Abstract
Lymphocyte antigen-plasminogen activator urokinase receptor domain-containing protein 6B (Lypd6b) is a newly identified molecule associated with neuromodulation. However, the role of Lypd6b in regulating the tumor microenvironment and its impact on CD8+ T cell-mediated antitumor immunity remain unknown. Here, we observe that Lypd6b expression is increased significantly in colorectal cancer (CRC) tumor tissues compared to normal tissues. Lypd6b is mainly expressed in CD8+ T cells in tumor tissues. Lypd6b knockout (Lypd6b−/−) mice are resistant to AOM/DSS-induced tumorigenesis. Furthermore, global deficiency or CD8+ cell deficiency of Lypd6b inhibits MC38 or CMT-93 tumor growth and promotes the infiltration of CD8+ T cells. Mechanistically, Lypd6b deficiency promotes activation and function of CD8+ T cells in anti-tumor response with increased glycolysis and reduced oxidative phosphorylation in a PI3K/mTOR/LDHA pathway-dependent manner. Notably, Lypd6b deficient CD8+ T cells have a more potent antitumor effect when combined with anti-PD1 antibody. Thus, Lypd6b as a negative regulator for T cell immunity promotes CRC development, providing a molecular target with therapeutic potential in CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common malignant neoplasm and the second most common cause of cancer mortality worldwide1. The risk factors for CRC include inherited and environmental factors, which influence the mode of disease presentation2,3. Chronic inflammation plays an important role in the development of CRC. Patients with inflammatory bowel diseases (IBDs) have an increased risk of developing colitis-associated colorectal cancer (CAC)4.
T cells play an essential role in anti-tumor immunity. Tumor infiltration of CD8+ effector T cells has been attributed to improved outcome of immunotherapy in patients with CRC5. Recent interest has focused on the administration of negative checkpoint regulators (NCRs), such as the cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and programmed death protein-1 (PD-1), for cancer immunotherapy6,7. In addition, inhibiting other recently discovered NCRs, such as V-domain Ig-containing suppressor of T cell activation (VISTA)8, suppressor of cytokine signaling (SOCS)9, and dendritic cell-associated heparan sulfate proteoglycan-integrin ligand (DC-HIL)10 promotes CD8+ T cell activation. Although immune checkpoint inhibitors (ICIs) benefit most patients with cancer, adverse effects, such as colitis, rash, pruritus, hepatic, and pneumonitis may occur following treatment with ICIs11. Therefore, identifying additional NCRs with fewer side effects has important therapeutic implications.
Lymphocyte antigen-plasminogen activator urokinase receptor domain-containing protein 6B (Lypd6b) belongs to the LY6/plasminogen activator urokinase receptor (uPAR) superfamily12. LYPD6B, also known as LYPD7, is first identified and cloned in 2009. It is expressed in the testis, prostate, stomach, lung, and cerebral cortex13,14. Additionally, Chrna5+ super-responders contain a population of acetylcholine super-responders, which express a unique complement of GPI-anchored lynx prototoxin genes including Lypd6b15. LYPD6B inhibits nAChR-mediated currents by α3β4 nAChRs16. Moreover, it is shown to be associated with autism and intellectual disability through a chromosome microarray analysis of patients17. In recent years, Viktória Koroknai et al. revealed predominantly hypermethylated genes, including LYPD6B, are associated with the invasive phenotype of malignant melanoma18. The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database analyses reveal that nine genes, including MYOCD and LYPD6B, are upregulated in gastric adenocarcinoma19. However, the function of Lypd6b in tumorigenesis has not been reported, and whether Lypd6b is involved in anti-tumor T cell immunity remains unclear.
In this study, we identified that Lypd6b exerts an anti-tumor effect in CRC. Moreover, Lypd6b deletion in CD8+ T cells enhances glycolysis while reducing oxidative phosphorylation (OXPHOS) in a PI3K/mTOR/LDHA pathway-dependent manner. Collectively, our findings demonstrate that Lypd6b functions as a critical regulator of CD8+ T cell metabolism during antitumor immunity in CRC, thereby representing a potential target for immunotherapeutic strategies.
Results
LYPD6B expression is significantly upregulated in CRC
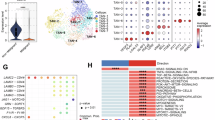
To determine the relationship between LYPD6B expression and tumor development, the TIMER 2.0 database was analyzed. LYPD6B expression was increased in tumor tissues of cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), etc., but decreased in tumor tissues of glioblastoma multiforme (GBM), kidney Chromophobe (KICH), etc. compared with normal tissues (Fig. 1A). In addition, the cancers with the highest LYPD6B gene mutation rates were uterine corpus endometrial carcinoma (UCEC) (19/531), stomach adenocarcinoma (STAD) (5/439), and COAD (4/406) (Fig. 1B). CRC is the third most prevalent malignant tumor with the second highest mortality rate in the world1. Using the UNLCAN (Fig. 1C, Supplementary Fig. 1A) and GEO (Fig. 1D) databases, we further confirmed that LYPD6B expression was higher in CRC tumor tissues compared with that in normal tissues. Similar results were obtained from transcriptome sequencing analysis of 154 patient CRC tissues (Fig. 1E).
