Abstract
Chimeric Antigen Receptor (CAR) T cells have induced remarkable clinical responses in patients with hematological cancers. However, CAR T-cell therapies against solid tumors have not elicited similar outcomes since immunosuppressive barriers in the tumor microenvironment attenuate anti-tumor activity. Here, we describe a multifaceted approach to engineer allogeneic CAR T-cells resistant to both biochemical (hypoxia-adenosinergic) and immunological (PD-L1 and TGF-β) inhibitory signaling using an adenine base editor and a CRISPR-Cas12b nuclease. The resulting EGFR-targeting CAR T-cell product comprised a combination of six gene edits designed to evade allorejection (B2M, CIITA), prevent graft-versus-host disease (CD3E) and overcome biochemical (ADORA2A) and immunological (PDCD1, TGFBR2) barriers in solid tumor microenvironment of subcutaneously grown EGFR+ human lung tumor xenografts. This combinatorial genetic disruption enhances CAR T cell effector function and anti-tumor efficacy leading to improved tumor elimination and survival in xenograft and humanized mouse solid tumor models. Our strategy confers CAR T cells resistance to multiple clinically relevant inhibitory signaling pathways that are amplified in hypoxic tumor areas and may improve the therapeutic potential of CAR T-cells against solid tumors.
Similar content being viewed by others
Introduction
Chimeric antigen receptor T (CAR-T) cell therapies have elicited remarkable clinical responses in treating hematological malignancies1,2. However, their effectiveness in treating solid tumors remains limited due to the hostile and hypoxic, immune-suppressive tumor microenvironment (TME)3,4. Tumor hypoxia, resulting from rapid tumor growth and irregular angiogenesis5,6,7,8, stabilizes hypoxia-inducible factor 1-alpha (HIF-1α) thereby promoting pro-tumoral and immune-suppressive transcriptional profiles and HIF-driven surface expression of adenosine (Ado) generating enzymes (e.g., CD38, CD39, CD73) leading to an adenosine-rich TME9,10,11,12. Extracellular adenosine activates Gs-protein coupled adenosine A2A receptors (A2AR) expressed on T cells that trigger the formation of intracellular cAMP and subsequent downstream PKA-mediated signaling events that culminate in potent suppression of T cell effector function13,14,15,16,17,18. In addition to this ‘hypoxia-adenosinergic’ biochemical suppression, tumor hypoxia augments immunological barriers and immunosuppressive pathways that may simultaneously converge to inhibit CAR T-cell functionality. For instance, hypoxia/HIF-1α increases the expression of PD-L1 and TGF-β19,20 that signal through PD-1 and TGF-βRI/RII, respectively, to attenuate T cell activity21,22. Prior attempts to improve CAR T-cell efficacy that were limited to single gene edits involved in these pathways have demonstrated promising preclinical efficacy23,24,25,26,27,28,29,30, but such unidimensional approaches may ultimately lead to immune evasion via the induction of alternative suppressive pathways in the heterogenous TME of solid tumors.
However, the development of a CAR T-cell product resistant to several distinct, major inhibitory pathways may offer multiple advantages in responding to intra-/inter-patient tumor heterogeneity and overcoming therapy resistance. Here, we leveraged base editing to address current challenges in developing multiplex gene-edited CAR T-cell products. Base editors (BE) have enabled precise base pair changes to disrupt gene expression without inducing double-strand DNA breaks or karyotypic abnormalities31,32,33 characteristic of current CRISPR nuclease systems when used in multiplex settings34,35. While BEs have been utilized to generate clinical-stage CD7-directed CAR T-cells (NCT05885464)36, the number of accessible BE target sites containing neighboring NGG protospacer adjacent motifs (PAMs) may be restricted for any given gene. Thus, to broaden the range of targeting loci and develop a higher-order gene-edited CAR T-cell product, it may be advantageous to combine base editing with a CRISPR nuclease. To this end, we complexed our adenine base editor (ABE) with a Cas12b nuclease37, allowing for the elimination of biochemical, immunological, and allogeneic barriers resulting in a novel 6-plex combination of a gene-edited solid tumor CAR T-cell product (termed Stealth-TKO CAR T-cells). This combinatorial engineering strategy represents an important technical advance that—to our knowledge—has achieved the highest number of simultaneous genetic edits in CAR T-cells without hindering efficacy.
Our preclinical data investigating the efficacy of these Stealth-TKO CAR T-cells may offer clinical promise as they are (i) conceptually ‘off-the-shelf’ by overcoming challenges associated with autologous CAR T-cell manufacturing and patient accessibility, (ii) safe by avoiding graft-versus-host disease (GvHD), and (iii) effective against solid tumors due to the genetic ablation of multiple, critical T cell inhibitory pathways that are prominent in solid tumors. Stealth-TKO CAR T-cells are shown to be resistant to adenosine (A2AR-KO), PD-L1 (PD-1-KO) and TGF-β (TGFBR2-KO), addressing the convergence of biochemical and immunological negative regulators within the TME. Furthermore, to generate allogeneic CAR T-cells that may avoid GvHD and host immunologic rejection, we introduced three additional ‘Stealth’ base edits by removing the T cell receptor (CD3E-KO), and HLA class-I (B2M-KO) and HLA class-II (CIITA-KO), respectively. This approach has been confirmed by Li et al. demonstrating enhanced resistance to allorejection by genetic disruption of the T cell receptor and HLA class-I/II in CAR T-cells38. We show that Stealth-TKO CAR T-cells exhibited enhanced production of major anti-tumor cytokines and cytotoxic capacities in in vitro suppression assays mimicking the hostile TME. Moreover, Stealth-TKO CAR-T demonstrated improved anti-tumor efficacy against solid tumors in stringent humanized xenograft murine systems, underscoring the power of multiplex gene editing to putatively overcome TME-associated immunosuppressive pathways.
Results
Base editor targeting and editing of ADORA2A to generate adenosine-resistant CAR T-cells
The accumulation of extracellular adenosine (Ado) in the solid tumor microenvironment is a powerful biochemical barrier inhibiting anti-tumor T cell responses39,40,41. Therefore, we engineered Ado-resistant CAR T-cells by using base editing to ablate functional expression of the adenosine A2A receptor (A2AR) encoded by ADORA2A. Single guide RNAs (sgRNAs) were designed to home adenine (ABE) and cytosine (CBE) base editors to ADORA2A and alter protein expression by mutating either the gene start codon, conserved intron-exon mRNA splice motifs, or by installing premature termination codons. These genomic changes were accomplished by base editor-mediated chemical modification of an adenine base to a guanine (ABE) or a cytosine base to a thymine (CBE) without the generation of double-stranded DNA breaks (Fig. 1a). To identify an sgRNA and base editor combination that mediates optimal ADORA2A editing, mRNAs encoding ABE or CBE were paired with a corresponding sgRNA designed to target loci spanning the entire ADORA2A gene and electroporated into activated primary human T cells (Fig. 1b, c; Supplementary Table 1). This sgRNA screen identified an ABE-sgRNA (TSBTx2043) complex targeting an intron-exon splice junction that achieved a mean on-target genomic editing efficiency of 88% (Fig. 1d). Subsequently, we electroporated TSBTx2043 into primary human T cells expressing a second-generation, epidermal growth factor receptor (EGFR)-specific, 4-1BB co-stimulated CAR construct (EGFR CAR) (Fig. 1e). The introduction of the ADORA2A base edit (A2AR-KO) had no impact on overall CAR expression (Fig. 1f) demonstrating base editor compatibility with the CAR T-cell manufacturing process. The CAR T-cell target antigen was selected since EGFR is widely expressed across multiple cancer types that form solid tumors, including lung, breast, pancreatic, prostate, and renal cancers where immune checkpoint blockade, inhibitors of the adenosine pathway are currently being tested in clinical studies. In addition, EGFR has been established clinically as a generally safe and effective target.
a Schematic of adenine (ABE) and cytosine (CBE) base editor/DNA complexes, showing precise A-to-G (left) and C-to-T (right) base pair conversion. Mutated Cas9 protein (gray), single guide RNA (sgRNA, magenta), deaminase (left, pink and right, purple), uracil DNA glycosylase (UGI, green). b Diagram of the human ADORA2A gene with sgRNA targeting loci (arrows) across both coding exons, with targeted nucleotide for base editing at the intron-exon junction (A, red) downstream of the protospacer adjacent motif (PAM, blue). c Target genomic editing efficiency determined by next-generation sequencing (NGS) of each sgRNA-base editor pairing (ABE, light gray and CBE white) in activated primary human T cells. d Cumulative frequency of ADORA2A on-target genomic editing efficiency of top candidate sgRNA (TSBTx2043) complexed with ABE (n = 8 independent biological donors). e Illustration of a second-generation, anti-EGFR Chimeric Antigen Receptor T cell (CAR-T) construct expressed under an MND promoter (purple). Single-chain variable fragment targeting EGFR (blue), CD8α hinge and transmembrane domain (gray), 4-1BB (yellow) and CD3ζ (green) co-stimulation. f Representative flow cytometry plots depicting CAR expression of untransduced (UTD) T cells, unedited (CAR) and A2AR-KO CAR T-cells detected with anti-CAR idiotype antibody. For all data, symbols and error bars reflect mean ± SD of individual biological replicates, except (c) where symbols represent technical replicates.
