in liquid nitrogen and stored at −80 °C until use.
Cryo-EM sample preparation and data acquisition
The hPLD3 samples (WT or HA+ssDNA) were adjusted to a concentration of 1.0 mg ml–1, For sample preparation in the presence of ssDNA, 40 μM of 49-mer ssDNA (poly(T) or poly(A)) was mixed with protein samples in buffer C (20 mM MES-NaOH buffer (pH 5.5), 150 mM NaCl) containing 0.1 mM sodium vanadate. Three-microliter aliquots of samples were placed onto a freshly glow-discharged Quantifoil holey carbon grids (R1.2/1.3, Cu, 300 mesh). After 3 s of blotting in 100% humidity at 6 °C, the grid was plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). For hPLD4, hPLD4(WT) was adjusted to concentrations of 4 mg ml–1. For sample preparation of hPLD4 in the presence of ssDNA, 40 μM of 49-mer ssDNA was mixed with 1 mg ml-1 protein samples in buffer C (20 mM MES-NaOH buffer (pH 5.5) containing 150 mM NaCl) for 3 min on ice and immediately placed onto a freshly glow-discharged Quantifoil holey carbon grids (R1.2/1.3, Cu, 300 mesh). After 2.5 s of blotting in 100% humidity at 6 °C, the grid was plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific). Cryo-EM data collection was performed using a Titan Krios G4 microscope (Thermo Fisher Scientific), running at 300 kV and equipped with a Gatan Quantum-LS Energy filter (GIF) and a Gatan K3 camera in electron-counting mode, at the Cryo-EM facility of the University of Tokyo, Japan. Imaging was performed at a nominal magnification of ×105,000, which corresponded to a calibrated pixel size of 0.83 Å px–1. Each movie was recorded for 5.0 s and subdivided into 64 frames at the specimen. Cryo-EM data were analyzed using CryoSPARC v434. Raw movie stacks were motion-corrected using patch motion correction. The CTF parameters were determined using patch CTF estimation. The data processing workflow is summarized in Supplementary Fig. 3-7. Particles were picked using the blob picker or template picker. Multiple rounds of 2D classification, ab initio reconstruction, and heterogeneous refinement were performed to select particle stacks for final 3D map reconstruction using nonuniform refinement35. The map was binned to a pixel size of 1.09 Å for hPLD4_apo while the other maps were not binned. The final resolution was estimated by gold-standard Fourier shell correlation (FSC) between the two independently refined half maps (FSC = 0.143)36. The atomic model was manually accomplished using the Coot program37. The built model was refined through alternating cycles using the Coot and PHENIX programs38. The refinement statistics are summarized in Supplementary Table 1.
Structure and sequence comparison
Multiple sequence alignments of the PLD family proteins were performed using CLUSTAL Omega. Structure figures were prepared using the PyMOL Molecular Graphics System, Schrödinger, L., & DeLano, W or UCSF Chimera39.
Electrophoretic mobility shift assay
A ssDNA (5’-ACGTTCCTGATGCTAAGTATGCACTTCATCGTCAAGCAATGCTATGCAT, Substrate-I) was used. Varying concentrations of hPLD3 (0, 2.5, 5.0, 10, 20 μM) were incubated with 5 μM of Substrate-I at 4 °C for 1 h. Sodium vanadate was added two equimolar amounts of hPLD3 when indicated. After incubation for 45-60 minutes, the mixtures were subjected to electrophoresis in Tris-Acetate buffer prepared at the indicated pH.
End-labeled fluorescence-quenched oligonucleotide (EFQO) assays
A reaction mixture was prepared by combining a purified protein sample at a concentration of 5 ng μl-1 with 0.5 μM of 5’FAM-ssDNA-BHQ1 (FASMAC) with the sequence TCCATGACGTTCCTGAtgct (ODN1668) in which lower case indicates a phosphorothioate linkage in a volume of 100 μl using a buffer solution containing 20 mM MES-NaOH at pH 5.5 and 150 mM NaCl. The fluorescence emission, detected at wavelengths between 500-550 nm following excitation at 485 nm, was continuously measured using Glomax Discover microplate reader (Promega). The measurements were taken at room temperature over a duration of 100 minutes at 1 minute intervals. Activity was calculated based on the initial rate, which was estimated by fitting a linear regression to the data from 1 to 11 minutes.