A Expression of LYPD6B in different cancers in TIMER 2.0. The red box highlights the expression data for COAD in both tumor and normal tissue. **P < 0.01, ***P < 0.001. B Ranking of LYPD6B mutation rate in TIMER 2.0. UALCAN database C (Normal: n = 41; Tumor: n = 286), GEO database D (n = 9), and transcriptome sequencing analysis E (n = 154) were used to analyze the expression of LYPD6B. **P < 0.01, ***P < 0.001. F–K Immunohistochemical (IHC) staining for LYPD6B in tissues from 30 colorectal cancer (CRC) patients. F Representative IHC images of median LYPD6B expression level (scale bar: 50 and 100 μm). The black rectangle indicates the region selected for higher-magnification viewing. G Quantification of LYPD6B intensity score (n = 30 per group). ***P < 0.001. H Representative LYPD6B staining (scale bar: 50 μm). I Percentage of the cases expressing LYPD6B. J Tumor depth in CRC patients with different LYPD6B expression (n = 30 per group). *P < 0.05, where the shaded region represents the lower through upper quartile, the whiskers represent the minimum and maximum values, the center line represents the median and the center point represents the mean. K Percentage of tissues with different LYPD6B expression and different tumor grades. L–P C57BL/6 mice were used to establish DSS-induced colitis (n = 3 per group) and AOM/DSS-induced CAC (n = 5 per group). The data are representative of three independent experiments. L, M Schematic representation of colitis and CAC model. N, O Protein levels of Lypd6b in DSS-induced colon tissues. ***P < 0.001. P, Q Immunofluorescence (IF) staining of Lypd6b and CD8 in colon tissues (scale bar: 100 μm). **P < 0.01, ***P < 0.001. R, S MC38 cells were inoculated into the flanks of Lypd6b gene expressing EGFP (Lypd6b-EGFP) mice (n = 3 per group). R Flow cytometry was used to analyze the percentage of multiple immune cells in Lypd6b-EGFP+ cells of MC38 tumor tissue, and S statistical results. T IF staining of Lypd6b-EGFP+ in mouse CRC tumor tissue, immune cells (red), EGFP (green), and DAPI (blue) nuclear staining (scale: 100 μm) and U statistical results. No number, HPF high-power field. All statistical tests in (A–U) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, we analyzed cancer tissues and paracancerous tissues from 30 patients with CRC using immunohistochemical (IHC) staining (Fig. 1F). LYPD6B expression in tumor tissues was higher compared with that in paracancerous tissues (Fig. 1G). In addition, by analyzing the various expression densities of LYPD6B in CRC tissues, we found that approximately 76.67% (23/30) of the cases exhibited strong LYPD6B expression, while 3.33% (1/30) and 20.00% (6/30) of the cases showed either weak or moderate expression, respectively (Fig. 1H and I). However, only 26.67% (8/30) of cases were strong for LYPD6B expression in paracancerous tissues. Moreover, patients with high LYPD6B expression had an increasing tumor depth (Fig. 1J). Also, LYPD6B was strongly expressed in more than 50% of CRC cases with different tumor grades (Fig. 1K). These results indicate that LYPD6B is abnormally expressed in various human cancers and upregulated in CRC.
Lypd6b is predominantly expressed in CD8+ and CD4+ T cells as well as macrophages in CRC
Next, we characterized the cell-specific expression of Lypd6b in human CRC tissues. Compared with normal tissues, double immunofluorescence (IF) staining revealed that LYPD6B expression was increased in multiple cell types within CRC tissues, with predominant expression observed in CD8+ T cells, CD4+ T cells (Supplementary Fig. 2A) and tumor cells in CRC tissues. In addition, a small subset of CD68+ and B220+ cells exhibited LYPD6B expression (Supplementary Fig. 2B).
To further explore the role of Lypd6b in CRC, we established DSS-induced acute colitis and AOM/DSS-induced CAC mouse models (Fig. 1L and M). Consistently, western blot and IF staining analysis revealed that the expression of Lypd6b in colitis or tumor tissues was significantly higher compared with that in normal tissues (Fig. 1N–Q). Double staining further confirmed that Lypd6b was highly expressed in CD8+ T cells (Fig. 1P and Q), CD4+ T cells, and macrophages (Supplementary Fig. 3A and B) in mouse colitis and tumor tissues.
We next constructed Lypd6b gene expressing EGFP (encoding Lypd6b-GFP protein) mice (Supplementary Fig. 4A and B) to detect Lypd6b expression in immune cells with the MC38 tumor. The flow cytometry results revealed that the highest percentage of Lypd6b+ cells in tumor tissues were CD8+ T cells (Fig. 1R and S). In addition, the IF staining results showed that GFP (labeling Lypd6b) was mainly expressed in CD8+ T cells, followed by CD4+ T cells (Fig. 1T and U), and CD11b+ cells (Supplementary Fig. 4C and D). The results were consistent with those obtained from the bioGPS database (Supplementary Fig. 4E). Taken together, Lypd6b expression in CRC is upregulated and closely associated with CRC progression. Given its predominant expression in CD8+ and CD4+ T cells, Lypd6b may function as an immunoregulatory molecule.
Lypd6b deficiency aggravates DSS-induced colitis and decreases AOM/DSS-induced colorectal tumorigenesis
IBD is widely accepted as a risk factor for CRC4. To determine the role of Lypd6b in colitis and CRC, we established Lypd6b knockout (Lypd6b−/−) mice (Supplementary Fig. 5A). The efficiency of Lypd6b deletion was verified by western blot analysis (Supplementary Fig. 5B) and IF staining for Lypd6b expression (Supplementary Fig. 5C). In addition, flow cytometry results revealed that there were no differences in DCs, macrophages, NK cells, B cells, CD4+ T cells, CD8+ T cells, and regulatory T cells (Tregs) in the spleen, peripheral blood, bone marrow, or lymph nodes in both wild type (WT) C57BL/6 and Lypd6b−/− mice fed with a normal diet (Supplementary Fig. 6A–D), indicating that Lypd6b deficiency has no obvious effects on the hematopoietic cells assessed in four central and peripheral mouse hematopoietic organs.
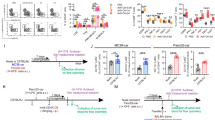
To define the role of Lypd6b in colitis-induced inflammation, Lypd6b−/− and WT mice were fed with 3% DSS to induce colitis (Fig. 2A). Ten days later, the mouse colon tissues were removed and photographed. Lypd6b−/− mice were significantly more susceptible to DSS treatment compared with the WT mice, as evidenced by increased weight loss on days 8, 9, and 10 (Fig. 2B), and severe shrinking of colon length (Fig. 2C and D). However, there were no differences in body weight or colon length between normal water-drinking Lypd6b−/− mice and the controls. The percentages of Gr1+CD11b+ cells (Supplementary Fig. 7A), CD8+ and CD4+ T cells (Supplementary Fig. 7B) in spleens (Supplementary Fig. 8A and B), and CD8+ and CD4+ T cells in colon tissues (Fig. 2E and F) from Lypd6b−/− mice were all significantly increased compared with that in WT mice analyzed by flow cytometry. In addition, Hematoxylin and eosin (H&E) staining of colitis tissues on day 10 of induction revealed worse damage and pronounced inflammatory cell infiltration in Lypd6b−/− mice compared with that in WT mice (Fig. 2G). Furthermore, CD8+, CD4+, CD11b+, F4/80+, and Gr1+ cells were increased in the colon tissues of Lypd6b−/− mice compared with control mice. There were no differences in colon immune cell infiltration between normal water-drinking Lypd6b−/− and WT mice (Fig. 2G and H). Moreover, the mRNA levels of several inflammatory cytokines and chemokines such as IL-2, IL-10, IFN-γ, IL-6, CCL5, and TGF-β were upregulated in colitis tissues of Lypd6b−/− mice compared with those in WT mice (Fig. 2I). In addition, IFN-γ and IL-2 production in CD8+ and CD4+ T cells from Lypd6b−/− mice was significantly higher compared with that in WT mice (Supplementary Fig. 7C), whereas Lypd6b deletion did not enhance TNF production in CD8+ or CD4+ T cells (Fig. 2J-L). These results demonstrate that Lypd6b deficiency exacerbates colitis by promoting immune cell infiltration and the inflammatory response.
