Abstract
Transient receptor potential (TRP) ion channels form heteromers through combinatorial associations of distinct subunits, contributing to the diversity of TRP channel functions. Among them, TRPC5, which forms a heteromer with TRPC1, represents an attractive pharmaceutical target for treating anxiety and depression. Here, we present the cryo-electron microscopy structure of the human TRPC1/C5 heteromer, composed of one TRPC1 subunit and three TRPC5 subunits. The incorporation of TRPC1 into the heteromer disrupts the C4 symmetry of the TRPC5 homotetramer, resulting in a distinct ion conduction pathway characterized by an asymmetrically constricted selectivity filter and an asymmetric lower gate. The TRPC1/C5 heteromer displays recognizable structural features compared to the TRPC1/C4 heteromer, including a noncanonically tilted coiled-coil domain and a distinct intersubunit interactions. Furthermore, we elucidate the structures of human TRPC5 bound to the TRPC1/4/5-specific agonist, (−)-Englerin A. Our findings establish a foundation for exploring the diversity of heteromeric TRP channels and pave the way for targeting TRPC1/C5 as a therapeutic strategy.
Similar content being viewed by others
Introduction
Transient receptor potential (TRP) channels assemble as heteromers through combinatorial associations of distinct subunits, contributing to diverse functionalities, including specific activation and inhibition mechanisms, electrophysiological properties, and ligand specificities1,2,3,4. Targeting tissue-specific heteromeric TRP channels with selective drugs holds promise as a therapeutic approach with reduced side effects, necessitating investigations into the structure-function relationships of heteromeric ion channels.
The transient receptor potential canonical (TRPC) subfamily includes TRPC1, TRPC4, and TRPC5, which form both homomers and heteromers in a tissue-specific manner5,6,7. While TRPC4 and TRPC5 alone show specific expression patterns primarily in the gastrointestinal tract and brain, respectively, TRPC1 is ubiquitously expressed across various tissues and is recognized as a negative regulator by forming heteromers with TRPC4 and/or TRPC5 (TRPC1/C4, TRPC1/C5, or TRPC1/C4/C5)8,9,10,11. Heteromerization with TRPC4 and/or TRPC5 enables the TRPC1 subunit to undergo membrane trafficking to the plasma membrane12. The resulting heteromers display distinct properties compared to homomeric TRPC4 and TRPC5 channels, including altered current-voltage (I–V) relationships, reduced calcium permeability (PCa/PNa), and different monovalent cation permeability13.
Notable distinctions between TRPC1/C4 and TRPC1/C5 heteromers lie in their desensitization mechanisms and phosphatidylinositol 4,5-bisphosphate (PIP2) affinity14,15. TRPC1/C4 desensitization is mediated by calcium and PIP2 depletion, whereas TRPC1/C5 desensitization involves a mechanism comprising protein kinase C (PKC) phosphorylation and PIP2 depletion. Regarding PIP2 sensitivity, TRPC4 exhibits a higher PIP2 affinity compared to TRPC516. Interestingly, TRPC1 modulates PIP2 sensitivity in opposite directions: it reduces TRPC4’s PIP2 sensitivity while enhancing that of TRPC5. This differential modulation results in comparable PIP2 sensitivities between TRPC1/C4 and TRPC1/C5 heteromers. In addition, TRPC1/C4 and TRPC1/C5 heteromers exhibit distinct activation mechanisms through interactions with specific Gα subtypes; they are activated by direct interaction with activated Gαq, whereas TRPC4 and TRPC5 homomers are activated by Gαi14,17,18. Notably, the cryo-electron microscopy (cryo-EM) structure of TRPC5 in complex with Gαi3 was recently resolved at a 4:4 stoichiometry in lipid nanodiscs, showing that Gαi3 binds to the ankyrin repeat domain (ARD) of TRPC518. Altogether, to further our understanding of the structure-function relationships in TRPC channels, structural studies of various TRPC heteromers are highly needed.
Recently, we resolved the cryo-EM structure of the human TRPC1/C4 heteromer in both the apo and antagonist-bound states, revealing an asymmetrically constricted selectivity filter and an asymmetric lower gate13. This provided insight into the structure-function relationship underlying the reduced calcium permeability and unique monovalent cation selectivity of the TRPC1/C4 heteromer19,20. However, the structural architecture and drug-binding sites of the TRPC1/C5 heteromer remain unresolved, although a recent high-resolution proteomics study of native rodent brain tissue identified a 1:3 stoichiometry for this complex6. Moreover, elucidating the molecular basis for the distinct physiological properties of different heteromeric assemblies (TRPC1/C4, TRPC1/C5, or TRPC1/C4/C5) in diverse cellular contexts is essential for advancing our mechanistic understanding of TRPC heteromer diversity1,11.
From a pharmacological perspective, TRPC heteromers represent distinct therapeutic targets, as evidenced by the observation that activators often exhibit subunit composition-dependent potency. For instance, (−)-Englerin A (EA), a selective and potent agonist of TRPC1/4/5 channels widely used to probe this subgroup, exhibits higher affinity for homomeric channels than for heteromeric assemblies14,21. Similarly, the xanthine derivative AM237 robustly activates TRPC5 homomers at nanomolar concentrations but fails to activate—and instead inhibits—the EA-activated TRPC1/C5 heteromers22. Although EA has played a pivotal role in advancing TRPC1/4/5 research, structural evidence for its binding remains unavailable, in contrast to several xanthine-based compounds whose channel interactions have been structurally resolved13,23,24,25. Another example is riluzole, an approved treatment for amyotrophic lateral sclerosis26, which acts as an agonist for both TRPC1/C5 heteromers and TRPC5 homomers, engaging TRPC5 through an extracellular Ca2+-dependent mechanism without affecting other closely related TRPC channels27,28. These findings highlight the functional divergence between TRPC homomers and heteromers and emphasize the importance of understanding how distinct activators interact with each complex to inform therapeutic strategies.
Here, we present the cryo-EM structures of the human TRPC1/C5 heteromer and the ternary complex of the TRPC5 homomer with EA and riluzole, supported by electrophysiological data. We provide structural evidence for the asymmetric architecture of the TRPC1/C5 heteromer, featuring a 1:3 stoichiometry and compare its unique features to those of the TRPC1/C4 heteromer. Our findings provide a basis for advancing our structural understanding of heteromeric TRPC channels and their unique, finely-tuned functions.
Results
Structure determination of human TRPC1/C5 heteromer
To analyze the TRPC1/C5 heteromeric channel through single-particle cryo-EM, we co-expressed full-length human TRPC1α and C-terminally truncated human TRPC5 (1–765) (hereafter referred to simply as TRPC1 and TRPC5) in HEK293S GnTI− cells. The TRPC1/C5 heteromer was solubilized in digitonin, purified using FLAG affinity chromatography, reconstituted into lipid nanodiscs, and subsequently purified using Strep affinity chromatography (Supplementary Fig. 1a). After final purification by size-exclusion chromatography, we analyzed the peak fractions using SDS-PAGE, which revealed co-elution of both TRPC1 and TRPC5, confirming the formation of a stable TRPC1/C5 heteromer. Due to similar molecular weights of the constructs for TRPC1 (123.6 kDa) and TRPC5 (119.7 kDa) subunits, we carefully monitored each purification step using in-gel fluorescence.
Primarily, we aimed to purify only the TRPC1/C5 heteromer by performing tandem affinity purification, with distinct epitope tags attached to TRPC1 and TRPC5, respectively. Using our previously determined structure of the TRPC1/C4 heteromer as a reference, we examined the TRPC1/C5 heteromer map by observing a weak ARD density, the absence of a calcium ion in the voltage-sensing-like domains (VSLD), and the lack of cholesteryl hemisuccinate (CHS) in the intraprotomer region formed around pre-S1, S1, S4, and the TRP helices (Fig. 1a and Supplementary Fig. 2a)13. However, during particle classification, we have also identified a co-existence of TRPC5 homomer (~ 14.4% of the initial particle selection), which was further resolved in two conformations (Class 1 and Class 2) (Supplementary Fig. 1b–e and Supplementary Table 1). This conformational diversity aligns with the previously characterized TRPC5 homomer structure in the same nanodiscs18. To confidently assign each 3D volume as either heteromer or homomer, we examined the positions of 10 amino acids with distinctive side chains in each of the four protomers within the heteromer or homomer (Supplementary Figs. 2c and 3). We therefore suggest that TRPC5 homomers are present in our TRPC1/C5 dataset, potentially due to unknown factors, including possible interactions between TRPC1/C5 heteromers and TRPC5 homomers, although this has yet to be confirmed.