Adenosine-resistant CAR T-cells demonstrate improved effector function in vitro
Next, we determined whether ADORA2A disruption prevents A2AR signaling in EGFR-specific CAR T-cells containing the ADORA2A base edit. A2AR-KO and unedited CAR T-cells were treated with the adenosine analogue 2-chloroadenosine (cADO) and evaluated for the level of phosphorylated cAMP Response Element-Binding Protein (pCREB), a critical downstream mediator of the adenosine-A2AR signaling pathway42. Notably, treatment with cADO increased the level of pCREB in unedited CAR T-cells, whereas the level of pCREB in A2AR-KO CAR T-cells did not deviate from the untreated control (Fig. 2a, b) indicating A2AR-KO attenuates proximal Ado-mediated signaling. We then investigated whether A2AR-KO alleviates Ado-induced immunosuppression by culturing CAR T-cells with the EGFR+ lung cancer cell line, H226. After in vitro tumor stimulation in the presence of cADO, A2AR-KO CAR T-cells maintained high levels of pro-inflammatory cytokine secretion including IFN-γ, IL-2, TNF-α, and GM-CSF. In contrast, cADO potently suppressed cytokine production of unedited CAR T-cells relative to untreated CAR T-cells in multiple healthy human donor T cells (Fig. 2c–g). The observations of increased cytokine production of A2AR-KO CAR T-cells relative to unedited CAR T-cells were confirmed using a separate EGFR-expressing cell line, A549 (Supplementary Fig. 1). Additional mechanistic assays demonstrated that A2AR-KO CAR T-cells also exhibited enhanced cytolytic capacities. Only the A2AR-KO CAR-T were able to eradicate H226 tumor spheroids in the presence of cADO, whereas unedited CAR T-cells and untransduced (UTD) control T cells failed to exert tumor control (Fig. 2h and Supplementary Fig. 2). Together, these data demonstrate that base editor-mediated disruption of ADORA2A in CAR T-cells confers a functional resistance to Ado-mediated immunosuppression in vitro.
a Flow cytometry histogram plots (a) and MFI quantification (b) of phosphorylated CREB (pCREB) in untransduced (UTD) T cells, unedited (CAR) and A2AR-edited CAR T-cells (A2AR-KO) treated with 2-chloroadenosine (cADO, blue) compared to an untreated control (DMSO), (Kruskal–Wallis, n = 4 independent biological donors, mean ± SD. P*** = 0.0005, n.s. = 0.212). c–f IFN-γ (c), IL-2 (d), TNF-α (e) and GM-CSF (f) production by UTD T cells (white), unedited (gray) or A2AR-KO (red) CAR T-cells measured by ELISA 48 h post-stimulation with H226 tumor cells in the presence (+) or absence (−) of cADO (Kruskal–Wallis test, n = 4 independent biological donors, mean ± SD. P*** = 0.00013, n.s. = 0.21, P** = 0.0014, n.s. = 0.383, P* = 0.014, n.s. = 0.083 P** = 0.0013, n.s. = 0.28). g Magnitude of GM-CSF, IFN-γ, IL2, TNF-α secretion by unedited (gray) and A2AR-KO (red) CAR T-cells in the presence of cADO normalized to respective untreated CAR T-cells (Two-sided Mann–Whitney t-test, n = 4 independent biological donors, mean ± SD. P*** = 0.0007, P*** = 0.0004, P*** = 0.0003, P** = 0.0037). h Incucyte live imaging tumor spheroid cytotoxicity of UTD T cells (white), unedited (gray) and A2AR-KO (red) CAR T-cells in the presence of cADO. Cytotoxicity was measured as a decrease in cumulative tumor GFP+ area over time (n = 3 average of technical replicates, mean ± SD). For all data, symbols and error bars reflect mean ± SD of individual biological replicates, except (h) where symbols represent technical replicates. b–g Kruskal–Wallis test performed to calculate statistical significance, g Mann–Whitney t-test performed to calculate statistical significance, ****P < 0.0001, ***P < 0.001, **P < 0.01.
A2AR-KO CAR T-cells overcome an immunosuppressive TME to control tumor progression in vivo
Adenosine accumulation in the TME is driven primarily by hypoxia/HIF and hypoxia-responsive gene expression43,44,45. Therefore, to determine whether A2AR-KO CAR T-cells were resistant to adenosine-mediated suppression in solid tumors, we established a preclinical H226 xenograft tumor model that recapitulates the hypoxic and adenosine-rich TME. Immunofluorescent staining confirmed widespread hypoxia and expression of the critical adenosine-generating ectoenzyme CD73 in H226 tumors resected from immunocompromised NCG mice (Fig. 3a, b). We then identified varying levels of hypoxia within the xenograft tumors based on the fluorescence intensity of hypoxia-sensitive pimonidazole (Hypoxyprobe) staining (Fig. 3c) and performed whole transcriptomic analysis using the Nanostring GeoMx digital spatial transcriptomics platform. Gene expression profiles of these regions were compared to the MGI Hypoxia Pathway Gene Ontology database, and notably, we observed induction of hypoxia-regulated genes, including ALDOA, CA9, ENO1, LDHA, NDRG1, TGFB1 and VEGF proportional to the intensity of Hypoxyprobe staining (Fig. 3d). These data support the utility of this in vivo system to mimic biochemical immune-suppressive features of a hypoxic and adenosine-rich TME.
a Flow cytometry staining of H226 tumor cells in vitro for CD73 expression. Isotype control (gray), anti-CD73 antibody (pink). b Immunofluorescent micrographs of H226 tumors resected from NCG mice 46 days post-implantation. Nucleated cells (DAPI, blue), hypoxia (Hypoxyprobe, green) and CD73 (pink). Representative images from four individual tumors from 10 to 20 different cutting surfaces. c Quantification of hypoxia in various tumor regions within resected H226 tumors from NCG mice determined by mean fluorescence intensity (MFI) of Hypoxyprobe. Representative image of a resected tumor section; quantification was performed across 6 independent slides (3 tumors per slide from individual mice) with an average of 13.5 regions of interest (ROI) analyzed per slide. d Spatial transcriptomics gene expression analysis from hypoxic regions in (c) (white = low hypoxia, light green = medium hypoxia, dark green = high hypoxia). Boxplots show the median (line), interquartile range (box), and whiskers extending to values within 1.5× the IQR. e 2 × 106 UTD T cells (white, n = 5 individual mice) or unedited (gray, n = 5 individual mice) and A2AR-KO (red, n = 5 individual mice) CAR T-cells injected I.V. into H226 tumor-bearing NCG mice. Group average of tumor volumes measured via calipers over time (Two-sided Mann–Whitney t-test, n = group average of individual mice, mean ± SEM, P** = 0.0079, P** = 0.0072). f Cumulative tumor burden, calculated as area under the curve, from (e) (Two-sided Mann–Whitney t-test, n = average of individual mice as above, mean ± SD, n.s. = 0.0556, P** = 0.00379). g 2 × 106 UTD T cells (white, n = 5 individual mice) or unedited (gray, n = 5 individual mice) and A2AR-KO (red, n = 5 individual mice) CAR T-cells injected I.V. into A549 tumor-bearing NCG mice. Group average of tumor volumes measured via calipers over time (Graph represents group mean ± SD, P** = 0.0072). h Cumulative tumor burden, calculated as area under the curve, from (g). (Two-sided Mann–Whitney t-test, n = average of individual mice as above, mean ± SD, n.s. = 0.490, P** = 0.0037). For all data, symbols and error bars reflect individual biological replicates and group mean ± S.E.M. e–h Mann–Whitney t-test performed to calculate statistical significance, **P < 0.01, *P < 0.05.
Using this model, we investigated whether elimination of A2AR enhances efficacy of CAR T-cell therapy in vivo. H226 tumor-bearing mice were treated with three dose levels of A2AR-KO CAR T-cells, unedited CAR T-cells or UTD T cells. At medium and high dose levels, both A2AR-KO and unedited CAR T-cells controlled tumor outgrowth, but A2AR-KO CAR T-cell-treated mice exhibited lower peak tumor volume and accelerated kinetics of tumor regression (Supplementary Fig. 3). Notably, at the lowest evaluated dose only A2AR-KO CAR T-cells eradicated tumors (Fig. 3e, f) and reduced cumulative tumor burden (Supplementary Fig. 3), while unedited CAR T-cells failed to control tumor. Of note, a slight decrease in average tumor size from all groups occurred at day ~60 post tumor implant, which could indicate an initial sign of graft-versus-tumor (GVT) induced by the native T cell receptor (TCR). Therefore, to determine the role of A2AR elimination in the activity of the CAR-signaling alone, and to exclude contributions of the TCR, we used base editing to eliminate the T cell receptor (CD3E-KO), on A2AR KO, control, and UTD CAR T-cells. In Fig. 3g, h, demonstrates in a separate tumor model (A549) that the improved tumor regression induced by A2AR-KO CAR T cells is mediated by the effects of the elimination of adenosine signaling on CAR activity, and not the native TCR.
These findings confirm in multiple tumor models that A2AR-KO CAR T-cells overcome an immunosuppressive TME and exhibit CAR-mediated improved anti-tumor efficacy compared to unedited CAR T-cells. Additionally, these data confirm and extend previous observations of the therapeutic benefit of A2AR-gene deletion in other models of adoptive cell transfer, including other CAR T-cell constructs9,10,23,30.
CAR T-cells are susceptible to biochemical and immunological barriers of solid tumors
While solid tumor hypoxia has been shown to augment immune suppression by increasing CD39/CD73-mediated generation of extracellular adenosine, numerous additional immune-inhibitory and tumor protecting pathways are also promoted by hypoxic signaling. Indeed, solid tumor microenvironments are often comprised of multiple immunosuppressive pathways including biochemical, metabolic, and immunological barriers that prevent anti-tumor T cell function46,47. Therefore, elimination of biochemical barriers alone, such as adenosinergic signaling, may not be sufficient to induce complete and durable tumor remissions48 (NCT02740985). Using digital spatial transcriptomics, we evaluated hypoxic regions in H226 xenografts and identified additional mechanisms of immunosuppression. Of note, regions of increasing hypoxia were associated with elevated levels of transforming growth factor beta 1 (TGF-β1) mRNA (Fig. 3c, d), which was then confirmed by detecting in vitro secretion of TGF-β by H226 tumor cells (Supplementary Fig. 4). In addition, flow cytometric analysis of cell surface proteins involved in T cell inhibitory pathways revealed that H226 tumor cells highly express PD-L1 (Supplementary Fig. 4). Both PD-L1 and TGF-β protein expression were confirmed by immunofluorescent staining of resected H226 tumors from NCG mice (Fig. 4a). Thus, it is expected that CAR T cells are suppressed by both biochemical and immunological barriers in the solid TME. The hypoxia-driven promotion of such negative regulators that facilitate tumor evasion may explain the lack of therapeutic efficacy of CAR T-cells against solid tumors compared to hematological malignancies.