A–L Groups of WT and Lypd6b−/− mice (n = 5 mice per group) were subjected to the DSS-induced colitis model. Data are representative of three independent experiments. A Schematic representation of DSS-induced colitis model. B Body weight changes. **P < 0.01, ***P < 0.001. C, D Colon photos and lengths of the mice after DSS treatment. ***P < 0.001. E, F FACS identified CD8+ and CD4+ cells in colon specimens. *P < 0.05, ***P < 0.001, ns not significant. G H&E, CD8, CD4, CD11b, F4/80, and Gr1 staining in colon specimens and H statistical analysis. *P < 0.05, **P < 0.01. I mRNA levels of cytokines and chemokines in DSS-induced colon tissue. *P < 0.05, **P < 0.01. J, K IFN-γ, IL-2, and TNF secretion of CD8+ and CD4+ T cells were detected by FACS and L statistical analysis. *P < 0.05, **P < 0.01, ns not significant. Data are representative of three independent experiments. (n = 4 mice per group). M–T WT and Lypd6b−/− mice (n = 10 per group) were subjected to the AOM/DSS-induced CAC model. Data are representative of three independent experiments. M Schematic representation of the AOM/DSS-induced CRC model. The black arrow indicates the administration of a 10 mg/g dose of AOM, based on each mouse’s body weight, with this day designated as Day 0. N Representative images of colon specimens, O colon length, P tumor number per mouse, Q tumor size, and R number of tumors greater than 3 mm in WT and Lypd6b−/− mice are shown. O–R Data are from 3 independent biological samples per group. *P < 0.05. **P < 0.01. S H&E, CD8, CD4, F4/80, and Ki67 staining with colon specimens (scale bar: 50 and 100 μm) and T statistical analysis. **P < 0.01, ***P < 0.001, ns not significant. All statistical tests in (B–T) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, we examined the role of Lypd6b during intestinal tumorigenesis with AOM/DSS induction in Lypd6b−/− mice. The mice received a single intraperitoneal injection of AOM followed by DSS treatment for three cycles and were monitored for 120 days and euthanized (Fig. 2M). In contrast to DSS-induced colitis, Lypd6b−/− mice exhibited a longer colon length (Fig. 2N and O). In addition, Lypd6b−/− mice showed significantly decreased tumor numbers and tumor sizes compared with WT mice (Fig. 2P–R). Pathologic analysis also confirmed decreased tumor growth in Lypd6b−/− mice (Fig. 2S). In addition, reduced tumorigenesis was accompanied by increased CD8+ T cells and decreased F4/80+ cells, Ki67+ cells in Lypd6b−/− mice (Fig. 2T), indicating that Lypd6b deficiency decreases colorectal tumorigenesis and promotes the infiltration of CD8+ T cells.
Lypd6b deficiency inhibits CRC tumor growth and promotes the infiltration of CD8+ T cells
CD8+ T cells exert dural action by effectively killing tumors and facilitating inflammation that leads to tumorigenesis20. To further investigate the function of Lypd6b in anti-tumor CD8+ T cell responses in CRC, MC38 cells were injected subcutaneously into Lypd6b−/− mice and the control mice on a C57BL/6 background. Compared with WT mice, tumor growth was slower in Lypd6b−/− mice (Fig. 3A–C). Proliferating (Ki67+) cells in tumors were significantly decreased in the Lypd6b−/− mice,
A–J MC38 cells were inoculated into the flanks of WT and Lypd6b−/− mice (n = 5 mice per group). Data are representative of at least three independent experiments. A Growth curves of tumor volume. *P < 0.05. B Representative tumor images and C tumor weight. **P < 0.01. D Representative staining of Ki67 in tumor tissues (scale bars: 100 μm). E, F Tumor-infiltrating CD11b+Gr1+ cells, G, H CD8+ and CD4+ cells were identified by FACS. *P < 0.05, ***P < 0.001. I, J IFN-γ and IL-2-secreting CD8+ T cells in tumors were detected by FACS and statistical analysis. **P < 0.01, ***P < 0.001. K–P CMT-93 cells were inoculated into the flanks of WT and Lypd6b−/− mice (n = 5 mice per group). Data are representative of three independent experiments. K Tumor growth curves. ***P < 0.001. L Representative tumor images. M Tumor weight. ***P < 0.001. N–P Tumor-infiltrating CD8+ and CD4+ T cells were identified by FACS. ***P < 0.001, ns not significant. Q–T RNA sequencing analysis of differentially expressed genes (DEGs) between CD8+ T cells sorted in the MC38 tumor from WT and Lypd6b−/− mice. (n = 3 mice per group). Q Venn diagrams and R histogram of DEGs, the threshold is P < 0.05, and the absolute value of log2 Ratio≥0. S Heatmap view and T fold change of the most significant DEGs related to T cell activation. *P < 0.01, **P < 0.01. U, V Activation status evidenced by CD69 expression was quantified by FACS. *P < 0.05, ns not significant. Data are representative of three independent experiments. (n = 3 mice per group). W–Z RT-PCR analysis of Lag3, Tigit, Gzmb, and Ifng expression of CD8+ T cells. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. Data are representative of three independent experiments. (n = 3 mice per group). All statistical tests in (A–Z) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
indicating the effective inhibition of tumor cell proliferation in vivo by Lypd6b knockout (Fig. 3D). In addition, tumor-infiltrating myeloid cells were significantly decreased in Lypd6b−/− mice (Fig. 3E and F), whereas the percentages of CD8+ and CD4+ T cells were increased in tumors of Lypd6b−/− mice. This difference was more apparent in CD8+ T cells as determined by flow cytometry (Fig. 3G and H). The results were further confirmed by IF staining (Supplementary Fig. 9A and B). Additionally, the production of IFN-γ by CD8+ and CD4+ T cells was increased in Lypd6b−/− mice. IL-2 levels in CD8+ T cells were markedly increased following Lypd6b depletion (Fig. 3I and J), whereas no obvious difference was observed in CD4+ T cells (Supplementary Fig. 9C and D), indicating that Lypd6b is essential for the regulation of anti-tumor immunity by CD8+ T cells.
In addition, Lypd6b deficiency also inhibited CMT-93 tumor growth in Lypd6b−/− mice compared with WT mice (Fig. 3K–M). The percentage of CD8+ T cells was significantly increased in tumors (Fig. 3N and O) and spleens (Supplementary Fig. 10A) of Lypd6b−/− mice, whereas no difference in CD4+ T cell infiltration was observed. The results further confirmed the inhibitory effect of Lypd6b deficiency on tumor growth, which might largely rely on CD8+ T cells.