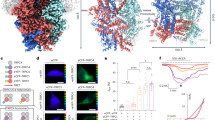
a, b Cryo-EM density map (a) and atomic model (b) of the TRPC1/C5 heteromer viewed from the side (left) and the top (right). c Schematic illustrations of the concatemer constructs used for electrophysiology experiments (TRPC5–TRPC5, TRPC5–TRPC1, and TRPC1–TRPC1) are presented, together with the possible tetramer combinations under the specified expression conditions, shown in separate boxes. The linker sequences are AELKLRILQSTVP for the TRPC5–TRPC5 concatemer, SAGSGGSGSLIEPG for the TRPC5–TRPC1 concatemer, and GSGGSGSLIS for the TRPC1–TRPC1 concatemer. d Representative (−)-Englerin A (EA)-induced current traces (left; inward and outward traces represent current responses from ± 100 mV, respectively, from every ramp pulse) and I–V relationships (right) obtained from the time point marked by a triangle are displayed using the same color scheme in (c). e Summary of the statistical data for inward and outward current responses induced by EA (positive data points represent current amplitudes at + 100 mV; negative data points represent current amplitudes at − 100 mV). f Diagrammatic representation of potential TRPC channel assemblies. ECFP or EYFP tags were attached to the amino termini of the channels (left). On the right, the tetrameric configurations of ECFP- or EYFP-tagged TRPC1 with TRPC5 (top), ECFP-tagged TRPC1 with EYFP-tagged TRPC5 (middle), and ECFP- or EYFP-tagged TRPC5 with TRPC5 are shown (bottom). g Quantitative summary of the FRET efficiency (EFRET) measurements obtained through the donor-centric method across all tested subunit combinations (n = 12, ECFP + EYFP; n = 9, ECFP–TRPC1 + EYFP–TRPC1; n = 9, TRPC5 + ECFP–TRPC1 + EYFP–TRPC1; n = 12, ECFP–TRPC1 + EYFP–TRPC5; n = 15, ECFP–TRPC5 + EYFP–TRPC5; n = 22, TRPC5 + ECFP–TRPC5 + EYFP–TRPC5). n represents a number of independent cells. Comparisons between groups were conducted using a two-sided Student’s t test. Bars represent the mean ± s.e.m. **p < 0.01, ****p < 0.0001.
Validation of asymmetric TRPC1/C5 stoichiometry in vivo
To validate the 1:3 stoichiometry of TRPC1/C5 heteromers in cells, we generated three dimeric concatemers (TRPC5–TRPC5, TRPC5–TRPC1, and TRPC1–TRPC1) (Fig. 1c) and expressed them individually or in combination in HEK293-T cells. Electrophysiological recordings in response to the agonist EA revealed that only co-expression of TRPC5–TRPC1 with TRPC5–TRPC5 produced functional channels, characterized by outward-rectifying currents distinct from the doubly rectifying currents observed for TRPC5–TRPC5 alone (Fig. 1d, e). In contrast, no currents were detected from TRPC5–TRPC1 or TRPC1–TRPC1 alone or together, suggesting that assemblies with 2:2, 3:1, or 4:0 stoichiometries do not give rise to functional channels. These results support the formation of heterotetramers composed of one TRPC1 and three TRPC5 subunits.
To further corroborate this stoichiometry and exclude potential contributions from TRPC5 homomers, we conducted comprehensive Förster resonance energy transfer (FRET) experiments using ECFP- and EYFP-tagged TRPC subunits (Fig. 1f). To ensure robust quantification, we employed the acceptor photobleaching (donor dequenching FRET) method29,30 (Fig. 1g), and analyses were performed using whole-cell regions of interest to minimize selection bias and ensure representative measurements across the plasma membrane.
Our experiments revealed that co-expression of EYFP–TRPC5 with ECFP-tagged TRPC1 or TRPC5 produced robust FRET signals, consistent with the formation of TRPC1/C5 heteromers and the incorporation of multiple TRPC5 subunits into tetrameric complexes (Fig. 1g). Moreover, when untagged TRPC5 was co-expressed at 70% of the total transfected DNA, the FRET efficiency between ECFP- and EYFP-tagged TRPC5 decreased considerably, demonstrating that untagged subunits compete for incorporation into tetramers30.
In contrast, co-expression of ECFP–TRPC1 and EYFP–TRPC1 with untagged TRPC5 did not yield significant FRET signals compared to the negative control, indicating that the tetramers contain at most one TRPC1 subunit. In addition, expression of ECFP–TRPC1 and EYFP–TRPC1 alone produced no detectable FRET signal, confirming that TRPC1 does not form homomers. Together, these electrophysiology and FRET data consistently support the selective incorporation of a single TRPC1 subunit into functional TRPC heteromeric channels.
Overall architecture of the heteromeric TRPC1/C5
Despite the similar tetrameric architecture and comparable dimensions (100 Å × 100 Å × 120 Å) observed in both the TRPC1/C5 heteromer and the TRPC5 homomer, the heteromer displays recognizable structural characteristics resulting from the incorporation of TRPC1 (Fig. 1b). Superimposition of the two structures revealed discrepancies across the tetrameric assembly (Fig. 2a, b). For clarity, we designated the TRPC5 subunits in the heteromer as TRPC5B, TRPC5C, and TRPC5D, according to their relative position in a clockwise direction from the TRPC1 subunit, as viewed from the extracellular region (Fig. 1b).
a Structural superposition of the global transmembrane domain of TRPC1/C5 heteromer and TRPC5 homomer, as viewed from the extracellular region. Voltage-sensing-like domain is highlighted at each edge to emphasize the movements with red arrows. Translations of the outer helices are indicated in angstroms (Å). b Single protomer overlap of TRPC1 with TRPC5 within TRPC1/C5 heteromer and TRPC5 homomer. ARD, ankyrin repeat domain; HLH, helix–loop–helix; PH, pore helix; CH, connecting helix; CCD, coiled-coil domain. TRPC1 displays an outward tilting of the S2–S3 linker, which also results in a tilted position of the S2 helix. On the right, movements of the S2 and S3 helices are highlighted within the dotted box, showing both tilting and translational shifts. c Intracellular view of TRPC1/C5 heteromer (bottom) depicted with quadrilaterals connecting residues on the S6 helix. For comparison, the TRPC5 homomer (top) is drawn alongside. d Top view of the TRPC1/C5 heteromer and TRPC5 homomer (left) and a zoomed-in view of the selectivity filter (right). Densities near the selectivity pore, along with pore-forming or neighboring residues in each subunit, are shown. e Sequence alignment of the selectivity filter-forming region of TRPC1 and TRPC5 in different species (human, Homo sapiens; mouse, Mus musculus; rat, Rattus norvegicus; bovine, Bos taurus). f Ion conduction pathways of each TRPC5 homomer and TRPC1/C5 heteromer, calculated using MOLE54, are shown as black density, highlighting the asymmetric pathway in the heteromer. Residues at the selectivity filter are highlighted by their backbone traces, and those lining the pore constriction site are also shown. g Densities of the pore constriction site and their corresponding residues. H646 in the TRPC1 subunit tightly constricts the pore in addition to N625 in the TRPC5 subunits. h Sequence alignment of the S6 helix demonstrating the differences in residues conserved across species between TRPC1 and TRPC5. i Electrostatic potential surface (scale in kcal mol−1e−1) of the heteromer and the homomer was calculated using ChimeraX56. j Densities of the pore cavity with its corresponding residues are shown: K639 (TRPC1) and N618 (TRPC5). k Pore radius calculation using HOLE55.
Firstly, the global tetrameric arrangement of the TRPC1/C5 heteromer deviated from that of the TRPC5 homomer (Fig. 2a). When aligned by TRPC1, a clockwise rotation of the TRPC1 subunit and an anticlockwise rotation of TRPC5B,C,D were observed. Furthermore, alignment of individual protomers—TRPC1, TRPC5C, and the TRPC5 homomer—revealed differences in the tilt of the S2 helix and the S2–S3 linker (Fig. 2b). This global asymmetry was also evident in the pore-forming S6 helices, as shown by quadrilateral lines connecting A622 (A643 in TRPC1) (Fig. 2c).
