Abstract
In response to drought stress, land plants close their stomata to minimize transpiration. This action precedes a gradual accumulation of the stress hormone abscisic acid (ABA) that enhances plant drought tolerance. However, the molecular mechanisms that cause the time lag between the onset of stomatal closure and ABA accumulation and coordinate these two phases remain unexplained. Here, we found that Arabidopsis thaliana loss-of-function CLAVATA3/ENDOSPERM SURROUNDING REGION 5 (CLE5) mutants are less tolerant to drought. The CLE5 dodecapeptide (CLE5p) acts as a local signal to induce stomatal closure by binding to the LEUCINE-RICH REPEAT RECEPTOR-LIKE KINASE (LRR-RLK) receptor complex, BARELY ANY MERISTEM 1 (BAM1)–GUARD CELL HYDROGEN PEROXIDE-RESISTANT 1 (GHR1), in guard cells. The BAM1–GHR1–CLE5p module directly phosphorylates two SNF1-related protein kinases, OPEN STOMATA1 (SRK2E) and SRK2D, the central regulators of drought responses in plants, to regulate stomatal movement and drought-responsive gene expression without stimulating ABA biosynthesis or ROS accumulation. Our findings mark a critical step in understanding how plants promptly counteract environmental stresses. The CLEp–LRR-RLK signalling components are highly conserved across plant phyla, suggesting that peptide-mediated rapid stomatal closure is a widespread survival strategy and can be exploited to generate drought-resistant crops.
Similar content being viewed by others
Introduction
Drought stunts plant growth and limits crop yield worldwide. To cope with this, terrestrial plants have evolved sophisticated strategies with an array of morphological, physiological, and biochemical adaptations1,2,3,4,5. Stomata are specialized pores located on the leaf surface; they control the evaporation of water absorbed from the soil via transpiration. Therefore, closing stomata is the most effective strategy for land plants to prevent water loss upon drought stress.
Decades of research have illuminated the central importance of the stress hormone abscisic acid (ABA) in actively regulating drought-induced stomatal closure6,7. Synthesized ABA is captured by PYRABACTIN RESISTANCE 1 (PYR)/PYR1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR, which binds to and inhibits clade A PROTEIN PHOSPHATASE TYPE 2Cs (PP2Cs), thereby releasing SNF1-related protein kinase 2s (SnRK2s)7,8,9,10. Activation of subclass III-type SnRK2s, such as SRK2E/OPEN STOMATA1/SnRK2.6 (SRK2E) and SRK2D/SnRK2.2 (SRK2D), by phosphorylation plays important roles in the regulation of stomatal movement and ABA signalling to counteract dehydration7,8,10,11,12. However, accumulating evidence suggests that desiccation stress-induced stomatal closure occurs prior to a gradual increase of the endogenous ABA level in leaves1,2,3,4. This implies that there are yet-unknown signalling pathways responsible for activating the initial response to desiccation stress without stimulating the canonical ABA pathway.
In this study, we identified a local signal transduction module in guard cells, in which the CLE5 peptide (CLE5p) is perceived by the BARELY ANY MERISTEM 1 (BAM1)-GUARD CELL HYDROGEN PEROXIDE-RESISTANT 1 (GHR1) receptor complex under drought stress. This interaction directly activates SnRK2 kinases, leading to rapid stomatal closure prior to ABA accumulation. These findings reveal a previously unrecognized mechanism that regulates the early drought response in plants.
Results
CLE5 is required for rapid drought response
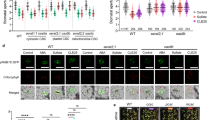
Peptide signalling modules are widely used in animals to transmit information. In plants, CLAVATA3/ENDOSPERM SURROUNDING REGION (CLE) peptides and their LEUCINE-RICH REPEAT RECEPTOR-LIKE KINASE (LRR-RLK) receptors were originally identified as key players in developmental processes13,14,15,16,17. Recently, a root-derived CLE peptide, CLE25p, has been found to be transported from roots to leaves, where it promotes drought stress resistance by stimulating ABA biosynthesis, thus representing a slower response18,19. To explore whether the CLE peptides are involved in rapid drought responses, we subjected a range of cle loss-of-function mutants to short-term dehydration stress and evaluated their sensitivity. Among these lines, CLE5 transfer DNA insertional (cle5-1) and its CRISPR/CAS-9 (CRISPR-cle5) loss-of function mutant leaves exhibited significant water loss within 1 h, while their stomatal density and index were equivalent to wild type leaves (Fig. 1a, b and Supplementary Fig. 1, 2c-e). CLE5 was broadly expressed in leaves and rapidly up-regulated in guard cells within 30 min after the dehydration stress (Fig. 1c, d and Supplementary Fig. 2a). The drought sensitivity phenotype displayed in the cle5 mutants was fully complemented by introducing a CLE5 genomic fragment (Fig. 1a, b and Supplementary Fig. 2b-e).
a Wild type, cle5-1, and pCLE5::CLE5/cle5-1 leaves were detached for 1.5 h. Scale bar: 1 cm. b Water loss in detached wild type, cle5-1, and pCLE5::CLE5/cle5-1 leaves at the indicated time points. c GUS (β-glucuronidase) staining of pCLE5::GUS in leaves (stomata) of eight-day-old seedlings before (left) and after (right) dehydration stress for 30 min. Scale bar: 10 µm. d qRT-PCR analysis of CLE5 expression in eight-day-old wild type leaves under 0, 5, and 15 min dehydration. e RNA-seq analysis of transcriptomic changes in CLE5ox shoots induced by β-estradiol for 2.5 h. Data in the volcano plot represent differentially expressed genes (DEG): red and blue points mark significantly up- and down-regulated genes, respectively (Supplementary Data 1). f Significantly enriched Gene Ontology (GO) terms of the CLE5-induced up-regulated genes in shoots (Supplementary Data 2). g qRT-PCR analysis of NCED3 expression in CLE5ox seedlings treated with Mock or 1 µM β-estradiol (Est) for 2.5 h. h Endogenous ABA level in eight-day-old CLE5ox shoots treated with Mock or 1 µM β-estradiol (Est) for 2.5 h. i Time-resolved stomatal conductance analysis of ABA (2 μM) responses in wild type, cle5-1, pCLE5::CLE5/cle5-1, and pGC1 (stomata-specific promoter)::CLE5/cle5-1 mutants: data represent the mean ± s.d.; ABA was added to the transpiration stream as indicated by the black arrowhead. DW: dry weight. Biological replicates (N) and sample size per replicate (n) for (b, d, g, and h) are listed in Supplementary Data 4; P values presented in (b, g, and h) were calculated with two-tailed unpaired t-test; Adjusted P values (Padj) in d were analyzed by one-way ANOVA followed by a Tukey Honest Significant Differences test; the experiments in a and b were repeated four times, with similar results. n.s., not significant. See also Supplementary Figs. 1 to 4.
To explore the primary gene regulatory network underlying this CLE5-mediated drought resistance, we generated an inducible CLE5 overexpression system (CLE5ox) using the β-estradiol promoter and conducted RNA-sequencing (RNA-seq) analysis. Upon CLE5 induction, 143 genes were significantly up-regulated (including CLE5), and these genes were enriched in the Gene Ontology categories of ABA response and water deprivation (Fig. 1e, f and Supplementary Data 1-2). Both RNA-seq and quantitative real-time PCR (qRT-PCR) analysis showed that CLE5 induction up-regulated the expression of drought-induced ABA-responsive genes, including RESPONSIVE TO DESICCATION 29B (RD29B) and RESPONSIVE TO ABA 18 (RAB18; Fig. 1e, and Supplementary Fig. 2f). Intriguingly, the expression of genes involved in ABA biosynthesis (9-CIS-EPOXYCAROTENOID DIOXYGENASE 3; NCED3 and NGATHA1), perception (ABA-INSENSITIVE 1; ABI1), and other stress-induced ABA-responsive genes (SALT OVERLY SENSITIVE 1; SOS1 and SALT OVERLY SENSITIVE 2; SOS2 (salt stress), DEHYDRATION RESPONSE ELEMENT B1A; DREB1A (osmotic and cold stress)) was barely affected by overexpressing CLE5 (Fig. 1g and Supplementary Fig. 2f and Supplementary Data 1)8,20,21. Consistent with these transcriptomic data, the endogenous ABA level in the induced CLE5ox leaves was similar to that in the mock control (Fig. 1h), suggesting that CLE5 peptide (CLE5p) signalling does not affect ABA biosynthesis in leaves.
CLE5p acts as a local signal to induce stomatal closure
Given that CLE5 expression is up-regulated in guard cells upon dehydration stress, we investigated whether stomatal movement is affected by CLE5p. To dissect CLE5p-mediated signal transduction in leaves, we characterized the mature form of CLE5p in planta. Structure analysis of secreted CLE5p with nanoscale liquid chromatography coupled to tandem mass spectrometry (nano LC-MS/MS) revealed the highest eluent peak at 798.4 m/z, suggesting that a 12-amino-acid CLE5 tri-arabinosylated at the 7th hydroxyproline residue ([Ara3]CLE5p) is the major form (Supplementary Fig. 3a). Since the tri-arabinosylated CLAVATA3 peptide (CLV3p), the closest paralog of CLE5p, shows higher biological activity than the non-arabinosylated form22, we evaluated the activities of CLE5p and [Ara3]CLE5p by comparing the stomatal response to the application of these peptides. Application of ~100-fold less concentrated [Ara3]CLE5p than CLE5p was sufficient to close stomata, indicating that [Ara3]CLE5p has higher biological activity in triggering stomatal closure in nature (Supplementary Fig. 3b). Moreover, the application of [Ara3]CLE5p to both wild type and cle5 mutant leaves triggered stomatal closure (Supplementary Fig. 3c), suggesting that CLE5p itself is the inducer.
