Abstract
The human ClpXP complex (hClpXP) orchestrates mitochondrial protein quality control through targeted degradation of misfolded and unnecessary proteins. While bacterial ClpXP systems are well characterized, the assembly and regulation of human ClpXP remain poorly understood. In this study, we elucidate the complete assembly pathway of hClpXP through high-resolution cryo-electron microscopy (cryo-EM) structures. Our findings confirm that hClpP exists as a single-ring heptamer in isolation and reveal a previously undocumented initial assembly complex in which hexameric hClpX first engages with heptameric hClpP. We further demonstrate how this interaction drives substantial conformational rearrangements that facilitate the formation of tetradecameric hClpP within the fully assembled complex. Notably, we characterize a unique eukaryotic sequence in hClpX, termed the E-loop, which plays a critical role in stabilizing hexamer assembly and maintaining ATPase activity. Additionally, we show that peptide binding at the hClpP active site triggers further structural changes essential for achieving full proteolytic competence. Together, these structures provide unprecedented mechanistic insights into the stepwise assembly and activation of hClpXP, significantly advancing our understanding of this essential mitochondrial protein degradation machinery.
Similar content being viewed by others
Introduction
Mitochondria are busy signaling hubs where a multitude of cellular processes converge, including oxidative phosphorylation, calcium homeostasis, redox signaling, and apoptosis1,2,3. Maintaining internal protein homeostasis (proteostasis) involves a sophisticated and tightly controlled protein quality control network that ensures proper import, folding, and turnover of proteins4. In humans, disruptions in this network are associated with aging and can lead to a wide range of diseases, including cancer, neurological disorders, diabetes, and skin disorders5. The ClpXP complex is one of the key ATP-dependent unfoldase-proteases involved in this protein quality control network, specifically targeting and degrading misfolded, damaged, or surplus proteins in the matrix of mitochondria. While all other mitochondrial proteases are encoded by a single gene, ClpXP is an assembly of two distinct proteins: ClpX, an ATPase associated with diverse cellular activities (AAA+) with unfoldase activity, and ClpP, a conserved serine protease. Together, these components work in concert to effectively degrade damaged proteins, as well as regulating essential cellular functions, including heme biosynthesis6,7, mitochondrial DNA nucleoid distribution8, and reactive oxygen species (ROS) production9.
Despite extensive biochemical and structural characterization of unicellular ClpXP systems10,11,12,13,14, many of the molecular details underlying human ClpXP (hClpXP) assembly and proteolytic activation are uncharacterized. ClpXP systems are broadly conserved across all forms of life, but the specific requirements of eukaryotic proteostasis have driven ClpX to evolve distinct regulatory components. For example, hClpX features a unique eukaryotic-specific insertion within its ATPase domain, previously termed the E-domain15 (Supplementary Figs. 1a, 2a). The function of this insertion was unknown, but its positioning at the distal end of hClpX suggests a potential role in substrate recognition16 (Supplementary Fig. 1d). Additionally, hClpP possesses an additional 28 amino acids at its C-terminus, termed the C-terminal extension (Supplementary Fig. 2b). Deleting this extension promotes tighter interaction between hClpP and hClpX17, but the precise underlying mechanism remains unclear.
The hClpP protease is also evolutionarily divergent. Bacterial ClpP assembles as a tetradecamer composed of two stacked heptameric rings that enclose 14 proteolytic active sites within the chamber18, but this is not necessarily the case for hClpP. Some studies suggest that hClpP exists as a tetradecamer19,20,21, whereas others indicate that ClpP assembles as a stable heptamer and only forms a tetradecamer upon binding with hClpX17,22. Studies employing small molecule activators of hClpP have provided some insights into its mechanism of activation21,23,24, but in the absence of a direct structural comparison between apo hClpP and hClpX-bound hClpP, the precise mechanism of hClpP activation remains elusive.
In this study, we use cryo-electron microscopy (cryo-EM) to investigate the assembly pathway and activation mechanisms of the hClpXP complex, revealing previously unobserved assemblies that distinguish hClpXP from its bacterial homologs. Our studies indicate that unbound hClpP natively adopts a single-ring asymmetric heptamer, which interacts with hClpX to form an initial assembly complex. Interaction with substrate drives formation of the fully assembled hClpXP complex, which we show transitions from a proteolytically inactive conformation to a proteolytically competent state when the peptide interacts with the protease site. Our hClpXP structures also reveal the architecture of the E-domain (hereafter referred to as the E-loop), which we show to be involved in nucleotide sensing and hClpX hexamer stabilization. Together, these findings highlight the distinct allosteric mechanisms that have evolved for hClpXP to meet the complex demands of the mitochondrial environment.
Results
Purification of the hClpXP sample for cryo-EM structural studies
We first aimed to compare the structure of the hClpXP complex to the numerous prior structures of the unicellular homologs to identify evolutionarily conserved or divergent features. However, generating a stable hClpXP complex for cryo-EM studies required substantial optimization. To structurally stabilize the hClpX construct for high-resolution structure determination, we removed the N-terminal zinc-binding domain16,25 and the C-terminal extension, both predicted to be disordered by AlphaFold (version 2) (Supplementary Fig. 1c, d). We also introduced a Walker B motif mutation (E359A), previously shown in bacterial ClpX and other AAA+ translocases, to slow ATP hydrolysis and promote hexamer stability11,12,13. This optimized construct is hereafter referred to as hClpXI146-∆CTE-E359A.
While the zinc-binding domain has been characterized in bacterial ClpX and shown not to affect the structural conformation of the assembly14,26, the functional relevance of the C-terminal extension in hClpX remains unclear. To investigate its potential role in ATPase activity, we performed an NADH-coupled ATPase assay comparing hClpXI146 variants with or without the C-terminal extension. Our data indicate that the C-terminal extension does not influence ATP binding affinity but decreases ATPase activity and the degradation of unfolded substrates in the presence of hClpP (Supplementary Fig. 3a,b). While removal of the hClpX C-terminal extension reduces its enzymatic activity and helps stabilize the oligomer for cryo-EM characterization, the underlying mechanisms by which this modification alters activity remain to be elucidated through future studies.
Although our construct optimization improved the stability and yield of hClpX, we did not observe formation of stable co-assembly with wild-type hClpP (Supplementary Fig. 4a). Thus, we next sought to enhance complex formation through hClpP construct optimization. Previous studies showed that removing the C-terminal extension (CTE)17,19 and introducing a proteolytically inactivating S153A mutation13 stabilize ClpXP complex assembly. Accordingly, we generated an hClpP∆CTE-S153A construct, which exhibited improved binding to hClpX, as observed by negative stain EM (Supplementary Figs. 4a, b, 5c). Titration experiments comparing wild-type hClpP and hClpP∆CTE suggested that both proteins bind and activate hClpX similarly, but hClpP∆CTE exhibits a modestly higher substrate turnover rate, indicating that the C-terminal extension negatively regulates hClpP activity (Supplementary Fig. 3c). To further stabilize the complex for high-resolution single-particle cryo-EM analysis, we applied disuccinimidyl suberate (DSS) crosslinking27, which substantially increased the number of intact hClpXP particles (Supplementary Fig. 4c). Notably, DSS does not crosslink hClpP alone (Supplementary Fig. 4e). For simplicity, we refer to this optimized, crosslinked complex (hClpXI146-∆CTE-E359A/hClpP∆CTE-S153A/DSS-X-linked) as hClpXPX-linked hereafter.
Cryo-EM structure of hClpXP reveals a conserved substrate translocation mechanism by hClpX
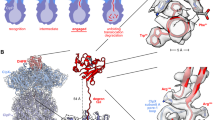
We determined the structure of the hClpXPX-linked complex at an overall resolution of ~3 Å (Fig. 1a, Supplementary Fig. 6 and Supplementary Table 1). The reconstruction is generally reminiscent of previously determined bacterial ClpXP structures, including a co-purified peptide bound within the central pore of the hClpX unfoldase, a common feature of recombinantly expressed AAA+ proteins containing a Walker B mutation12,28,29. The hexameric assembly of hClpX adopts a shallow spiral conformation consistent with bacterial ClpX and other AAA+ unfoldases11,12,14, and we labeled the subunits of hClpX in alphabetical order (A, B, C, D, E, and F) with chain A at the uppermost position of the spiral and continuing downwards, and chain F identified as the displaced “seam” subunit disengaged from the peptide substrate (Fig. 1b). Also consistent with other AAA+ unfoldases was density corresponding to an ATP nucleotide with a coordinated magnesium cofactor observed in the binding pockets of chains A through D, where the ATP β- and γ- phosphates are stabilized by interactions with K299 (Walker A motif) and R562 (sensor-II) of the same subunit, as well as R499 (arginine finger) from the adjacent protomer (Supplementary Fig. 7). The nucleotide density in lowest subunit of the spiral, chain E, was representative of ADP and lacked observable magnesium density (Supplementary Fig. 7b). Further, the arginine finger from the neighboring F subunit is shifted away to produce a more open nucleotide-binding pocket. No nucleotide density was resolved in the seam subunit F.
a Cryo-EM map of the hClpXPX-linked complex with a substrate peptide bound. Each subunit of the hClpX homo-hexamer is colored distinctly. b Atomic structural model of hClpXP. The substrate peptide within the hClpX central pore is modeled as poly-alanine. The hClpX subunits are labeled A-F with the nucleotide states indicated. Chain F, the seam subunit, is disengaged from the substrate. c Top view of the hClpP map with the hClpX LGF loops (shown as helices) occupying the hydrophobic pocket. An empty LGF loop binding pocket is located between chains D and E. d Cut-away view of the hClpXPX-linked 3D reconstruction highlighting several features: pore-1 loops, nucleotides, LGF loops, the hClpP N-terminal β-hairpins, and the active site. The inset shows Y327 from the pore-1 loops forming a spiral staircase around the substrate peptide backbone.