Secondly, the selectivity filter is constricted by two additional residues (FSLAH) in the TRPC1 subunit, compared to the TRPC5 subunit (FGL) (Fig. 2d, e). These cause a protrusion of the loop towards the pore axis, with TRPC1’s L601 creating both a constricted selectivity filter and a hydrophobic surface (Fig. 2f). Of note, L601 has been previously identified as a key determinant of monovalent cation selectivity13. Thirdly, the incorporation of TRPC1 results in the substitution of the lower gate’s constriction residue N625 with H646 (Fig. 2g, h). This substitution further narrows the pore at the lower gate, a feature similarly observed in the TRPC1/C4 and shown to influence its cation selectivity13. Lastly, TRPC1’s K639, which faces the ion conduction pathway, is unique compared to the neutral N618 in the corresponding region of TRPC5 subunits (Fig. 2i, j). This lysine residue creates a positive charge in the pore, reducing calcium permeability in TRPC1/C413. Taken together, the incorporation of TRPC1 creates asymmetry in various regions of TRPC1/C5, including the selectivity filter, constriction gate, and central cavity, resulting in a narrower and more asymmetric pathway compared to the homomeric channel (Fig. 2k).
Dynamic structural features of the cytosolic domains in heteromeric TRPC1/C5
We next examined the structural differences between TRPC1/C5 and TRPC1/C4, two heteromeric TRPC channels that have been structurally characterized. While TRPC1 shares ~47% sequence identity with TRPC5, TRPC4 and TRPC5 share ~ 77%. Despite high sequence homology, the apo-state structures of TRPC1/C5 and TRPC1/C4 display marked differences, particularly in the intracellular regions.
Before further analysis, it is noteworthy that our previously determined structure of human TRPC5 in nanodiscs revealed two distinct conformations, referred to as Class 1 and Class 2, based on the arrangement of the cytosolic domain18. These classes are defined by the proximity between the ARD and coiled-coil domain (CCD): in Class 1, the ARD is distant (> 10 Å) from both its own and adjacent CCDs, whereas in Class 2, the connecting helix (CH) forms a loop that brings the CCD closer to its ARD, with the ARD rotating toward the CCD to facilitate tighter interaction (< 10 Å). Interestingly, within the TRPC5 homomeric structures in the TRPC1/C5 heteromer dataset (Supplementary Fig. 1e), Class 1 matches the previous reported Class 1, but Class 2 shows a unique ARD–CCD interaction (Fig. 3a). Instead of forming a loop at the CH–CCD junction, the CCD tilts toward the ARD of an adjacent protomer, creating a close ARD–CCD proximity.
a Ankyrin repeat domain (ARD) and coiled-coil domain (CCD) conformation of the TRPC5 homomer from the TRPC1/C5 heteromer dataset (left). Class 1 shows more extended arrangement, whereas Class 2 exhibits a clockwise ARD rotation and CCD tilt, resulting in reduced ARD–CCD distance with the adjacent subunit. b In the TRPC1/C4 heteromer (PDB: 8WPL), the CCD adopts a straight conformation. All subunits (one TRPC1 and three TRPC4 subunits) show close ARD–CCD proximity. c In the TRPC1/C5 heteromer, the ARD–CCD axis is non-linear, with the CCD tilted toward the TRPC1 and TRPC5D subunits. Alignment across chains reveals varied ARD–CCD distances, highlighted in angstroms (Å).
According to this classification, the previously resolved TRPC1/C4 heteromer (PDB: 8WPL) corresponds to Class 2, characterized by a CH loop and close ARD–CCD proximity (Fig. 3b)13. In contrast, the TRPC1/C5 heteromer exhibits a markedly bent CCD orientation, directed toward the ARD of the TRPC1 subunit (Fig. 3c). Notably, whereas previously reported TRPC channels adopt a uniform conformational state—either Class 1 or Class 2—across all four subunits, the TRPC1/C5 heteromer shows a mixed conformation: the TRPC1 subunit adopts a Class 2 conformation, while the three TRPC5 subunits adopt Class 1 conformations (Supplementary Fig. 4).
This observation supports the notion that TRPC5 preferentially adopts a Class 1 conformation in lipid nanodiscs, whereas TRPC1 and TRPC4 tend to maintain the Class 2 conformation, regardless of the reconstitution method. Collectively, these observations reveal that the dynamic nature of the ARD–CCD interface in TRPC channels is modulated by subunit composition, contributing to their conformational versatility within a conserved architecture.
During the preparation of our manuscript, a preprint article describing chimeric human TRPC1 and mouse TRPC5 in detergent conditions was uploaded31. Although the overall architecture around the pore-forming regions appears similar, several distinct features observed in our study—including the bending of the CCD, the distal positioning of the helix–loop–helix (HLH), and the assembly of TRPC5 homomers—were not reported in the preprint. While further validation is warranted, these differences are likely attributable to the distinct membrane environments, particularly the use of lipid nanodiscs in our study, which play a critical role in shaping TRPC5 structural dynamics.
Distinct intersubunit interactions in TRPC5 and TRPC1/C5
To investigate the molecular basis of heteromer formation, we analyzed the intersubunit interactions within the TRPC1/C5 heteromer, which are distinct from those in the TRPC5 homomer (Fig. 4). First, at the outer pore entrance region, K560 in TRPC5 is replaced by Q578 in TRPC1 (Fig. 4a). In TRPC5 homomer, K560, together with E559, establishes a complementary electrostatic interaction (Fig. 4b). This substitution with an electroneutral residue in TRPC1/C5 weakens the interaction with the adjacent subunit and introduces an asymmetric charge distribution at the pore entrance. The significance of this residue was previously demonstrated, as the K560N mutation in human TRPC5 reduces EA-induced currents32. Second, the interprotomer interactions around the domain-swapped regions also differ, as we designated each junction as Views I–IV (Fig. 4c). At the TRPC5–TRPC5 junction, R466 in the S3 helix forms a hydrogen bond with Y542 in the adjacent subunit’s S5 helix (Fig. 4d, top). As shown in View II, the TRPC1 subunit contains R486 at the corresponding position, preserving the interaction between TRPC5 (S5 helix) and TRPC1 (S3 helix) (Fig. 4e). However, in View I, the equivalent position of Y542 in TRPC5 is replaced by G562 in TRPC1, disrupting this interaction. This loss further results in the absence of density in the outer pore loop preceding the pore helix (PH) in TRPC1, a region that remains clearly visible in TRPC5 subunits.
a, b Electrostatic charge representation of the pore entrance region of TRPC1/C5 heteromer (a) and TRPC5 homomer (b), calculated in ChimeraX56 (scale in kcal mol−1e−1). The TRPC1/C5 heteromer includes neutral Q578, which corresponds to positively charged K560 in the equivalent region of TRPC5. c Cryo-EM structure of the TRPC1/C5 heteromer, with dotted boxes (left) and a schematic structure (right) highlighting intersubunit junctions expanded in (d–i). d Close-up of the S3–S5 intersubunit interface (top) and intraprotomeric region with cholesteryl hemisuccinate (CHS, yellow) comprising the S1 helix, S4–S5 linker, and pre-S1 domain (bottom) in the TRPC5 homomer. e Equivalent S3–S5 intersubunit interactions in the TRPC1/C5 heteromer, shown across Views I–IV. In View I, Y542 of TRPC5 is substituted by G562 in TRPC1, leading to a loss of electrostatic interaction. f Corresponding intraprotomeric region in the TRPC1/C5 heteromer, with lipid-like densities visualized in transparent yellow. Notably, in View II, the CHS-like density is absent. g Close-up of the helix–loop–helix (HLH) region in the TRPC5 homomer. h HLH region in the TRPC1/C5 heteromer, shown in Views I and II. In View I, the loss of loop and interaction is indicated by a dotted circle. i HLH region in the previously resolved TRPC1/C4 heteromer (PDB: 8WPL), shown in the same orientations for comparison.
Third, the lipid-like density in the intraprotomer region, formed around the pre-S1 helix, S1 helix, S4 helix and TRP domain, also differs in TRPC1. CHS is commonly modeled in this region of many TRP channels (Fig. 4d, bottom). Similarly, the densities in TRPC5 subunits closely resemble CHS, exhibiting hydrophobic interactions with the VSLD and a hydrogen bond with Y316 and its succinate head. However, in View II, highlighting the intraprotomer region of TRPC1, the CHS-like density is absent and is possibly replaced by a phospholipid, which constitutes the majority of the lipids used to reconstitute the nanodiscs (Fig. 4f). This substitution is likely due to the loss of an oxygen atom caused by the introduction of F336 in place of Y316, while the VSLD retains predominantly hydrophobic residues. Notably, this residue in mouse TRPC5 has been shown to be critical for channel stability, as the Y316A mutation resulted in a significant loss of channel function33.