Since ABA has been shown to induce rapid stomatal closure23, we investigated stomatal conductance upon ABA treatment in cle5-1 and tissue-specific CLE5-expressing leaves (Fig. 1i and Supplementary Fig. 3d). Wild type and the complemented pCLE5::CLE5/cle5-1 leaves began to close their stomata promptly after ABA application, whereas the leaves from cle5-1 mutants were insensitive to ABA treatment (Fig. 1i). Interestingly, stomata-specific induction of CLE5 (pGC1::CLE5/cle5-1) was sufficient to elicit ABA-induced stomatal closure (Fig. 1i).
Together with the previous RNA-seq analysis, these results suggest that CLE5p acts as a local signal, stimulating neither ABA biosynthesis nor ABA perception to trigger stomatal closure and altering the expression of drought-induced ABA-responsive genes. This action is different from the root-derived CLE25p, which induces stomatal closure by modulating ABA accumulation18.
CLE5p-mediated stomatal closure is not dependent on ROS production
Since overexpressing CLE5 also up-regulates the expression of genes that respond to oxidative stress (Fig. 1f) and reactive oxygen species (ROS) are important signals involved in controlling stomatal movement24, we investigated whether the CLE5p-induced stomatal closure requires ROS-related pathways. First, we performed 3,3′-diaminobenzidine (DAB) staining of CLE5p-treated detached leaves and observed no hydrogen peroxide (H2O2) accumulation, whereas ABA treatment induced H2O2 accumulation (Supplementary Fig. 4a)24. Next, we quantified endogenous oxidant levels in living guard cells using the dye CM-H2DCFDA (5-(and-6)-chloromethyl-2’,7’-dichlorodihydrofluorescein diacetate). No significant differences in fluorescence intensity were observed between the wild type and cle5-1 mutant cells, or during stomatal closure upon CLE5p treatment, suggesting that ROS levels in guard cells are not stimulated by CLE5p (Supplementary Fig. 4b).
Furthermore, we observed that CLE5p treatment of the leaves from the RESPIRATORY BURST OXIDASE HOMOLOG D and F (rbohDrbohF) double mutant, which is known to be NADPH oxidase-deficient in guard cells, induced stomatal closure (Supplementary Fig. 4c)24. The stomata of cle5-1 and bam1-3 (a mutant defective in CLE5p perception in guard cells described in the next section) mutants showed similar sensitivities to H2O2-induced closure as the wild type (Supplementary Fig. 4d). Taken together, these results imply that ROS signals are not involved in CLE5p-mediated stomatal closure.
BAM1–GHR1 is the CLE5p receptor in guard cells
To identify the receptor(s) that perceive CLE5p in guard cells, we screened our LRR-RLK mutant pools by exogenously applying CLE5p to mutant seedlings25. Because most CLE peptides have been shown to induce root growth inhibition13,14,15,16,17, we first searched for mutants that were insensitive to growth inhibition by prolonged exposure to CLE5p. Insensitive mutants with loss-of-function of BARELY ANY MERISTEM 1 (BAM1) and GUARD CELL HYDROGEN PEROXIDE-RESISTANT 1 (GHR1)26,27 genes were further selected based on these genes’ expression in stomatal cells (Supplementary Fig. 5a-d). Neither single bam1-3 and ghr1 nor double bam1-3ghr1 mutants showed significant growth defects like wild type caused by CLE5p treatment (Supplementary Fig. 5a-c).
Since activation of LRR-RLKs usually relies on ligand-induced heterodimerization with a shape-complementary co-receptor through their ectodomains16,28, we asked whether CLE5p, BAM1, and GHR1 form a complex. We first performed co-immunoprecipitation and bimolecular fluorescence complementation assays using full-length BAM1 and GHR1 proteins (Supplementary Fig. 6) and revealed that GHR1 interacts with BAM1, but not with BAM3, in planta. In accordance with this molecular analysis, bam3 seedlings, similar to the wild type, were sensitive to the CLE5p-induced root growth inhibition (Supplementary Fig. 5b).
Next, we performed a thermal shift assay (TSA) to investigate the interaction of CLE5p with BAM1 or GHR1. Compared to the GST (glutathione-S-transferase) control, the melting temperatures of the BAM1–CLE5p and GHR1–CLE5p combinations were significantly shifted in a CLE5p dose-dependent manner, with a higher shift in the BAM1–CLE5p combination (Supplementary Fig. 7). Furthermore, we investigated the binding affinity of CLE5p to BAM1 and GHR1 receptors using an isothermal titration calorimetry (ITC) assay and computational modelling. ITC analysis showed that CLE5p binds to both receptors, with dissociation constant (Κd) values of 2.52 µM for BAM1 and 7.39 µM for GHR1 (Fig. 2a). The thermodynamic parameters of enthalpy factor (ΔH) and entropy factor (-T∆S) were similar between BAM1 and GHR1, suggesting a similar binding mechanism with differences in affinity. A computational docking study (Supplementary Fig. 8) showed that the CLE5p interface score was lower when it forms a complex with BAM1 (−60.5 REU) than with GHR1 (−45.2 REU). Together with the TSA result, these data suggest a higher binding affinity of CLE5p to BAM1.
a Isothermal titration calorimetry (ITC) profiles for interactions of CLE5p with the BAM1 (left) and GHR1 (right) extracellular domain. The dissociation constant (Κd) ± fitting errors, binding stoichiometries (N), and thermal parameters (ΔG Gibbs free energy, ΔH enthalpy, and -TΔS entropy) are indicated. The experiments were repeated twice independently, with similar results. b Representative images of stomata in wild type and bam1-3ghr1 leaves, treated with Mock or 1 µM [Ara3]CLE5p for 1 h. Scale bar, 10 μm. c Statistics of stomatal aperture, measured as the ratio of pore width to length. d qRT-PCR analysis of RD29B (ABA response) and NCED3 (ABA biosynthesis) expression in wild type, bam1-3, and bam1-3ghr1 seedlings treated with Mock or 1 µM [Ara3]CLE5p for 1 h. Biological replicates (N) and sample size per replicate (n) for a, b, c, and d are listed in Supplementary Data 4; P values were calculated with two-tailed unpaired t-test. n.s., not significant. See also Supplementary Fig. 5–8.
We then assessed whether BAM1–GHR1–CLE5p signalling is required for stomatal closure by exogenously applying [Ara3]CLE5p to wild type and bam1-3ghr1 mutant leaves. [Ara3]CLE5p application induced stomatal closure and increased RD29B expression in wild type leaves, but not in two different bam1 mutant alleles (bam1-3 and bam1-4) or bam1-3ghr1 leaves (Fig. 2b, c, and Supplementary Fig. 5e). As described earlier, overexpression of CLE5 did not alter the expression of genes involved in ABA biosynthesis or the endogenous ABA content in leaves (Fig. 1e–h). Consistent with this, NCED3 expression level was unaffected by [Ara3]CLE5p treatment (Fig. 2d). Taken together, our results pinpoint the role of BAM1–GHR1–CLE5p signalling in activating stomatal closure, without stimulating ABA biosynthesis.
The BAM1–GHR1–CLE5p module activates SRK2E
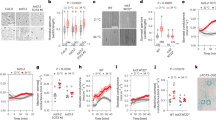
To further dissect the CLE5p signalling pathway, we searched for downstream components that could be phosphorylated by the BAM1–GHR1–CLE5p complex. Unlike other catalytic LRR-RLKs (including BAM1), GHR1 is considered as a pseudokinase due to the absence of a C-terminal cytoplasmic kinase domain (Supplementary Fig. 6b)26. Therefore, it is thought to act as a scaffold for yet-to-be-identified regulators to induce stomatal closure, which partially depends on the phosphorylation status of SRK2E29,30. We thus reasoned that the BAM1–GHR1–CLE5p complex in stomata might phosphorylate SnRK2 kinases. To test this, we performed a protein mobility shift assay by applying [Ara3]CLE5p to Arabidopsis leaf protoplasts transfected with 35S::SRK2E-GFP and 35S::SRK2D-GFP, which are involved in drought-induced ABA signalling, for up to 30 min31. In both cases, a distinct band shift was detected in the [Ara3]CLE5p- or ABA-treated wild type protoplasts, and this band was diminished by phosphatase treatment (Fig. 3a, Supplementary Fig. 9a). In contrast, no such mobility shift was observed in bam1-3, ghr1, or bam1-3ghr1 protoplasts (Fig. 3a, Supplementary Fig. 9a). These findings indicate that the BAM1–GHR1–CLE5p module mediates the phosphorylation of SRK2D and SRK2E, the key regulators involved in drought-induced ABA signalling31.
a Short-term [Ara3]CLE5p application is sufficient to phosphorylate SRK2E-GFP (P-SRK2E) in vivo. Wild type, bam1-3, ghr1, and bam1-3ghr1 protoplasts expressing 35S::SRK2E-GFP were treated with or without 1 μM [Ara3]CLE5p for 30 min. Peptide-untreated protoplasts (-) or 1 μM ABA-treated protoplasts (ABA) were used as a negative or positive control, respectively. SRK2E-GFP transfected protoplasts were first treated with 1 μM [Ara3]CLE5p for 30 min and then incubated with 1 μM λ-phosphatase (λ-PPase) for 30 min. SRK2E-GFP proteins were detected by anti-GFP antibody (αGFP). Phosphorylated (P-) and non-phosphorylated isoforms are indicated. Tubulin was used as the internal control. The samples derive from the same experiment and that blots (αGFP and αTubulin) were processed in parallel. b Structure of SRK2E and its major phosphorylation sites catalysed by the BAM1 kinase domain identified from an LC-MS/MS spectrum. Red highlights the phosphorylated resides within the activation domain (T-loop). c In vitro kinase assay. The recombinant GST-BAM1 kinase domain (GST-BAM1(KD)), MBP-SRK2E(D140A), MBP-SRK2E(G33R/D140A), and MBP-SRK2E (S175A/T176A/T179A, Triple A) proteins were tested. d Kinase activity of GST-BAM1(KD) to the MBP-SRK2Es (D140A, G33R/D140A, Triple A). histone III was used as the substrate. e, f Time-resolved stomatal conductance analysis of ABA (2 μM) responses in wild type, bam1-3, ghr1, and bam1-3ghr1 (e) or srk2e, 35S::SRK2E(Triple A)/srk2e, and 35S::SRK2E/srk2e leaves (f): data represent the mean ± s.d. from at least three biological replicates (Supplementary Data 4). Coomassie brilliant blue (CBB) staining in (a, c, and d) was used as the loading control; The experiments in (a, c, and d) were repeated at least in duplicate, with similar results. See also Supplementary Fig. 9.