a Immunofluorescent staining of H226 tumors resected from NCG mice 46 days post-tumor implantation. Nucleated cells (DAPI, blue), PD-L1 (green), TGF-β (purple). b IL2 production by UTD T cells (white) and A2AR-KO CAR T-cells (red) measured by ELISA 48 h post-stimulation with recombinant human EGFR and PD-L1 protein conjugated beads incorporating increasing amounts of PD-L1 protein (n = 3 technical replicates, mean ± SD). c IFN-γ, IL-2, TNF-α and GM-CSF production by A2AR-KO CAR T-cells 48 h post-stimulation with H226 tumor cells in the presence of exogenous TGF-β normalized to DMSO treated control (n = 3 independent biological donors). d Flow cytometry plot indicating PD-1 surface expression on unedited (dotted line) and PD1-KO (solid line) CAR T-cells 24 h post-stimulation with phorbol myristate acetate. e IL2 production by UTD T cells (white), unedited (light gray) and PD1-KO (dark gray) CAR T-cells measured 48 h post-stimulation with recombinant human EGFR and PD-L1 conjugated beads (n = 3 average of technical replicates, mean ± SD). f Flow cytometry plot of phosphorylated SMAD2/3 (pSMAD) in unedited (black) or TGFβRII-KO (green) CAR T-cells treated with exogenous TGF-β and unedited CAR T-cells treated with DMSO (dashed line). g IFN-γ, IL-2, TNF-α and GM-CSF production by unedited (light gray) and TGFβRII-KO (green) CAR T-cells 48 h post-stimulation with H226 tumors in the presence of exogenous TGF-β. All functional assays were performed in duplicate (Two-sided Mann–Whitney t-test, n = 5 biological replicates from independent experiments, mean ± SD, P*** = 0.0079, n.s. = 0.31, P*** = 0.0079, n.s. = 0.31, P*** = 0.0079, n.s. = 0.69, P*** = 0.0079, n.s. = 0.69). b, e Symbols and error bars represent mean ± SD of technical replicates, c symbols represent independent biological replicates, and error bars represent mean ± group SD, g symbols represent five individual biological donors in technical duplicate. g Mann–Whitney t-test performed to calculate statistical significance, ***P < 0.001,**P < 0.01, *P < 0.05.
Therefore, we tested whether A2AR-KO CAR T-cells were still susceptible to PD-L1- or TGF-β-mediated inhibition. Upon in vitro stimulation in the presence of PD-L1, A2AR-KO CAR T-cells secreted lower amounts of IL-2 inversely proportional to the amount of PD-L1 inhibition (Fig. 4b). Similarly, the addition of exogenous TGF-β suppressed A2AR-KO CAR T-cell secretion of multiple pro-inflammatory cytokines including IFN-γ, IL-2, TNF-α and GM-CSF (Fig. 4c, Supplementary Fig. 5). These data indicate that A2AR-KO CAR T-cells are still potently suppressed by immunological negative regulators, and further suggest that PD-L1 and TGF-β, for which pharmacologic inhibitors are clinically approved, may synergize with adenosine in solid tumors to suppress anti-tumor CAR T-cell responses in vivo.
Multiplex gene-editing confers resistance to biochemical and immunological negative regulators
Since solid tumor environments are heterogeneous, CAR T cells must be poised to respond in any environmental condition. Single-gene editing approaches may ultimately prove ineffective in solid tumors due to immune evasion via the induction of alternative suppressive pathways. Indeed, A2AR-KO CAR T-cells retained sensitivity to clinically relevant immunological negative regulators such as PD-L1 and TGF-β. Therefore, we sought to simultaneously confer CAR T-cells resistance to both biochemical and immunological barriers that are augmented in hypoxic tumors.
To this end, we utilized our previously described BE sgRNA targeting PDCD1, which encodes PD-1, the receptor responsible for PD-L1-mediated inhibition36. PD1-KO CAR T-cells did not upregulate PD-1 cell surface expression (Fig. 4d) and resisted PD-L1-mediated suppression of IL-2 secretion after in vitro antigen stimulation in multiple healthy human donor T cells (Fig. 4e; Supplementary Fig. 6). Next, we evaluated sgRNAs spanning TGFBR2 (Supplementary Table 1), the gene encoding TGF-β receptor 2 (TGFβRII) and screened each sgRNA-ABE complex in primary human T cells. None of the evaluated sgRNAs drastically attenuated downstream TGF-β-mediated signaling indicated by the phosphorylation of SMAD2/3 (Supplementary Fig. 7). Therefore, we designed CRISPR-Cas12b nuclease sgRNAs, and with its unique ATTN protospacer adjacent motif (PAM) sequence, were able to target alternative loci within TGFBR2 compared to ABE (Supplementary Table 1). In doing so, we identified a Cas12b-sgRNA pairing that completely abrogated pSMAD2/3 relative to unedited control T cells (Fig. 4f), and after in vitro tumor stimulation, TGFβRII-KO CAR T-cells maintained secretion of effector cytokines despite the presence of exogenous TGF-β (Fig. 4g; Supplementary Fig. 8).
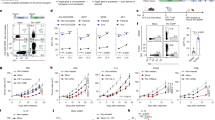
Next, we co-introduced these three genetic edits, termed triple knock-out (TKO), into EGFR-specific CAR T-cells to simultaneously overcome Adenosine, PD-L1 and TGF-β tumor-associated inhibitory pathways (Fig. 5a). TKO CAR T-cells were generated by combining ADORA2A-, PDCD1-, and TGFBR2-specific sgRNAs into a single electroporation reaction with mRNAs encoding ABE and Cas12b, which achieved genomic on-target editing efficiencies of 93% ± 2.5%, 95% ± 1.7%, and 92% ± 3.8%, respectively (Fig. 5b). To determine whether multiplex gene editing confers functional resistance to these biochemical and immunological inhibitory pathways we developed a triple-suppression assay, where TKO CAR T-cells were tumor stimulated in an in vitro system that contemporaneously mimics Ado, PD-L1 and TGF-β inhibition in the TME. TKO CAR T-cells resisted the suppressive effects of both biochemical (Ado) and immunological (PD-L1 and TGF-β) mechanisms as evidenced by their ability maintain elevated levels of the pro-inflammatory cytokines IL-2, IFN-γ, TNF-α, GM-CSF. In stark contrast, the anti-tumor activity of unedited CAR T-cells, and A2AR-KO CAR T-cells was attenuated by these inhibitory pathways (Fig. 5c). Using the same in vitro suppression system, only the TKO CAR T-cells fully eradicated H226 tumor spheroids, exhibiting superior anti-tumor activity compared to A2AR-KO CAR T-cells (Fig. 5d). These findings demonstrate that multiplex gene editing simultaneously confers TKO CAR T-cells functional resistance to multiple immunosuppressive pathways in vitro.
a Schematic of triple knock out (TKO) EGFR-specific CAR T-cells comprising three genomic edits targeting ADORA2A (A2AR-KO), PDCD1 (PD-1-KO) and TGFBR2 (TGFβRII-KO). b Frequency of ADORA2A, PDCD1, and TGFBR2 on-target genomic editing efficiencies in unedited and TKO CAR T-cells quantified by NGS (n = 4 independent biological donors, mean ± SD). c IFN-γ, IL-2, TNF-α, and GM-CSF production by A2AR-KO (red) and TKO (blue) CAR T-cells measured by ELISA 48 h post-stimulation with H226 tumors in the presence of triple suppression comprising exogenous cADO, PD-L1 and TGF-β proteins normalized to a DMSO treated control (Kruskal–Wallis t-test, n = 4 independent biological donors, mean ± SD. P* = 0.021, P* = 0.021, P** = 0.0028, P* = 0.02). d Incucyte-based live imaging tumor spheroid cytotoxicity of tumor alone (white) and tumor cultured with A2AR-KO (red) or TKO (blue) CAR T-cells in the presence of triple suppression. Cytotoxicity was measured as a decrease in total tumor GFP+ area over time (n = 3 average of technical replicates, mean ± SD). e 2 × 106 ZsGreen+ (gray, n = 5 individual mice), A2AR-KO (red, n = 7 individual mice) or TKO (blue, n = 7 individual mice) CAR T-cells, or untreated mice (tumor alone, white, n = 5 individual mice)were infused into NCG mice when tumor volume reached an average volume of 150 mm3. Longitudinal tumor volume (e), cumulative tumor burden (Kruskal–Wallis test average of individual mice, mean ± SEM, P** = 0.0033, P** = 0.0035, P** = 0.0071) (f) (calculated as area under the curve) (Kruskal–Wallis, symbols represent individual mice, mean ± SD P** = 0.0023, P** = 0.0025, P** = 0.0025), and survival curves (g) (log-rank test, lines represent group average, P* = 0.029), are shown. h Immunofluorescent detection of ZsGreen+ unedited (left), A2AR-KO (center) and TKO (right) CAR T-cells within H226 tumors resected 7 days post-CAR-T infusion. Nucleated cells (DAPI, blue), ZsGreen+ CAR T-cells (green). i Quantification of CAR-T infiltration from (h) (Kruskal–Wallis, n = 4 biological replicates, mean ± SD, P** = 0.0009, P* = 0.045). For (b, c), symbols represent biological donor replicates and error bars reflect mean ± SD. d Symbols represent technical replicates ± SD, e, f, i symbols represent individual mice and error bars represent S.E.M. c, e, f, i Kruskal–Wallis and g log-rank tests performed to calculate statistical significance, ***P < 0.001, **P < 0.01, *P < 0.05.
TKO CAR T-cells overcome immune-suppression to eliminate solid tumor xenografts
To determine if TKO CAR T-cells overcome the convergence of these negative regulators in vivo, we compared their potency to A2AR-KO CAR T-cells and unedited CAR T-cells at subtherapeutic dose levels in H226 tumor-bearing NCG mice. As expected, unedited CAR T-cells demonstrated limited capacity to control tumor outgrowth, while A2AR-KO CAR T-cells mitigated tumor progression but did not clear tumor. However, TKO CAR T-cells durably eradicated established tumors (Fig. 5e; Supplementary Fig. 9), decreased cumulative tumor burden (Fig. 5f), and improved survival of treated mice (Fig. 5g).