Finally, the HLH domain displayed dissimilar interactions. Although no residues differed, this region exhibited changes in densities and side-chain orientations. At the junction of TRPC5–TRPC5, Q309 in HLH 8 forms a hydrogen bond with two serine residues, S189 and S193, in the adjacent subunit’s HLH 2 (Fig. 4g). While this interaction is conserved in TRPC1/C5 heteromer, the electrostatic interaction between R323 in the HLH 8–pre-S1 loop and E236 (E253 in TRPC1) in the HLH 4–HLH 5 loop is disrupted (Fig. 4h). In View I of the TRPC1/C5 heteromer, the loop containing R323 in the TRPC5D subunit lacks resolved density, suggesting structural flexibility. Consistently, the side chain of E253 in TRPC1 is oriented away from the predicted position of R323, preventing the formation of a potential electrostatic interaction between these two residues. In contrast, this interaction in the previously determined TRPC1/C4 structure (PDB: 8WPL)13 was similar to that observed in homomeric interaction, involving both hydrogen bonding and electrostatic interactions (Fig. 4i). Overall, TRPC1/C5 heteromer exhibits weaker interfaces at the S3 helix–S5 helix and HLH regions, reflecting distinct subunit interactions and lipid-binding properties.
Binding modes of (−)-Englerin A and riluzole in TRPC5
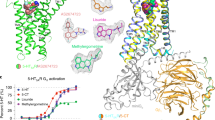
To investigate the binding mode of EA in TRPC5, we initiated structural determination of TRPC5 in complex with EA. Despite multiple attempts, we were unable to resolve EA-bound structures of either the TRPC5 homomer or the TRPC1/C5 heteromer. To overcome this, we adopted a dual-agonist strategy targeting distinct binding sites to achieve a synergistic activation effect—an approach previously shown to be effective for TRPM8 and TRPM3 channels34,35. In this context, riluzole was identified as a suitable co-agonist, as it exhibited a clear synergistic effect with EA on TRPC5 activation (Fig. 5a). Co-application of both ligands elicited a greater response than the sum of their individual effects (Fig. 5b). Utilizing this strategy, we successfully determined the EA-bound TRPC5 structure, which revealed well-resolved densities for both ligands (Fig. 5d).
a A representative current trace of TRPC5 at − 60 mV upon 20 μM riluzole (beige), 30 nM (−)-Englerin A (EA, pink), and both agonists (green), with I–V relationships from peak points indicated by triangles shown as an inset. b Statistical data of current responses at − 60 mV induced by each agonist combination (n = 11, riluzole; n = 7, EA; n = 7, riluzole + EA). c Summary of dose-dependent responses of TRPC channels with WT or mutant TRPC1/C5 subunits (n = 4–9 for each EA concentration condition, currents obtained at − 60 mV. Detailed data are available in the Source Data file). d Cryo-EM map of TRPC5 homomer (Class 1) in complex with EA and riluzole. The agonist binding regions are highlighted. e–g Close-up view of the interprotomer cavity of the EA-bound TRPC5 (e), apo TRPC5 (f), or TRPC1/C5 heteromer (g). F520 (TRPC5) or F540 (TRPC1) is indicated by the dotted circle. h Close-up view of the VSLD of riluzole-bound TRPC5 (left) and TRPC1 subunit in TRPC1/C5 heteromer (right). i Pore radius calculation using HOLE55 to compare the effect of agonists binding to TRPC5. j Sequence alignment of the interprotomer site-forming region of human TRPC1–7. n represents a number of independent cells. Comparisons between groups were conducted using two-sided Student’s t test. Bars represent the mean ± s.e.m. **p < 0.01.
Consistent with the previously described TRPC5 homomeric channel, 3D classification focused on the cytosolic domain revealed that the TRPC5 adopts two distinct conformational states (Class 1 and Class 2), resolved at 2.43 Å and 3.03 Å, respectively (Supplementary Fig. 5 and Supplementary Table 2). In addition, two distinct non-protein densities were clearly identified: riluzole near the Ca2+ binding site of the VSLD and EA within the interprotomer cavity formed by the S5 helix, PH, and S6 helix of the adjacent subunit (Fig. 5d). Despite the binding of two agonists interacting at distinct sites, the most constricted pore radii are less than 1 Å in both classes, indicating the closed conformations (Fig. 5i).
The density within the interprotomer cavity of the pore domain, originally occupied by diacylglycerol (DAG) in apo TRPC518,24, was assigned to EA based on its distinct size and shape, closely matching EA (Fig. 5e, f). EA forms extensive interactions with TRPC5, spanning from its head to tail. The indole ring of W577 forms a hydrogen bond with EA’s glycoloyl head alcohol group, while Q573 forms a polar contact with the same oxygen atom. A602 on the adjacent S6 helix makes a van der Waals interaction with this oxygen, while F576 on the PH and T607 on the adjacent S6 helix interact hydrophobically with EA’s guaiane core. L521 on the S5 helix and V610 on the adjacent subunit hydrophobically interact with EA’s cinnamoyl tail (Fig. 5e). In addition, EA’s cinnamoyl tail engages with F520 on the S5 helix, a residue that does not interact with DAG in apo TRPC5 (Fig. 5e, f). Intriguingly, Y524 on the S5 helix forms a parallel π–π stacking interaction with the phenyl ring of EA’s cinnamoyl tail, contrasting with antagonist-bound TRPC1/4/5 structures, where Y524 or equivalent residues adopt perpendicular orientations (e.g., Pico145) or show no interaction (e.g., HC-070)13,23,24. Similarly, photoswitchable TRPC4/5 agonists align parallel with Y524, unlike their antagonistic forms25, suggesting an agonist-specific role.
Whole-cell patch clamp recordings on single mutants targeting these interacting residues supported the interaction between EA and TRPC5 (Fig. 5c). The F520A and Y524A mutants were nearly unresponsive to EA, despite proper membrane expression (Supplementary Fig. 6), indicating that these residues are critical for EA binding or channel activation (Fig. 5c). Meanwhile, the Q573A and W577A mutants required higher EA concentrations to achieve full activation compared to WT channel, which is consistent with previous observations from intracellular calcium measurements23.
The second non-protein density in the VSLD near the Ca2+-binding site was identified as riluzole based on its position, size, and shape, consistent with the known structure of riluzole-bound TRPC5 (Fig. 5h)28. Riluzole forms π–π interactions with Y374 on the S1 helix and F414 on the S2 helix, along with hydrogen bonds to D439 on the S3 helix and S495 on the S4 helix28. The binding mode of riluzole in both classes closely matched that of the previously reported structure, with VSLDs exhibiting Cα root mean square deviation (RMSD) less than 0.7 Å, indicating nearly identical conformations (Supplementary Fig. 7)28. While riluzole acts as an agonist for TRPC5 homomers and TRPC1/C5 heteromers, TRPC1’s corresponding residues differ significantly, including M434, S459, and F515, which are less capable of π–π stacking or hydrogen bonding (Fig. 5h). These differences, combined with the absence of Ca2+ in TRPC1’s VSLD, likely reduce riluzole sensitivity in the TRPC1 subunit within the TRPC1/C5 heteromer.
Channel selectivity of (−)-Englerin A within the TRPC subfamily
To elucidate the structural determinants of EA selectivity within the TRPC subfamily, we performed a structure-guided functional analysis using swap-mutant channels. The EA-interacting residues in TRPC5 are highly conserved in TRPC4 but differ in TRPC1 (Fig. 5j). In particular, TRPC5’s Y524 and Q573, key components of the EA-binding site, are replaced by F544 and F592 in TRPC1 (Fig. 5e, g). To assess their roles in EA binding, we generated corresponding swap mutants.
Swapping these residues significantly impacted EA affinity (Fig. 5c). The Y524F/Q573F double mutant of TRPC5 (TRPC5YQ/FF) exhibited a more than 30-fold increase in the half-maximal effective concentration (EC50) for EA compared to the WT TRPC5 homomer, even exceeding that of the WT TRPC1/C5 heteromer. Conversely, the TRPC1/C5 heteromer containing the TRPC1 F544Y/F592Q mutant (TRPC1FF/YQ) showed increased EA sensitivity relative to the WT TRPC1/C5 heteromer. These changes primarily arise because F592 in TRPC1 cannot form a polar interaction with EA’s alcohol group, in contrast to Q573 in TRPC5, resulting in weaker EA binding. This explains why the TRPC5YQ/FF homomer exhibits even lower EA sensitivity than the WT TRPC1/C5 heteromer, which retains three EA-interacting glutamines.