Since phosphorylation of SnRK2 kinase within the T-loop is known to be essential for its activation32, we undertook LC-MS/MS analysis using recombinant BAM1 and GST proteins to identify phosphorylation site(s) of SRK2E that are targeted by the BAM1–GHR1–CLE5p module. The GST-fused kinase domain of BAM1 (hereafter GST-BAM1(KD)) phosphorylated S175, T176, and T179 residues of SRK2E, located within its T-loop (from D160 to E184; Fig. 3b, and Supplementary Fig. 9b). Previous studies have shown that the ATP-γ-phosphate proton acceptor site at Asp140 is pivotal for SRK2E activity, while the Gly-to-Arg G33R mutation suppresses its activity to a kinase-dead level33,34. As shown in Fig. 3c, GST-BAM1(KD) effectively phosphorylated these SRK2E kinase-dead variants (D140A and G33R/D140A), whereas the phosphorylation signal was reduced when using the SRK2E mutations on S175A/T176A/T179A sites, hereafter SRK2E (Triple A). We also examined the kinase activity of BAM1 using an in-gel kinase assay, and revealed that GST-BAM1(KD) significantly enhanced the activity of maltose-binding protein (MBP)-SRK2E, while MBP-SRK2E (Triple A), like inactive SRK2E(D140A), showed no detectable activity (Fig. 3d). Taking these findings together, the cytosolic kinase domain of BAM1 protein directly activates SRK2E kinase by phosphorylating three residues (S175/T176/T179) within the activation T-loop.
To address the in vivo physiological significance of the direct SRK2E phosphorylation by BAM1, we next investigated whether the BAM1–GHR1–CLE5p-directed phosphorylation of SRK2E is required for rapid stomatal closure in vivo. We first monitored stomatal conductance in bam1-3, ghr1, and bam1-3ghr1 mutant leaves treated with 2 µM ABA (Fig. 3e)23. In contrast to wild type and similar to the cle5 mutants (Supplementary Fig. 3d), all tested mutant leaves were insensitive to the ABA-induced reduction of stomatal conductance, suggesting that the BAM1–GHR1–CLE5p module is required for ABA-induced stomatal closure (Fig. 3e). We then monitored stomatal conductance in 35S::SRK2E/srk2e and 35S::SRK2E(Triple A)/srk2e leaves treated with 2 µM ABA (Fig. 3f). Compared to the steady-state stomatal conductance in the ABA-treated loss-of-function srk2e leaves, stomatal conductance in 35S::SRK2E/srk2e leaves decreased rapidly, indicating that the expression of 35S::SRK2E in srk2e leaves fully complements SRK2E function in mediating stomatal movement. In contrast, stomatal conductance of 35S::SRK2E(Triple A)/srk2e leaves was insensitive to the ABA treatment, indicating that BAM1-catalysed phosphorylation is crucial for SRK2E function in ABA-induced rapid stomatal closure.
The BAM1–GHR1–CLE5p/SRK2E module regulates rapid drought responses
We showed that CLE5p is required for the drought stress response in leaves and that the BAM1–GHR1–CLE5p module induces stomatal closure without altering the expression of key genes involved in ABA biosynthesis (Figs. 1g and 2d). Moreover, in response to dehydration, stomatal closure is achieved prior to the boost of ABA accumulation1,2,3,4,7,18,19. Therefore, we asked whether ABA and canonical ABA signalling is required for drought-induced BAM1–GHR1–CLE5p/SRK2E-mediated stomatal closure.
First, we performed the dehydration assay on bam1 mutant alleles (bam1-3 and bam1-4), ghr1, bam1-3ghr1, and bam1-3ghr1cle5-1 mutant leaves. These mutant leaves showed significantly greater water loss than the wild type (Fig. 4a and Supplementary Fig. 10a), similar to the cle5 leaves (Fig. 1a, b and Supplementary Fig. 2b–d). Next, we measured the change of ABA level in cle5-1 and bam1-3ghr1 shoots upon drought. Endogenous ABA content in the receptor mutant was comparable to that in the wild type and cle5-1 (Fig. 4b). Once those plants were exposed to drought, ABA accumulation and the expression of an ABA biosynthesis gene (NCED3) were both markedly induced (Fig. 4b and Supplementary Fig. 10b), suggesting that the BAM1–GHR1–CLE5p module is not involved in drought-induced ABA accumulation. However, induced expression of the dehydration stress marker genes RD29B and RAB18 was observed only in the wild type leaves upon 1 h dehydration stress (Supplementary Fig. 10c, d). Furthermore, the application of ABA to cle5-1 and bam1-3ghr1 mutant leaves failed to induce stomatal closure in 30 min (Supplementary Fig. 10e). These results suggest that ABA accumulation cannot mask the BAM1–GHR1–CLE5p-mediated drought responses.
a Images of wild type, bam1-3, ghr1, bam1-3ghr1, and bam1-3ghr1cle5-1 leaves detached for 1.5 h. Scale bar: 1 cm. b Endogenous ABA level in wild type, cle5-1, and bam1-3ghr1 shoots after 2.5 h dehydration stress. Different letters above the columns represent a two-way ANOVA analysis followed by a Tukey Honest Significant Differences test; Pr( > F) for the treatment = 1.8e-08, Pr( > F) for the genotype = 0.695, Pr( > F) for the treatment:genotype interaction = 0.977. DW: dry weight. c Ten-day-old wild type and srk2e seedlings were exposed to drought stress for 20 min, and 20 µg of their crude extracts were subjected to an in-gel kinase reaction with histone III as the substrate. d Short-term application of 1 μM [Ara3]CLE5p to wild type, srk2e, and nced3 leaves for 30 min. Left: representative images of stomata. Scale bars, 5 μm. Right: statistics of stomatal aperture, measured as the ratio of pore width to length (W/L). e qRT-PCR analysis of RD29B and RAB18 (drought stress-responsive genes) expression in wild type and ABA-insensitive (abi1-1) seedlings treated with Mock or 1 μM [Ara3]CLE5p for 1 h. f Schematic representation of the BAM1–GHR1–CLE5p/SRK2E signal transduction module. Upon drought stress, CLE5p is captured by the BAM1–GHR1 receptor complex, located on the plasma membrane of guard cells. This module phosphorylates SRK2E and SRK2D kinases to induce stomatal closure and activate the expression of drought stress-responsive genes without stimulating the canonical ABA pathway. Biological replicates (N) and sample size per replicate (n) for (b, c, and e) are listed in Supplementary Data 4; P values in (c, e) were calculated with two-tailed unpaired t-test; the experiments in a and e were repeated three times, with similar results. n.s., not significant. See also Supplementary Fig. 10.
Consistent with the previous finding that srk2e mutants cannot endure a rapid humidity decrease31, we found that SRK2E in wild type plants was already activated under short-term dehydration ( < 30 min), during which ABA accumulation is not likely to be sufficient (Fig. 4c, and Supplementary Fig. 10f)1,2,3,4. Therefore, we examined whether SRK2E or ABA biosynthesis is required for the drought responses operated by the CLE5p signalling. In contrast to wild type plants, SRK2E was not activated in srk2e, cle5-1, bam1-3, ghr1 or bam1-3ghr1 mutants under the short-term dehydration (Fig. 4c, Supplementary Fig. 10f). Moreover, we found that [Ara3]CLE5p application caused stomatal closure within 30 min in wild type and nced3 mutant leaves, but not in srk2e mutant leaves (Fig. 4d). Taken together, our results suggest that stomatal closure triggered by CLE5p signalling requires at least prompt SRK2E activity, but not ABA biosynthesis.