To further investigate the mechanism of improved anti-tumor efficacy of TKO CAR T-cells against solid tumors in vivo, we performed histological analyses of the TME of mice treated with CAR T-cells. One week post CAR T-cell infusion, tumors were excised, and tumor infiltration of CAR T-cells was quantified by immune-fluorescence imaging. Approximately two times as many A2AR-KO CAR T cells were observed infiltrating tumors compared to control CAR T cells. However, more than four times as many TKO CAR T-cells were identified in the TME relative to control CAR T cells, indicating enhanced capacity to penetrate and locally proliferate within the immunosuppressive TME (Fig. 5h, i). These data indicate that CAR T-cells resistant to biochemical inhibition alone are insufficient to overcome additional suppression in the TME and highlight the power of multiplex gene editing to generate a CAR T-cell product that is simultaneously resistant to multiple TME-associated immunosuppressive features in vivo.
Data from Figs. 4 and 5 indicated that A2AR-KO CAR T-cells were still susceptible to immunological negative regulators (PD-L1, TGF-β) in the TME. To test the reverse, whether CAR T cells lacking major immunological barriers are still inhibited by biochemical barriers, we engineered CAR T cells with TGFβRII-KO and PD1-KO (double knockout, DKO) (Fig. 6a). In vitro assays confirmed that DKO CAR T-cells remain susceptible to adenosine-mediated biochemical suppression since the production of critical anti-tumor cytokines IFN-γ and IL-2 were substantially reduced in the presence of adenosine (Fig. 6b, c and Supplementary Fig. 10). Moreover, in vitro 3-D spheroid cytotoxicity assay demonstrated that TKO and A2AR-KO CAR T-cells were able to completely eliminate H226 tumor spheroids in the presence of adenosine (Fig. 6d). While DKO CAR T-cells exhibited improved cytolytic capacity relative to control, they were incapable of clearing tumor spheroids. Taken together, these data suggest that even after clinically approved blockade of immunological negative regulators (e.g., PD1, TGFβ), biochemical barriers (e.g., hypoxia/HIF → CD73/CD39 → adenosine → A2AR) may still suppress CART cells within the TME of solid tumors.
a Schematic of double knock out (DKO) EGFR-specific CAR T-cells comprising two genomic edits targeting PDCD1 (PD-1-KO) and TGFBR2 (TGFβRII-KO). b IFN-γ and IL-2 production by DKO CAR T-cells measured by ELISA 48 h post-stimulation with H226 tumors in the presence of a DMSO control (gray) or exogenous cADO (orange), (Kruskal–Wallis t-test, n = 4, two technical replicates of two biological donors). c Reduction of IFN-γ and IL-2 levels normalized to a DMSO treated control from (b) (n = 4, two technical replicates of two independent biological donors, mean ± SD). d Incucyte-based live imaging of GFP + H226 tumor spheroid cytotoxicity tumor of tumor alone (black) and tumor cultured with A2AR-KO (red), DKO (orange) and TKO (blue) CAR T-cells in the presence of cADO. Cytotoxicity was measured as a decrease in total tumor GFP+ area over time (n = 3 technical replicates, mean ± SD). e Magnitude of IFN-γ, IL-2 and TNF-α secretion by A2AR-KO (red), DKO (orange) and TKO (blue) CAR T-cells in the presence of triple suppression comprising exogenous cADO, PD-L1, and TGF-β proteins normalized to respective untreated CAR T-cell controls, (Kruskal–Wallis t-test, n = 4, two technical replicates of two donors). f 2 × 106 or 0.5 × 106 DKO (orange, n = 9 individual mice), TKO (blue, n = 9 individual mice) or UTD (gray, n = 6 individual mice) CAR T-cells were infused into H226 tumor-bearing NCG mice when tumor volume reached an average volume of 300 mm3 (Two-sided unpaired t-test, average of individual mice, Day49 P* = 0.0125, Day73 P* = 0.0464). g Longitudinal analysis of mice bearing palpable H226 tumors from (f). Black bars indicate mice with remaining tumors and white bars indicate tumor-free mice (Two-sided Fisher’s exact test, n = 36 individual mice, P* = 0.0354). h 2 × 106 DKO (orange, n = 9 individual mice), TKO (blue, n = 9 individual mice) or UTD (gray, n = 6 individual mice) CAR T-cells were infused into A549 tumor-bearing NCG mice when tumor volume reached an average volume of 300 mm3 (Two-sided unpaired t-test, n representing individual mice, Day10 P*** = 0.0004, Day14 P*** = 0.0001, Day17 P*** = 0.0001, Day21 P*** = 0.0001, Day24 P** = 0.001). Right panel shows rapid growth kinetics of individual UTD CAR T-cell treated mice. i A549 cumulative tumor burden calculated as area under the curve (Two-sided unpaired t-test, n = 8 representing individual mice, P*** = 0.0004). For (b, c, e), symbols represent biological replicates from two different donors, error bars represent S.D., d symbols represent technical replicates, error bars represent S.D. and (f–i) where symbols represent group average of individual mice and error bars are S.E.M. f Kruskal–Wallis and h Mann–Whitney tests performed to calculate statistical significance, ***P < 0.001, *P < 0.05.
We further tested this hypothesis by comparing the therapeutic efficacy of TKO versus DKO CAR T-cells against H226 tumors in vivo. In our initial assays, a dose of 2 × 106 TKO or DKO CAR T cells was sufficient to eliminate tumors in all mice bearing H226 tumors, albeit more rapidly in the TKO CAR T-cell treated mice (Supplementary Fig. 11a). However, 5 out of 10 mice treated with DKO CAR T-cells relapsed and tumors resumed outgrowth. By contrast, mice treated with TKO CAR T-cells exhibited complete tumor elimination for the duration of the assay (100 days post-CAR T-cell infusion), except for one mouse with a lesion that had the appearance of remnant scar tissue (Supplementary Fig. 11a). We repeated this experiment using CAR T-cell products derived from a different donor and tested their therapeutic responses upon infusing 2 × 106 and 0.5 × 106 TKO and DKO CAR T-cells into H226 tumor-bearing mice. TKO CAR T-cells exhibited greater tumor control than DKO CAR T-cells at both dose levels (Fig. 6f) and resulted in a higher proportion of tumor-free mice at study endpoint (Fig. 6g). Of note, at these doses, unedited control CAR T-cells do not delay tumor outgrowth, and tumors progress with the same kinetics as untreated mice. In longitudinal assessment of tumor-bearing mice treated with the higher dose (2 × 106), 67% (6/9) of mice treated with TKO CAR T-cells demonstrated durable and long-term tumor elimination. By contrast, only 22% (2/9) of mice treated with DKO CAR T-cells exhibited complete tumor control, with several mice demonstrating tumor-growth relapse (Supplementary Fig. 11b, left).
To confirm and extend these findings in a separate tumor model, we tested the therapeutic efficacy of TKO versus DKO CAR T-cells against A549 tumors. In this model, TKO CAR T-cell-treated mice demonstrated improved therapeutic responses as indicated by accelerated kinetics of tumor regression (Fig. 6h) and reduced cumulative tumor burden (Fig. 6i) compared to mice that received DKO CAR T-cells. Moreover, all mice treated with TKO CAR T-cells achieved durable and long-lasting tumor elimination with no signs of tumor-relapse for the duration of the assay. By contrast, 8 out of the 9 DKO CAR T-cell-treated mice took over twice as long (72-days post infusion) to control tumor, with one mouse unable to clear tumor (Supplementary Fig. 11b, right).
The goal of our unique multiplex editing strategy was to generate a CAR T-cell product that is broadly applicable to heterogenous tumor subtypes that often have different immune-suppressive barriers due to tumor heterogeneity. Here we show that all three edits (A2AR, PD1, TGFβRII) afford the strongest and most durable tumor remission in these models. Taken together, data from Figs. 5 and 6 suggest that for the maximal therapeutic benefit observed with TKO CAR T-cells, elimination of both biochemical and immunological barriers are necessary in these models.
Stealth TKO CAR T-cells resist allorejection in immunocompetent mice and potently reject tumors
A limitation of tumor-engrafted immunocompromised mice is the paucity of immunologically relevant human cell types that contribute to forming the TME. Therefore, we wanted to investigate the functionality of TKO CAR T-cells in a small-animal model that reconstitutes a human immune system. To do so, we utilized humanized NCG (huNCG) mice that are generated via adoptive transfer of human cord blood-derived CD34+ hematopoietic stem cells into recipient mice. huNCG mice developed peripheral CD4+ and CD8+ T cells, B cells, myeloid cells, and a limited pool of mature NK cells (Supplementary Fig. 12). Importantly, CD4+FoxP3+ regulatory T cells and CD68+ tumor-associated macrophages were detected in resected tumor sections from H226 bearing huNCG mice (Supplementary Fig. 13) indicating that these mice recapitulate a tumor milieu resembling TMEs described in humans49,50,51,52.