Interestingly, the specificity of EA for TRPC1/4/5 over TRPC3/6/7 can be attributed to differences in the binding pocket. In TRPC3/6/7, Q573 is not conserved, and the preceding L572 is substituted by a phenylalanine, likely hindering EA accommodation (Fig. 5j). Consistent with this, substituting these residues of TRPC5 individually with TRPC3/6/7 counterparts greatly diminished EA sensitivity (Supplementary Fig. 8c). Conversely, we tested whether introducing the corresponding TRPC5 residues into TRPC6 could confer EA responsiveness. However, the F675L/K676Q double mutant in TRPC6 neither restored EA binding nor preserved DAG activation (Supplementary Fig. 8d), consistent with previous studies36,37 highlighting the essential role of these residues in DAG-mediated gating. Together, these swap-mutant analyses show that EA exhibits specificity for TRPC1/4/5 channels—despite binding to the interprotomer cavity, a ligand-binding site shared across the TRPC family—with particular preference for TRPC5 and TRPC421,37,38.
Discussion
We determined the structure of the human TRPC1/C5 heteromeric channel, providing additional insights into the structural characterization of heteromeric TRP channels. Recently, our group also reported the structure of the human TRPC1/C4 heteromeric channel, highlighting the structural and functional changes induced by the incorporation of a single TRPC1 subunit13. A comparison of the TRPC1/C4 and TRPC1/C5 structures, despite the ~77% sequence homology between TRPC4 and TRPC5, revealed noticeable differences in the apo-state, particularly in the intracellular regions. These differences are largely attributed to the dynamic ARD–CCD interface. In addition, we characterized the binding mode of the TRPC1/4/5-specific agonist EA to TRPC5, identifying key residues critical for its specificity. While the binding of EA to the DAG site had been previously proposed based on intracellular calcium measurements of TRPC5 F576A and W577A mutants by D. J. Wright and colleagues23, direct structural evidence was lacking. Our study not only confirmed the EA binding site but also provided structural and functional evidence for EA’s specificity toward TRPC1/4/5 channels and its selectivity for TRPC homomers over heteromers (Fig. 5 and Supplementary Fig. 8).
The TRPC1/C5 channel plays a significant role in brain function, establishing it as an emerging pharmacological target1,39,40. Despite their low abundance—approximately 500 times less than AMPA-type glutamate receptors6 and metabotropic GABA (GABAB) receptors41—TRPC1/4/5 channels have a notable impact on the brain. Recent studies have spotlighted TRPC5’s involvement in neurological disorders, including autism, depression, anxiety, and obesity, underscoring its clinical relevance42. In addition, epitope-based brain extractions indicate that heteromeric TRPC1/4/5 channels are more prevalent than homomeric forms, suggesting that homomers are less common in native brain tissues6. Intriguingly, this study further revealed that only a single TRPC1 subunit incorporates into each tetramer, resulting in a 1:3 stoichiometry with either TRPC4 or TRPC5. The ubiquitous expression of TRPC1 further supports the dominance of heteromeric TRPC1/4/5 channels in native tissues, with their dysfunction linked to broader physiological and organismal issues. Functionally, the TRPC1/C5 heteromer is closely associated with homomeric TRPC5, particularly in its role in neurological conditions43,44. Notably, in Huntington’s disease, TRPC1/C5 exerts a neuroprotective effect by preventing toxic calcium influx mediated by homomeric TRPC5, thereby slowing disease progression45.
In this context, a molecular understanding of how TRPC5 homomers and heteromers are modulated by agonists and antagonists is crucial. A recent study suggested that lysophosphatidylcholine (LPC) activates TRPC5 channels by directly binding to the EA binding site, independently of Ca2+ or PIP2, indicating that this site may accommodate multiple lipophilic modulators46. This finding raises a critical question: whether EA-mediated activation operates through a similar Ca2+- and PIP2-independent mechanism or cooperates with these physiological modulators to achieve channel gating, which remains a key question for future investigation.
Consistent with the functional importance of this binding pocket, direct mutagenesis of residues within the EA-binding site in our study markedly reduced agonist sensitivity, providing experimental validation of its role in channel activation (Fig. 5c and Supplementary Fig. 8c). Collectively, these findings suggest that TRPC channel activation profiles are fine-tuned by the specific arrangement of residues and ligands within the interprotomer cavity, which serves as a binding site for EA and other lipophilic ligands but also contains the highly conserved LFW motif (residues 575–577 in TRPC5), known to play a critical role in channel gating across the TRPC family12,20,47.
Beyond the significant insights provided in this study, including the structures of human TRPC1/C5 and human TRPC5 bound to potent agonists (EA and riluzole), the pore is observed in a closed conformation. As no open-state, intermediate, or sensitized structures have yet been resolved for TRPC1/4/5 channels, further investigations into the open-state structure are necessary to uncover the activation mechanism. Nevertheless, we believe that the structure of TRPC1/C5 and the identification of the EA binding mode establish a solid framework for advancing the molecular understanding of TRPC1/4/5 channels, paving the way for pharmacological developments aimed at treating anxiety and depression.
Methods
Expression and purification of the human TRPC1/C5 heteromer and TRPC5 homomer in lipid nanodiscs
Both TRPC5 and TRPC1 used in this study were cloned into in-house-modified pEG BacMam vectors. To achieve high expression levels and stability, a C-terminally truncated human TRPC5 (1–765) sequence was inserted into a vector containing a C-terminal EGFP and FLAG tag. For full-length TRPC1 (1–793), the same vector used in a previous study, which includes an N-terminal Twin-StrepII tag and mCherry followed by an HRV-3C protease site, was used13. The molecular cloning was performed using the following primers:
hTRPC5 forward, 5′-CGCGGAATTCGCCACCATGGCCCAACTGT-3′
hTRPC5 reverse, 5′-GAGAACTTCAAGTCTAGATTTTCTATTTCCCAAGAGGTCAAGCACTTCAT-3′
The constructs were expressed by baculovirus-mediated transduction of human embryonic kidney (HEK) 293S GnTI− (N-acetylglucosaminyltransferase I-negative, ATCC) suspension cells. Baculovirus was amplified to the P3 stage using Spodoptera frugiperda (Sf9) insect cells according to the manufacturer’s protocol. For each batch of baculovirus, the expression ratio was screened to achieve similar levels of expression between EGFP and mCherry. The determined ratio of each P3 baculovirus was added to the HEK293S GnTI− suspension at a cell density of 3.0 million cells ml−1 at 37 °C. After 16–18 h, 10 mM sodium butyrate was added to the culture, then the temperature was adjusted to 30 °C. After 48 h, the cell pellet was harvested following a PBS wash and stored at − 80 °C for future purification.
All purifications were conducted at 4 °C or on ice. The cell pellet from 2.4 L was resuspended in buffer A (100 mM Tris-HCl, pH 8.0, 150 mM NaCl) with 4 μg ml−1 leupeptin, 4 μM pepstatin, 4 μM aprotinin, and 1 mM phenylmethylsulfonyl fluoride. After 30 min, 1% (w/v) digitonin was added, and the mixture rotated for 2 h at 4 °C. After solubilization, the cell lysate was centrifuged at 100,000 × g for 1 hour. The cleared lysate was mixed with anti-FLAG resin (Wako) for 3 h with rotation at 4 °C. After binding, the resin was collected using a gravity-flow column (Bio-Rad), and washed with 50 column volumes (CV) of buffer A supplemented with 0.1% digitonin. Protein was eluted with 5 CV of buffer containing 0.2 mg ml−1 FLAG peptide. The eluates were checked via SDS-PAGE analysis and in-gel fluorescence to confirm the presence of TRPC1 co-elution.
For the nanodisc reconstitution, MSP2N2 scaffold protein and soybean polar lipid extracts dissolved in 0.5% digitonin and 0.05% cholesteryl hemisuccinate (CHS) were used at a molar ratio of 1:3:200, and Bio-Beads SM-2 (Bio-Rad) was used to remove digitonin. Subsequently, nanodisc-reconstituted TRPC1/C5 and TRPC5 were applied to Strep-Tactin resin and incubated overnight. After washing with buffer A supplemented with 1 mM DTT, TRPC1/C5 was eluted using 5 mM d-desthiobiotin. The elutions were analyzed using SDS-PAGE, and the fractions containing TRPC1/C5 were concentrated using a 100 kDa molecular weight cut-off (MWCO) centrifugal filter (Millipore). In-house-purified HRV-3C was added to the concentrated fractions to remove tags, and the protein was further purified using SuperoseTM 6 increase 10/300 GL column (Cytiva) pre-equilibrated with buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM DTT. The peak fractions containing TRPC1/C5 were further concentrated to 1.1 mg ml−1 for cryo-EM analysis.
For the preparation of TRPC5 in complex with EA and riluzole, the flow-through (FT) fraction from the Strep affinity purification step during TRPC1/C5 purification was used. This FT contained TRPC5 reconstituted in MSP2N2 lipid nanodiscs. To remove residual FLAG peptides, the FT was buffer-exchanged by repeated cycles of concentration and dilution (10 mL to 1 mL, three times) using a 100 kDa MWCO centrifugal filter. A second FLAG affinity purification was then performed, followed by size-exclusion chromatography under the same buffer conditions as described above. The resulting peak fractions were concentrated to 4.0 mg ml−1 for cryo-EM analysis.