In the canonical ABA pathway, group A PP2C members interact physically with SnRK2s to inactivate their kinase activity, thereby repressing downstream ABA responses35,36. This repression is counteracted by the PYR/PYL/RCAR ABA receptors in response to ABA7,8,9,10. However, even though ABA accumulation and ABA biosynthesis gene expression were not stimulated by CLE5p (Figs. 1g, h, 2d, 4b), the BAM1–GHR1–CLE5p module activated SRK2E and the expression of a subset of drought-responsive genes (Figs. 2d, 3a, d, and Supplementary Fig. 10b–d, f). Therefore, we assessed whether ABA perception is required for the action of the BAM1–GHR1–CLE5p module by applying [Ara3]CLE5p to abi1-1 mutant leaves, in which the G180D mutation in the ABI1 protein leads to a constitutive inactivation of SnRK2s10,31,37. We showed that [Ara3]CLE5p treatment partially masks the dominant negative role of abi1-1 protein in ABA perception to activate drought-responsive gene expression (Fig. 4e), suggesting that the BAM1–GHR1–CLE5p module partially bypasses ABI1-mediated ABA perception to induce drought responses (Fig. 4f)7,8,38.
Discussion
Understanding signalling mechanisms that specifically respond to environmental stimuli has the potential to yield new technologies for optimizing plant growth in ever-changing environments. To date, many hormonal and intracellular molecular mechanisms underlying stress responses have been identified, but the regulatory mechanisms that act upstream of these intracellular signalling pathways remain enigmatic. Here, we revealed the BAM1–GHR1–CLE5p/SRK2E signalling module as a local player to initiate primary drought responses without stimulating ABA biosynthesis or ROS production. Interestingly, modelling the association of CLE5p with the BAM1–GHR1 complex showed a lower interface score (−73.04 REU) compared to its binding to either BAM1 or GHR1 alone (Supplementary Fig. 8), suggesting that the binding affinity of CLE5p for its receptor complex is vital for activating downstream signalling pathways.
Beyond drought stress, SRK2E proteins have been reported to play a central role in the response to other environmental stresses29,30,39,40,41,42,43, such as oxidative, osmotic, and cold stresses. Our experiments showed that the BAM1–GHR1–CLE5p module phosphorylates three residues within the T-loop region of SRK2E for its activation. However, since more phosphorylation sites are predicted in the active centre of SRK2E, the degree of phosphorylation in SRK2E may change under various environmental stresses, thereby fine-tuning its biological activity and substrate specificity. At least one of the residues (S175) in SRK2E protein phosphorylated by the BAM1–GHR1–CLE5p signalling module has been reported to be dephosphorylated by PP2Cs, including ABI135,36, and the CLE5p signal is able to partially overcome the dominant effect on the abi1-1 protein (Fig. 4e). These data suggest that CLE5p and ABA synthesis pathways may compete to activate or inhibit SnRK2 function, thereby regulating SnRK2 activities in a post-translational manner.
A variety of guard cell-expressed ion channels and membrane-localized receptor proteins have been reported to function in drought responses and stomatal closure44,45,46. We observed that the water loss in srk2e leaves is more severe than in cle5-1 or bam1-3ghr1 mutant leaves. Therefore, we speculate that other peptide molecules and functional analogues of the receptors may act together to induce drought responses. The interaction of the BAM1–GHR1–CLE5p module with other signalling cascades, such as RAPIDLY ACCELERATED FIBROSARCOMA-like (Raf-like) kinase and MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) pathways, may jointly activate SRK2E to confer or boost resistance against drought and other stresses29,30,39,40,41,42,43. Several ion channels, such as SLOW ANION CHANNEL-ASSOCIATED 1 (SLAC1), QUICK-ACTIVATING ANION CHANNEL 1 (QUAC1/ALMT12), and POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1 (KAT1), may also jointly induce stomatal closure together with the BAM1–GHR1–CLE5p module33,45,47. Unraveling the interplay of these networks is a major challenge to fully understand the nature of plant responses to environmental changes.
In the future, identification of the downstream targets catalysed by SRK2E and SRK2D via the CLE5 signalling module will provide a more complete depiction of the rapid response to drought. In addition, how the BAM1–GHR1–CLE5p signalling module is initially activated remains an interesting open question. Since CLE peptides and SnRK2 signalling components are broadly conserved in plants48,49,50, our study illuminates the exciting question of how specific combinations of signalling peptide(s) and SnRK2(s) form versatile signal transduction networks that help plants to resist environmental stresses. Further insights into these signalling networks will facilitate a better design of breeding strategies to improve stress tolerance in crops.
Methods
Plant materials and growth conditions
Other than in stomatal conductance measurement (Figs. 1i, 3e, 3f, and Supplementary Fig. 3d), Arabidopsis thaliana (Arabidopsis) was grown under long-day conditions (16/8-h photoperiod) at 22 °C. Nicotiana benthamiana was grown under short-day conditions (10/14-h photoperiod) at 22 °C and used for BiFC analysis and co-immunoprecipitation assay (see below). For intact leaf gas exchange analysis using Arabidopsis leaves, plants were grown under short-day conditions (10/14-h photoperiod) at 22 °C. All seeds were sterilized with bleach solution (10% commercial bleach, 0.02% Triton X-100) for 10 min, washed three times with sterile water, and first cold-imbibed for three days in the dark at 4 °C. The T-DNA mutants in Supplementary Data 3 were all obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org) or GABI Kat (https://www.gabi-kat.de). The rbohDrbohF double mutant 51 was a kind gift from K. Kuchitsu of Tokyo University of Science. We established homozygous lines by genotyping PCR (Supplementary Data 3). bam1-3ghr1 double mutants were generated by crossing. For CLE5 induction experiments as well as RNA-seq analysis, 8-d.a.g. (days after germination) seedlings harbouring the pMDC7-CLE5 construct were placed on 1/2 Murashige and Skoog (1/2MS) agarose plates containing either 1 µM β-estradiol (FUJIFILM) or solvent as sample or mock, respectively. For the peptide application test, Arabidopsis seedlings were transferred to 1/2MS plates containing 0.75 µM CLE5 peptide (CLE5p) or 0.1 µM [Ara3]CLE5 peptide ([Ara3]CLE5p) for the indicated times.
Plasmid DNA constructs and transformation
PCR fragments encompassing the functional CLE5, BAM1, BAM3, and GHR1 coding sequences as well as their promoter regions were designed with reference to previous publications (Supplementary Data 3)27,52,53,54. For the CLE5 complementation test, a 4-kb genomic region spanning 3.2 kb upstream of the translation start and the 3′UTR (denoted as the full-length CLE5 genomic region) was cloned by PCR and used to generate the pCLE5::CLE5 transgene (Supplementary Data 3). To generate a complemented plant that specifically expresses CLE5 in stomatal guard cells, the 1.73-kb promoter region of GC1 was amplified by PCR and inserted into the pGWB504 vector in-frame with the CLE5 coding region (pGWB504::pGC1) (Supplementary Data 3). PCR fragments of coding sequences were all inserted into pENTR/D-TOPO vectors according to the GATEWAY manufacturer’s instructions (Invitrogen). After sequencing, each clone was recombined with pMDC163 (GUS reporter vector), pBGYN (EYFP + NLS tag fusion vector), R4pGWB501 (complementation test), pGWB11 (C-terminal FLAG tag fusion vector), pGWB20 (C-terminal 10×Myc tag fusion vector), pGWB504::pGC1 (stomatal-specific expression vector), and pMDC7 (ß-estradiol-inducible overexpression vector) using LR Clonase (Invitrogen), respectively. The CRISPR-cle5 mutant was generated using the CRISPR/Cas9 system, as described elsewhere55. The target sequence was determined using CRISPRdirect software56. Cloning was performed according to Fauser et al.55. Two 35S::SRK2–GFP constructs (35S::SRK2D–GFP and 35S::SRK2E–GFP) were generated according to previous reports31,57. All primer information used in this study is provided in Supplementary Data 3, and all primers were synthesized by eurofins scientific (https://www.eurofins.com/). All constructs were verified by sequencing. Arabidopsis plants were transformed via flower dipping using Agrobacterium tumefaciens (strain C58MP90)58.
Site-directed mutagenesis and recombinant protein purification
For in vitro protein purification, pMALc4x-SRK2Es were constructed by inserting the coding region of SRK2E. Site-directed mutagenesis was performed by inverse PCR, which resulted in the production of pMALc4x-SRK2E(G33R), pMALc4x-SRK2E(D140A), pMALc4x-SRK2E(G33R/D140A), and pMALc4x-SRK2E(S175A/T176A/T179A, Triple A). The kinase domain sequence of BAM1 was amplified and cloned into pDONR221 vector using BP Clonase (Invitrogen), and subsequently inserted into the pDEST15 binary vector using LR Clonase (Invitrogen). Primer sequences are listed in Supplementary Data 3. All recombinant proteins were batch-purified according to manufacturers’ protocols (for GST proteins, Cytiva; for MBP proteins, New England Biolabs).