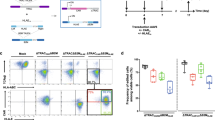
Additionally, successful engraftment of adoptively transferred allogeneic donor-derived CAR T-cells into a genetically dissimilar lymphoreplete host relies on incorporating a cloaking strategy to evade the host immune system. Given that huNCG mice primarily reconstitute a human T cell compartment, we used ABE to simultaneously introduce two ‘Stealth’ gene edits into allogeneic CAR T-cells targeting B2M (beta-2-microglobulin) and CIITA (class-II transcriptional activator) to disrupt HLA class-I and HLA class-II surface expression to prevent recipient CD8+ and CD4+ T cell-mediated rejection, respectively53. Additionally, these HLA-deficient CAR T-cells were edited at CD3E to ablate surface expression of the endogenous T cell receptor complex to prevent graft-versus-host disease (Fig. 7a)53. To determine whether genetic disruption of B2M and CIITA protect allogeneic Stealth CAR T-cells from immune-mediated rejection, we co-transferred Stealth and unedited (HLA+) CAR T-cells into recipient huNCG mice, and the persistence of these cell products was monitored by longitudinally sampling peripheral blood. Unedited CAR T-cells were readily eliminated from peripheral blood within 2-weeks post-infusion, whereas Stealth CAR T-cells resisted allorejection and persisted for the duration of the study (Fig. 7b, c).
a Flow cytometry histograms indicating surface expression of endogenous TCR, HLA-class I (HLA-ABC) and HLA-class II (HLA-DR) on unedited (gray) and Stealth (green) T cells. b, c 5 × 106 unedited (gray, n = 4 individual mice) and Stealth (green, n = 5 individual mice) T cells were co-infused into humanized NCG mice. Flow cytometry plots indicate longitudinal frequency of peripheral T cells from within the same mouse at 1-, 6-, 13- and 20-days post-infusion (b) and cumulative concentration of peripheral T cells 20 days post-infusion (c) (Mann–Whitney t-test, n= average of individual mice as above, mean ± SD P** = 0.0079). d Schematic of Sealth-TKO EGFR-specific CAR T-cells comprising six genomic edits targeting ADORA2A (A2AR-KO), PDCD1 (PD-1-KO), TGFBRII (TGFBR2-KO), CD3E (TCR-KO), B2M (HLA-Class I-KO), and CIITA (HLA-Class II-KO). TKO edits colored red and Stealth edits colored gray. e Frequency of on-target genomic editing efficiencies at six genomic loci in Unedited (black) and Stealth-TKO (purple) CAR T-cells quantified by NGS (Symbols represent individual biological donors from individual experiments). f–h 4 × 106 Stealth CAR T-cells (gray, n = 8 individual mice) and Stealth-TKO CAR T-cells (purple, n = 8 individual mice) were infused into humanized NCG mice when tumor volume reached an average volume of 150 mm3. Longitudinal tumor volume (f) (Mann–Whitney t-test, n = 8 average of individual mice, mean ± SEM, P* = 0.018, P** = 0.003), cumulative tumor burden (g), (Mann–Whitney t-test, n = 8 average of individual mice, mean ± SD, P*** = 0.0007, P** = 0.003) and survival curves (h), (log-rank test, graph lines represent group mean, P* = 0.012) are shown. For all data, symbols represent individual mice or T cell donors, and error bars reflect mean ± SD, except (f) where error bars represent S.E.M. c,f, g Mann–Whitney, and h log-rank test performed to calculate statistical significance, ***P < 0.001, **P < 0.01, *P < 0.05.
After determining that Stealth CAR T-cells successfully engraft into huNCG mice, we engineered TKO CAR T-cells with CD3E, B2M and CIITA base edits (Fig. 7d). The resulting 6-plex gene edited cell product, termed Stealth-TKO CAR T-cells, maintained a high degree of on-target genomic editing efficiency at all loci (Fig. 7e) without any discernible negative impact on cell viability or overall expansion, which correlated to our prior observations in multiplex base editing experiments36. Upon infusion into H226 tumor-bearing huNCG mice, Stealth-TKO CAR T-cells potently eliminated tumor at multiple dose levels (Fig. 7f; Supplementary Fig. 14). In contrast, the tumor growth kinetics in mice treated with CAR T-cells containing only Stealth edits mirrored untreated control mice (Fig. 7g). Moreover, the superior anti-tumor responses mediated by Stealth-TKO CAR T-cells significantly reduced cumulative tumor burden and extended the long-term survival of tumor-bearing mice (Fig. 7g, h). The ability of Stealth-TKO CAR T-cells to evade allorejection and mediate potent anti-tumor activity despite a complex TME within a lymphoreplete host highlights the utility of this unique combinatorial multiplex gene editing strategy to maximize the therapeutic efficacy of CAR T-cell therapies against solid tumors.
Discussion
Overcoming multiple immunosuppressive barriers acting on CAR T-cells within the TME likely requires a multifaceted genome engineering strategy. To this end, we employed a unique combination of our Base Editing platform31 with a CRISPR nuclease38 to manufacture allogeneic CAR T-cells resistant to biochemical and immunological negative regulators. These data emphasize the importance of understanding CAR T-cell-TME interactions, as well as the therapeutic advantages of simultaneously blocking multiple immunosuppressive pathways in the heterogenous tumor microenvironments.
Solid tumors promote hypoxic and adenosine-rich microenvironments that can be ameliorated by pharmacological approaches54,55,56,57. While initial Phase II (NCT05024097) and Phase III (NCT05221840) clinical results are promising, inhibitors of the adenosinergic pathway may not be completely effective due to the presence of alternative suppressive mechanisms and the paucity of anti-tumor T cells in the TME. To ensure sufficient numbers of tumor-reactive T cells, we developed CAR T-cells for adoptive cell therapy that are resistant to adenosine-mediated immunosuppression. We selected EGFR as a target antigen since it has been used preclinically and clinically to test cancer therapies with small molecules, biologics and CAR T-cells. EGFR is also widely expressed across multiple solid tumor types, including lung, breast, pancreatic, prostate, and renal cancers where immune checkpoint blockade and inhibitors of the adenosine pathway are currently being tested in clinical trials. While our study focused on solid tumor xenografts from EGFR-expressing lung carcinoma cell lines, it is likely that such an engineering approach would be applicable to other CAR target antigens and tumor types since hypoxia/adenosine, the PD1:PD-L1 axis, and TGF-β are common features of many solid cancers.
This study demonstrated the first use of base editing in CAR T-cells to genetically ablate ADORA2A, the gene encoding A2AR, which is the major receptor that binds extracellular adenosine initiating inhibitory programming. Ablation of functional A2AR in CAR T-cells attenuated downstream phosphorylation of CREB, increased production of pro-inflammatory cytokines and improved tumor elimination. This confirms and extends previous observations by Beavis et al. using CRISPR-Cas9 mediated elimination30. However, A2AR-KO CAR T-cells remained sensitive to immunological negative regulators such as PD-L1 and TGF-β. Likewise, CAR T-cells deficient in both PD-1 and TGF-βRII expression were highly suppressed by adenosinergic signaling. These data support the notion that i) non-redundant inhibitory mechanisms within the TME act in concert to inhibit anti-tumor responses, and ii) engineering CAR T-cells to overcome a single pathway may be insufficient to engender durable remissions.
A key conceptual advance of this study is in tailoring our engineering strategy to target specific negative immune regulators in solid tumor types. In this case, we eliminated major biochemical (adenosine) and immunological (PD-L1 and TGF-β) negative regulators that are prominent in the hypoxic tumor areas that are a common feature of virtually all solid tumors. This engineering approach potently enhanced the therapeutic efficacy and durability of responses of CAR T-cells against solid tumors. Recent research has identified additional negative regulators within the TME56,57,58,59,60 and is likely to reveal more target genes for interrogation, ultimately requiring a complex genome engineering strategy to generate higher-order multiplex gene-edited CAR T-cells. To meet the challenge of complex genome engineering, we combined an adenine base editor (ABE) and Cas12b, a nuclease with a distinct ATTN protospacer adjacent motif (PAM) sequence dissimilar to the NGG PAM used by ABE37. By utilizing editors with distinct PAM restriction, this unique combination acts complementarily to expand the range of accessible target sites up to six genes reported here and provides future avenues for nuclease-driven gene knock-in. This technical advance in engineering strategy resulted in the highest number of simultaneous genomic edits to date in a CAR T-cell platform with no measurable negative impact on CAR T-cell viability and provided proof-of-concept for the rational design of CAR T-cell products with a synergistic combination of gene edits that confer potent anti-tumor efficacy.
Although single-gene editing strategies have previously been applied, particularly in murine models, to disrupt immune-suppressive pathways, such approaches are likely insufficient in solid tumors, where immune evasion occurs through multiple mechanisms (Fig. 4a, b; Supplementary Fig. 10). In this study, we leveraged the flexibility of multiplex gene editing to simultaneously target the dominant negative regulators reinforced by the hypoxic TME. We propose that the key limitation of CAR T-cells in solid tumors, compared with their success in hematological malignancies, is the array of immune-suppressive barriers driven by hypoxia. Hypoxia, a hallmark of virtually all solid tumors, promotes aggressiveness, metastasis, and therapy resistance in part through the induction of extracellular adenosine, PD-L1, and TGF-β. Our goal, therefore, was to engineer a CAR T-cell product with multiplex edits capable of overcoming suppression in these hostile, therapy-resistant niches. Importantly, this work does not aim to rank the relative contributions of adenosine, PD-L1, and TGF-β; indeed, the impact of each pathway is likely to vary across tumor models and TME contexts. Rather, we emphasize that solid TMEs are heterogeneous, and CAR T-cells must be broadly fortified to withstand diverse suppressive pressures. Notably, the selected targets represent distinct, non-redundant pathways of T-cell inhibition. Collectively, our findings demonstrate that even in the absence of major immunological barriers such as PD-1 and TGF-β, CAR T-cells remain vulnerable to biochemical suppression (Fig. 6).
Moreover, we have demonstrated the power of multiplex gene editing in successfully incorporating additional edits to manufacture a conceptually clinically relevant, allogeneic CAR T-cell product. CAR T-cells derived from an allogeneic donor hold promise in solving challenges associated with autologous, patient-derived products including manufacturing failure and the urgency to reduce patient wait-time to treatment61,62,63,64. Thus, a critical aspect of this study applied ‘Stealth’ gene edits to TKO CAR T-cells to prevent GvHD (CD3E-KO) and evade T cell-mediated rejection caused by HLA haplotype mismatch between patient and allogeneic CAR T-cell donor (B2M-KO and CIITA-KO). Disrupting these target genes permitted stable engraftment of allogeneic CAR T-cells into lymphoreplete humanized mice resulting in superior solid tumor clearance. However, it is important to note that stem cell-engrafted humanized mice often exhibit incomplete immune reconstitution, e.g., mature NK cells64, suggesting that additional modifications such as genetic ablation of NK cell activating receptors65 or overexpression of the invariant HLA-E inhibitory receptor66 may be necessary to truly render allogeneic CAR T-cells hypoimmunogenic.
While adverse events are also an important consideration with any CAR T-cell therapy, in these studies we did not observe any such indications with the Stealth-TKO CAR T-cells in mice. There may be inherent safety built into approaches that target surface receptors that rely on external factors as opposed to disrupting cell-intrinsic negative regulators of T cell function (i.e., regnase, roquin, rasa2, etc.) which function independent of environmental cues. In addition, the deletion of the TCR likely prevents aberrant off-tissue immune responses that could potentially be exacerbated by TKO edits. However, if safety concerns arose, the multiplex base editing strategy employed here would be amenable to the incorporation of an additional safety switch that would mitigate such risks.