Gel-based nuclease activity assay
A reaction mixture was created by combining a purified protein sample at a concentration of 20 nM for PLD3 or 40 nM for PLD4 with 2.5 μM of substrates. The mixture was then adjusted to the specified pH and incubated at 37 °C for a designated period. Following the incubation, the mixtures were heated at 95 °C for 2 minutes to halt the nuclease activity, and subsequently analyzed by electrophoresis in Tris-Borate-EDTA buffer.
Analysis of TLR9-dependent NFκB activation
TLR9 and Unc93B1 genes were amplified from mouse genomic DNA via PCR, while wild-type and mutant forms of hPLD3 were amplified from cDNA obtained from RIKEN. The TLR9 and Unc93B1 cDNAs were subsequently cloned into retroviral pMXpuro vectors provided by Dr. T. Kitamura. Both wild-type and mutant hPLD3 cDNAs were cloned into pMX-IRES-BFP vectors. Cloning procedures were performed using the NEBuilder HiFi DNA Assembly Cloning Kit (New England BioLabs) and the Rapid DNA Ligation Kit (Roche Applied Science). For retroviral packaging, the pMX vectors were transfected into Plat-E packaging cells using FuGENE 6 (Roche Applied Science). Following 24 hours of incubation, viral supernatants were collected. Ba/F3 cells harboring an NF-κB GFP reporter plasmid were then infected with these viral suspensions in the presence of DOTAP (Roche Applied Science). To standardize hPLD3 expression levels, BFP-positive cells were isolated using FACS Aria (BD Bioscience). Post-stimulation with CpG B, cell analysis was conducted using an ID7000 spectral flow cytometer (Sony Biotechnology). The method of normalization involved subtracting the MFI of unstimulated samples from the MFI of stimulated samples.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or Supplementary Information. Additional data and resources related to this paper may be requested from the authors. The cryo-EM maps and related structure coordinates have been deposited in the EMDB and PDB under accession codes, EMD-64921 and PDB:9VBG [https://doi.org/10.2210/pdb9VBG/pdb] for hPLD3_apo, EMD-64922 and PDB:9VBH [https://doi.org/10.2210/pdb9VBH/pdb] for hPLD3_poly(T), EMD-64923 and PDB:9VBI [https://doi.org/10.2210/pdb9VBI/pdb] for hPLD3_poly(A), EMD-64924 and PDB:9VBJ [https://doi.org/10.2210/pdb9VBJ/pdb] for hPLD4_apo, EMD-64925 and PDB:9VBK [https://doi.org/10.2210/pdb9VBK/pdb] for hPLD4 in the presence of poly(T), respectively. The previously determined structural coordinates used in this study are 8V06, 8V07 and 8Q1X. Source data are provided as a Source Data file. Source data are provided with this paper.
References
Bartok, E. & Hartmann, G. Immune Sensing Mechanisms that Discriminate Self from Altered Self and Foreign Nucleic Acids. Immunity 53, 54–77 (2020).
Okude, H., Ori, D. & Kawai, T. Signaling Through Nucleic Acid Sensors and Their Roles in Inflammatory Diseases. Front Immunol. 11, 625833 (2020).
Miyake, K. et al. Nucleic Acid Sensing by Toll-Like Receptors in the Endosomal Compartment. Front Immunol. 13, 941931 (2022).
Ablasser, A. & Hur, S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 21, 17–29 (2020).
Chan, M. P. et al. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat. Commun. 6, 5853 (2015).
Rodero, M. P. et al. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8, 2176 (2017).
Yoshida, H., Okabe, Y., Kawane, K., Fukuyama, H. & Nagata, S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat. Immunol. 6, 49–56 (2005).
Ahn, J., Gutman, D., Saijo, S. & Barber, G. N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA 109, 19386–19391 (2012).
Ostendorf, T. et al. Immune Sensing of Synthetic, Bacterial, and Protozoan RNA by Toll-like Receptor 8 Requires Coordinated Processing by RNase T2 and RNase 2. Immunity 52, 591–605.e596 (2020).
Greulich, W. et al. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 179, 1264–1275.e1213 (2019).
Liu, K. et al. Skewed endosomal RNA responses from TLR7 to TLR3 in RNase T2-deficient macrophages. Int Immunol. 33, 479–490 (2021).
Gavin, A. L. et al. PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing. Nat. Immunol. 19, 942–953 (2018).
Gavin, A. L. et al. Cleavage of DNA and RNA by PLD3 and PLD4 limits autoinflammatory triggering by multiple sensors. Nat. Commun. 12, 5874 (2021).
Akizuki, S. et al. PLD4 is a genetic determinant to systemic lupus erythematosus and involved in murine autoimmune phenotypes. Ann. Rheum. Dis. 78, 509–518 (2019).