In vitro phosphorylation assays and in-gel kinase assays
Phosphorylation assays were performed as described in Soma et al.59 with slight modifications. Kinase (0.5 µg) and substrate (1.5 µg) were incubated for 30 min at room temperature in 12.5 µl of 1× kinase buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 2.5 mM CaCl2, 1× protease inhibitor cocktail (Roche), 10 mM NaF, and 5 mM β-glycerophosphate with 20 µCi of [γ−32P]ATP. The reaction was stopped by adding 4 × SDS-PAGE loading buffer and heating at 95 °C for 5 min. Proteins were separated by SDS-PAGE and phosphorylation of the substrate was detected by applying a phosphor imaging screen to the dried gel using a Typhoon FLA9000 Phosphor Imager (GE Healthcare). For in-gel assays, total proteins were extracted from 10-day-old Arabidopsis seedlings that were untreated (Mock) or subjected to dehydration stress (50–55% humidity) for 20 min. The seedlings were then ground to a fine powder in liquid nitrogen and homogenized at 4 °C in extraction buffer (100 mM HEPES-HCl (pH 7.5), 5 mM EDTA, 5 mM EGTA, 0.5% Triton X-100, 150 mM NaCl, 0.5 mM DTT, 10 mM NaF, 5 mM Na3VO4, 5 mM β-glycerophosphate, and complete ULTRA protease inhibitor cocktail (Sigma)). The resultant crude extracts were centrifuged at 15,300 g and 4 °C for 30 min to remove debris. The supernatants were collected and 20 µg per sample was separated by SDS-PAGE in a polyacrylamide gel containing 200 ng of histone III (Sigma). For recombinant MBP-SRK2E proteins, 0.2 µg of purified proteins were loaded. The gel was washed three times in buffer (25 mM Tris-HCl (pH 8.0), 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/ml BSA, 0.1% (v/v) Triton X-100), and further incubated in renaturation buffer (25 mM Tris-HCl (pH 8.0), 1 mM DTT, 0.1 mM Na3VO4, 5 mM NaF) overnight at 4 °C. The in-gel assay followed a published method60. The resultant gel was dried and analysed using a Typhoon FLA9000 Phosphor Imager.
Nano LC-MS/MS and data analysis
Full-length recombinant maltose binding protein (MBP)-SRK2E protein purified from Escherichia coli was subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. An in vitro phosphorylation reaction was performed in the same kinase reaction buffer as above, except with 100 µM cold ATP. Samples were separated by SDS-PAGE, the selected protein bands were excised from the gel, and subsequently in-gel digestion was performed61. The resultant peptides were analysed by nano LC-MS/MS using the UltiMate 3000 Nano LC system (ThermoFisher Scientific) coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific) with a nanoelectrospray ionization source. After injection, the peptides were trapped on a 5 × 0.3 mm ID trap column packed with 5 µm C18 resin and separated at a flow rate of 500 nl/min using a 5–40% buffer B gradient over 100 min on a NANO-HPLC capillary column C18 (0.1 × 125 mm, Nikkyo Technos). The composition of LC buffer A was 0.5% (v/v) acetic acid in water and that of LC buffer B was 80% (v/v) acetonitrile, 0.5% (v/v) acetic acid. Survey full scan MS spectra were collected from 350 to 1800 m/z in the Orbitrap with a resolution of 70,000 and an AGC target of 3E6. For the MS/MS experiment, the 10 most intense multiplied charged precursors (z ≥ 2) were accumulated to a 1E5 target value and fragmented in the collision cell by higher-energy collisional dissociation (HCD). The precursor isolation width was 2.0 m/z. The HCD normalized collision energy was 27%. The maximum injection time was set to 60 ms and dynamic exclusion was set to 10 s. The obtained MS and MS/MS data were analyzed using Proteome Discoverer 2.5.0.400 (Thermo Fisher Scientific). Database searches were performed against the TAIR10 Arabidopsis protein database, including the SRK2E protein sequence and the common Repository of Adventitious Proteins (cRAP) contaminant database (http://www.thegpm.org/crap/). Search parameters were as follows: enzyme specificity, trypsin; maximum of two missed cleavages; minimum and maximum peptide lengths, 6 and 144 amino acids, respectively; variable modifications, phosphorylation (Ser/Thr/Tyr) and oxidation (Met); static modification, carbamidomethylation (Cys); precursor mass tolerance, ±10 ppm; fragment mass tolerance, ±0.02 Da. Peptide-spectrum matches (PSMs) were filtered to 1% false discovery rate (FDR) using the Percolator algorithm, corresponding to high-confidence identifications. Phosphorylation site localization probabilities were calculated using the ptmRS node integrated in Proteome Discoverer. Precursor quantification was based on precursor ion intensities.
RNA preparation and transcriptome analysis
Total RNA was isolated from Arabidopsis seedlings using an RNeasy Plant Mini kit (QIAGEN) following the manufacturer’s instructions. A NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) was used to prepare cDNA libraries according to the manufacturer’s instructions. RNA-seq was performed on the Illumina NextSeq500 platform (Illumina) in single-end mode62. The actual read length obtained was 86 bp. Raw reads containing adapter sequences were trimmed using bcl2fastq (Illumina), and nucleotides with low quality (QV < 25) were masked by N using an in-house script. Reads were aligned against TAIR10 reference genome by STAR (v2.7.10b). Counts per gene were computed with featureCount (v2.0.1). Differential expression analysis was performed with R (v4.2.2) script tool DESeq2 (v3.16). TAIR10 reference genome sequence and Araport11 annotation can be downloaded from: (https://www.arabidopsis.org/). Differentially expressed genes were defined with the adjust P-value (padj) <0.05 and |log2FC (fold change)| ≥ 1. Adjust P values shown in Fig. 1e, Supplementary Data 1 and Supplementary Data 2 (sheet “sigUP_regulated_Genes”) were calculated via R script tool DESeq2 (v3.16) using Benjamini-Hochberg method to control the false discovery rate (FDR). Gene Ontology enrichment analysis was performed using DAVID63. P values shown in Fig. 1f and Supplementary Data 2 (sheet “GOup_CLE5_shoot”) were calculated via DAVID using Expression Analysis Systematic Explorer (EASE) score, a modified Fisher’s Exact Test that assesses the statistical significance of gene-term enrichment. Adjust P values using FDR estimate, Benjamini-Hochberg method and Bonferroni correction were also provided in the Supplementary Data 2 (sheet “GOup_CLE5_shoot”). For quantitative real-time PCR (RT-qPCR) analysis, 1 µg of total RNA was reverse transcribed using Superscript III (Invitrogen) with oligo-dT primer and subsequently amplified using LightCycler SYBR Green Master (Roche) according to the manufacturer’s guidelines. Expression data were normalized against the average expression values of the reference gene UBQ10 (AT4G05320). Each assay was performed on at least three biological replicates, and similar results were obtained. All primer information for RT-qPCR is listed in Supplementary Data 3.
Stomatal conductance measurements
Gas exchange measurements were performed using an LI-6800 instrument (LI-COR). Sample preparation is described elsewhere23. Detached leaves were placed in the gas exchange chamber and equilibrated until the stomatal conductance value became stable. The values of stomatal conductance to water (gsw) were recorded under the following conditions: the LED light source was set as 200 µmol m−2 s−1 (90% red and 10% blue), gas exchange temperature was 22 °C, relative humidity in the chamber was set at 75%, airflow was set to 200 rpm, and the CO2 gas concentration setting was kept at 400 ppm. The indicated ABA treatments were conducted 10 min after recording started. All data represent n ≥ 3 per genotype and were normalized to the average of the first 10 min gsw values.
Bimolecular fluorescence complementation (BiFC) assay
The functional coding sequences of BAM1, BAM3, and GHR127,52,53,54 were amplified by PCR, cloned into pENTR, and recombined into the vectors pBGYN and pBGCN (Funakoshi) by Gateway LR (ThermoFisher Scientific) to make C-terminal-fused YFP constructs of each protein (Supplementary Fig. 6b). These proteins were transiently expressed in N. benthamiana epidermis cells under short-day conditions (10/14-h photoperiod at 22 °C). Leaves were infiltrated with A. tumefaciens (C58MP90) that had previously been transformed with appropriate plant expression vectors according to established protocols64. After incubation for two days at 22 °C, leaf discs were excised from the infiltrated leaves and observed under an FV1200 confocal microscope (Olympus). Fluorescence signals for YFP (excitation 514 nm, emission 517–569 nm) and PI (excitation 536 nm, emission 617 nm) were detected. The BiFC assays were performed on more than four individual plants for independent transfected lines with two repetitions, and similar expression patterns were obtained. Primer information is listed in Supplementary Data 3.
Histochemical GUS staining
GUS staining and fixation were performed according to the method of Willemsen et al.65. For the dehydration assay, 10-d.a.g. seedlings were transferred to Whatman filter paper (GE Healthcare) for the indicated time. They were then immediately fixed in cold 90% acetone for 1 h, rinsed with water, immersed in staining solution, and vacuum-infiltrated for 10 min in staining solution [1 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide), 0.5 mM K3Fe(CN)6, 0.5 mM K2Fe(CN)6, 1% Triton X-100, 10 mM EDTA, and 100 mM NaPO4 (pH 7.2)] followed by incubation at 37 °C. Pictures were taken with Nomarski Optics (BX53; Olympus).
Peptide structure analysis
CLE5 cDNA was obtained by RT-qPCR using total RNA isolated from Arabidopsis roots and specific primers (Supplementary Data 3). The amplified fragment was cloned into pMDC7 using Gateway LR Clonase II (Invitrogen). The construct was introduced into wild type Col-0 by Agrobacterium-mediated transformation. Arabidopsis seeds ( ~ 100) containing the β-estradiol-inducible CLE5 gene were sown directly in 100 ml of B5 liquid medium containing 1% sucrose and incubated at 22 °C without shaking under continuous light. After 14 days of culture, expression of the transgene was induced by addition of 75 µM β-estradiol for 24 h. Peptides were extracted from the culture medium using o-chlorophenol and then performing acetone precipitation66. Fractions containing total secreted peptide were adjusted to a total volume of 500 µl with 0.1% trifluoroacetic acid. Nano LC-MS analysis was performed using a DiNa-M splitless nano HPLC system (KYA Technologies) connected to an LTQ Orbitrap XL mass spectrometer (ThermoFisher Scientific). A 5 µl aliquot of peptide was loaded onto a C18 trap column (0.5 mm i.d. × 1 mm cartridge; KYA Technologies), which was then washed with 10 µl of 0.1% formic acid. Peptides were subsequently eluted from this precolumn and separated on a MonoCap C18 Fast-flow nano-column (100 µm i.d. × 150 mm; GL Sciences) using a gradient of 2–50% acetonitrile containing 0.1% formic acid for 30 min at a flow rate of 500 nl/min. Tandem mass spectra were obtained by scanning the mass range from m/z 350 to m/z 1,500 using data-dependent acquisition methods with HCD fragmentation. The data were analysed with Proteome Discoverer 1.3 software using the SEQUEST search engine (ThermoFisher Scientific).