Lypd6b deficiency promotes the activation and function of CD8+ T cells during the anti-tumor response
To gain insight into Lypd6b function in anti-tumor immunity, RNA-sequencing was performed on CD8+ T cells in MC38 tumors from WT and Lypd6b−/− mice. We identified 539 and 592 genes unique to the Lypd6b−/− and WT groups, respectively (Fig. 3Q). Furthermore, 134 differentially expressed genes (DEGs) were identified, of which 105 were upregulated and 29 were downregulated in Lypd6b−/− mice compared with WT mice (fold change [FC] > 0) (Fig. 3R). Heat maps of the DEGs showed that the expression of Nkg7, Thy1, Gzmb, TNFRSF9, and CD3 in Lypd6b−/− mice was significantly up-regulated, and most of these genes were related to T cell activation (Supplementary Fig. 11A and B). Subsequently, an examination of T cell activation-related genes in a heat map revealed the significant upregulation of Thy1, Gzmb, Cd3e, Cd8a, Cd6, Cd5, and Gzmk in CD8+ T cells of Lypd6b−/− mice (Fig. 3S and T).
Then the effects of Lypd6b on T cell proliferation and activation were verified. Splenic T cells were isolated from WT and Lypd6b−/− mice and labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE). Fluorescence-activated cell sorting (FACS) analysis revealed that Lypd6b knockout (Lypd6b-KO) T cells showed increased proliferation compared with that of WT T cells, particularly CD8+ T cells, following anti-CD3 and anti-CD28 stimulation (Supplementary Fig. 7D, and Supplementary Fig. 11C). In addition, the T cell activation marker (CD69+) of CD8+ T cells but not CD4+ T cells in tumors was significantly increased in Lypd6b−/− mice (Fig. 3U and V). Conversely, the percentages of TIM3+PD1+CD8+ T cells were significantly decreased in tumors from Lypd6b−/− mice compared with those in controls (Supplementary Fig. 11D and E). We also observed decreased mRNA levels of T cell exhaustion-related genes such as Lag3 and Tigit (Fig. 3W and X) and increased mRNA levels of T cell activation-related genes such as Gzmb and Ifng in activated Lypd6b-KO CD8+ T cells (Fig. 3Y and Z). Also, Lypd6b deficiency markedly augmented the secretion of IL-2 and IFN-γ in activated CD8+ T cells as determined by ELISA (Supplementary Fig. 11F). These results indicate that Lypd6b−/− mice have increased anti-tumor immunity by fostering CD8+ T cell proliferation, activation, and effector functions.
Lypd6b deficiency in CD8+ T cells inhibits CRC growth
To investigate whether the anti-tumor function of Lypd6b deficiency is mediated through CD8+ T cells, we performed an adoptive CD8+ T cell transfer experiment using nude mice as recipients. Nude mice received adoptive transfer of activated WT CD8+ T cells and Lypd6b-KO CD8+ T cells, followed to establish the MC38 tumor model (Fig. 4A). MC38 tumors in nude mice with over-transferred Lypd6b-KO CD8+ T cells grew more slowly (Fig. 4B) and the tumors were smaller than those in the control group (Fig. 4C–E). These results indicated that Lypd6b-KO CD8+ T cells had a stronger anti-tumor effect than WT CD8+ T cells.
A–E 5 × 106 CD8+ T cells from the spleens of WT and Lypd6b−/− mice were sorted and injected into 8-week-old nude mice (n = 5 mice per group) by tail vein injection. The second day, 7 × 105 MC38 cells were injected subcutaneously into the nude mice to construct the MC38 tumor model. Data are representative of three independent experiments. A Diagram of adoptive CD8+ T cell transfer experiment, created in BioRender. Li, B. (2024) https://BioRender.com/fibcpif. B Tumor volume growth curve, **P < 0.01. C Representative nude mouse images. D Representative tumor photographs. E Tumor weight, ***P < 0.001. F Genotyping of Lypd6bfl/fl and CD8Lypd6b−/− mice using PCR assay. (G-O) Groups of Lypd6bfl/fl and CD8Lypd6b−/− mice (n = 5 mice per group) were subjected to the MC38 tumor model. Data are representative of three independent experiments. G Tumor growth curves. ***P < 0.001. H Representative tumor images. I Tumor weight. ***P < 0.001. J Tumor-infiltrating CD11b+, Gr1+ cells (K) and CD4+, CD8+ T cells were identified by FACS. *P < 0.05, ***P < 0.001. L, M IFN-γ and IL-2 levels of CD8+ T cells in MC38 tumors of Lypd6bfl/fl and CD8Lypd6b−/− mice were detected by FACS. **P < 0.01. N, O Quantification of TIM3+ and PD1+ in MC38 tumor-infiltrating CD8+ and CD4+ T cells from Lypd6bfl/fl and CD8Lypd6b−/− mice by FACS. *P < 0.05, ns no significant. All statistical tests in (A–O) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We next constructed the Lypd6bflox/flox (Lypd6bfl/fl) transgenic mice (Supplementary Fig. 12A and B) and crossed them with CD8-Cre mice. The obtained CD8-Cre Lypd6bfl/fl conditional knockout mice were named as CD8Lypd6b−/− mice (Fig. 4F). Compared with Lypd6bfl/fl mice, CD8Lypd6b−/− mice showed significantly slower MC38 tumor growth and reduced tumor weights (Fig. 4G–I). In addition, the percentages of spleen (Supplementary Fig. 13A) and tumor-infiltrating (Fig. 4J) myeloid cells were reduced in CD8Lypd6b−/− mice. The proportions of CD8+ T cells were significantly increased in both spleens (Supplementary Fig. 13B) and tumors (Fig. 4K) of CD8Lypd6b−/− mice, and the CD4+ T cell proportions were slightly increased. These results demonstrated that Lypd6b deficiency in CD8+ T cells is a key factor in suppressing tumor growth and promotes the formation of a tumor immunosuppressive environment in CRC.
In addition, compared with Lypd6bfl/fl mice, IFN-γ and IL-2 levels were significantly increased in tumor CD8+ T cells in CD8Lypd6b−/− mice (Fig. 4L and M), whereas changes in IFN-γ and IL-2 levels in CD4+ T cells were not significant (Supplementary Fig. 13C and D). We also found a lower percentage of TIM3+PD1+CD8+ T cells in spleens of Lypd6bfl/fl mice (Supplementary Fig. 13E and F) and tumors (Fig. 4N and O, and Supplementary Fig. 7E). Taken together, these results suggest that Lypd6b deficiency in CD8+ T cells promotes CD8+ T cell activation and effector function, which in turn promotes anti-tumor immunity.