As previously observed in other AAA+ proteins, the nucleotide state of each protomer correlates with the extent of the subunit’s interaction with the incoming substrate within the central ATPase channel30,31,32. The pore-1 loop of each nucleotide-bound hClpX subunit in the right-handed spiral engages the translocating peptide via a conserved GYVG sequence, so that the aromatic-hydrophobic motif intercalates into the substrate backbone (Fig. 1d). This organization maintains grip on the incoming polypeptide to facilitate translocation towards the peptidase chamber using a putative hand-over-hand mechanism11,12,31,32,33.
ClpX attaches to ClpP via loops containing a conserved Ile/Leu-Gly-Phe (IGF or LGF) motif, which binds to the hydrophobic pockets formed at the inter-protomer ClpP interface11. Our hClpXP structure shows how F441 of the hClpX LGF loop forms conserved hydrophobic interactions with residues Y118, W146, Y138, and L104 in the hydrophobic pocket of hClpP (Supplementary Fig. 8). Prior cryo-EM studies of bacterial ClpXP systems have shown that one of the hydrophobic binding pockets in ClpP is empty due to the six-to-seven protomer mismatch between the unfoldase and protease. However, this empty pocket is sometimes situated between the clefts occupied by the IGF loops of ClpX subunits E and F11,12,14 while in others it is located between subunits D and E34,35. It was proposed that the ATP hydrolysis-driven motions of the ATPase subunits coordinate repositioning of the ClpX I/LGF loops as well as the stability of the N-terminal β-hairpins comprising the ClpP axial channel to facilitate substrate translocation11. However, recent studies show that the crosslinked ClpAP complex remains capable of degrading unstructured substrates, suggesting that a rotational motion may not be required for proteolysis36. Our crosslinked hClpXP abolished FITC-casein degradation activity, which may result from impaired substrate unfolding and translocation caused by the promiscuity of the DSS crosslinker (Supplementary Fig. 9a). The pore-2 loops of ClpX are also thought to coordinate substrate translocation through interactions with the axial ClpP β-hairpins that vary based on ClpX nucleotide state11,12,13,37. In our hClpXPX-linked reconstruction, we observe an empty pocket between hClpX chains D and E (Fig. 1c), and although our pore-2 loops are not resolved, we observe that the pore-2-adjacent residue D374 from hClpX subunit E interacts with R68 at the top of a hClpP β-hairpin (Supplementary Fig. 10). This observation is similar to the Neisseria meningitides ClpXP structure, where only a single ClpX protomer (subunit E) interacts with the arginine residue from ClpP β-hairpin at the same position11, suggesting a conserved ClpXP communication and substrate translocation mechanism.
E-domain of hClpX senses nucleotide states and promotes hexamer formation
A notable distinguishing feature of our hClpXPX-linked structure is the presence of the E-loop, a eukaryotic-specific insertion of unknown function within the AAA+ module (Fig. 2a). Within the hClpX hexamer, the E-loop extends from the large AAA+ domain of one subunit over the adjacent nucleotide binding pocket and interacts with the neighboring subunit at the interface of the large and small AAA+ domains. Despite these numerous interactions, the E-loop is generally flexible and is only partially resolved. The interface between the large and small AAA+ domains is distinct for each subunit within translocating AAA+ unfoldases due to the staircase organization of the hexamer30,31,32, thereby impacting E-loop interactions. Accordingly, the resolvability of the E-loop density varies among different subunits, indicating that the register of the subunit within the hexamer impacts the E-loop stability (Supplementary Fig. 11b). The least-resolved E-loop is that of the seam subunit, chain F, which extends over the ADP-bound pocket of chain E. Proceeding from the top of the ATPase spiral to the bottom, the E-loop arching across each of the preceding nucleotide binding pockets becomes increasingly ordered, with the E-loop extending over the lower-most ATP pocket being the best-resolved of the loops. Given the location of the E-loop, and that E285 within the loop is within hydrogen bonding distance of both the nucleotide ribose and R565 in the binding pocket (Fig. 2a and Supplementary Fig. 11a, b), we surmised that the E-loop may play a role in the ATPase mechanochemical cycle.
a The hClpX hexamer is shown as a surface representation, highlighting the E-loop (shown as licorice/ovals style) of one hClpX subunit extends away to interact with the neighboring subunit. The inset illustrates the network of interactions between the E-loop of chain E and chain D, emphasizing that E285 from chain E is within hydrogen bonding distance of the ATP nucleotide. b NADH/ATP-coupled ATPase assays were performed. The negative controls include reactions without enzymes and the Walker B mutant—hClpXE395A. Removal of the E-loop from hClpX caused a 2-fold decrease in hClpX ATPase activity. The E285A mutation has a similar deleterious effect on hClpX ATPase activity as the E-loop deletion. Data are presented as bar graphs showing mean values with error bars representing standard deviation from three independent replicates. c Removal of the E-loop from hClpX or introduction of the E285A mutation resulted in a 10X reduction in the FITC-casein degradation activity of the hClpXP complex. Data are presented as bar graphs showing mean values with error bars showing standard deviation from three independent replicates. Source data are provided as a Source Data file.
To test this hypothesis, we removed the E-loop (residues 230-304) from hClpX (hClpXdE) and evaluated its impact on ATPase activity and substrate degradation. Deletion of the E-loop decreased ATP affinity by 4-fold and decreased ATPase activity by more than half (Fig. 2b, Supplementary Fig. 3a, b). Removing the E-loop also impaired the ability of hClpXP to degrade substrates, as indicated by a 11-fold reduction in FITC-casein degradation (Fig. 2c). Negative stain EM analysis of this truncation mutant revealed an inability of the hClpXdE to form hexamers, instead assembling into higher-order oligomers, including heptameric, octameric, and nonameric assemblies in the absence of exogenous ATP (Supplementary Fig. 11 c, d). Although the addition of ATP and MgCl2 facilitated the formation of some loosely organized hexamers, we also observed heptamers and unclosed ring assemblies. Given that the ClpXdE construct shows impaired substrate degradation (Supplementary Fig. 3b), these data collectively suggest that the E-loop is critical for organizing hClpX into a hexamer that is competent to engage and translocate substrate. Notably, site-specifically mutating the nucleotide-interacting residue E285 within the E-loop to alanine was as deleterious to hClpXP enzymatic activities as removal of the E-loop (Fig. 2b,c), but the mutation did not negatively affect hClpX hexamer formation (Supplementary Fig. 11e). Given our observed four-fold decrease in ATP binding affinity of the E285A mutation (equivalent to the hClpXdE construct, Supplementary Fig. 3a, b) we posit that the E285 residue plays a key role in hClpX ATPase and substrate translocation activities by sensing and communicating changes in nucleotide states between neighboring subunits.