Cryo-EM sample preparation and data acquisition
For grid preparation, 4 μl of purified TRPC1/C5 heteromer was applied onto glow-discharged Quantifoil 1.2/1.3 300-mesh Cu holey carbon grids (Electron Microscopy Sciences; EMS). For TRPC5 homomer with agonists, 1 mM (−)-Englerin A (EA) (PhytoLab) and 0.5 mM riluzole (Sigma-Aldrich) dissolved in dimethyl sulfoxide were added and incubated for 30 min prior to grid preparation. All samples were blotted at 4 °C with 100% humidity for a blot time of 7 s, and plunged into liquid ethane using Vitrobot Mark IV (FEI).
For TRPC1/C5 heteromer datasets, cryo-EM data collection was performed using a Krios G4 (Thermo Fisher Scientific) at 300 kV accelerated voltage at the Institute for Basic Science (IBS). The movies were recorded using a BioQuantum K3 detector (Gatan Inc.) in counting mode with EPU automated data-acquisition software. For TRPC1/C5, a total of 8,046 movies (51 frames) were collected with a pixel size of 0.828 Å/pixel and a defocus range of − 1.0 to − 2.0 μm. Each movie was acquired with a total dose of 66.8 e−/Å2.
For the TRPC5 homomer with EA and rliuzole, cryo-EM data collection was performed using a Krios G4 equipped with SelectrisX and Falcon 4i detector (Thermo Fisher Scientific) at the National Center for Inter-university Research Facilities (NCIRF), Seoul National University. The movies were recorded with a slit width of 10 eV in counting mode with EPU automated data-acquisition software. Total of 6,858 movies (40 frames) with a pixel size of 0.95 Å/pixel and a defocus range of − 1.0 to − 2.0 μm. Each movie with a total dose of 44.3 e−/Å2.
Cryo-EM data processing
For the TRPC1/C5 heteromer, a total of 8046 movies were subjected to patch motion correction and CTF estimation in CryoSPARC version 4.1.148. 7181 movies were selected according to a CTF cut-off value of 10 Å. After initial templates were made via several 2D classifications, the particles were subjected to Topaz training and extraction, resulting in 289,935 particles49. The resulting particles were 3D reconstructed using an ab initio approach, which was further classified using 3D classification with 4 classes and a target resolution of 3 Å. At this stage of processing, one optimal tetrameric 3D volume was further classified. The optimal tetrameric 3D volume was subjected to another round of 3D classification with the same parameters. Only two classes emerged: one representing the TRPC1/C5 heteromer and the other representing the TRPC5 homomer. At this stage, no symmetry was applied to the 3D structures, and amino acid side chains were analyzed for a total of eight protomers. The TRPC5 homomer map in C1 symmetry had a gold-standard Fourier shell correlation (GSFSC) resolution of 3.04 Å. The particles representing TRPC1/C5 heteromer were further subjected to reference-based motion correction and non-uniform refinement, resulting in GSFSC resolution of 2.7 Å. For the TRPC5 homomer, the volume was imposed with C4 symmetry during non-uniform refinement, and further 3D classification was carried out with a mask comprising ARD and CCD regions. Based on the distance between ARD and CCD, similar classes were combined to carry out reference-based motion correction and non-uniform refinement. The final 3D volume had GSFSC resolution of 2.62 Å and 2.90 Å for Class 1 and Class 2, respectively.
For TRPC5 in complex with EA and riluzole, a total of 6,858 movies were processed for patch motion correction and CTF estimation in CryoSPARC version 4.1.148. Several rounds of 2D classifications were performed to separately select particles corresponding to side and bottom views; after duplicate removal, 459,895 particles were retained for ab initio reconstruction (C1), non-uniform refinement (C4), and heterogeneous refinement (C4). Among the four resulting classes, the two with the best resolutions were selected, and the particles were further divided by 3D classification using masks focused either on the ARD and CCD regions or on the transmembrane domain. As a result, 270,318 particles were assigned to the Class 1 conformation with a GSFSC resolution of 2.43 Å, and 22,419 particles to the Class 2 conformation with a GSFSC resolution of 3.03 Å. All 3D volumes at this stage showed clear densities for both EA and riluzole.
Model building, refinement and validation
For the TRPC1/C5 heteromer, TRPC1 from the previously deposited TRPC1/C4 heteromer (PDB: 8WPL) was used as the initial model. For the three TRPC5 subunits in the TRPC1/C5 heteromer and TRPC5 homomers, the previously deposited human TRPC5 (PDB: 7X6C, 8GVW) was used as the initial model. All models were manually adjusted in Coot version 0.9.4.150. The models for EA and riluzole were generated using the eLBOW module in Phenix51,52, and manually docked into the densities and adjusted in Coot. The models were further refined by real-space refinement in Phenix52, with the MolProbity53 tool for validation. Pore radius was calculated using MOLE54 and HOLE55. All figures were prepared using UCSF ChimeraX56.
Electrophysiology
Human embryonic kidney (HEK) 293-T cells (ATCC) were cultured following the manufacturer’s guidelines. For whole-cell patch clamp recordings, cells were plated in 12-well plates and transfected at the subsequent day using the transfection reagent X-tremeGene 9 (Roche Molecular Biochemicals) according to the provided protocol. Plasmid co-expression was achieved with a standard 1:1 transfection ratio, unless specified differently. Following 24–48 h of incubation, cells were undergone trypsinization and were transferred to a small chamber on the stage of an inverted microscope (TE2000S or Eclipse Ti, Nikon), allowing 10 min for cell attachment before electrophysiological recording. Whole-cell patch clamp recordings were performed using borosilicate patch pipettes (Harvard Apparatus) with 2–4 MΩ pipette resistance. Patch clamp recordings utilized an Axopatch 200B amplifier (Molecular Devices), with data acquisition managed through pCLAMP software version 11.1 and a Digidata 1550B interface (Molecular Devices). Ramp pulse protocols consisted of 550 ms duration sweeps from − 120 mV to + 100 mV, delivered at 0.4 mV ms−1 resolution. The holding membrane potential was maintained at − 60 mV, with ramp pulses applied every 10 s. Data were sampled at 2–5 kHz and filtered using a low-pass Bessel filter with a 2 kHz cut-off frequency.
Standard recordings employed Normal Tyrode solution, containing (in mM): 135 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 Glucose, 10 HEPES (pH 7.4, adjusted with NaOH). Bath solutions were continuously perfused at 1–2 ml min−1. The pipette solution contained (in mM): 140 CsCl, 0.5 EGTA, 10 HEPES, 3 Mg-ATP, 0.2 Tris-GTP (pH 7.2, adjusted with CsOH), unless otherwise mentioned. For TRPC6 channel recordings, intracellular solution was prepared by replacing 0.5 mM EGTA with 10 mM BAPTA, while modifying the concentration of CaCl2 to 2.9 mM to achieve a free calcium concentration of 100 nM (calculated using WebMaxC Extended Calculator (UC Davis Pharmacology))57. Recordings were conducted at room temperature (18–20 °C), with data analysis performed using pCLAMP (v.11.1, Molecular Devices), OriginPro (2023b, OriginLab), and Prism (9.3, GraphPad) software. All reagents were sourced from Sigma Aldrich.
Molecular cloning and mutagenesis
Plasmid constructs of human TRPC1α inserted into the pcDNA3.1 vector and human TRPC1α tagged with CFP at the amino terminus were kindly donated by Drs. M. Mederos y Schnitzler and V. Luis, respectively. Plasmid constructs of human and mouse TRPC5 in the pEGFP-N1 vector were kindly donated by Drs. S. Kaneko and Y. Mori, respectively. Wild-type and mutant human TRPC5 channels were utilized in a C-terminally truncated form (1–765 amino acids), unless otherwise mentioned. The plasmid construct of human TRPC6 in the p3xFLAG-CMV-10 vector was kindly donated by Dr. K. P. Lee. Site-directed mutagenesis was performed using custom-designed missense primers and the PrimeSTAR HS DNA polymerase system (Takara Bio). DNA sequencing confirmed the accuracy of all channel construct sequences. Dimeric concatemers were generated as previously mentioned58. In this study, we used the TRPC5–TRPC1 concatemer linked by the sequence (SAGSGGSGSLIEPG), the TRPC5–TRPC5 concatemer linked by the sequence (AELKLRILQSTVP), and the TRPC1–TRPC1 concatemer linked by the sequence (GSGGSGSLIS).