Lypd6b deficiency enhances the activation of CD8+ T cells through increased glycolysis and reduced OXPHOS
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis based on RNA sequencing data of CD8+ T cells revealed that Lypd6b deficiency primarily affects glycolysis /gluconeogenesis and carbon metabolism (Fig. 5A). In addition, Gene set enrichment analysis (GSEA) from the DEGs in Lypd6b-KO CD8+ T cells revealed the enrichment of metabolism pathways (Fig. 5B). Moreover, we found that the DEGs related to glycolysis such as Hif1α, Mtor, Hk1, G6pdx, Slc2a3, Hk2, and Atp6V0A2, were significantly up-regulated in Lypd6b-KO CD8+ T cells compared with that in WT cells (Fig. 5C).
A KEGG analysis of DEGs. B GSEA results showed metabolism pathways enriched in Lypd6b-KO CD8+ T cells. C Heatmap view of the DEGs related to T cell metabolism. Splenic CD8+ T cells were treated with anti-CD3 and anti-CD28 for 48 h, D glucose consumption, E lactate production, F ATP levels, G, H 2-NBDG, and I LDH levels were detected. *P < 0.05, **P < 0.01, ***P < 0.001. J The mRNA level of glycolysis-related genes. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. K Schematic of LDHA reaction. L NAD+/NADH ratio of CD8+ T cells. ***P < 0.001. D–L Data are representative of three independent experiments. (n = 3 mice per group). M, N The extracellular acidification rate (ECAR) and O, P oxygen consumption rate (OCR) using the Seahorse XFe96 BioAnalyzer. *P < 0.05, **P < 0.01, ***P < 0.001. M–P Data are representative of three independent experiments. (n = 5 mice per group). Q Quantification of mean fluorescence intensity for TMRE and R Mito Tracker Red. **P < 0.01.***P < 0.001. MFI mean fluorescence intensities. Q, R Data are representative of three independent experiments. (n = 3 mice per group). All statistical tests in (D–R) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Glycolysis plays an important role in the enhanced activation and function of CD8+ T cells21. Therefore, we analyzed the effects of Lypd6b on glycolysis in CD8+ T cells in vitro. Glucose consumption, lactate, and ATP production were increased in Lypd6b-KO CD8+ T cells (Fig. 5D–F). The mean fluorescence intensities (MFI) of 2-(N-[7-nitrobenz-2-oxa-1,3-diazol-4-ylamino)-2-deoxyglucose (2-NBDG) in Lypd6b-KO CD8+ T cells was stronger compared with that in WT CD8+ T cells (Fig. 5G and H, and Supplementary Fig. 7F), indicating increased glucose uptake in CD8+ T cells from Lypd6b−/− mice. Furthermore, LDH levels were increased in Lypd6b-KO CD8+ T cells (Fig. 5I). The mRNA levels of glycolytic genes including Glut1, Hk2, Pkm2, Gapdh, and Ldha were also significantly increased in CD8+ T cells from Lypd6b−/− mice (Fig. 5J). Of these, LDHA levels were markedly increased. LDHA catalyzes the interconversion of pyruvate to lactate and is accompanied by NAD+ regeneration (Fig. 5K)22. The NAD+/NADH ratios increased in Lypd6b-KO CD8+ T cells, indicating the LDHA activity in CD8+ T cells was enhanced following Lypd6b knockout (Fig. 5L).
To further investigate the role of Lypd6b in CD8+ T cell metabolic homeostasis, glycolysis and OXPHOS were determined by measuring extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) using a seahorse XF96 analyzer. Lypd6b-KO CD8+ T cells exhibited increased levels of glycolysis, glycolytic capacity, and glycolytic reserve (Fig. 5M and N). In contrast, basal respiration, maximal respiration, and spare respiration capacity were attenuated in Lypd6b-KO CD8+ T cells compared with WT CD8+ T cells (Fig. 5O and P), indicating Lypd6b-KO CD8+ T cells rely more on glycolysis and less on OXPHOS for energy production compared with that in WT CD8+ T cells. Mitochondrial metabolic reprogramming toward aerobic glycolysis facilitates the reprogramming of naïve T cells into specialized effector T cells23. Consequently, the mitochondrial membrane potential (MMP) and mitochondrial number in CD8+ T cells were evaluated using two specific fluorescent dyes tetramethylrhodamine ethyl ester (TMRE) and Mito Tracker Red, respectively. FACS analysis revealed that the MMP (Fig. 5Q) and number of mitochondria (Fig. 5R) decreased significantly in Lypd6b-KO CD8+ T cells. These results indicate that Lypd6b deficiency promotes LDHA activity in CD8+ T cells, resulting in increased glycolytic flux and attenuated OXPHOS.
Lypd6b deficiency regulates CD8+ T cells metabolism through PI3K/mTOR/LDHA pathway
T cells depend on mTOR signaling to integrate immune signals and metabolic cues for proper maintenance and activation24. LDHA has an essential role in T cell metabolic programming25. In addition, mTOR is a downstream target of phosphatidylinositol 3 kinase (PI3K) and regulates glycolysis26. mTOR pathways were enriched in Lypd6b-KO CD8+ T cells from the RNA sequence analysis (Fig. 6A). The phosphorylated PI3K (p-PI3K), p-mTOR, and LDHA protein levels were significantly increased in both DSS-induced colon tissue (Fig. 6B and C) and CD8+ T cells (Fig. 6D and E) from Lypd6b−/− mice.