Similar to unicellular ClpX systems, ATP binding is important for hClpX to form hexamers that are competent for substrate translocation16 (Supplementary Fig. 11c). However, our data indicate that the E-loop serves as a critical structural feature required for proper hexameric assembly of hClpX and effective ATP-driven substrate translocation. Given that the E-loop is specific to eukaryotes, these findings suggest an evolutionary adaptation of hClpX to incorporate a more complex set of structural requirements for stabilizing hexamer formation and sensing nucleotide states during substrate translocation in mitochondria.
hClpP assembles as an asymmetric heptamer whose C-terminal extension negatively regulates its activity
Having probed the evolutionarily distinct features of the assembled hClpXP complex, we next aimed to investigate the mechanism of hClpXP assembly. It is well-established that bacterial ClpP proteins form stable tetradecamers composed of two stacked heptamers, but there are conflicting results regarding whether hClpP assembles as a heptamer or tetradecamer in the absence of hClpX in solution17,20,21,22. When we expressed and purified our wild-type hClpP construct, we observed that it eluted from size exclusion chromatography at a mass corresponding to a heptameric assembly (Supplementary Fig. 5a). We further confirmed that hClpP predominantly exists as a heptamer in solution using mass photometry and negative-stain EM (Supplementary Fig. 5b, c) and next aimed to determine a high-resolution cryo-EM structure. We observed that the apo hClpP heptamer exhibits a strong preferred axial orientation when vitrified on cryo-EM grids (Supplementary Fig. 12a, b), which was alleviated by the addition of fluorinated fos-choline-8 detergent (Supplementary Fig. 12c). Although the majority of imaged hClpP oligomers were heptameric according to 2D classification, there was a minor population of tetradecameric hClpP, possibly induced by the high concentration of hClpP applied to the cryo-EM grid (15 mg/ml). Our 2D averages also showed that the tetradecamers were not well-resolved (circled in red in Supplementary Fig. 12d), indicative of the flexibility of transient inter-heptamer interactions. These data indicate that the hClpP heptamers may not readily form stable tetradecamers in cellular conditions and that docking of the hClpX LGF loop in the ClpP hydrophobic pocket plays a role in promoting assembly of the hClpP tetradecamer.
Despite challenges from reduced apparent particle concentration and ice thickness, further processing of our fluorinated fos-choline-8 dataset allowed us to obtain a heptameric hClpP reconstruction with a reported overall resolution of ~3.5 Å (Supplementary Fig. 12 and Supplementary Table 1), which was sufficient to generate an atomic model (Fig. 3a, b). Surprisingly, we found that the hClpP heptamer is not symmetric as expected, but rather assembles into an asymmetric oligomer in the absence of hClpX (Supplementary Fig. 13a, c). A salient feature of the asymmetric heptamer is the ordering of three of the N-terminal loops into β-hairpins, forming a partial wall around the axial hClpP pore, while these loops from the other four subunits are disordered (Fig. 3b, Supplementary Fig. 14). We surmise that this distinct asymmetric organization of the N-terminal loops forming a partial wall at the ClpP pore impacts hClpX recruitment, although further biophysical studies will be required to elucidate the possible regulatory mechanism of this unexpected structural feature.
a 3D reconstruction and b atomic model of C1 refined apo hClpP. The N-terminal loops of three hClpP subunits form well-ordered β-hairpins and are pointed upwards, while the N-terminal loops of the remaining subunits are disordered. The inset highlighted with a magenta box shows one of the three well-ordered N-terminal β-hairpins c Overlay of the apo hClpP (shown in burlywood) and the top part of the hClpX-bound hClpP tetradecamer (shown in grey). The insets highlight conformational changes in three different regions of hClpP upon binding to hClpX: (1) inward movement of the handle region; (2) a downward movement of the C-terminal extension; (3) outward movement of the N-terminal loops from the central pore as well as the ordering of the N-terminal loops in the subunits that originally had disordered loops.
hClpP contains an unstructured 28-residue C-terminal extension (Supplementary Fig. 2b) that is unresolved in our hClpP reconstructions. We wondered how the removal of this disordered region from ClpP increases affinity for ClpX and increases substrate degradation17 (Supplementary Fig. 3c, 4b, 15c). Prior studies of the C-terminal extension in ClpP homologs from Mycobacterium smegmatis38 and Listeria monocyotenes13 suggest that this segment may be positioned to block the interaction between the ClpX loops and their respective hydrophobic binding pockets in ClpP. In our apo hClpP structure, we noted that the two proline residues preceding the C-terminal extension are well-resolved, stably positioned atop the LGF loop binding pocket (Supplementary Fig. 15 a). The location of these proline residues confirms that the C-terminal extensions are located in the vicinity of the hClpP LGF loop binding pockets, where they could potentially shield the pockets from hClpX interaction. Further, these prolines residues are positioned so that they would sterically hinder the binding of LGF loops (Supplementary Fig. 15a). We noted that these residues are shifted away from the LGF binding pocket in our hClpXP structure to accommodate the interactions necessary to form the heterocomplex (Supplementary Fig. 15b). These structural and biochemical observations suggest that the C-terminal extension negatively regulates the enzymatic activity of hClpXP.
hClpP switches from an asymmetric heptamer to a symmetric tetradecamer upon interaction with hClpX
In addition to displacing the C-terminal extension of hClpP, hClpX binding is associated with a large-scale reorganization of the hClpP heptamer. Superimposing our structures of the apo hClpP heptamer and one heptameric ring of the hClpX-bound hClpP reveals several notable conformational changes (Fig. 3c). Upon hClpX binding, the four N-terminal loops that were disordered in our asymmetric heptameric hClpP structure adopt the canonical β-hairpin fold, concurrent with expansion of the axial pore to allow unfolded substrates to access the proteolytic chamber.
Notably, we also observe a ~15° axial rotation of the subunits away from the hClpX hexamer as the hClpP heptamer transitions from an asymmetric heptamer to a symmetric assembly (Fig. 3c). The rotation of the hClpP subunits upon hClpX binding results in an inward movement of the handle regions to facilitate critical interactions at the hClpP-hClpP interface, promoting a stable tetradecamer conformation (Supplementary Fig. 16a). In the hClpX-bound hClpP tetradecameric structure, the conserved oligomerization sensors E225 and R226 from one ClpP heptamer are positioned in proximity to form salt bridges with R226 and E225 from the opposing ClpP heptamer (Supplementary Fig. 16a, b). When we introduced the E225A and R226A mutations to hClpP, these mutations destabilize, but do not entirely abolish, hClpP tetradecamer formation in the presence of hClpX, as assessed by negative stain EM (Supplementary Fig. 4d). However, these mutations completely abolished hClpXP’s ability to degrade FITC-casein substrates, indicating that the oligomeric sensors are essential for the enzymatic activity of hClpP (Supplementary Fig. 9b). We also observe several other residues in the handle region, including E196, K202, and Y206, that interact with residues from the opposing subunit to maintain the tetradacameric state of hClpP (Supplementary Fig. 16b). Additionally, the catalytic residue D227 contributes to stabilizing the tetradecamer by forming a salt bridge with Q194. These interactions were also recently observed in a tetradecameric cryoEM structure of hClpP determined while this manuscript was under preparation39.
hClpXP forms a substrate-free initial assembly complex
To confirm that our substrate-bound ClpXP structure was not impacted by chemical crosslinking and construct engineering, we set out to determine a structure of the uncrosslinked hClpXP complex using the wild-type proteins (Supplementary Fig. 1b, 17, Supplementary Table 1). Although our negative stain images did not show assembled complexes, our observed activity of the purified components indicated that complex formation was nonetheless occurring. We posited that the low (nanomolar) concentrations of protein used for our negative staining may be promoting complex dissociation. Thus, we incubated hClpX and hClpP together at high concentration of 3 mg/ml in the presence of ATPγS and froze grids of this sample for cryoEM imaging. Although hClpX and hClpP were mostly dissociated, a large dataset of cryoEM micrographs yielded sufficient hClpXP complexes to obtain moderate-resolution maps of the hClpXP complex, with hClpX and hClpP locally refined to resolutions of ~4.5 Å and ~3.5 Å, respectively (Supplementary Fig. 17, Supplementary Table 1). We generated a model of uncrosslinked hClpXP by rigid-body fitting the crosslinked hClpX and hClpP models into the composite map of uncrosslinked hClpX and hClpP, followed by Phenix real-space refinement (Supplementary Fig. 18a). An overlay of the uncrosslinked and crosslinked models revealed an RMSD of 0.5 Å, indicating that the two structures are nearly identical within the limits of the map resolution. While the hClpP tetradecamer shares a consistent conformation, we observe subtle shifts in hClpX subunits (Supplementary Fig. 18b). Overall, this analysis suggests that the cross-linked hClpXP structures closely represent the uncross-linked full-length hClpXP structure.