Synthesis of [Ara3]CLE5 peptide
Asp(tBu)-Pro-Gln(Trt)-His(Trt)-His(Trt)-PEG-resin was prepared on an automated peptide synthesizer (Initiator+Alstra). A mixture of Fmoc-[AcAra3]Hyp–OH (6.0 mg, 5.8 µmol)22, 1-hydroxybenzotriazole (2.7 mg, 20 µmol), O-benzotriazole-N,N,N’,N’-tetramethyluronium hexafluorophosphate (7.6 mg, 20 µmol) and N,N-diisopropylethylamine dissolved in dry N-methylpyrrolidone (200 µl) was added to the peptide-resin (25 µmol) pre-swollen with dry N-methylpyrrolidone (50 µl). The mixture was stirred for 2 h at room temperature. Peptide-resin was recovered by filtration, and the remaining N-terminal amino acids were added to the peptide using the peptide synthesizer. The synthesized peptide was deprotected and cleaved from the resin using trifluoroacetic acid/water (95:5 v/v) (1 ml) for 30 min and then precipitated by adding cold ether (10 ml) for 5 min at −20 °C. The precipitate was washed twice with cold ether, dissolved in water, and lyophilized. Crude peptide was dissolved in dry methanol (3 ml) and treated with sodium methoxide (28% solution, 60 µl) at room temperature for 1 h. The reaction was terminated by adding acetic acid (60 µl). HPLC purification using an amide column (TSK-gel amide-80; TOSOH) gave analytically pure [Ara3]CLE5 (3.7 mg).
Co-immunoprecipitation Experiments in N. benthamiana
Fully expanded six-week-old N. benthamiana leaves were co-infiltrated with Agrobacterium cultures bearing 35S::GHR1-FLAG, 35S::BAM1-10xMyc, and 35S::BAM3-10xMyc plasmids in the appropriate combinations64. After two days, leaves were harvested and homogenized with extraction buffer [20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, 2.5 mM EDTA, 0.1% Nonidet P-40, 1 mM PMSF, and protease inhibitor cocktail (Sigma)]. Crude extracts were cleared by centrifugation at 15,300 g for 10 min at 4 °C, and 1 ml of the supernatant was subsequently incubated with 50 µl anti-FLAG agarose beads (#A2220; Sigma) and rotated at 4 °C for 3 h. Beads were washed three times with 1 ml extraction buffer containing protease inhibitor. Immunoprecipitates were mixed with 25 µl 4 × SDS-PAGE loading dye and eluted by heating for 5 min at 95 °C. Anti-FLAG (#F1804; Sigma) and anti-cMyc (#M4439; Sigma) antibodies were used for immunodetection and anti-mouse antibody conjugated with HRP (#W4021; Promega) was used as the secondary antibody. Three independent biological replications were performed, with similar results.
LRR-RLK screening
As a method of receptor exploration, we initially examined the sensitivity of roots treated with a high concentration of CLE5 peptide (CLE5p) to inhibit growth. The lrr-rlk mutants25 were germinated on a 1/2MS agar plate for 3 days before transfer to a 1/2MS agar plate containing 0.75 µM CLE5p, and primary root length of each mutant was measured after 3 days of peptide treatment. To compare the biological activity of two CLE5 peptides (CLE5p and [Ara3]CLE5p), three-day-old wild type seedlings were transferred onto a 1/2MS agar plate containing the indicated amount of CLE5p and grown for a further 4 days. Primary root length was measured by ImageJ software (https://imagej.nih.gov/ij/). Of the candidates from this primary screening, we then examined expression patterns at the cellular level using GUS staining and the eFP browser 2.0 (https://bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html) to narrow down the receptors expressed in stomatal guard cells, and finally targeted receptor mutants that did not cause stomatal closure upon CLE5 peptide treatment.
Protein extraction and Western Blotting in Arabidopsis
The preparation and transfection of Arabidopsis mesophyll protoplasts have been described previously (see http://genetics.mgh.harvard.edu/sheenweb/). Protoplasts (6 × 104) were transfected with 10 μg of plasmid DNA and incubated at 21 °C for 18 h in W5 solution under continuous illumination. Arabidopsis mesophyll protoplasts expressing 35S::SnRK2s–GFP (SRK2D or SRK2E) were treated with 1 µM [Ara3]CLE5p or 1 µM ABA for the indicated times, and then immediately frozen in liquid nitrogen, ground using a multi-beads shocker (QIAGEN), and lysed in protein extraction buffer [50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 0.1% Triton X-100, 1 mM DTT, 10% glycerol, protease inhibitor cocktail (Merck)]. For phosphatase treatment, the proteins were extracted with extraction buffer [100 mM HEPES-KOH, pH 7.5, 10% glycerol, 0.5% PVP, protease inhibitor cocktail (Merck)] and incubated with λ-phosphatase (New England Biolabs) at 30 °C for 30 min in the presence/absence of phosphatase inhibitor cocktail (50 mM NaF and 20 mM NaVO3). The resultant homogenized cell lysates were centrifuged at 15,300 g for 10 min at 4 °C, separated using the Phos-Tag SDS-PAGE system (FUJIFILM), transferred to a PVDF membrane (Bio-Rad), and blotted following the manufacturer’s instructions67. Western blotting was performed with anti-GFP antibody (Invitrogen, A-11122) as primary antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (Santa Cruz, sc-2004) as secondary antibody. Tubulin (Agrisera, AS10 680) was used as the internal control. Immunodetection was performed using the Pierce ECL detection reagent (ThermoFisher Scientific).
Dehydration assay and water loss measurement
To determine water loss rate, aerial parts of 3-week-old plants were detached, dried on Whatman filter paper (GE Healthcare) at 22 °C under normal light conditions at the indicated time points, and then weighed. The experiment was repeated independently three times. For the dehydration leaf assay, fully expanded true leaves were detached and placed on Whatman filter paper at room temperature (humidity 50–55%) for up to 2.5 h. For short-time dehydration stress treatment, 12-d.a.g. seedlings grown in 1/2MS plates with 1% (w/v) sucrose were placed on Parafilm for 1.5 h and total RNA was then extracted for RT-qPCR analysis. Three independent experiments were performed, and similar results were obtained.
Stomatal aperture measurement
The abaxial side of Arabidopsis leaves from 3-to-4-week-old plants of each genotype was peeled away manually and floated on stomatal opening buffer (5 mM mesbistrispropane, pH 6.5, 50 mM KCl, 0.1 mM CaCl2) under normal light conditions at least for 2 h to induce stomatal opening68. Samples were then treated with 2 µM ABA or 1 µM [Ara3]CLE5p or 0.1 mM H2O2 as indicated. After stomatal apertures were recorded with a light microscope (BX53; Olympus), their width and length were measured using ImageJ software, which allowed calculation of the aperture ratio (width/length). At least three independent experiments (n = 15/time point) were performed per genotype, and similar results were obtained.
ABA quantification
ABA was extracted and semi-purified as previously described69,70 in biological triplicate for each sample. ABA was quantified with an ultra-high performance-liquid chromatography (UHPLC)-electrospray interface-quadrupole-orbitrap mass spectrometer (UHPLC/Q-Exactive; ThermoFisher Scientific)70,71 with an ODS column (AQUITY UPLC BEH C18 1.7 µm, 2.1 × 100 mm; Waters).
Thermal shift assay (TSA)
The LRR repeat coding sequences of BAM1 and GHR1 were cloned into pET42b expression vector, designed to produce an N-terminal GST fusion. Primer information is in Supplementary Data 3. Purification steps followed the manufacturer’s protocol (Cytiba). The purity of the resulting recombinant protein was assessed by CBB staining and Western blotting. The TSA was performed using the CFX Connect real time detection system (Bio-Rad). Protein unfolding was monitored by measuring the fluorescence of SYPRO Orange dye (Bio-Rad). Samples contained 30–50 µM GST-fused protein and the CLE5p concentration was varied from 0–0.5 µM in a total volume of 20 µl. We used 96-well PCR plates for the assay. The buffer solution contained 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, pH 7.4. Each reaction was run in duplicate or triplicate and repeated on at least two different plates. Data were analysed using CFX Maestro Software, and melting temperature (Tm) values were calculated according to the manufacturer’s protocol (Bio-Rad); ΔTm was determined from the first derivative of the melt curve with and without CLE5p.
ROS measurements
Histochemical detection of H2O2 in situ was performed by DAB staining as described by Daudi et al.72. Specifically, four-week-old Arabidopsis rosette leaves were treated with 100 µM CLE5p or 1 µM ABA for 2 h and immersed in freshly prepared 100 µg/ml 3,3’-diaminobenzine (DAB, Sigma-Aldrich) solution in the dark for 8 h. Stained leaves were fixed and bleached with a solution of ethanol/acetic acid/glycerol (3:1:1), rinsed with distilled water, and then photographed on a white background. ROS in stomatal guard cells were detected using H2DCF-DA (Sigma-Aldrich) as described elsewhere73. Briefly, leaf epidermis peels were prepared from four-week-old Arabidopsis plants and incubated for 3 h in 30 mM KCl, 10 mM MES-KOH, pH 6.15, after which 20 µM H2DCF-DA was added and the solution incubated for a further 30 min. Excess dye was removed by washing three times with distilled water. The pre-treated epidermal strips were incubated with or without 1 µM [Ara3]CLE5p for 30 min. Fluorescence was captured by a confocal microscope (Zeiss LSM 800 Confocal Laser Scanning Microscope) operated with Zen software (version 13), using an excitation wavelength of 488 nm. Fluorescence intensity of guard cells was quantified using ImageJ.