This study provides rationale and precedent for the use of combinatorial gene editing to manufacture an allogeneic CAR T-cell product that maximizes therapeutic efficacy by overcoming orthogonal immunosuppressive barriers in the solid TME. While it must be considered that an inflection point may be reached whereby increasing the number of edits decreases overall CAR T-cell viability or functionality, our data indicate that this was not the case with the 6-plex edited Stealth-TKO CAR T-cells. By employing a unique Base Editor/CRISPR nuclease combination, we focused on eliminating biochemical and immunological barriers that are common to many solid TMEs in an effort to enhance the therapeutic capacity of CAR T-cells. Thus, the engineering approach described here may unlock the potential of solid tumor targeting CAR T-cells.
Methods
Tumor cell lines
NCI-H226 (CRL-5826), A549 (CCL-185), NCI-H460 (HTB-177), MDA-MB-231 (HTB-26), SKOV3 (HTB-77), MCF7 (HTB-22), A-375 (CRL-1619) cells were obtained from the American Type Culture Collection (ATCC) where STR profiling was performed. Cell lines were cross-checked with the database of cross-contaminated or otherwise misidentified cell lines maintained by the International Cell Line Authentication Committee and not found on this list. NCI-H226 and NCI-H460 cells were grown in RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) and A549, MDA-MB-231, SKOV3, MCF7 and A375 cells were grown in DMEM medium (Gibco) supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2 incubator. For in vivo studies, Mice were inoculated with 5 × 106 tumor cells resuspended in 20% Matrigel Matrix (Corning), randomized into different groups and tumors measured in a blinded manner 2–3 times per week using vernier calipers. Mice were sacrificed when tumor volume reached 2000 mm3 in accordance with the guidelines and approval of the Institutional Animal Care and Use Committee at Northeastern University and the Charles River Accelerator and Development Lab.
Mouse strains and study approval
For all immune compromised xenograft models, female 6–8-week-old NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG, strain code: 572) coisogenic immunodeficient mice were obtained from Charles River Laboratories (CRL). For all humanized mouse models, human umbilical cord blood-derived CD34+ humanized coisogenic NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl female mice (huNCG, strain code: 695) mice were obtained from CRL. NCG mice were engrafted with Hu-CD34+ stem cells at CRL at 6–8 weeks old followed by 14–16-week engraftment. All animals were housed in a specific pathogen–free environment under controlled conditions and received food and water ad libitum according to the National Institutes of Health (NIH) guidelines. All animal experiments were conducted in accordance with the guidelines and approval of the Institutional Animal Care and Use Committee at Northeastern University and the Charles River Accelerator and Development Lab.
Flow cytometry
Surface expression of anti-EGFR CARs was detected by staining with an anti-Cetuximab-AF647 idiotype at 5 µL/test (Clone: 2259C, R&D Systems). T cell editing efficiency, T cell phenotyping, T cell signaling, tumor cell phenotyping and mouse blood phenotyping were evaluated with the following anti-human antibodies at 2 µL/test: CD155 (SKII.4, Biolegend), CD19 (HIB19, Biolegend), CD2 (RPA-2.10, Biolegend), CD3 (UCHT1, Biolegend), CD33 (P67.6, Biolegend), CD38 (HIT2, Biolegend), CD39 (A1, Biolegend), CD4 (OKT4, Biolegend), CD45 (2D1, Biolegend), CD47 (CC2C6, Biolegend), CD56 (NCAM, Biolegend), CD68 (FA11, Biolegend), CD73 (AD2, Biolegend), CD8 (SK1, Biolegend), CD80 (B7-1, BD Biosciences), CD86 (IT2.2, Biolegend), EGFR (AY13, Biolegend), FasL (NOK-1, Biolegend), HLA-ABC (W6/32, Biolegend), PD1 (A17188B, Biolegend), PDL1 (29E.2A.3, Biolegend), TCR (IP26, Biolegend). CD73 (AD2, Cell Signaling), PDL1 (D8T4X, Cell Signaling), Phospho-CREB (87G3, Cell Signaling), HLA-DR (L203.rMAb, BD Biosciences), Phospho-SMAD2/3 (027-670, BD Biosciences), TGFβ1 (Polyclonal, Bioss). Antibodies for Mass Cytometry all supplied from Fluidigm: Alpha-SMA (1A4), Collagen-1 (polyclonal), E-cadherin (2.4e + 11), Histone H3 (D1H2), Vimentin (D21H3), Granzyme B (EPR20129-217), Ki-67 (B56), PD-1 (EPR4877(2)), PD-L1 (Sp142), CD20 (H1), CD3 (polyclonal), CD4 (EPR6855), CD45RO (UCHL1), CD68 (KP1), CD8α (CD8/144B), FoxP3 (PCH101), Pan-keratin (C11). For detection of surface proteins, cells were stained for 20 min at 4 °C in the dark. For intracellular staining, cells were stimulated with agonist and then fixed with pre-warmed Cytofix (BD Biosciences) for 15 min at 37 °C, then permeabilized with Perm III buffer (BD Biosciences) for 30 min at −20 °C. Fix and permeabilized cells were then stained with antibodies for 1 h at room temperature in the dark. All samples were washed with PBS prior to analysis on LSR Fortessa (BD Biosciences) flow cytometer and data analyzed using FlowJo v.10.9 software.
Generation of CAR T-cells
CD4+ and CD8+ cells were positively selected from a de-identified healthy human donor apheresis (Charles River Laboratories) using anti-CD4 and anti-CD8 microbeads (Miltenyi) following the manufacturer’s protocol. Post isolation, cells were washed and cryopreserved in a mixture of Plasma-Lyte A (Hanna Pharmaceutical), 2% HSA (Access Biologicals) and CS10 (BioLife Solutions). Cryopreserved cells were thawed and activated with anti-CD3/anti-CD28 TransAct (Miltenyi) 1:100 v/v in OpTmizer media (Thermo Fisher) containing 2% GlutaMAX (Thermo Fisher), 2.5% Immune cell serum replacement (Thermo Fisher), 100 IU/mL rhIL-2, 2 ng/mL rhIL-7 and 0.4 ng/mL rhIL-15 (R&D Systems). T cells were transduced 24 h post-activation with lentivirus encoding an anti-EGFR CAR with 41bb/CD3ζ signaling domains (Flash Therapeutics). Forty-eight hours post activation, T cells were electroporated with ABE8.20 base editor mRNA, Cas12b nuclease mRNA and 5’/3’ end modified target sgRNAs (Agilent) with 4-D Nucleofector (Lonza). Post-electroporation, cells were transferred to GRex culture flasks (Wilson Wolf) and expanded at 37 °C in a 5% CO2 incubator. Verification of editing was confirmed via flow cytometry and next-generation sequencing. All control CAR T cells were run through the same electroporation protocol as the edited CAR T cells to control for any effects on T cell health, viability or change in effector functions. This was demonstrated to be the most relevant editing control in previous studies using base editors with respect to CAR T-cell viability. Each experiment also used multiple donor replicates to account for donor-to-donor variability in CAR T-cell expansion, transduction, editing efficiency and functional anti-cancer responses. These donors were combined and averaged in presented in vitro data. Due to the size and complexity of the in vivo experiments, a single donor was utilized per experiment. CAR T cells were expanded in a 10-day expansion process optimized to routinely yield 1−3e9 multiplex edited CAR+ T cells, sufficient numbers to fulfill the needs of these large in vivo experiments. In this case, no pooling of donors was required. CAR T cells were administered in a blinded manner. For in vitro assays, 2–4 unique donors were used and each experiment performed in duplicate or triplicate. In vivo experiments employed T cells from different donors. For immunodetection, CAR T-cells in some studies were transduced with ZsGreen sequence (Addgene) cloned into a lentiviral backbone. ZsGreen was selected due to its high fluorescence intensity compared to GFP, which enhances detection sensitivity allowing CAR T-cells to be readily detected by native ZsGreen fluorescence.
Next-generation sequencing
Genomic DNA was extracted from cell pellets with QuickExtract DNA Extraction Solution (Lucigen) via manufacturer’s protocol. Subsequently, 2 μl of genomic DNA was added to a 25 μl PCR reaction containing Phusion U Green Multiplex PCR Master Mix (Thermo Fisher) and 0.5 μM of each forward and reverse primer for each target site. Following initial PCR amplification, PCR products were barcoded using Illumina barcoding primer pairs. DNA was sequenced on an Illumina MiSeq instrument according to the manufacturer’s protocol. NGS data was analyzed by first performing Illumina demultiplexing, then read trimming and filtering, followed by alignment of all reads to the reference amplicon sequence prior to quantification generation of alignment statistics and editing rates.
A2AR signaling assay
CAR T-cells were placed at 37 °C in a 1% O2 incubator for 48 h. Following hypoxic pretreatment, cells were washed in PBS and counted prior to assay. 1 × 106 cells were plated per well of 96-well round-bottom plate. Cells were treated with 30 µM 2-chloroadenosine (cADO, Sigma) or equivalent volume of DMSO for 1 h at 37 °C. Subsequently, cells were stained and analyzed as described above.
TGFβR signaling assay
CAR T-cells were rested in media without serum replacement overnight at 37 °C. Then, cells were washed in PBS and counted prior to assay. 1 × 106 cells were plated per well of 96-well round-bottom plate. Cells were treated with 10 ng/mL of TGF-β1 (Peprotech) or equivalent volume of DMSO for 20 min at 37 °C. Cells were then stained and analyzed as described above.
Cytokine release assay
1 × 105 tumor cells and 5 × 104 CAR+ cells were plated per well of 96-well flat bottom plate. DMSO, 10 µM cADO or 10 ng/mL TGF-β1 were added, and cell co-culture was incubated for 48 h. At assay endpoint, plates were spun down, and supernatants taken for ELISA analysis on Ella platform (Protein Simple) via manufacturer’s protocol.