During our analysis of this dataset, we were surprised to identify a population of particles corresponding to a single hClpX hexamer bound to a single hClpP heptamer (Fig. 4a, Supplementary Fig. 17). This organization is distinct from all previously characterized ClpXP systems, where ClpX exclusively associates with tetradecameric ClpP. This organization was absent in our crosslinked hClpXP dataset, suggesting that it represents an intermediate in the hClpXP-substrate assembly pathway. Notably, the ATPase domains of hClpX in this stacked hClpX-hClpP complex adopt a more open configuration compared to substrate-bound structure, and the central pore is devoid of substrate (Fig. 4d). Further, the arrangement of individual hClpX subunits is relatively flat, a notable departure from the conventional staircase conformation used by hClpX and other AAA+ ATPases to facilitate substrate peptide unfolding and translocation (Fig. 4d). The heptameric hClpP in this identified hClpXP complex exhibits slight asymmetry, representing an intermediate state between asymmetric hClpP heptamer and the symmetric hClpX bound hClpP tetradecamer (Supplementary Fig.13b). These characteristics indicate this hClpXP complex represents an early stage in the assembly process, poised for subsequent substrate engagement and nucleotide exchange. Based on these observations, we term this structure the “initial assembly” complex (hClpXPIA).
a atomic model of hClpXPIA and hClpXPX-linked docked into low-resolution cryo-EM maps of hClpXP initial assembly and uncrosslinked hClpXP complex, respectively. b The LGF loop from hClpX chain E in the intermediate state is situated in the hydrophobic pocket formed between hClpP subunits I and J, while the loop shifts to the neighboring pocket formed between subunits I and H in the hClpXP complex. c The ATPase and protease rings rotate approximately 10 degrees relative to one another during the transition from the initial assembly state to the fully assembled complex. d The ATPase ring of the hClpXPIA state adopts a wider, more open conformation that does not contain substrate, compared to the substrate-bound hClpX. Further, the staircase organization of the ATPase domains is not as prominent in hClpXPIA, with substrate binding inducing chains F and A move slightly upward, and chains D and E to move slightly downward to form the canonical substrate-bound staircase.
Comparison of the substrate-free hClpXPIA complex with the substrate-bound hClpXP structure enables us to define the structural rearrangements associated with substrate engagement. One of the most striking rearrangements is a ~10 degree rotation of the hClpX relative to hClpP upon substrate binding (Fig. 4c), which prompted us to examine the interactions between the hClpX LGF loops and the hydrophobic pockets of the hClpP heptamer. During this inter-oligomeric rotation, the hClpX LGF loops remain positioned in their respective hClpP pockets with the exception of the LGF loop from the “E” subunit, which shifts from the binding pocket formed by hClpP chains I and J to the neighboring pocket (Fig. 4a,b). It is proposed that the ClpX I/LGF loops dynamically engage and disengage from one ClpP binding pocket to the next during substrate processing, and the clockwise movement of this LGF loop is consistent with the directionality associated with hClpX enzymatic activity11. This repositioning of the LGF loop during the transition from initial assembly complex to fully assembled complex suggests a mechanistic link between substrate engagement, ATPase activation, and formation of the hClpP tetradecamer.
Our hClpXPX-linked structure showed that the “E” subunit of hClpX interacts closely with an N-terminal β-hairpin of hClpP (Supplementary Fig. 10), and our isolated hClpP heptamer structure also presented an asymmetric arrangement of N-terminal β-hairpins, with some more stably directed upward (Fig. 3b, Supplementary Fig. 14). This asymmetric arrangement of hClpP β-hairpins may play a role in the initial recruitment of hClpX by providing recognition elements that establish a preferred asymmetric distribution of LGF loops in the hydrophobic hClpP binding pockets. Such a directed assembly mechanism could establish an energetically favorable substrate recruitment complex, poised to reposition the first LGF loop to the next binding pocket upon substrate engagement as the first step of substrate processing.
Substrate binding is required for hClpP activation
Prior structural studies of bacterial ClpP revealed three distinct states of the tetradecameric assembly, defined as extended, compact, and compressed18. The extended state is considered to be the proteolytically active conformation, whereas the compact and compressed states are regarded as transition states posited to facilitate peptide exit10,18. However, in seeming contradiction to these results from unicellular systems, the crystal structure of apo hClpP revealed a tetradecamer in an extended state19, while crystal structures of hClpP bound to small molecule activators showed tetradecamers in a compact state21,23,24,40. While it is possible that hClpP has evolved a fundamentally distinct proteolytic mechanism, it is also conceivable that these structural conformations were not representative of their corresponding proteolytic states due to crystallization conditions. We thus aimed to clarify these structure-activity relationships with cryo-EM analyses.
Prior single particle cryo-EM studies of bacterial ClpXP complexes indicate that ClpX binding promotes ClpP to adopt the proteolytically competent extended conformation10. Given the level of structural conservation observed between bacterial ClpXP and our hClpXP complexes, we expected this feature to be preserved. Intriguingly, our hClpXP complex, despite being poised for degradation with a substrate peptide in the hClpX central pore, contains hClpP in a compact conformation that is presumed to be proteolytically inactive (Fig. 5a). In this inactive conformation, the catalytic histidine is orientated away from the nucleophilic serine such that the necessary hydrogen bond network required for catalysis cannot be achieved41.
a Atomic models of the compact hClpX-bound hClpP and b the extended bortezomib-bound hClpP tetradecamers are shown as secondary structure cartoons, with insets highlighting the active site conformations represented as ribbons with salient residues as sticks. To determine our hClpX-bound hClpP structure, S153 was mutated to alanine, but is shown in (a) as a serine for visualization purposes. In the proteolytically inactive conformation, the imidazole of H178 is positioned too far from S153 for peptide hydrolysis. Conversely, in the bortezomib-bound hClpP, the active site adopts a proteolytically active conformation, with H178 shifting towards S153 in proximity for catalysis.
We previously observed an analogous discrepancy between proteolytically competent conformations in our comparison of the bacterial and human Lon protease30. Substrate interaction with the AAA+ module of bacterial Lon promotes rearrangement of the protease component to an active conformation but is insufficient for protease activation in human LONP1. We previously showed that the proteolytically active conformation of LONP1 could be obtained by introducing the peptide mimetic bortezomib30, and we surmised that hClpP activation may similarly require substrate peptide interaction with the peptidase to promote the active/extended conformation. To test this, we used negative staining and found that covalent binding of bortezomib indeed promotes heptameric hClpPs to assemble into tetradecamers (Supplementary Fig. 4f). We next determined a cryo-EM structure of bortezomib-bound hClpP to an overall resolution of ~2.3 Å, which was sufficient to resolve the covalent linkage between bortezomib and the catalytic S153 residue at the protease site (Fig. 5b and Supplementary Fig.19, Supplementary Table 1). Notably, bortezomib binding induced hClpP to adopt an active, extended tetradecamer, confirming our hypothesis (Fig. 5b).
The similarity between our bortezomib-bound hClpP structure and the previous X-ray crystal structure of apo hClpP may be explained by the presence of an unknown density linked to the catalytic serine residue in the crystal structure, which may have contributed to adoption of the active conformation19. The bortezomib density in our structure also resembles that of a putative tripeptide identified in the protease active site of Thermus thermophilus ClpP (TtClpP), which also crystallized in the active conformation42. Analogous to what has been described for Thermus thermophilus ClpP (TtClpP)42 and Mycobacterium tuberculosis ClpP1ClpP243 (MtClp1ClpP2), bortezomib mimics peptide binding at the active sites, interacting with the handle region to stabilize an extended/active conformation. Upon bortezomib binding, the catalytic H178 residue shifts towards S153 while the neighboring Q179 residue shifts equatorially to establish an interaction network stabilizing the extended conformation (Supplementary Fig. 16d,e,f). The previously described “oligomerization sensors10,” corresponding to E225 and R226 in hClpP, contribute to the stabilization of the handle region in the extended state by interacting with N187 and T189 residues, which are located in a loop that is disordered in the compact state. These additional structural observations—coupled with our degradation assay on the E225A and R226A constructs—further confirm the conserved role of these residues in hClpP (Supplementary Fig. 9b, 16f).
Collectively, our structural data clarify the interpretation of prior crystallographic studies and demonstrate a role for substrate peptide interaction with the proteolytic active site in protease activation in various species. The conformational impact of substrate interaction is also consistent with findings from another cryo-EM study of hClpP that was performed while this manuscript was under preparation39.
Discussion
ClpXP is a conserved protease that is critical for proteostasis in organisms belonging to all the kingdoms of life. While previous studies have shed light on the mechanisms that regulate substrate processing in bacterial ClpXP, it was unclear if human ClpXP employed these same mechanisms in the mitochondria. The specific proteostatic demands of the mitochondrial environment are distinct from those of unicellular organisms, and our findings reveal both conserved and evolutionarily distinct regulatory features that govern hClpXP function. While hClpXP shares a similar mechanism of substrate unfolding and degradation to its bacterial homologs, the assembly and activation of this mitochondrial protease are unique, involving a multi-tiered regulatory mechanism.