Isothermal titration calorimetry (ITC) assay
Experiments were performed using a MicroCal ITC200 calorimeter (Malvern Instruments, UK) at 37 °C. A syringe was used to inject 2 μl of 1 mM CLE5p sequentially into a 200 μl titration cell containing protein samples. To ensure rapid mixing, injections were performed with a time interval of 180 s between successive injections while maintaining a reference output of 5 µcal/s, and the solution in the sample cell was continuously stirred at 800 rpm. Recombinant GST-BAM1 and GST-GHR1 proteins prepared as described in the TSA were subjected to gel-filtration in titration buffer (20 mM Tris-HCl pH 7.5, 25 mM NaCl, and 5% glycerol). The binding stoichiometry (N), Gibbs free energy (ΔG), enthalpy (ΔH), entropy (-TΔS), and dissociation constant (Κd) changes were calculated from the titration of 0.1 mM GST-BAM1 and 0.1 mM GST-GHR1 with 1 mM CLE5p in titration buffer. GST protein alone was used as a negative control. Data were analysed using the One Set of Sites Model Fitting with the Origin software package (MicroCal ITC program) provided by the manufacturer.
Docking simulations
A structural model of the leucine-rich repeat (LRR) domain of GHR1 and BAM1 was built using the SWISS-MODEL homology-modelling server74. To identify the possible binding sites of the peptide to each receptor, blind docking (i.e., no explicit descriptions of binding site residues) was performed with HPEPDOCK75. Given a slightly modified CLE5p peptide sequence (standard proline instead of hydroxyproline at residues 4 and 7) and the BAM1 and GHR1 LRR domain models obtained above, rigid-body docking of 1000 peptide conformations, generated from the MODPEP program, over the receptor surface was performed. Supplementary Fig. 8a–c illustrates that the binding region between the modified CLE5p and BAM1 or GHR1 LRR domains was close to the homologous TDR/TDIF peptide complex interface76. In the case of GHR1, more variability was observed, but a few poses still indicated a conserved interacting region.
Given the blind docking results and the likelihood that CLE peptide receptors share a conserved ligand recognition mechanism77, to obtain more detailed structural models of CLE5p bound to BAM1 and GHR, we performed template-based modelling starting from the structure of TDR bound to the TDIF peptide (TDIFp) (PDB ID:5GIJ)76. First, BAM1 and GHR1 were aligned with the TDR–TDIFp complex, and the CLE5p sequence was threaded onto TDIFp. Placing the amino acid sequence of CLE5p at the TDIFp coordinate maintained the overall structure of the peptide and its orientation to the homologous receptor binding site. Such an approach was described by Alam et al. (2016)78 to identify peptides that bind to a histone deacetylase (HDAC8). These initial coarse-grained CLE5p-LRR domain models were energy-minimized and refined using the Rosetta FlexPepDock refinement protocol79, designed for cases where coarse-grained models of the complex are available. During this process, the peptide backbone and its rigid-body orientation for the receptor protein were optimized along with the orientation of the peptide and receptor side chains. Backbone minimization of the receptor was also carried out during the refinement of the peptide–receptor interface. The procedure started with low-resolution refinement (centroid mode) and generated 500 unique models. Subsequently, the top model underwent further high-resolution refinement, producing an additional 500 models. All refinements were performed using the Rosetta Energy Function in the Rosetta modelling suite (v3.13)80.
To construct the BAM1–GHR1 complex model, protein–protein docking of BAM1 and GHR1 was performed using HDOCK in blind dock mode (i.e., no explicit descriptions of binding site residues of receptors)81, and the complex was optimized with FlexPepDock. Protein–protein docking of the BAM1–GHR1 complex showed that the peptide-binding region of BAM1 forms a protein–protein interface with GHR1. Therefore, to obtain a model of the BAM1–GHR1 complex bound with CLE5p, BAM1 (in complex with GHR1) was first aligned to TDR bound to TDIFp (PDB ID:5GIJ). The CLE5p sequence was threaded onto the TDIFp coordinates, followed by energy minimization of the side chain positions of the interface residues of the BAM1–GHR1 complex and CLE5p. Optimization was performed using the FlexPepDock protocol. For each FlexPepDock refinement protocol, the Rosetta Energy Unit (REU) interface score was used to rank the models. This score is calculated by subtracting the monomeric energy from the complex energy. Lower interface scores indicate a more favorable and stable peptide receptor complex.
Statistical analyses and reproducibility
Biological replicates (N), sample size per biological replicate (n) and P values can be found in figures, figure legends, Supplementary Data 4 and 5 or Source Data. Box-and-whisker plots overlaid with a dot plot (Figs. 1b, d, g, h, 2c, d, 4b, d and e, and Supplementary Fig. 1b, 2c–f, 3b, c, 4a–d, 5b, c, e, 7c and 10a–e), volcano plot (Fig. 1e), dot plot (Fig. 1f), bar chart overlaid with a dot plot (Fig. 2a) and violin plots overlaid with a box and a dot plot (Supplementary Fig. 1d and 1e) were generated using R (v4.4.1) and R studio (v2024.4.2.764); line graphs (Figs. 1i, 3e, f, and Supplementary Fig. 3d) were generated using Microsoft Excel 2016. All statistical graphs were edited using Adobe Illustrator CC 2018. The centre line in a box-and-whisker plot overlaid with a dot plot indicates the sample median; the bottom and top edge of the box represents the first quartile (Q1, 25th percentile) and third quartile (Q3, 75th percentile) of the dataset, respectively; the lower and upper whisker represents the minimum and maximum values (excluding outliers) of the dataset, respectively; each dot represents the value per sample or the mean value per biological replicate. In a violin plot overlaid with a box and dot plot, an error bar represents the lower and upper whisker showing the minimum and maximum values (excluding outliers) of the dataset, each dot represents the value per sample. Error bars in Figs. 1i, 2a, 3e and 3f, and Supplementary Fig. 3d are standard division (s.d.). Statistical significance was calculated with a two-tailed Student’s t-test (P values), or one-way ANOVA analysis (Fig. 1d) followed by a Tukey Honest Significant Differences (HSD) test (adjust P values). For two-way ANOVA analysis (Fig. 4b, and Supplementary Fig. 5c) followed by a HSD test, Pr ( > F) that represents the P-value associated with the F-statistic for each factor and their interaction was calculated using R (v4.4.1). All samples were randomly assigned to experimental groups, and all experiments were blinded during data acquisition and analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequence data from this article can be found in the GenBank/EMBL data libraries (https://www.ncbi.nlm.nih.gov/genbank/) under the following accession numbers: NCED3, AT3G14440; RD29B, AT5G52300; RAB18, AT5G66400; ABI1, AT4G26080; CLE5, AT2G31083; BAM1, AT3G23920; BAM3, AT4G17090; GHR1, AT4G20940; SRK2D/SnRK2.2, AT3G50500; SRK2E/SnRK2.6/OST1, AT4G33950; SRK2G/SnRK2.1, AT5G08590. Data are available in the main text or the supplementary materials. The RNA-seq data generated in this study have been deposited in the DDBJ under accession no. DRA011039 (https://ddbj.nig.ac.jp/search/entry/bioproject/PRJDB10736). The LC-MS/MS data generated in this study have been deposited in jPOST repository database (project ID, JPST002362, accession ID, PXD046831) (https://repository.jpostdb.org/entry/JPST002362). Source data are provided with this paper. All lines used in this study will be provided upon signature of an appropriate material transfer agreement. Source data are provided with this paper.
References
Huber, A. E., Melcher, P. J., Piñeros, M. A., Setter, T. L. & Bauerle, T. L. Signal coordination before, during and after stomatal closure in response to drought stress. N. Phytol. 224, 675–688 (2019).
Tombesi, S. et al. Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 5, 1–12 (2015).
Trejo, C. L. & Davies, W. J. Drought-induced closure of Phaseolus vulgaris L. stomata precedes leaf water deficit and any increase in xylem ABA concentration. J. Exp. Bot. 42, 1507–1516 (1991).
Tanaka, Y. et al. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 138, 2337–2343 (2005).
Basu, S., Ramegowda, V., Kumar, A. & Pereira, A. Plant adaptation to drought stress. F1000Research https://doi.org/10.12688/f1000research.7678.1. (2016).
Brodribb, T. J. & McAdam, S. A. Passive origins of stomatal control in vascular plants. Science 331, 582–585 (2011).
Sah, S. K., Reddy, K. R. & Li, J. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7, 571 (2016).
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R. & Abrams, S. R. Abscisic acid: emergence of 15 a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 (2010).
Fujii, H. et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664 (2009).
Umezawa, T. et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106, 17588–17593 (2009).
Lind, C. et al. Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr. Biol. 25, 928–935 (2015).
Ng, L. M. et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. USA 108, 21259–21264 (2011).
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M. & Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 (2000).
Pallakies, H. & Simon, R. The CLE40 and CRN/CLV2 signaling pathways antagonistically control root meristem growth in Arabidopsis. Mol. Plant 7, 1619–1636 (2014).