PD-1 induction assay
1 × 106 cells were plated per well of 96-well round-bottom plate. Cells were treated with 1:1000 diluted Cell Stimulation cocktail (Biolegend) overnight at 37 °C. Next, cells were washed with PBS, and stained with anti-PD1 antibody (Biolegend). Data acquired on LSR Fortessa (BD Biosciences) and analyzed via FlowJo.
PD-L1 bead suppression assay
Saturating amounts of rhEGFR-Biotin and rhPD-L1-Biotin (ACRO Biosystems) were coated onto separate Streptavidin-coated Dyna Beads (Thermo Fisher). Protein-labeled beads were washed twice in PBS and resuspended in cell culture media at 6.5 × 108 beads/mL. 1 × 106 CAR T-cells were plated per well of 96-well flat bottom plate. 2 µL of EGFR-coated beads and either 2 µL, 5 µL or 10 µL of PD-L1-coated beads were added to each well and incubated for 48 h. At assay endpoint, plates were spun down, and supernatants taken for ELISA analysis on Ella platform via manufacturer’s protocol.
Cytotoxicity assay
H226 and A549 lung carcinoma cell lines were stably transduced with lentivirus encoding GFP. 1.5 × 104 GFP+ tumor cells were plated in ultra-low attachment 96-well plates (Corning) and placed at 37 °C for 72 h to allow for spheroid formation. After 72 h, 7 × 103 CAR+ cells were plated in tumor wells with or without 10 µM cADO, 10 ng/mL TGF-β1 or 10 µL PD-L1 coated beads. Cytotoxicity was quantified as reduction of GFP area via imagining in Incucyte S3 Live-Cell Analysis System (Sartorius). All cytotoxicity assays were done in technical triplicate and each iteration of the cytotoxicity assay was done using a different donor. These assays were also repeated a minimum of 3 times.
In vitro tumor cell TGF-β production assay
1 × 106 NCI-H226, A549, H460, MDA-MB-231, SKOV3, MCF7 or A375 tumor cells were plated in 24-well tissue culture plate in 1 mL of media. After 24 h, supernatants were taken for ELISA analysis on Ella platform via manufacturer’s protocol.
CAR T-cell efficacy in vivo
5 × 106 H226 cells were resuspended in 200 μL of 20% Matrigel Matrix (Corning) and injected subcutaneously into hind limb of NCG or huNCG mice followed by randomization. Tumor size was measured via calipers 2–3x per week in a blinded manner. Once tumors grew to an average volume of 150 mm3, 2–8 × 106 CAR+ cells were injected via tail vein in 200 µL HBSS in a blinded manner. Weekly submandibular bleeds were used to track human cells within the blood over the course of the experiment. Prior to sacrificing the animals, mice were injected with 80 mg/kg of Hypoxyprobe (pimonidazole HCl, 4.3.11.3, Hypoxyprobe) solution via tail vein. Sixty minutes post Hypoxyprobe injection, the mice were sacrificed, and tumors resected. Tumors were washed in PBS and then flash frozen in OCT blocks and stored at −80 °C.
Allogeneic CAR-T in vivo persistence assay
CD2+CD3+HLA-ABC+HLA-DR+ (unedited) and CD2+CD3-HLA-ABC-HLA-DR- (Stealth) T cells expressing a CD4-based CAR were generated as previously described67. 5 × 106 CAR+ T cells of each population were co-injected intravenously into huNCG mice in a blinded manner. Blood was collected via puncture of the submandibular vein 1-day post-infusion and weekly thereafter until 20 days post-infusion, into K2 EDTA coated microvette tubes (Sarstedt Inc) and stained as described above and analyzed on MACSQuant 16 (Miltenyi) flow cytometer.
Tissue sectioning and immunohistochemistry
Tumors were excised from mice, washed in PBS, flash frozen into OCT blocks and stored at −80 °C until further processing. Blocks were removed from freezer and placed in pre-chilled NX70 CryoStat (ThermoFisher). 5-μm tissue sections were prepared from 10 to 20 different cutting surfaces and mounted onto polysine-coated slides and air-dried for 45–60 min. Sections were fixed in a 1:1 mixture of acetone and methanol for 10 min and subsequently air-dried for 10 min. Next, hydrophobic barrier PAP pen was applied to the edges of the sections and IHC buffer (PBS, 0.5% BSA, 0.1% Tween-20) added for 20 min. Then, sections were blocked with Fc block in IHC buffer for 10 min followed by immunostaining of antibody cocktail in IHC buffer for 3 h at room temperature. Slides were then washed three times with IHC buffer for 5 min, stained with DAPI, coverslips applied with fluoromount and imagined with an Olympus IX83 inverted microscope and analyzed using the Olympus CellSense software.
Spatial transcriptomics
Tumors were excised and prepared as described above. Tissues were mounted on slides, prepared and stained per Nanostring protocol. Briefly, tissue sections were fixed with 10% NBF overnight. Next, slides were washed in PBS, rehydrated in ethanol, treated with proteinase K and hybridized with RNA probes from the Nanostring Human Whole Transcriptome Atlas panel overnight at 37 °C. Next, slides were washed and stained with nuclear dye Syto-83 or DAPI (Thermo Fisher), PD-L1 (Cell Signaling), TGFβ1 (Bioss), Hypoxyprobe, or CD73 (Cell Signaling) for 1 h at room temperature. Slides then washed and imaged on GeoMX DSP (Nanostring). RNA collected and run on MiSeq (Illumina). Standard quality control checks assessing imaging, binding density, positive control linearity and limit of detection were performed per Nanostring protocol. The mRNA expressed below background were filtered from the analysis using cutoffs of mean plus two standard deviations of negative controls and only probes with counts greater than the background threshold were included in the analysis.
Mass cytometry
Tissue sections were thawed to room temperature and then baked at 60 °C for one hour. The tissue was washed twice in PBS followed by four 5-min washes in ethanol (50%, 70%, 100% x2), followed by a DEPC water wash. Antigen retrieval was done in 1X Tris EDTA (pH 9.0) at 99 °C for 20 min using a Hamilton beach steamer. Slides were washed in PBS; tissue was encircled using a PAP pen and allowed to dry for 5 min. Tissue was blocked in 3% BSA in PBS for 45 min at room temperature in a hydration chamber. Antibody cocktail was prepared in 0.5% BSA in PBS, pipetted onto slide and incubated overnight at 4 °C in a hydration chamber. The tissue was then washed twice in 0.2% Triton X-100, followed by two washes in PBS for 8 min with slow agitation. The tissue was stained with Intercalator-Ir solution for 30 min at room temperature in a hydration chamber. The slides were washed in DEPC water for 5 min and air-dried for 20-min at room temperature. Tissue slides were stored in a slide box at 4 °C until acquired on the CyTOF Hyperion (Standard BioTools).
Statistical methods
All analyses were performed with GraphPad Prism software. Data was presented as mean ± S.D. or S.E.M. with statistically significant P-values determined by non-parametric Mann–Whitney t-test or Kruskal–Wallis or one-way ANOVA as indicated in figure legends. All experiments were conducted a minimum of three times with similar results. All in vivo studies were blinded to group allocation during data collection and analysis.
Sex as a biological variable
Female mice were used for the tumor immunology assays described in this study since they exhibit more consistent immune responses in these models. Findings are not expected to be relevant to one sex more than the other given the nature of the immunologic pathways that our murine model was designed to investigate. The use of the female sex mice was solely for the purpose of achieving consistency in immunologic responses measured.
Data availability
Source data for all figures are provided as a Source data file and published alongside the paper. Analyzed sequencing data and summary metadata generated in this study are included within the article and the Source data file. Spatial transcriptomics sequencing data generated in this study has been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under BioProject accession number PRJNA1299304. The next-generation sequencing was performed at Beam Therapeutics as part of a sponsored research agreement with Northeastern University and all raw sequencing files are maintained at Beam Therapeutics. Data can be made available upon request with clear scientific justification. Requests should be directed to Colby Maldini (cmaldini@wistar.org). Data transfer will be facilitated within 30 days, and access will remain available for 30 days thereafter.
References
Cappell, K. M. & Kochenderfer, J. N. Long-term outcomes following CAR T cell therapy: what we know so far. Nat. Rev. Clin. Oncol. 20, 359–371 (2023).
Leick, M. B., Maus, M. V. & Frigault, M. J. Clinical perspective: treatment of aggressive B cell lymphomas with FDA-approved CAR T-cell therapies. Mol. Ther. 29, 433–441 (2021).
Martinez, M. & Moon, E. K. CAR T cells for solid tumors: new strategies for finding, infiltrating, and surviving in the tumor microenvironment. Front. Immunol. 10, 128 (2019).
Newick, K., Moon, E. & Albelda, S. M. Chimeric antigen receptor T-cell therapy for solid tumors. Mol. Ther. Oncolytics 3, 16006 (2016).
Wang, Q. et al. Role of tumor microenvironment in cancer progression and therapeutic strategy. Cancer Med. 12, 11149–11165 (2023).
Muz, B., Puente, P., de la, Azab, F. & Azab, A. K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 3, 83–92 (2015).
Hatfield, S. M. et al. Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1α-dependent and extracellular adenosine-mediated tumor protection. J. Mol. Med. 92, 1283–1292 (2014).
Kojima, H. et al. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1α-deficient chimeric mice. Proc. Natl Acad. Sci. USA 99, 2170–2174 (2002).
Sitkovsky, M. V. T regulatory cells: hypoxia-adenosinergic suppression and re-direction of the immune response. Trends Immunol. 30, 102–108 (2009).
Hatfield, S. M. et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci. Transl. Med. 7, 277ra30–277ra30 (2015).
Li, Y., Patel, S. P., Roszik, J. & Qin, Y. Hypoxia-driven immunosuppressive metabolites in the tumor microenvironment: new approaches for combinational immunotherapy. Front Immunol. 9, 1591 (2018).
Hatfield, S. M. & Sitkovsky, M. V. Antihypoxic oxygenation agents with respiratory hyperoxia to improve cancer immunotherapy. J. Clin. Investig. 130, 5629–5637 (2020).
Sek, K. et al. Targeting adenosine receptor signaling in cancer immunotherapy. Int. J. Mol. Sci. 19, 3837 (2018).
Ohta, A. & Sitkovsky, M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature 414, 916–920 (2001).