Among the most striking of our observations is that, unlike previously characterized ClpP homologs that assemble as a symmetric tetradecamer, hClpP assembles as an asymmetric heptamer. In most bacterial species, such as N. meningitidis and E. coli, the ClpP protease sites are sequestered within the tetradecameric chamber of ClpP, where flexible N-terminal loops restrict substrate access until ClpX binds, whereupon N-terminal gates of ClpP allow unfolded substrate peptides to enter and be degraded44,45. In contrast, the proteolytic active sites of the hClpP heptamer are exposed to the mitochondrial matrix. However, the catalytic residues of the protease in the heptamer are positioned such that they are incompatible for proteolysis, preventing non-specific degradation of proteins, representing the first layer of regulation in a multi-tier regulatory system involved in hClpXP-mediated substrate degradation. Both hClpX-induced conformational rearrangements, as well as interactions with another heptamer, are necessary to achieve proteolytic competence. The presence of a heptameric ClpP was also observed in M. tuberculosis (Mt), where MtClpP forms a heterocomplex consisting of MtClpP1 and MtClpP2. Each MtClpP is predominantly heptameric in isolation, and both forms are required to assemble tetradecamers43. Similar to our hClpP, MtClpP1 and MtClpP2 heptamers are catalytically inactive despite having proteolytic sites exposed to the solvent43. Such tight control of protease activity may have evolved in bacteria such as M. tuberculosis to regulate proteolytic function during prolonged dormancies within host cells46,47,48. Mitochondrial hClpP, which must rapidly respond to stress stimuli49,50, likely integrates a similarly sophisticated assembly and activation pathway to ensure specific proteolytic activities.
The discovery of an initial assembly complex between the substrate-free hClpX hexamer and a single hClpP heptamer ring provides insights into how the proteolytic apparatus is assembled and activated in a controlled, stepwise manner. This intermediary configuration likely serves as a regulatory checkpoint to prevent premature substrate processing, ensuring that the complex only achieves full proteolytic competence upon substrate engagement. The observation that hClpX induces partial symmetrization of the hClpP heptamer during this initial interaction, followed by complete symmetrization upon substrate binding and tetradecameric assembly, highlights the intricate allosteric communication that exists between these components. This multi-step assembly process likely provides the mitochondrial system with an additional regulatory control point that may be absent in prokaryotic homologs, reflecting the complex proteostatic demands of the eukaryotic mitochondrial environment.
Based on our collective findings, it is likely that two hClpXP initial assembly complexes coordinate to form the functional tetradecamer upon hClpX interaction with substrate. However, in vitro hClpX hexamerization and hClpXP assembly for high-resolution structural studies required substantial construct and condition optimization, suggesting that this assembly pathway is unlikely to be spontaneous in the mitochondrial matrix and likely only occurs in response to specific stimuli. The hClpX NTD contains a conserved C4-type zinc-finger motif known to be involved in adaptor binding and substrate recruitment16,25. Removing the NTD from hClpX enhanced its stability and facilitated hexamer formation, suggesting that the NTD may inhibit hClpX oligomerization unless it is bound to a mitochondrial adaptor protein or specific substrate. For example, interaction between the hClpX NTD and adaptor protein polymerase delta-interacting protein (PDIP38) was shown to limit hClpX turnover by LONP1 in mitochondria25. PDIP38 may interact with the hClpX NTD to promote hexamerization and ClpXP assembly, explaining the stabilizing role this adaptor protein has on hClpX25. PDIP38 is a nucleoid factor; it has been previously proposed that hClpX associates with nucleoid condensates50,51. It is plausible that PDIP38 facilitates hClpX oligomerization as a means of tightly regulating its activity in the nucleoid for maintenance of mitoribosome assembly50. Analogously, it was observed in B. subtilis that the adaptor MecA regulates ClpC activity by facilitating its oligomerization in the presence of nucleotides52.
The next layer of regulation involves the interaction between hClpX and hClpP, which also requires construct design and crosslinking to stabilize in vitro, suggesting that specific conditions are required for hClpXP complex assembly in the mitochondrion. Similar to hClpX hexamerization, hClpXP formation may also have substrate or cofactor requirement to ensure degradation of appropriate substrates. The more transient nature of the hClpX-hClpP interaction compared to bacterial homologs may be associated with the role hClpX plays as a chaperone in the mitochondrial matrix. For example, hClpX activates ALA synthase (ALAS) by partially unfolding the protein to facilitate binding of a cofactor, heme6, yet under heme-repleting conditions, ALAS becomes targeted for hClpXP-mediated degradation53. To balance these dual chaperoning and unfoldase roles, regulatory checkpoints such as displacement of the hClpP C-terminal extension have likely evolved to enable substrate sensing and molecular decisions regarding unfolding, chaperoning, and degradation to be carried out effectively.
Unlike previously characterized ClpXP orthologs, the assembly of the hClpXP complex involves an intermediate step in which hClpX is bound to a single hClpP heptamer. In this initial assembly complex (hClpXPIA), the binding of hClpX promotes the symmetrization of an asymmetric apo hClpP heptamer. However, in this initial complex, the protease ring remains slightly asymmetric until the full hClpXP complex is formed (Supplementary Fig. 13b). A detailed structural comparison between hClpXPIA and the fully assembled hClpXP complex reveals that substrate engagement acts as another regulatory step in the assembly process. Only upon engagement can hClpXP undergo the conformational rearrangement necessary for the formation of a complete hClpXP complex. Previous studies on bacterial ClpXP have shown that the N-terminal domain of ClpP interacts with ClpX to facilitate communication between the two proteins13,54. The interaction between hClpX E subunit and hClpP N-terminus, as well as the movement of the hClpX E subunit during transition from the intermediate state to a fully assembled complex, further supports allosteric communication between hClpX and the N-terminal domain of hClpP. It is likely that the intermediate structure was not observed in our hClpXP X-linked dataset due to the use the hClpX walker-B mutant, which traps the substrate peptide in the central pore12,28,29, thereby stabilizing the fully assembled complex.
Once fully assembled, the hClpXP complex bears a conserved organization to unicellular ClpXP structures that likely operates through a conserved mechanochemical cycle. However, the hClpX AAA+ domain has integrated an E-loop that is critical for stabilizing the hexamer and carrying out the ATP hydrolysis associated with substrate unfolding and translocation. Unique motifs or insertions that facilitate inter-subunit communication or nucleotide sensing are found throughout the different classes of AAA+ modules. For example, AAA+ enzymes of the classical clade, such as YME1 and AFG3L2, harbor an inter-subunit signaling (ISS) motif that detects the nucleotide state of the adjacent subunit28,31. ClpX is a member of the HCLR clade, which all contain a presensor-1 β-hairpin (PS1βH) that has been shown in other members of the clade to be crucial for AAA+ function, such as sensing nucleotide states32 or stabilizing the pore-1 loop during substrate translocation30. However, the PS1βH sequence is highly divergent (Supplementary Fig. 2a) and seems to play varying functional roles in different AAA+ members of the PS1βH superclade55. It is plausible that the nucleotide-sensing role of PS1βH in hClpX has been evolutionarily transferred to the E-loop. Additionally, this addition to the hClpX AAA+ module may help fine-tune the rate of ATP hydrolysis or inter-subunit coordination in response to different substrates under varying mitochondrial conditions29.
Finally, in contrast to most bacterial species, hClpX binding is insufficient to induce the active conformation of hClpP. While hClpX binding drives hClpP to form compact, symmetric tetradecamers, these tetradecamers remain catalytically inactive until the binding of substrate peptide at the catalytic site. To date, all crystal structures of small-molecule-activator-bound hClpP have been resolved in a compact state and are largely interpreted as transition states due to a limited understanding of the hClpXP activation mechanism21,24,56. In light of our findings, it seems unlikely that the compact state is a transition state, but rather the resting state of the hClpP tetradecamer. Similarly, it has been reported that substrate-binding pockets act as an additional allosteric site for M. tuberculosis ClpP1P2 (MtClpP1P2)43 and TtCpP42.
Our study highlights key regulatory adaptations that hClpXP has evolved to maintain mitochondrial proteostasis, which likely enhance the capacity of hClpXP to rapidly and efficiently respond to the dynamic mitochondrial environment. Consequently, our findings pinpoint regulatory facets that should be carefully considered in future investigations that seek to characterize the diverse roles of hClpXP within the mitochondrial space. We anticipate that our insights into assembly and activation of the hClpXP complex will lay the groundwork for new studies exploring how these mechanisms are used to maintain mitochondrial proteostasis and cellular health.