Terao, C. et al. PLD4 as a novel susceptibility gene for systemic sclerosis in a Japanese population. Arthritis Rheum. 65, 472–480 (2013).
Okada, Y. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet 44, 511–516 (2012).
Cruchaga, C. et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 505, 550–554 (2014).
Nibbeling, E. A. R. et al. Exome sequencing and network analysis identifies shared mechanisms underlying spinocerebellar ataxia. Brain 140, 2860–2878 (2017).
Liu, Y. H. et al. Case Report: A Homozygous Mutation (p.Y62X) of. Front Aging Neurosci. 13, 671296 (2021).
Van Acker, Z. P. et al. Phospholipase D3 degrades mitochondrial DNA to regulate nucleotide signaling and APP metabolism. Nat. Commun. 14, 2847 (2023).
Bérouti, M. et al. Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation. Immunity https://doi.org/10.1016/j.immuni.2024.04.010 (2024).
Uesugi, Y. & Hatanaka, T. Phospholipase D mechanism using Streptomyces PLD. Biochim Biophys. Acta 1791, 962–969 (2009).
Yang, W. Nucleases: diversity of structure, function and mechanism. Q Rev. Biophys. 44, 1–93 (2011).
Stanacev, N. Z. & Stuhne-Sekalec, L. On the mechanism of enzymatic phosphatidylation. Biosynthesis of cardiolipin catalyzed by phospholipase D. Biochim Biophys. Acta 210, 350–352 (1970).
Stuckey, J. A. & Dixon, J. E. Crystal structure of a phospholipase D family member. Nat. Struct. Biol. 6, 278–284 (1999).
Gottlin, E. B., Rudolph, A. E., Zhao, Y., Matthews, H. R. & Dixon, J. E. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc. Natl. Acad. Sci. USA 95, 9202–9207 (1998).
Roske, Y. et al. Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5’ exonuclease-mediated nucleic acid degradation. Nucleic Acids Res https://doi.org/10.1093/nar/gkad1114 (2023).
Yuan, M. et al. Structural and mechanistic insights into disease-associated endolysosomal exonucleases PLD3 and PLD4. Structure https://doi.org/10.1016/j.str.2024.02.019 (2024).
Gonzalez, A. C. et al. Unconventional Trafficking of Mammalian Phospholipase D3 to Lysosomes. Cell Rep. 22, 1040–1053 (2018).
Cappel, C., Gonzalez, A. C. & Damme, M. Quantification and characterization of the 5’ exonuclease activity of the lysosomal nuclease PLD3 by a novel cell-based assay. J. Biol. Chem. 296, 100152 (2021).
Grazulis, S. et al. (2005) Proc Natl Acad Sci USA 102, 15797 .
Davies, D. R., Interthal, H., Champoux, J. J. & Hol, W. G. Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide. Chem. Biol. 10, 139–147 (2003).
Flett, F. J. et al. Structural basis for DNA 3’-end processing by human tyrosyl-DNA phosphodiesterase 1. Nat. Commun. 9, 24 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. Biol. Crystallogr 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank Masahide Kikkawa and Yoichi Sakamaki for the management and support of the Graduate School of Medicine cryo-EM facility at the University of Tokyo. This work was supported by a Grant-in-Aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology Grant Nos. 22K06110, 25K09523 (Y.H.), 22H02556, 23K18211 (U.O.), 22H05184(T.S., K.M.) and 23H00366 (T.S.). CREST, JST (grant no. JPMJCR21E4; T.S.、K.M.), the Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) from the Japan Agency of Medical Research and Development (AMED) (Grand No. JP21am0101115; support No. 1570, 620 1846, 1848).
Author information
Authors and Affiliations
Contributions
Y.H. and W.E. designed the experiments. Y.H. and W.E. prepared the protein samples. Y.H. and U.O. conducted cryo-EM data collection. Y.H., W.E. and U.O. processed the cryo-EM data and built the structural model. Y.H., W.E., R.S. and K.M performed the biochemical assays. Y.H. and U.O. performed the computational work. Y.H. and T.S. composed and wrote the manuscript with the assistance of all the authors. Y.H. and T.S. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no Competing interests.
Peer review
Peer review information
Nature Communications thanks David Teran and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hirano, Y., Ezaki, W., Sato, R. et al. Mechanistic insights into single-stranded DNA degradation by lysosomal exonucleases PLD3 and PLD4 from structural snapshots. Nat Commun 16, 11431 (2025). https://doi.org/10.1038/s41467-025-66261-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66261-2