Live-cell image acquisition and Förster Resonance Energy Transfer (FRET) measurements
HEK293-T cells were grown on 35 mm coverslip-bottom dishes for imaging and measurement of FRET efficiency. For FRET measurements, three different types of cDNA-containing plasmids were co-transfected at a 9:1:3 ratio (fluorescent untagged human TRPC5: ECFP–TRPC5 or ECFP–TRPC1α: EYFP–TRPC5 or EYFP–TRPC1α). Live cells were imaged on an Eclipse Ti inverted microscope (Nikon) equipped with an S Fluor 100 × oil objective lens (Nikon). The fluorescence intensities required for calculating donor dequenching FRET efficiencies were acquired from a pE-4000 LED light source (CoolLED) to three-cube FRET (excitation, dichroic mirror, filter) through a fixed collimator: CFP (ET 435/20 nm, ET CFP/YFP/mCherry beam splitter, ET 470/24 nm, Chroma); and YFP (ET 500/20 nm, same dichroic, ET 535/30 nm, Chroma). After identifying target cells in the field of view, excitation wavelengths for YFP and CFP were applied sequentially. Switching between these excitation settings typically took under 200 ms, a duration dictated mainly by the exposure time rather than the control speed limit of the light source, under 0.5 ms. Emitted fluorescence from each fluorophore was selectively acquired through a DV2 dual-view beam splitter (Teledyne Photometrics) with a filter pair of ET470/24 and ET535/30 (Chroma). Split images were acquired using a cooled 3 MHz (14 bit) sCMOS (optiMOS) camera (Teledyne Photometrics) at 100–300 ms exposure without binning, managed through MetaMorph 7.10 (Molecular Devices) software. The acquired images were aligned using an in-house program written in MATLAB (MathWorks), and FRET-gradient maps were rendered using MetaMorph 7.10.
For epifluorescence imaging to validate the membrane trafficking of EA binding site mutants, which do not respond to the highest concentration of EA treatment, EGFP-tagged TRPC5 mutant channels were transfected into HEK293-T cells. Before the acquisition, cells were fixed with 0.5% formaldehyde for 15 mins, and then stained with 0.5 μg ml−1 of WGA-Alexa Fluor 633 (Invitrogen) for 10 mins. For the acquisition, most settings were identical to those used for FRET acquisition, but the excitation wavelength for the membrane marker was changed to 580 nm, and the DV2 dual-view beam splitter was equipped with a filter pair of ET510/80 m and ET630/75 m (Chroma).
Computation of FRET efficiency (E FRET)
For acceptor photobleaching experiments, donor dequenching was measured by selectively photobleaching EYFP acceptors while monitoring the recovery of ECFP donor fluorescence. For donor dequenching measurements, the FRET efficiency (EFRET), which incorporates both the intrinsic efficiency (E) and the bound donor fraction (Db), was calculated as:
Here, SCUBE(SPECIMEN) denotes intensity measurements using the CFP filter cubes on cells expressing both donor and acceptor (DA). Thus, SCFP(DA)before and SCFP(DA)after represent donor fluorescence intensities before and after acceptor photobleaching, respectively. We confirmed in preliminary experiments that acceptor emission could be sufficiently photobleached under our imaging system settings, decreasing to less than 5% of the initial intensity and thereby ensuring complete elimination of energy transfer. Acceptor photobleaching was performed by illuminating the sample with maximum LED intensity for 2 min.
All measurements were performed on regions of interest encompassing entire cells. Statistical analyses were performed on data from independent transfections, with n values representing the number of individual cells analyzed. Competition experiments utilized transfection ratios optimized to achieve a substantial excess of untagged competitors while maintaining detectable fluorescence signals from tagged subunits. Overall computations were implemented via a custom MATLAB script (Mathworks).
Statistics
Data are presented as means ± standard error of the mean (SEM). Comparisons between groups were conducted using two-sided Student’s t test, with statistical significance set at P < 0.05. Precise p-values are documented in the source data file. Statistical analyses were performed using OriginPro (2023b, OriginLab).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density maps and corresponding atomic coordinates for TRPC1/C5, TRPC5 Class 1, TRPC5 Class 2, TRPC5 with EA and riluzole Class 1, and TRPC5 with EA and riluzole Class 2 have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) under EMDB accession codes EMD-62343 (TRPC1/C5), EMD-62344 (TRPC5 Class 1), EMD-62345 (TRPC5 Class 2), EMD-63533 [https://www.ebi.ac.uk/emdb/EMD-63533] (TRPC5 with EA and riluzole Class 1), and EMD-63534 [https://www.ebi.ac.uk/emdb/EMD-63534] (TRPC5 with EA and riluzole Class 2), and under PDB accession codes 9KHI (TRPC1/C5), 9KHJ (TRPC5 Class 1), 9KHK (TRPC5 Class 2), 9LZY (TRPC5 with EA and riluzole Class 1), and 9LZZ (TRPC5 with EA and riluzole Class 2), respectively. The in-house image alignment code used in this study have been deposited in the Zenodo OpenAIRE database under accession code 17539483. The source data underlying Fig. 1e, g, 5b, c, Supplementary Fig. 8c and d are provided as a Source Data file. Uncropped scans of all gels in the Supplementary Figs. are provided at the end of the Supplementary Information. Source data are provided in this paper.
References
Strübing, C., Krapivinsky, G., Krapivinsky, L. & Clapham, D. E. TRPC1 and TRPC5 form a novel cation channel in ümammalian brain. Neuron 29, 645–655 (2001).
Schaefer, M. Homo- and heteromeric assembly of TRP channel subunits. Pflüg. Arch. 451, 35–42 (2005).
Cheng, W., Sun, C. & Zheng, J. Heteromerization of TRP channel subunits: extending functional diversity. Protein Cell 1, 802–810 (2010).
Cheng, W. et al. Heteromeric heat-sensitive transient receptor potential channels exhibit distinct temperature and chemical response. J. Biol. Chem. 287, 7279–7288 (2012).
Bröker-Lai, J. et al. Heteromeric channels formed by TRPC1, TRPC4 and TRPC5 define hippocampal synaptic transmission and working memory. EMBO J. 36, 2770–2789 (2017).
Kollewe, A. et al. Subunit composition, molecular environment, and activation of native TRPC channels encoded by their interactomes. Neuron 110, 4162–4175 (2022).
Phelan, K. D. et al. Heteromeric canonical transient receptor potential 1 and 4 channels play a critical role in epileptiform burst firing and seizure-induced neurodegeneration. Mol. Pharmacol. 81, 384–392 (2012).
Wes, P. D. et al. TRPC1, a human homolog of a Drosophila store-operated channel. Proc. Natl. Acad. Sci. USA 92, 9652–9656 (1995).
Okada, T. et al. Molecular cloning and functional characterization of a novel receptor-activated TRP Ca2+ channel from mouse brain. J. Biol. Chem. 273, 10279–10287 (1998).
Torihashi, S., Fujimoto, T., Trost, C. & Nakayama, S. Calcium oscillation linked to pacemaking of interstitial cells of Cajal: requirement of calcium influx and localization of TRP4 in caveolae. J. Biol. Chem. 277, 19191–19197 (2002).
Kim, J. et al. TRPC1 as a negative regulator for TRPC4 and TRPC5 channels. Pflüg. Arch. 471, 1045–1053 (2019).
Hofmann, T., Schaefer, M., Schultz, G. & Gudermann, T. Subunit composition of mammalian transient receptor potential channels in living cells. Proc. Natl. Acad. Sci. USA 99, 7461–7466 (2002).
Won, J. et al. Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel. Nat. Struct. Mol. Biol. 32, 326–338 (2025).
Myeong, J. et al. Dual action of the Gαq-PLCβ-PI(4,5)P2 pathway on TRPC1/4 and TRPC1/5 heterotetramers. Sci. Rep. 8, 12117 (2018).
Ko, J., Myeong, J., Kwak, M., Jeon, J.-H. & So, I. Identification of phospholipase C β downstream effect on transient receptor potential canonical 1/4, transient receptor potential canonical 1/5 channels. Korean J. Physiol. Pharmacol. 23, 357–366 (2019).
Ko, J., Myeong, J., Shin, Y.-C. & So, I. Differential PI(4,5)P2 sensitivities of TRPC4, C5 homomeric and TRPC1/4, C1/5 heteromeric channels. Sci. Rep. 9, 1849 (2019).
Myeong, J. et al. Close spatio-association of the transient receptor potential canonical 4 (TRPC4) channel with Gαi in TRPC4 activation process. Am. J. Physiol. Cell Physiol. 308, C879–C889 (2015).