Methods
Protein expression and purification
The genes encoding hClpX (residues 66-633) and hClpP (residues 58-277) were cloned into a PET28a expression vector containing a His6-SUMO tag at the N-terminus. Various hClpX variants (hClpXI146-dCTE, hClpXI146-dCTE-dE, hClpXI146-dCTE-E396Q, and hClpXI146-dCTE-E396A) and hClpP variants (hClpPdCTE and hClpPdCTE-S153A) were generated using the Q5 Site-Directed Mutagenesis Kit from New-England Biolabs. The hClpX and hClpP constructs were transformed into BL21 Rosetta competent cells and grown overnight in LB medium supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol. The overnight culture was then diluted 1:100 in fresh LB medium and incubated at 37 °C with a shaking speed of 225 rpm. When OD600 reached 0.6, protein expression was induced by adding isopropyl beta-D-thiogalactopyranoside (IPTG) at a final concentration of 0.5 mM, and the culture was shaken overnight at 16 °C. The following day, cells were harvested by centrifugation at 4500 rpm for 30 minutes, and the pellet was resuspended in lysis buffer (25 mM HEPES, pH 7.5, 100 mM KCl, 400 mM NaCl, 10% glycerol, 0.1 mM PMSF protease inhibitor). The cell suspension was sonicated, and the lysate was clarified by centrifugation at 35,000 rpm for 30 minutes. The supernatant containing the proteins was purified using Ni-NTA beads. The eluted proteins were cleaved with Ulp1 protease and dialyzed overnight in a buffer containing 25 mM HEPES, pH 7.5, 300 mM KCl, and 10% glycerol. To remove the His6-SUMO tag and Ulp1 protease, the cleaved proteins were passed through Ni-NTA beads again. The proteins were subsequently further purified using size exclusion chromatography (SEC) on a Superdex 200 10/300 GL column. For hClpX, the SEC buffer contains 25 mM HEPES pH 7.5, 300 mM KCl, 10% glycerol, 2 mM magnesium, and 1 mM DTT. hClpP was purified using SEC buffer containing 25 mM HEPES pH7.5, 150 mM KCl, 5% glycerol, and 1 mM DTT. Purified proteins were concentrated, aliquoted, flash frozen in liquid nitrogen, and stored at -80 °C.
Sample preparation for electron microscopy
For all three cryo-EM structures presented in this paper, 4 μl of sample was applied onto 300 mesh R1.2/1.3 UltraAuFoil Holey Gold grids (Quantifoil), which were plasma cleaned for 15 seconds using a Pelco glow discharge cleaning system (TED PELLA, INC.) with atmospheric gases at 15 mA. The grids were manually blotted using Whatman #1 blotting paper for 6 seconds before being plunged frozen in liquid ethane in a 4 °C cold room with 90% humidity (all subsequently described vitrification of samples were performed using the same methodology).
To prepare hClpXPX-linked sample, hClpXI146-dCTE-E396Q (50 μM) and hClpPdCTE-S153A (8 μM) were incubated in a buffer containing 25 mM HEPES (pH 7.5), 150 mM KCl, 5 mM ATP, and 5 mM MgCl2 for 5 minutes at room temperature. The sample was then crosslinked with 160 μM disuccinimidyl suberate (DSS) (Thermo Fisher A39267) for 30 minutes at room temperature, and the reaction was quenched with 40 μM Tris (pH 7.5). The crosslinked sample was concentrated and injected onto a Superose 6 increase 10/300 GL column in SEC buffer (25 mM HEPES, 150 mM KCl, 5% glycerol, 2 mM MgCl2, 2 mM ATP). Fractions corresponding to the crosslinked hClpXP complexes were collected and concentrated 15-fold. The concentrated sample was then applied to the cryo-EM grid and manually blotted before being plunged frozen in liquid ethane. For the uncrosslinked hClpXP sample, a similar concentration of full-length hClpX and hClpP (with the mitochondrial targeting sequence removed) was incubated in a buffer containing 25 mM HEPES (pH 7.5), 150 mM KCl, 5 mM ATPyS, and 5 mM MgCl2 for 30 minutes at room temperature. The sample was then applied to the cryoEM grid and manually blotted before being plunged frozen in liquid ethane.
For the apo hClpP cryo-EM, 4 μl of 15 mg/ml of hClpP (WT) in a buffer containing 25 mM HEPES, 150 mM KCl, 2 mM DTT, and a final 4 mM fluorinated fos-choline or 0.08% n-Decyl-beta-Maltoside (DM) detergent was applied onto the cryo-EM grid and immediately blotted before being plunged into liquid ethane. For the bortezomib-bound hClpP structure, a 10X molar excess of bortezomib (Sigma-Aldrich) was added to 15 mg/ml of hClpP and incubated on ice for 30 minutes. Just before applying the sample to the cryo-EM grid, detergent at a final 0.08% DM concentration was added to the sample to help mitigate the preferred orientation of hClpP in vitrified ice.
Cryo-EM data collection and processing
For all datasets, cryo-EM data were collected on a Thermo-Fisher Talos Arctica transmission electron microscope operating at 200 kV with parallel illumination conditions57,58. Micrographs were acquired with a Gatan K2 Summit direct electron detector with a total electron exposure of 50 e-/Å2. All subsequent image processing was performed using cryoSPARC59. Detailed processing workflows and dataset-specific parameters are provided in Supplementary Figs. 6, 12, 17, 19 and Supplementary Table 1.
For the hClpXPX-linked complex cryo-EM dataset, Leginon data collection software60 was used to collect 10,333 micrographs at 45 K nominal magnification (0.91 Å/pixel-1), with a nominal defocus range from −0.23 μM to −-3.3 μm. Stage movement was used to target the center of either 4 or 16 1.2 μm holes for focusing, and image shift was utilized to acquire high magnification images. The Appion image processing wrapper61 was used to run MotionCor262 for micrograph frame alignment and dose-weighting in real-time during data collection. Subsequent image processing was done in cryoSPARC. The contrast transfer function (CTF) parameters were estimated using patch CTF estimation (multi). Template-based particle picking, using representative 2D class averages generated from 1,175 particles selected with blob picking using a diameter of 200 Å to 320 Å, resulted in approximately 2.07 million particles. Particles were extracted from the motion-corrected and dose-weighted micrographs with a box size of 352 pixels. These particles were classified into 100 2D classes using default parameters, and 2D classes (containing 1,940,578 particles) displaying high-resolution secondary features were selected to generate four reference-free 3D Ab-initio models. The initial volumes generated from ab initio were used for heterogeneous refinement to classify the particles into four classes with C1 symmetry. The class containing well-resolved structural features, corresponding to 1,321,170 particles, was chosen for non-uniform refinement of hClpP with C1 symmetry, resulting in a final reconstruction with a reported GSFCS at 2.91 Å. For hClpX, the same particle set underwent additional 3D classification (six classes) with a mask corresponding to hClpX (O-EM learning rate init: 0.2, initialization mode: PCA; PCA/simple: particles per reconstruction: 750; PCA: number of reconstructions: 75; Force hard classification; class similarity: 0.1). Two good classes (513,188 particles total) were combined and subjected to a final round of non-uniform refinement with C1 symmetry. Subsequently, the 3D volume was re-aligned centering on hClpX, and the particles re-extracted with the centered coordinates were locally refined with a mask around hClpX, resulting in a final resolution of 3.11 Å based on the FSC at a cutoff of 0.143. A composite map was generated using individual high-resolution maps of hClpX and hClpP in ChimeraX.
For the uncrosslinked hClpXP complex cryo-EM dataset, Leginon data collection software60 was used to collect 3298 micrographs at 36 K nominal magnification (1.15 Å/pixel-1), with a nominal defocus range from -0.2 μM to -3.6 μm. Stage movement was used to target the center of either 4 or 16 1.2 μm holes for focusing, and image shift was utilized to acquire high magnification images. The Appion image processing wrapper61 was used to run MotionCor262 for micrograph frame alignment and dose-weighting in real-time during data collection. Subsequent image processing was done in cryoSPARC. The contrast transfer function (CTF) parameters were estimated using patch CTF estimation (multi). Template-based particle picking, using representative 2D class averages generated from 2,205 particles selected with blob picking using a diameter of 200 Å to 320 Å, resulted in approximately ~1.1 million particles. Particles were extracted from the motion-corrected and dose-weighted micrographs with a box size of 352 pixels (Fourier cropped to a box size of 176 pixels). These particles were classified into 100 2D classes using default parameters, and 2D classes (containing 70,623 particles) displaying high-resolution secondary features were selected to generate four reference-free 3D Ab-initio models. The initial volumes generated from ab initio were used for heterogeneous refinement to classify the particles into four classes with C1 symmetry. The class containing 26,401 particles of hClpXPIA was re-extracted with full box size and used as input for non-uniform refinement with C1 symmetry, resulting in a final reconstruction with a reported GSFCS at 4.4 Å. Focused-refinement of hClpXIA and hClpPIA using individual masks led to final resolution of 4.3 Å and 4.0 Å, respectively. The class containing 23,084 particles of hClpXP was re-extracted and chosen for homogenous refinement and symmetry expansion (D1), followed by local refinement of the top half of hClpXP and 3D classification (5 classes). The three classes of particles with good hClpX density were combined for local refinement, while the other 2 classes were combined for homogenous refinement. The particles from local refinement and homogenous refinement were then combined for local refinement with a hClpX mask, leading to a reconstruction of hClpX resolved to ~4.4 Å resolution. Further local refinement with a hClpP mask resulted in hClpP density wth an overall resolution of ~3.5 Å. A composite map of uncrosslinked hClpXP was generated using individually local refined maps of hClpX and hClpP in ChimeraX.