A GSEA results showed mTOR pathways enriched in Lypd6b-KO CD8+ T cells. B, C Protein levels of p-mTOR, mTOR, p-PI3K, PI3K, LDHA, and Lypd6b in DSS-induced colitis colons and D, E splenic CD8+ T cells. *P < 0.05, **P < 0.01, ***P < 0.001. B–E Data are representative of three independent experiments. (n = 3 mice per group). Splenic CD8+ T cells were treated with 25 nM rapamycin and 10 μM GSK2837808A for 48 h. F–I Glucose consumption, lactate production, ATP levels, and NAD+/NADH ratio in CD8+ T cells. **P < 0.01, ***P < 0.001. J, K LDHA and GAPDH mRNA levels were analyzed. **P < 0.01, ***P < 0.001, ns: not significant. Quantification of mean fluorescence intensity for L TMRE and M Mito Tracker Red. **P < 0.01, ***P < 0.001. N, O IFN-γ and IL−2 levels were measured by ELISA. *P < 0.05, **P < 0.01, ***P < 0.001. P, Q Protein of p-mTOR, mTOR, p-PI3K, PI3K, and LDHA in CD8+ T cells after 10 μM GSK2837808A and 10 μM LY294002 treatment. *P < 0.05, **P < 0.01. D–Q Data are representative of three independent experiments. (n = 3 mice per group). R Schematic of targets of PI3K, mTOR, and LDHA. created in BioRender. Li, B. (2024) https://BioRender.com/n3s6ltk. All statistical tests in B–P were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
The LDHA inhibitor GSK2837808A, and the mTOR inhibitor rapamycin effectively reversed increased glucose consumption, lactate production, ATP production, and NAD+/NADH ratio of CD8+ T cells with Lypd6b deficiency (Fig. 6F–I). Moreover, the mRNA levels of glycolytic enzymes, LDHA and GAPDH, in activated CD8+ T cells were attenuated following rapamycin treatment (Fig. 6J and K). In addition, the OXPHOS metabolic activities in CD8+ T cells were evaluated following GSK2837808A and rapamycin treatment. FACS analysis revealed that in the presence of GSK2837808A and rapamycin, severely reduced MMP was partially restored (Fig. 6L) and the mitochondrial number was increased in Lypd6b-KO CD8+ T cells (Fig. 6M). GSK2837808A and rapamycin also abolished IFN-γ and IL-2 levels in activated CD8+ T cells following Lypd6b knockout (Fig. 6N and O). These results indicate that mTOR and LDHA inhibition are sufficient to decrease glycolysis and IFN-γ and IL-2 secretion resulting from Lypd6b deficiency. CD8+ T cells were further treated with LY294002 (a PI3K inhibitor) and GSK2837808A. LY294002 inhibited the protein expression of p-mTOR and LDHA, whereas GSK2837808A had no effect on the expression of p-PI3K and p-mTOR (Fig. 6P and Q). These results indicate that Lypd6b deficiency promotes metabolic reprogramming and activation of CD8+ T cells by activating the PI3K/mTOR/LDHA pathway (Fig. 6R).
Lypd6b deficient CD8+ T cells suppress tumor growth and result in a marked antitumor effect when combined with oxaliplatin or anti-PD1 antibody
To determine whether CD8+ T cells contribute to the enhanced anti-tumor effect mediated by Lypd6b deficiency, Lypd6b−/− and WT mice with MC38 tumors were administrated an intraperitoneal injection of anti-CD8a to deplete CD8+ T cells (Fig. 7A). The effect of depleting CD8+ T cells in tumors was determined by a flow cytometry analysis of the spleen (Fig. 7B–D) and IF staining in the tumor (Supplementary Fig. 14A and B) from tumor-bearing mice. Both WT and Lypd6b−/− mice exhibited increased tumor size and weight following anti-CD8a antibody injection, indicating that anti-CD8a antibody treatment effectively reverses tumor growth inhibition caused by Lypd6b deficiency (Fig. 7E and G). In addition, we implemented the depletion of CD8+ T cells in WT and Lypd6b−/− mice with colitis via injection of CD8a neutralization antibody. The depletion of CD8+ T cells was confirmed through flow cytometry analysis in the spleen (Supplementary Fig. 15A and B). CD8a neutralization antibody treatment considerably inhibited colitis progression in Lypd6b−/− mice, as evidenced by a decrease in weight loss and an increase in colon length (Supplementary Fig. 15C–E).
A–G For CD8+ T cell depletion, 5 mg/kg anti-CD8a in PBS was injected intraperitoneally twice a week during the MC38 tumor growth (n = 5 mice per group). The arrow indicate that each mouse was injected with 7 × 105 MC38 cells. Data are representative of three independent experiments. A Schematic of anti-CD8a treatment tumor model. B–D FACS analysis of CD8+ T cells and CD4+ T cells of the spleen. ***P < 0.001, ns not significant. E Growth curves of tumor volume. *P < 0.05, **P < 0.01, ***P < 0.001. F Representative tumor images. G Tumor burdens. *P < 0.05, **P < 0.01, ***P < 0.001. H–N For oxaliplatin (OXP) treatment, 20 mg/kg OXP dissolved in 5% dextrose was injected intraperitoneally once every two days during the MC38 tumor growth (n = 5 mice per group). Data are representative of three independent experiments. H Schematic of the OXP treatment tumor model. The arrow indicate that each mouse was injected with 7 × 105 MC38 cells, marking this day as Day 0. I Growth curves of tumor volume. *P < 0.05, **P < 0.01, ***P < 0.001. J Representative tumor images. K Tumor burdens. *P < 0.05, **P < 0.01. L–N CD8 and CD4 staining in MC38 tumor tissues. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. O–U For anti-PD1 treatment, Lypd6bfl/fl and CD8Lypd6b−/− mice were injected intraperitoneally with 200 μg of anti-PD1 diluted in PBS every two days during MC38 tumor growth. The arrow indicate that each mouse was injected with 7 × 105 MC38 cells. Data are representative of three independent experiments. (n = 5 mice per group). O Schematic of anti-PD1 treatment tumor model. P Growth curves of tumor volume. *P < 0.05, **P < 0.01, ***P < 0.001. Q Representative tumor images. R Tumor burdens. *P < 0.05, **P < 0.01, ***P < 0.001. S FACS analysis of CD8+ and CD4+ T cells from the spleen of Lypd6bfl/fl and CD8Lypd6b−/− mice. T, U Statistics results. *P < 0.05, ***P < 0.001, ns not significant. V Spearman correlations between Lypd6b and immunostimulators in COAD samples from TISIDB databases. W Different expression of Lypd6b between responders and non-responders of different cancers based on TISIDB databases. X Expression of Lypd6b between responders and non-responders of COAD in Kaplan–Meier plotter databases. All statistical tests in (C–U) were carried out using two-tailed Student’s t tests. Multiple comparisons were made using two-way ANOVA with GraphPad Prism V8.0. Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Despite the promising anti-tumor effects of oxaliplatin (OXP) for the treatment of CRC, the occurrence of drug resistance remains challenges27. To determine whether Lypd6b deficiency affects CRC sensitivity to OXP, the mice with MC38 tumors were administered an intraperitoneal injection of OXP (Fig. 7H). As shown in Fig. 7I–K, tumor growth in Lypd6b−/− mice combined with OXP treatment was remarkably decreased compared with WT mice. Furthermore, the infiltration of CD8+ T cells in tumors from Lypd6b−/− mice treated with OXP was significantly enhanced compared with untreated mice (Fig. 7L and M), whereas no differences were observed in the infiltration of CD4+ T cells following OXP treatment (Fig. 7N). These results suggest that CD8+ T cells are major immune cells involved in anti-tumor immunity in MC38 tumors in Lypd6b−/− mice, and Lypd6b deficiency promotes MC38 tumor sensitivity to OXP.