Won, J. et al. Molecular architecture of the Gαi-bound TRPC5 ion channel. Nat. Commun. 14, 2550 (2023).
Storch, U., Forst, A.-L., Philipp, M., Gudermann, T. & Mederos y Schnitzler, M. Transient receptor potential channel 1 (TRPC1) reduces calcium permeability in heteromeric channel complexes. J. Biol. Chem. 287, 3530–3540 (2012).
Kim, J. et al. Isoform- and receptor-specific channel property of canonical transient receptor potential (TRPC)1/4 channels. Pflüg. Arch. 466, 491–504 (2014).
Akbulut, Y. et al. Englerin A is a potent and selective activator of TRPC4 and TRPC5 calcium channels. Angew. Chem. Int. Ed. Engl. 54, 3787–3791 (2015).
Minard, A. et al. Potent, selective, and subunit-dependent activation of TRPC5 channels by a xanthine derivative. Br. J. Pharmacol. 176, 3924–3938 (2019).
Wright, D. J. et al. Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site. Commun. Biol. 3, 704 (2020).
Song, K. et al. Structural basis for human TRPC5 channel inhibition by two distinct inhibitors. Elife 10, https://doi.org/10.7554/elife.63429 (2021).
Müller, M. et al. Ideal efficacy photoswitches for TRPC4/5 channels harness high potency for spatiotemporally-resolved control of TRPC function in live tissues. Preprint at https://doi.org/10.1101/2024.07.12.602451v1 (2024).
Miller, R. G., Mitchell, J. D. & Moore, D. H. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst. Rev. 2012, Cd001447 (2012).
Richter, J. M., Schaefer, M. & Hill, K. Riluzole activates TRPC5 channels independently of PLC activity. Br. J. Pharmacol. 171, 158–170 (2014).
Yang, Y., Wei, M. & Chen, L. Structural identification of riluzole-binding site on human TRPC5. Cell Discov. 8, 67 (2022).
Erickson, M. G., Alseikhan, B. A., Peterson, B. Z. & Yue, D. T. Preassociation of calmodulin with voltage-gated Ca2+ channels revealed by FRET in single living cells. Neuron 31, 973–985 (2001).
Amiri, H., Schultz, G. & Schaefer, M. FRET-based analysis of TRPC subunit stoichiometry. Cell Calcium 33, 463–470 (2003).
Chen, Y. et al. The cryo-EM structure of TRPC1 and TRPC5 heterotetramer. Preprint at https://doi.org/10.1101/2024.11.03.621705v1 (2024).
Jeong, S. et al. Englerin A-sensing charged residues for transient receptor potential canonical 5 channel activation. Korean J. Physiol. Pharmacol. 23, 191–201 (2019).
Duan, J. et al. Cryo-EM structure of TRPC5 at 2.8-Å resolution reveals unique and conserved structural elements essential for channel function. Sci. Adv. 5, eaaw7935 (2019).
Yin, Y. et al. Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP2. Science 378, eadd1268 (2022).
Yin, Y. et al. Molecular basis of neurosteroid and anticonvulsant regulation of TRPM3. Nat. Struct. Mol. Biol. 32, 828–840 (2025).
Lichtenegger, M. et al. An optically controlled probe identifies lipid-gating fenestrations within the TRPC3 channel. Nat. Chem. Biol. 14, 396–404 (2018).
Bai, Y. et al. Structural basis for pharmacological modulation of the TRPC6 channel. Elife 9, https://doi.org/10.7554/elife.53311 (2020).
Carson, C. et al. Englerin A agonizes the TRPC4/C5 cation channels to inhibit tumor cell line proliferation. PLoS ONE 10, e0127498 (2015).
Greka, A., Navarro, B., Oancea, E., Duggan, A. & Clapham, D. E. TRPC5 is a regulator of hippocampal neurite length and growth cone morphology. Nat. Neurosci. 6, 837–845 (2003).
Davare, M. A. et al. Transient receptor potential canonical 5 channels activate Ca2+/calmodulin kinase Iγ to promote axon formation in hippocampal neurons. J. Neurosci. 29, 9794–9808 (2009).
Schwenk, J. et al. Modular composition and dynamics of native GABAB receptors identified by high-resolution proteomics. Nat. Neurosci. 19, 233–242 (2016).
Li, Y. et al. Loss of transient receptor potential channel 5 causes obesity and postpartum depression. Cell 187, 4176–4192 (2024).
Phelan, K. D. et al. Canonical transient receptor channel 5 (TRPC5) and TRPC1/4 contribute to seizure and excitotoxicity by distinct cellular mechanisms. Mol. Pharmacol. 83, 429–438 (2013).
Selvaraj, S. et al. Neurotoxin-induced ER stress in mouse dopaminergic neurons involves downregulation of TRPC1 and inhibition of AKT/mTOR signaling. J. Clin. Invest. 122, 1354–1367 (2012).
Hong, C. et al. Increased TRPC5 glutathionylation contributes to striatal neuron loss in Huntington’s disease. Brain 138, 3030–3047 (2015).
Ptakova, A., Zimova, L., Barvik, I., Bon, R. S. & Vlachova, V. Functional determinants of lysophospholipid- and voltage-dependent regulation of TRPC5 channel. Cell. Mol. Life Sci. 81, 374 (2024).
Strübing, C., Krapivinsky, G., Krapivinsky, L. & Clapham, D. E. Formation of novel TRPC channels by complex subunit interactions in embryonic brain. J. Biol. Chem. 278, 39014–39019 (2003).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T., Kelley, K., Noble, A. J. & Berger, B. Topaz-Denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun. 11, 5208 (2020).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Moriarty, N. W., Grosse-Kunstleve, R. W. & Adams, P. D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pravda, L. et al. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–W373 (2018).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Bers, D. M., Patton, C. W. & Nuccitelli, R. A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol. 99, 1–26 (2010).
Kang, H. & So, I. Unique responses of the fixed stoichiometric TRPC1–TRPC5 concatemer to G proteins. Front. Physiol. 15, 1392980 (2024).
Acknowledgements
We thank Dr. Eric Gouaux (Vollum Institute, OR, United States) for the pEG BacMam vector; Drs. Michael Mederosy Schnitzler (LMU, Germany) and Luis Vaca (UNAM, Mexico) for the human TRPC1α cDNA-containing plasmid; Drs. Shuji Kaneko (OSU, OH, United States) and Yasuo Mori (Kyoto University, Japan) for the human and mouse TRPC-containing plasmids, respectively; and Dr. Kyu Pil Lee (CNU, Republic of Korea) for the human TRPC6 cDNA-containing plasmid; Dr. Bumhan Ryu (Research Solution Center, IBS, Republic of Korea); Dr. Hanseong Kim (National Center for Inter-university Research Facilities, SNU, Republic of Korea) for the assistance in cryo-EM data collection. This work was supported by Samsung Science & Technology Foundation and Research (SSTF-BA2101-13 to H.H.L.). This work was also supported by the National Research Foundation of Korea grants RS-2025-00514715 and 2021R1A4A2001857 (I.S.), by the Education and Research Encouragement Fund of Seoul National University Hospital (I.S.), by the Mid-Career Bridging Program through Seoul National University (800-20240545) (I.S.), by the Seoul National University Hospital Research Fund (03-2024-0440) (I.S.), by the BK21 FOUR education program scholarship (J.K. and H.K.), and by KBRI basic research program through Korea Brain Research Institute funded by Ministry of Science and ICT (25-BR-01-02 to B.-C.L.). Use of the cryo-EM facilities of the NEXUS consortium was supported by a National Research Foundation of Korea grant (RS-2024-00440289). The authors thank the National Center for Inter-university Research Facilities (NCIRF) for assistance with cryo-EM.
Author information
Authors and Affiliations
Contributions
S.-H.K., H.P., J.K., I.S., and H.H.L. conceived and designed the experiments. S.-H.K., H.P., J.K., H.K., J.W., and B.-C.L. performed the experiments. S.-H.K. performed the structural experiments and analyses for the heteromeric channel. H.P. and J.W. performed the structural experiments and analyses for the homomeric channel with agonists. J.K. and H.K. performed the electrophysiological recordings. B.-C.L. constructed concatemers. S.-H.K., H.P., J.K., H.K., J.W., I.S., and H.H.L. analyzed the data. S.-H.K., H.P., J.K., I.S., and H.H.L. wrote the manuscript. I.S. and H.H.L. directed the work. All the authors have edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, SH., Park, H., Kim, J. et al. Molecular architecture of the human TRPC1/C5 heteromeric channel. Nat Commun 17, 317 (2026). https://doi.org/10.1038/s41467-025-67024-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67024-9