For the apo hClpP (WT) structure, a total of 14,989 micrographs were collected at 150 K nominal magnification (0.94 Å/pixel-1) with a nominal defocus range from −0.3 μm to −3.5 μm using the EPU software. A maximum image shift of 2 µm was used to acquire high magnification images. Dose-weighting, motion correction, and CTF estimation were performed within cryoSPARC live. Template-based particle picking, using representative 2D class averages generated from 4,710 particles selected with blob picking (diameter of 70 Å to 100 Å), resulted in ~2.7 million particles. Two rounds of 2D classification into 100 class averages using default parameters were performed to remove junk particles, with 935,410 particles selected for Ab-initio and iterative hetero-refinement (four classes). The class with the best-resolved structural features (corresponding to 501,826 particles) was used for non-uniform refinement (C1 symmetry), yielding a hClpP reconstruction at 3.63 Å resolution. Subsequent global and local CTF refinement, and another round of non-uniform refinement,t slightly improved the resolution to 3.61 Å. 3D classification into 10 classes was performed, with particles from eight good classes used for the final round of non-uniform refinement (C1 symmetry), resulting in a final reported GSFCS resolution of 3.52 Å.
For the bortezomib-bound-hClpP dataset, 3972 micrographs were collected using EPU at 150 K nominal magnification (0.94 Å/pixel-1) with a nominal defocus range from -0.1 μm to -3.28 μm. Image shift was used to acquire high magnification images as for the prior dataset. Dose-weighting, motion correction, and CTF estimation were performed within cryoSPARC live. Blob picking of the CTF estimated images using a particle diameter of 100 Å to 150 Å resulted in ~1.9 million particles. Two rounds of 2D classification were performed to remove junk particles, leaving 545,529 particles for Ab-initio and subsequent hetero-refinements (4 classes). The class with the best-resolved structural features (corresponding to 224,782 particles) was used for non-uniform refinement (C1 symmetry), producing a hClpP reconstruction at 2.67 Å resolution. Further global and local CTF refinement slightly increased the resolution to 2.63 Å with C1 symmetry and 2.28 Å with D7 symmetry.
Atomic model building and refinement
For the hClpXPx-linked complex, the AlphaFold model of hClpX and the hClpP crystal structure (PDB 1TG6) were used as starting models. For the uncrosslinked hClpXP complex, the refined models from the crosslinked hClpXP were used as initial references. For the bortezomib-bound hClpP, the hClpP crystal structure (PDB 6BBA) served as a starting model, whereas for the hClpP apo structure, the hClpX-bound hClpP model was used. The models were docked into the 3D reconstruction using UCSF ChimeraX63. Manual model building and real-space structural refinement were performed with Coot64 and Phenix65, respectively. Phenix real-space refinement includes global minimization, rigid body, local grid search, atomic displacement parameters, and morphing for the first cycle. It was run for 100 iterations, five macro-cycles, with a target bonds RMSD of 0.01 and a target angles RMSD of 1.0. The refinement settings also include secondary structure restraints and Ramachandran restraints. ChimeraX plug-in ISOLDE66 was used to manually fix any Ramachandran outliers, rotamers, and clashes not fixed by Phenix. For the bortezomib-bound hClpP, a covalent linkage between bortezomib and hClpP S153 was made using Phenix. Figures for publication were prepared using UCSF ChimeraX. Supplementary Table 1 includes data collection, refinement, and validation statistics.
ATPase activity assay
The ATPase activity experiments of different hClpX variants were carried out in a black flat-bottom 96-well plate in a final volume of 100 μl. 0.3 μM of hClpX6 proteins were pre-incubated at 37 °C for 5 min in the activity buffer (50 mM HEPES pH 7.5, 75 mM KCl, 5 mM MgCl2, and 1 mM DTT, 1.6 mM NADH (Caymen Chemical; 606-69-8), 4 mM phosphoenolpyruvate (1PlusChem; 5541-93-5), 120 U/ml lactate dehydrogenase (Worthington Chemical; LS002755), 40 U/ml pyruvate kinase (Sigma; 10128155001). The reaction was initiated by adding 5 mM ATP. For ATP titration assays, increasing final concentrations (0.05 mM, 0.1 mM, 0.15 mM, 0.2 mM, 0.3 mM, 0.8 mM, 2 mM, 5 mM, and 8 mM) of ATP were added to initiate the reaction. Absorbance at 340 nm was measured at 37 °C using a TECAN plate reader.
Degradation assays
The ClpXP-mediated degradation of FITC-casein was carried out in a black flat-bottom 96-well plate in a final volume of 100 μl. 1 μM ClpX and 0.5 μM ClpP were pre-incubated at 37 °C for 5 min in activity buffer containing 50 mM HEPES pH 7.5, 75 mM KCl, 5 mM MgCl2, 5 mM ATP, and 1 mM DTT. 1 μM FITC-casein (Sigma Aldrich; C3777), which had also been pre-incubated at 37 °C, was added to initiate the degradation reaction. For substrate titration assays, increasing final concentrations (0.125 μM, 0.250 μM, 0.5 μM, 1 μM, 2 μM, 4 μM, 8 μM, 10 μM, and 16 μM) of FITC-casein was added to initiate the reaction. For the hClpP or hClpPdCTE titration assay, 0.5 μM of hClpX and increasing concentrations of hClpP or hClpPdCTE (0.03 μM, 0.06 μM, 0.125 μM, 0.25 μM, 0.5 μM) were used in the reaction. 2 μM FITC-casein was added to initiate the degradation reaction. An increase of fluorescence resulting from FITC molecules was measured (excitation 485 nm, emission 535 nm) using a TECAN plate reader.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM maps and associated atomic models generated in this study have been deposited to the Electron Microscopy Databank (EMDB) and the Protein Databank (PDB), respectively, with the following EMDB and PDB IDs: hClpXPx-linked (focused alignment of ClpX): EMD-47226 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD47226] and 9DVY; hClpXP x-linked (focused alignment of ClpP): EMD-47232 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD47232] and 9DW0; apo hClpP heptamer: EMD-47233 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD47233] and 9DW1; hClpP tetradecamer bound to bortezomib: EMD-47234 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD47234], 9DW3 [https://doi.org/10.2210/pdb9YKX/pdb]; hClpXPuncrosslinked (focused alignment of hClpX): EMD-71595 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD71595], 9YKX [https://www.rcsb.org/structure/unreleased/9YKX]; hClpXPuncrosslinked (focused alignment of hClpP): EMD-71596 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD71596], 9YKZ; hClpXPuncrosslinked composite map: EMD-73107 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD73107]; hClpXP initial assembly complex (focused alignment of hClpX): EMD-71425, 9P9V; hClpXP initial assembly complex (focused alignment of hClpP): EMD-71450 [https://www.ebi.ac.uk/pdbe/entry/emdb/EMD71450], 9PB1). Source data are provided with this paper.
References
Osellame, L. D., Blacker, T. S. & Duchen, M. R. Cellular and molecular mechanisms of mitochondrial function. Best. Pr. Res Clin. Endocrinol. Metab. 26, 711–723 (2012).
Kuznetsov, A. V. & Margreiter, R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J. Mol. Sci. 10, 1911–1929 (2009).
Tait, S.W. & Green, D.R. Mitochondrial regulation of cell death. Cold Spring Harb. Perspect. Biol. 5, a008706 (2013).
Jadiya, P. & Tomar, D. Mitochondrial protein quality control mechanisms. Genes 11, 563 (2020).
Javadov, S., Kozlov, A. V. & Camara, A. K. S. Mitochondria in health and diseases. Cells 9, 1177 (2020).
Kardon, J. R., Moroco, J. A., Engen, J. R. & Baker, T. A. Mitochondrial ClpX activates an essential biosynthetic enzyme through partial unfolding. Elife 9, e54387 (2020).
Kardon, J. R. et al. Mitochondrial ClpX Activates a Key Enzyme for Heme Biosynthesis and Erythropoiesis. Cell 161, 858–867 (2015).
Kasashima, K., Sumitani, M. & Endo, H. Maintenance of mitochondrial genome distribution by mitochondrial AAA+ protein ClpX. Exp. Cell Res 318, 2335–2343 (2012).
Seo, J. H. et al. The Mitochondrial Unfoldase-Peptidase Complex ClpXP Controls Bioenergetics Stress and Metastasis. PLoS Biol. 14, e1002507 (2016).
Mabanglo, M. F. & Houry, W. A. Recent structural insights into the mechanism of ClpP protease regulation by AAA+ chaperones and small molecules. J. Biol. Chem. 298, 101781 (2022).
Ripstein, Z. A., Vahidi, S., Houry, W. A., Rubinstein, J. L. & Kay, L. E. A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery. Elife 9, e52158 (2020).