Tumor immunotherapy, represented by “PD1/PD-L1”, has gained a lot of attention worldwide28. To confirm the contribution of Lypd6b to CRC immunotherapy, we treated Lypd6bfl/fl and CD8Lypd6b−/− mice with anti-PD1 antibody (Fig. 7O). Both Lypd6b deficiency in CD8+ T cells and anti-PD1 treatment inhibited MC38 tumor growth effectively. Additionally, Lypd6b deficiency in CD8+ T cells further augmented the response to anti-PD1 treatment (Fig. 7P–R). Flow cytometry analysis further revealed that Lypd6b deficiency in CD8+ T cells alongside anti-PD1 treatment significantly increased the infiltration of CD8+ T cells (Fig. 7S–U). Thus, targeting Lypd6b enhanced anti-PD1 therapeutic efficacy in CRC by promoting the filtration of CD8+ T cells.
A database analysis revealed that LYPD6B is negatively associated with immunostimulatory factors, such as CD28, and CD40 in COAD patient samples (Fig. 7V). Responders to PD1 treatment were primarily associated with low expression of LYPD6B (Fig. 7W). Also, LYPD6B expression in patients with COAD, who responded to immunotherapy, was lower compared with that in non-responders (Fig. 7X). These results indicate that COAD patients with low LYPD6B expression are more sensitive to immunotherapy, and LYPD6B may represent a target for CRC therapy.
Patients with CRC and LYPD6B-low expression exhibit increased CD8+ T cell infiltration and altered glycolysis
A single-cell RNA sequence (scRNA-seq) analysis of 16 CRC samples in the GEO database further delineates a functional role of LYPD6B in human CRC. The various clusters were annotated by canonical marker genes using the “Seurat” package. Integrative analysis of this scRNA-seq cohort revealed that LYPD6B mRNA expression in tumor tissues was higher compared with that in normal tissues (Fig. 8A). Distinct clusters corresponding to canonical markers of the indicated cell type were identified (Supplementary Fig. 16A and Fig. 8B). Furthermore, we found the LYPD6B expression in CRC (Supplementary Fig. 16B) and used the median LYPD6B expression level in all cells as a cutoff value to split the enrolled patients into LYPD6B-high and LYPD6B-low groups (Fig. 8C). Patients exhibiting low LYPD6B expression in tumors showed an increased presence of CD8+ and CD4+ T cells, indicating that decreased LYPD6B expression promotes a tumor-inhibiting immune environment (Fig. 8D, Supplementary Fig. 16C). Furthermore, a significantly negative correlation between LYPD6B expression and IL-2, IFN-γ (Fig. 8E) in CD8+ T cells was revealed. Also, LYPD6B expression was negatively correlated with glycolysis-related gene expression (Fig. 8F) and PIK3CA, MTOR, LDHA expression in CD8+ T cells (Fig. 8G).
A–G Single-cell data analysis based on the GSE245552 dataset. A LYPD6B mRNA levels in tumor and paired non-tumor tissues. B UMAP plot of all major cell types with cells colored by cell type. C–G Based on the average mRNA levels of the LYPD6B in all cells, these samples were divided into two groups: LYPD6B-high and LYPD6B-low groups (C), and the levels of LYPD6B expression were shown. D The proportions of CD8+ and CD4+ T cell subsets in two groups. E The IL-2, IFNG and F glycolysis-related gene mRNA levels in LYPD6B-high and LYPD6B-low groups within CD8+ T cells. G The PIK3CA, PIK3CB, MTOR, and LDHA mRNA levels in LYPD6B-high and LYPD6B-low groups within CD8+ T cells. H–J Correlations between the expression of LYPD6B and IFNG, IL-2, CD8A, CD69, HK2, PIK3CA in patient-derived tissues with CRC (n = 154). K MTOR or LDHA in patient-derived tissues with CRC (n = 154). L, M Comparison of the survival rate of CRC patients with LYPD6Blow and LYPD6Bhigh or LYPD6Blow/CD8high and LYPD6Blow/CD8low from Kaplan–Meier plotter and TIMER 2.0 databases, respectively. P < 0.05 was considered statistically significant. N Schematic diagram of Lypd6b regulates colon-associated CD8+ T cell metabolism, created in BioRender. Li, B. (2024) https://BioRender.com/uudywff. PN paired non-tumor, PT paired tumor.
Consistently, we performed RNA sequencing analysis of 154 CRC samples and found that LYPD6B expression was negatively correlated with IFNG and IL-2 expression (Fig. 8H). Additionally, LYPD6B expression was negatively correlated with CD8A, CD69, HK2, PIK3CA, MTOR, and LDHA expression in CRC patient samples (Fig. 8I–K). In addition, patients with CRC and high LYPD6B expression had shorter survival times following anti-PD1 treatment (Fig. 8L), while patients with CRC and LYPD6Blow/CD8high signature exhibited substantial survival benefits compared with those having-LYPD6Blow/CD8low signature (Fig. 8M) based on an analysis using the Kaplan–Meier Plotter and the Timer2.0 databases, respectively. Overall, these findings indicated an increased anti-tumor immune response in CRC patients with low LYPD6B expression and highlighted a negative correlation between the LYPD6B and IL-2, IFN-γ production, and glycolysis.
Discussion
This study revealed that Lypd6b is a negative regulator in the T cell anti-tumor response in CRC. Lypd6b deficiency inhibited colon carcinogenesis by increasing CD8+ T cell activity. In addition, Lypd6b deficiency promoted CD8+ T cell metabolic reprogramming through the PI3K/mTOR/LDHA signaling pathway (Fig. 8N).
CD8+ T cells kill tumor cells by secreting cytokines and inducing apoptosis29, NCRs such as lymphocyte activation gene 3 (LAG-3), TIM-3, and T cell immunoreceptor with Ig and ITIM domains (TIGIT) are associated with the activation and exhaustion of CD8+ T cells in anti-tumor immunity30,31. Similarly, our study is the first to demonstrate that Lypd6b acts as a potential immunomodulatory inhibitor in CRC. Lypd6b deficiency promotes the activation and anti-tumor effect of CD8+ T cells in CAC and transplanted tumor models (Figs. 2 and 3). In addition, Lypd6b−/− mice were more sensitive to DSS-induced colitis. This phenomenon may be related to the complex role of CD8+ T cells in inflammation and tumors, particularly pro-inflammation and tumor suppression32.