Fukuda, H. & Ohashi-Ito, K. Vascular tissue development in plants. Curr. Top. Dev. Biol. 131, 141–160 (2019).
Narasimhan, M. & Simon, R. Spatial range, temporal span, and promiscuity of CLE-RLK signaling. Front. Plant Sci. 26, 906087 (2022).
Xi, L., Wu, X. N., Gilbert, M. & Schulze, W. X. Classification and interactions of LRR receptors and co-receptors within the Arabidopsis plasma membrane - an overview. Front. Plant Sci. 16, 472 (2019).
Takahashi, F. et al. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238 (2018).
Christmann, A., Weiler, E. W., Steudle, E. & Grill, E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 52, 167–174 (2007).
Yoshida, T. et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49 (2015).
Xiong, L. & Zhu, J. K. Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36 (2003).
Shinohara, H. & Matsubayashi, Y. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54, 369–374 (2013).
Ceciliato, P. H. O. et al. Intact leaf gas exchange provides a robust method for measuring the kinetics of stomatal conductance responses to abscisic acid and other small molecules in Arabidopsis and grasses. Plant Methods 15, 38 (2019).
Kwak, J. M. et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633 (2003).
Hirakawa, Y. et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105, 15208–15213 (2008).
Sierla, M. et al. The receptor-like pseudokinase GHR1 is required for stomatal closure. Plant Cell 30, 2813–2837 (2018).
Hua, D. et al. A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24, 2546–2561 (2012).
Hohmann, U., Lau, K. & Hothorn, M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137 (2017).
Takahashi, Y. et al. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 12 (2020).
Lin, Z. et al. A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11, 613 (2020).
Yoshida, R. et al. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318 (2006).
Xie, T. et al. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J. Biol. Chem. 287, 794–802 (2012).
Geiger, D. et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106, 21425–21430 (2009).
Belin, C. et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 141, 1316–1327 (2006).
Vlad, F. et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184 (2009).
Soon, F. F. et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88 (2012).
Park, S. Y. et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 (2009).
Vishwakarma, K. et al. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161 (2017).
Soma, F. et al. Constitutively active B2 Raf-like kinases are required for drought-responsive gene expression upstream of ABA-activated SnRK2 kinases. Proc. Natl. Acad. Sci. USA 120, e2221863120 (2023).
Sun, Z. et al. RAF22, ABI1 and OST1 form a dynamic interactive network that optimizes plant growth and responses to drought stress in Arabidopsis. Mol. Plant 15, 1192–1210 (2022).
Kollist, H. et al. Rapid responses to abiotic stress: priming the landscape for the signal transduction network. Trends Plant Sci. 24, 25–37 (2019).
Lin, Z. et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 12, 2456 (2021).
Fàbregas, N., Yoshida, T. & Fernie, A. R. Role of Raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Commun. 11, 6184 (2020).
Hsu, P. K., Dubeaux, G., Takahashi, Y. & Schroeder, J. I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 105, 307–321 (2021).
Imes, D. et al. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 74, 372–382 (2013).
Chen, X. et al. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53–78 (2021).
Zhang, A. et al. S-type Anion Channels SLAC1 and SLAH3 Function as Essential Negative Regulators of Inward K+ Channels and Stomatal Opening in Arabidopsis. Plant Cell 28, 949–955 (2016).
Shinozawa, A. et al. SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2, 30 (2019).
Whitewoods, C. D. Evolution of CLE peptide signalling. Semin. Cell Dev. Biol. 109, 12–19 (2021).
Olsson, V. et al. Look closely, the beautiful may be small: precursor-derived peptides in plants. Annu. Rev. Plant Biol. 70, 153–186 (2019).
Torres, M. A., Dangl, J. L. & Jones, J. D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522 (2002).
Rodriguez-Villalon, A. et al. Molecular genetic framework for protophloem formation. Proc. Natl. Acad. Sci. USA 111, 11551–11556 (2014).
Guo, Y., Han, L., Hymes, M., Denver, R. & Clark, S. E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63, 889–900 (2010).
Wu, Y. et al. Genome-Wide Expression Pattern Analyses of the Arabidopsis Leucine-Rich Repeat Receptor-Like Kinases. Mol. Plant 9, 289–300 (2016).
Fauser, F., Schiml, S. & Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 (2014).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Soma, F. et al. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3, 16204 (2017).
Logemann, E., Birkenbihl, R. P., Ülker, B. & Somssich, I. E. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2, 16 (2006).
Soma, F., Takahashi, F., Suzuki, T., Shinozaki, K. & Yamaguchi-Shinozaki, K. Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 11, 1373 (2020).
Brandt, B. et al. Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. Elife 4, e03599 (2015).
Kano, K., Noda, S., Sato, S., Kuwata, K. & Mishiro-Sato, K. An efficient in-gel digestion method on small amounts of protein sample from large intact gel pieces. Sep. Sci. Plus, 6, 2200121–2200121 (2023).
Sakamoto, T., Sotta, N., Suzuki, T., Fujiwara, T. & Matsunaga, S. The 26S Proteasome is required for the maintenance of root apical meristem by modulating auxin and cytokinin responses under high-boron stress. Front Plant Sci. 10, 590 (2019).
Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Occhialini, A. Visualization of RMRs (Receptor Membrane RING-H2) dimerization in Nicotiana benthamiana leaves using a bimolecular fluorescence complementation (BiFC) assay. Methods Mol. Biol. 1789, 177–194 (2018).
Willemsen, V., Wolkenfelt, H., de Vrieze, G., Weisbeek, P. & Scheres, B. The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development 125, 521–531 (1998).
Ohyama, K., Ogawa, M. & Matsubayashi, Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160 (2008).
Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K. & Koike, T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell Proteom. 5, 749–757 (2006).
Kinoshita, T. et al. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660 (2001).
Kojima, M. et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214 (2009).
Kojima, M. & Sakakibara, H. Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 918, 151–164 (2012).
Shinozaki, Y. et al. Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83, 237–251 (2015).
Daudi, A. & O’Brien, J. A. Detection of hydrogen peroxide by DAB staining in arabidopsis leaves. Bio Protoc. 2, e263 (2012).
Murata, Y., Pei, Z. M., Mori, I. C. & Schroeder, J. Abscisic acid activation of plasma membrane Ca(2+) channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13, 2513–2523 (2001).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Zhou, P., Jin, B., Li, H. & Huang, S. Y. HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res 46, W443–W450 (2018).
Morita, J. et al. Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide. Nat. Commun. 7, 12383–12383 (2016).
Zhang, H., Lin, X., Han, Z., Qu, L. J. & Chai, J. Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res 26, 543–555 (2016).
Alam, N. et al. Structure-Based Identification of HDAC8 Non-histone Substrates. Structure 24, 458–468 (2016).
Raveh, B., London, N. & Schueler-Furman, O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins: Struct., Funct., Bioinforma. 78, 2029–2040 (2010).
Alford, R. F. et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J. Chem. Theory Comput. 13, 3031–3048 (2017).
Yan, Y., Tao, H., He, J. & Huang, S. Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Acknowledgements
We deeply thank B. Scheres and I. Blilou for critical reading of the manuscript. We also thank T. Kinoshita for suggestions for stomatal pore size measurement; R. Iwasaki and T. Anan for technical assistance with plant care and genotyping; and T. Shinagawa, A. Furuta, and T. Niwa for assistance with RNA-seq analysis. This work was funded by Grants-in-Aid from the MEXT KAKENHI (20H04882 to A.S., 15H05958 and 21H02500 to H.F.), Grants-in-Aid from the JSPS KAKENHI (19K05943, 22K05556, and 25K01977 to A.S., 16H06377 to H.F., 24H02266 to T.H. and 24H02260 to F.T.), This work was supported in part by FOCUS (Establishing Supercomputing Center of Excellence) and MEXT as “Program for Promoting Researches on the Supercomputer Fugaku” (Development and application of large-scale simulation-based inferences for biomolecules JPMXP1020230119) to F.T., and a Grant for Basic Science Research Projects from the Sumitomo Foundation, Toyoaki Scholarship, Fujimori Foundation, Shorai Foundation for Science and Technology, Nagase Science and Technology Foundations, and DAIKO FOUNDATION to A.S.
Author information
Authors and Affiliations
Contributions
A.S. wrote the original draft paper with substantial input from Y.D. and additional input from all other authors. A.S. and Y.D. took the lead in revising and editing the manuscript, with contributions from T.H., K.U.T., and H.F. A.S. designed and led the study together with Y.D., H.K., and V.S. M.K. helped to perform LI-6800 measurements. A.S., T.H., and V.S. performed ITC experiment. A.S., V.S., M.K., and F.T. performed docking simulation. Y.M. performed structural analysis of CLE5 in planta and synthesized the arabinosylated peptide. E.M.S. and K.K. performed structural analysis of MBP-SRK2E. A.S. and H.F. obtained funding and administrated for the study. Y.D. and T.S. performed RNA-seq analysis. M.K., Y.T., and H.S. analysed ABA content. F.S. and K.Y. prepared ABA-related experimental materials.
Corresponding authors
Ethics declarations
Competing interests
A.S. and Y.M. have filed a provisional patent application for the function of peptide module as a stomatal regulator reported in this study through Nagoya University, and declare no other competing interests. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shimotohno, A., Matsubayashi, Y., Du, Y. et al. Local peptide signalling induces stomatal closure under drought stress. Nat Commun 17, 177 (2026). https://doi.org/10.1038/s41467-025-66392-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66392-6