Fei, X. et al. Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate. Elife 9, e52774 (2020).
Gatsogiannis, C., Balogh, D., Merino, F., Sieber, S. A. & Raunser, S. Cryo-EM structure of the ClpXP protein degradation machinery. Nat. Struct. Mol. Biol. 26, 946–954 (2019).
Fei, X., Bell, T. A., Barkow, S. R., Baker, T. A. & Sauer, R. T. Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates. Elife 9, e61496 (2020).
Truscott, K. N., Lowth, B. R., Strack, P. R. & Dougan, D. A. Diverse functions of mitochondrial AAA+ proteins: protein activation, disaggregation, and degradation. Biochem. Cell Biol. 88, 97–108 (2010).
Lowth, B. R. et al. Substrate recognition and processing by a Walker B mutant of the human mitochondrial AAA+ protein CLPX. J. Struct. Biol. 179, 193–201 (2012).
Kang, S. G. et al. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J. Biol. Chem. 277, 21095–21102 (2002).
Liu, K., Ologbenla, A. & Houry, W. A. Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases. Crit. Rev. Biochem. Mol. Biol. 49, 400–412 (2014).
Kang, S. G., Maurizi, M. R., Thompson, M., Mueser, T. & Ahvazi, B. Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J. Struct. Biol. 148, 338–352 (2004).
Brodie, E. J., Zhan, H., Saiyed, T., Truscott, K. N. & Dougan, D. A. Perrault syndrome type 3 caused by diverse molecular defects in CLPP. Sci. Rep. 8, 12862 (2018).
Wong, K. S. et al. Acyldepsipeptide analogs dysregulate human mitochondrial ClpP Protease activity and cause apoptotic cell death. Cell Chem. Biol. 25, 1017–1030.e9 (2018).
Kang, S. G., Dimitrova, M. N., Ortega, J., Ginsburg, A. & Maurizi, M. R. Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 280, 35424–35432 (2005).
Wang, P. et al. Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma. Cell Chem. Biol. 29, 1396–1408.e8 (2022).
Ishizawa, J. et al. Mitochondrial ClpP-mediated proteolysis induces selective cancer cell lethality. Cancer Cell 35, 721–737.e9 (2019).
Strack, P. R. et al. Polymerase delta-interacting protein 38 (PDIP38) modulates the stability and activity of the mitochondrial AAA+ protease CLPXP. Commun. Biol. 3, 646 (2020).
Ghanbarpour, A. et al. A closed translocation channel in the substrate-free AAA+ ClpXP protease diminishes rogue degradation. Nat. Commun. 14, 7281 (2023).
Fux, A., Korotkov, V. S., Schneider, M., Antes, I. & Sieber, S. A. Chemical cross-linking enables drafting ClpXP proximity maps and taking snapshots of in situ interaction networks. Cell Chem. Biol. 26, 48–59 e7 (2019).
Puchades, C. et al. Unique structural features of the mitochondrial AAA+ Protease AFG3L2 reveal the molecular basis for activity in health and disease. Mol. Cell 75, 1073–1085.e6 (2019).
Puchades, C., Sandate, C. R. & Lander, G. C. The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol. 21, 43–58 (2020).
Shin, M. et al. Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Nat. Commun. 12, 3239 (2021).
Puchades, C. et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358, eaao0464 (2017).
de la Pena, A. H., Goodall, E. A., Gates, S. N., Lander, G. C. & Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725 (2018).
Shin, M. P. et al. Structural basis for distinct operational modes and protease activation in AAA protease Lon. Sci. Adv. 6, eaba8404 (2020).
Ghanbarpour, A., Fei, X., Baker, T. A., Davis, J. H. & Sauer, R. T. The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery. Proc. Natl. Acad. Sci. USA 120, e2219044120 (2023).
Ghanbarpour, A., Sauer, R. T. & Davis, J. H. A proteolytic AAA+ machine poised to unfold protein substrates. Nature Communications 15, 9681 (2024).
Kim, S., Zuromski, K. L., Bell, T. A., Sauer, R. T. & Baker, T. A. ClpAP proteolysis does not require rotation of the ClpA unfoldase relative to ClpP. Elife 9, e61451 (2020).
Martin, A., Baker, T. A. & Sauer, R. T. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell 29, 441–450 (2008).
Nagpal, J. et al. Molecular and structural insights into an asymmetric proteolytic complex (ClpP1P2) from Mycobacterium smegmatis. Sci. Rep. 9, 18019 (2019).
Goncalves, M. M. et al. Mechanism of allosteric activation in human mitochondrial ClpP protease. bioRxiv (2024).
Stahl, M. et al. Selective Activation of Human Caseinolytic Protease P (ClpP). Angew. Chem. Int Ed. Engl. 57, 14602–14607 (2018).
Malik, I. T. & Brotz-Oesterhelt, H. Conformational control of the bacterial Clp protease by natural product antibiotics. Nat. Prod. Rep. 34, 815–831 (2017).
Felix, J. et al. Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors. Mechanism of the allosteric activationof the ClpP protease machinery by substratesand active-site inhibitors. Sci. Adv. 5, eaaw3818 (2019).
Vahidi, S. et al. An allosteric switch regulates Mycobacterium tuberculosis ClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR. Proc. Natl. Acad. Sci. USA 117, 5895–5906 (2020).
Mabanglo, M. F. et al. ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores. Commun. Biol. 2, 410 (2019).
Baker, T. A. & Sauer, R. T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys. Acta 1823, 15–28 (2012).
Raju, R. M. et al. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 8, e1002511 (2012).
Schmitz, K. R. & Sauer, R. T. Substrate delivery by the AAA+ ClpX and ClpC1 unfoldases activates the mycobacterial ClpP1P2 peptidase. Mol. Microbiol. 93, 617–628 (2014).
Alhuwaider, A. A. H. & Dougan, D. A. AAA+ Machines of Protein Destruction in Mycobacteria. Front Mol. Biosci. 4, 49 (2017).
Dittmar, D. et al. Complementation studies with human ClpP in Bacillus subtilis. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118744 (2020).
Key, J., Gispert, S. & Auburger, G. Knockout Mouse Studies Show That Mitochondrial CLPP Peptidase and CLPX Unfoldase Act in Matrix Condensates near IMM, as Fast Stress Response in Protein Assemblies for Transcript Processing, Translation, and Heme Production. Genes 15, 694 (2024).
Key, J. et al. Inactivity of Peptidase ClpP Causes Primary Accumulation of Mitochondrial Disaggregase ClpX with Its Interacting Nucleoid Proteins, and of mtDNA. Cells 10, 3354 (2021).
Kirstein, J. et al. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491 (2006).
Cottle, T., Joh, L., Posner, C., DeCosta, A. & Kardon, J. R. An adaptor for feedback regulation of heme biosynthesis by the mitochondrial protease CLPXP. bioRxiv (2024).
Martin, A., Baker, T. A. & Sauer, R. T. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell 27, 41–52 (2007).
Burrows, P. C. et al. Functional roles of the pre-sensor I insertion sequence in an AAA+ bacterial enhancer binding protein. Mol. Microbiol. 73, 519–533 (2009).
Mabanglo, M. F. et al. Potent ClpP agonists with anticancer properties bind with improved structural complementarity and alter the mitochondrial N-terminome. Structure 31, 185–200.e10 (2023).
Herzik, M. A. J. Setting up parallel illumination on the Talos Artica for high resolution data collection. Methods Mol. Biol. 2215, 125–144 (2021).
Herzik, M., Wu, M. & Lander, G. Achieving better-than3-A-resolution by single-particle cryo-EM at 200-kev. Nat. Methods 14, 1075–1078 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 74, 519–530 (2018).
Acknowledgements
We thank the members of the Lander lab for helpful discussion. We thank Will Lessin at the Scripps Research Electron Microscopy Facility for microscopy support. We thank Charles Bowman and J.C. Ducom at Scripps Research High Performance Computing for computational support. This work is supported by a grant from the National Institutes of Health (NIH) NS095892 to G.C.L., and a National Science Foundation predoctoral fellowship to W.C. Cryo-EM data collection used equipment supported by NIH grant S10OD032467.
Author information
Authors and Affiliations
Contributions
J.Y. and G.C.L. initially conceptualized the project. W.C. and G.C.L. led the project; W.C. generated the constructs, purified the proteins, and performed all cryo-EM structure determination, model building, and refinement, and mechanistic interpretation with guidance from J.Y. and G.C.L.; W.C. and G.C.L. wrote the manuscript, with edits from J.Y.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, W., Lander, G.C. & Yang, J. Cryo-EM structures of human ClpXP reveal mechanisms of assembly and proteolytic activation. Nat Commun 17, 1064 (2026). https://doi.org/10.1038/s41467-025-67010-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67010-1