Wu, V. H. et al. The GPCR–Gαs–PKA signaling axis promotes T cell dysfunction and cancer immunotherapy failure. Nat. Immunol. 24, 1318–1330 (2023).
Ohta, A. et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl Acad. Sci. USA 103, 13132–13137 (2006).
Nakagawa, Y. et al. Effects of extracellular pH and hypoxia on the function and development of antigen-specific cytotoxic T lymphocytes. Immunol. Lett. 167, 72–86 (2015).
Kaczmarek, E. et al. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J. Biol. Chem. 271, 33116–33122 (1996).
Hu, M. et al. The regulation of immune checkpoints by the hypoxic tumor microenvironment. PeerJ 9, e11306 (2021).
Mallikarjuna, P., Zhou, Y. & Landström, M. The synergistic cooperation between TGF-β and hypoxia in cancer and fibrosis. Biomolecules 12, 635 (2022).
Massagué, J. TGFbeta in cancer. Cell 134, 215–230 (2008).
Ou, Z., Dou, X., Tang, N. & Liu, G. Pressure increases PD-L1 expression in A549 lung adenocarcinoma cells and causes resistance to anti-ROR1 CAR T cell-mediated cytotoxicity. Sci. Rep. 12, 6919 (2022).
Kjaergaard, J., Hatfield, S., Jones, G., Ohta, A. & Sitkovsky, M. A2A adenosine receptor gene deletion or synthetic A2A antagonist liberate tumor-reactive CD8+ T cells from tumor-induced immunosuppression. J. Immunol. 201, 782–791 (2018).
Tang, N. et al. TGFβ inhibition via CRISPR promotes the long-term efficacy of CAR T-cells against solid tumors. JCI Insight 5, e133977 (2020).
Masoumi, E. et al. Genetic and pharmacological targeting of A2a receptor improves function of anti-mesothelin CAR T cells. J. Exp. Clin. Cancer Res. 39, 49 (2020).
Giuffrida, L. et al. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances CAR T cell efficacy. Nat. Commun. 12, 3236 (2021).
Kurago, Z. et al. Inhibitors of the CD73-adenosinergic checkpoint as promising combinatory agents for conventional and advanced cancer immunotherapy. Front. Immunol. 14, 1212209 (2023).
McGowan, E. et al. PD-1 disrupted CAR T-cells in the treatment of solid tumors: promises and challenges. Biomed. Pharmacother. 121, 109625 (2020).
Hirayama, A. V. et al. Timing of PD-L1 blockade with durvalumab may affect outcomes of CD19 CAR T-cell therapy for relapsed/refractory large B-cell lymphoma. Blood 140, 7447–7449 (2022).
Beavis, P. A., Milenkovski, N., Stagg, J., Smyth, M. J. & Darcy, P. K. A2A blockade enhances anti-metastatic immune responses. OncoImmunology 2, e26705 (2013).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Webber, B. R. et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 10, 5222 (2019).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Samuelson, C. et al. Multiplex CRISPR/Cas9 genome editing in hematopoietic stem cells for fetal hemoglobin reinduction generates chromosomal translocations. Mol. Ther. Methods Clin. Dev. 23, 507–523 (2021).
Diorio, C. et al. Cytosine base editing enables quadruple-edited allogeneic CAR T cells for T-ALL. Blood 140, 619–629 (2022).
Strecker, J. et al. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 10, 212 (2019).
Li, W. et al. Simultaneous editing of TCR, HLA-I/II and HLA-E resulted in enhanced universal CAR-T resistance to allo-rejection. Front. Immunol. 13, 1052717 (2022).
Vigano, S. et al. Targeting adenosine in cancer immunotherapy to enhance T-cell function. Front. Immunol. 10, 925 (2019).
Xia, C., Yin, S., To, K. K. W. & Fu, L. CD39/CD73/A2AR pathway and cancer immunotherapy. Mol. Cancer 22, 44 (2023).
Sun, C., Wang, B. & Hao, S. Adenosine-A2A receptor pathway in cancer immunotherapy. Front. Immunol. 13, 837230 (2022).
Ghiringhelli, F., Bruchard, M., Chalmin, F. & Rébé, C. Production of adenosine by ectonucleotidases: a key factor in tumor immunoescape. J. Biomed. Biotechnol. 2012, 1–9 (2012).
Anderson, K. G., Stromnes, I. M. & Greenberg, P. D. Obstacles posed by the tumor microenvironment to T cell activity: a case for synergistic therapies. Cancer Cell 31, 311–325 (2017).
Chen, Y. et al. Tumor microenvironment characterization for assessment of recurrence and survival outcome in gastric cancer to predict chemotherapy and immunotherapy response. Front. Immunol. 13, 890922 (2022).
Yu, L., He, R. & Cui, Y. Characterization of tumor microenvironment and programmed death-related genes to identify molecular subtypes and drug resistance in pancreatic cancer. Front. Pharmacol. 14, 1146280 (2023).
Nanjireddy, P. M., Olejniczak, S. H. & Buxbaum, N. P. Targeting of chimeric antigen receptor T cell metabolism to improve therapeutic outcomes. Front. Immunol. 14, 1121565 (2023).
Verma, N. K. et al. Obstacles for T-lymphocytes in the tumour microenvironment: Therapeutic challenges, advances and opportunities beyond immune checkpoint. eBioMedicine 83, 104216 (2022).
Barrera, C. et al. Deep computational image analysis of immune cell niches reveals treatment-specific outcome associations in lung cancer. NPJ Precis. Oncol. 7, 52 (2023).
Zhang, J. et al. Comprehensive characterization of the tumor microenvironment for assessing immunotherapy outcome in patients with head and neck squamous cell carcinoma. Aging 12, 22509–22526 (2020).
Huo, M. et al. Tumor microenvironment characterization in head and neck cancer identifies prognostic and immunotherapeutically relevant gene signatures. Sci. Rep. 10, 11163 (2020).
Şenbabaoğlu, Y. et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 17, 231 (2016).
Sitkovsky, M. V. Lessons from the A2A adenosine receptor antagonist–enabled tumor regression and survival in patients with treatment-refractory renal cell cancer. Cancer Discov. 10, 16–19 (2020).
Bedoya, D., Dutoit, V. & Migliorini, D. Allogeneic CAR T cells: an alternative to overcome callenges of CAR T cell therapy in glioblastoma. Front. Immunol. 12, 640082 (2021).
Jacobson, K. A., Tosh, D. K., Jain, S. & Gao, Z.-G. Historical and current adenosine receptor agonists in preclinical and clinical development. Front Cell Neurosci. 13, 124 (2019).
Fong, L. et al. Adenosine 2A receptor blockade as an immunotherapy for treatment-refractory renal cell cancer. Cancer Discov. 10, 40–53 (2020).
Jin, D. et al. CD73 on tumor cells impairs antitumor T-cell responses: a novel mechanism of tumor-induced immune suppression. Cancer Res. 70, 2245–2255 (2010).
Brownlie, R. J., Wright, D., Zamoyska, R. & Salmond, R. J. Deletion of PTPN22 improves effector and memory CD8+ T cell responses to tumors. JCI Insight 5, e127847 (2019).
Wei, J. et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476 (2019).
Guo, X. et al. CBLB ablation with CRISPR/Cas9 enhances cytotoxicity of human placental stem cell-derived NK cells for cancer immunotherapy. J. Immunother. Cancer 9, e001975 (2021).
Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020).
Blache, U., Popp, G., Dünkel, A., Koehl, U. & Fricke, S. Potential solutions for manufacture of CAR T cells in cancer immunotherapy. Nat. Commun. 13, 5225 (2022).
Sun, W., Jiang, Z., Jiang, W. & Yang, R. Universal chimeric antigen receptor T cell therapy—the future of cell therapy: a review providing clinical evidence. Cancer Treat. Res. Commun. 33, 100638 (2022).
Caldwell, K. J., Gottschalk, S. & Talleur, A. C. Allogeneic CAR cell therapy—more than a pipe dream. Front. Immunol. 11, 618427 (2021).
Chuprin, J. et al. Humanized mouse models for immuno-oncology research. Nat. Rev. Clin. Oncol. 20, 192–206 (2023).
Jo, S. et al. Endowing universal CAR T-cell with immune-evasive properties using TALEN-gene editing. Nat. Commun. 13, 3453 (2022).
Maldini, C. R. et al. Dual CD4-based CAR T cells with distinct costimulatory domains mitigate HIV pathogenesis in vivo. Nat. Med. 26, 1776–1787 (2020).
Chester, C., Fritsch, K. & Kohrt, H. E. Natural killer cell immunomodulation: targeting activating, inhibitory, and co-stimulatory receptor signaling for cancer immunotherapy. Front. Immunol. 6, 601 (2015).
Acknowledgements
We would like to thank the members of the Beam Therapeutics Immunology team and the Hatfield Laboratory at Northeastern University that contributed to this collective work. Special acknowledgement for J. Decker and the Beam sequencing core, as well as R. Manoukian and the Beam imaging core for their technical expertise. B. Lutz for her expert guidance with spatial transcriptomics at Beam Therapeutics, and BioRender for schematic design. These studies were completed with collaborative research funding from Beam Therapeutics.
Author information
Authors and Affiliations
Contributions
R.M., C.M., and S.H. conceptualized the project and designed the experiments. R.M., S.H., R.C., N.R., K.D., S.C., B.C., Y.X., A.M., H.S., J.R., F.M., A.C., and G.C. performed experiments, analyzed results and interpreted data. R.M., C.M., and S.H. wrote the manuscript. M.V.S. provided institutional knowledge and infrastructure at the New England Inflammation and Tissue Protection Institute, provided constructive criticism, and participated in manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
R.M., A.M., H.S., J.R., F.M., A.C., C.M., and G.C. were employees of Beam Therapeutics when the work was conducted and are shareholders in the company. Beam Therapeutics has filed patent applications on this work. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sebastian Kobold and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Murray, R., Chowdhury, M.R., Botticello-Romero, N.R. et al. Multiplex gene-editing strategy to engineer allogeneic EGFR-targeting CAR T-cells with improved efficacy against solid tumors. Nat Commun 16, 11593 (2025). https://doi.org/10.1038/s41467-025-66737-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66737-1