Lypd6b is predominantly expressed in CD8+, CD4+, and E-cadherin+ cells, while other NCRs, such as B7-H1 (PD-L1), are expressed in B cells, T cells, macrophages, and DCs33. B7-DC (PD-L2) expression primarily occurs in lymphoid tissues and DCs34. In this study, we found that Lypd6b is expressed in several types of cells, including CD8+, CD4+ T cells, and macrophages. Lypd6b deficiency inhibited the growth of MC38 and CMT-93 tumors primarily through the anti-tumor effect of CD8+ T cells. Studies with adoptive T cell transfer and neutralizing CD8+ T cells further confirmed this result. MC38 tumor growth in CD8Lypd6b−/− mice further confirmed that Lypd6b deficiency in CD8+ T cells inhibited CRC growth (Fig. 4). However, the role of Lypd6b in other cell types in CRC remains unclear. Further studies using conditional knockout mice with Lypd6b deficiency should be conducted to determine the function of Lypd6b in tumors.
T cells exhibit distinct metabolic plasticity and activity at different stages of differentiation. Quiescent T cells provide energy through OXPHOS, while activated T cells rely on glycolysis35. Glycolysis is a strong driver of CD4+ T and CD8+ T cell differentiation and function36,37, and is primarily dependent on the mTOR, p38 MAPK, and ERK1/2 pathways38. Our study demonstrated that Lypd6b deficiency reduces OXPHOS and increases aerobic glycolysis of CD8+ T cells through the PI3K/mTOR/LDHA axis (Fig. 6). The effects of PI3K/mTOR and LDHA on metabolism have been reported. Enhanced glycolysis is maintained by PI3K signaling downstream of CD28 co-stimulation, and mTOR is a downstream molecule of PI3K and an important metabolic checkpoint39. In addition, LDHA is increased in activated T cells to promote aerobic glycolysis and IFN-γ secretion40. We discovered a previously unknown role of Lypd6b in regulating CD8+ T cell aerobic glycolysis. Future studies are needed to determine the effect of Lypd6b on other metabolic pathways, including lipid metabolism, amino acid metabolism, and glutamine metabolism in T cells.
Immunotherapy is considered a major treatment modality for the majority of solid cancers41. Inhibiting CTLA4 or PD1 signaling enhances the anti-tumor effect of T cells during CRC therapy42. Although anti-PD1 antibodies have advantages, such as high binding affinity and target specificity, several limitations, including tumor penetration and immune-related toxicities, impact their clinical utility43. Thus, exploring more effective treatments for patients with CRC is an urgent unmet need. In this study, we found that the expression of LYPD6B is increased in human CRC tissues. Patients with CRC and low LYPD6B expression are more sensitive to immunotherapy (Fig. 1 and Fig.8). In addition, Lypd6b deficiency enhances the CRC sensitivity to oxaliplatin in vivo (Fig. 6). Lypd6b deficiency in CD8+ T cells significantly enhances the anti-tumor effect of anti-PD1 antibody. Therefore, therapeutic strategies targeting Lypd6b inhibition may enhance anti-tumor immunity in CRC. Whether Lypd6b deficiency can promote anti-tumor responses in other cancers is unknown. Further studies are needed to determine the role of Lypd6b in other cancers and whether targeting Lypd6b enhances anti-tumor immunity.
Overall, our findings reveal an important negative regulatory role for Lypd6b in the anti-tumor CD8+ T cell response through metabolic regulation. Lypd6b deficiency promotes aerobic glycolysis of CD8+ T cells via the PI3K/mTOR/LDHA pathway and enhances CD8+ T cell anti-tumor immunity in CRC. Patients with CRC and low LYPD6B expression exhibit increased CD8+ T cell infiltration and altered glycolysis. Our findings suggest that Lypd6b is a target for cancer diagnosis and immunotherapy.
Methods
Cell lines
The murine colon cancer cell line MC38 and CMT-93 were purchased from iCell Bioscience, Inc. These cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum at 37 °C with 5% CO2.
Generation of Lypd6b gene expressing EGFP (Lypd6b-EGFP) Mice
The P2A-EGFP cassette was inserted before the stop codon of Lypd6b. Guide RNAs (gRNAs) were designed using the CRISPR guide-design web tool (https://crispr.dbcls.jp/), with the gRNA sequence being CCAgcgatgtaaagcacagaaac. A target vector was constructed containing a 1186 bp 5’homology arm and a 1102 bp 3’ homology arm. Mouse zygotes obtained by mating superovulated C57BL/6J females with males, followed by microinjected of a mixture consisting of Cas9 protein (50 ng/μl), sgRNA (30 ng/μL) and donor vector (10 ng/μL) into the male pronucleus of the fertilized oocytes. The injected zygotes were then transferred into pseudopregnant CD1 female mice, and viable adult mice were obtained. Mice were genotyped by PCR and confirmed by sequencing.
Generation of Lypd6b knockout (Lypd6b −/−) Mice
To generate the Lypd6b−/− mice, gRNAs were designed using the CRISPR guide-design web tool (https://crispr.dbcls.jp/). The selected gRNA sequences are as followes: gRNA1: CCCagcatcgactagctgtcccc; gRNA2: gcagggtgagtatccgagctTGG. Mouse zygotes were obtained by mating superovulated C57BL/6 J females with males, and then microinjected with a mixture of sgRNAs (30 ng/μL) and Cas9 mRNA (80 ng/μL). The treated zygotes were transferred into pseudopregnant CD1 female mice, yielding viable adult mice. Lypd6b−/− mice were genotyped by PCR and confirmed by sequencing.
Generation of Lypd6bflox/flox (Lypd6bfl/fl) Mice
Exon 7 of Lypd6b was selected to anchor with the LoxP sequence. gRNAs were designed using the CRISPR guide-design web tool (https://crispr.dbcls.jp/), with the two gRNA sequences being CCCagcatcgactagctgtcccc and tgctatagtcttacatggcaTGG. A target vector was constructed containing a 1057 bp 5’ homolology arm and a 1011 bp 3’homology arm. Mouse zygotes obtained by mating superovulated C57BL/6 J females with males, followed by microinjection of a mixture consisting of Cas9 protein (50 ng/e malsgRNA (24 ng/μL each, totaling 48 ng/μL) and donor vector (10 ng/μL) into the male pronucleus of fertilized oocytes. Injected zygotes were transferred into pseudopregnant CD1 female mice, and viable adult mice were obtained. Mice were genotyped using PCR and confirmed by sequencing.
Lypd6bflox/flox (Lypd6bfl/fl) mice on the C57BL/6 background were crossed with CD8-Cre (GemPharmatech Co., Ltd) transgenic mice on the C57BL/6 background. The resulting Lypd6bfl/+; CD8Cre+/− mice were then crossed with Lypd6bfl/fl mice to obtain Lypd6bfl/fl; CD8Cre+/+ mice for study subjects and their control littermates (Lypd6bfl/fl mice). gRNA for generate Lypd6b-EGFP, Lypd6bfl/fl, and Lypd6b−/− mice on a C57BL/6 background were listed in Supplementary Table 3. The sequence of primers used for genotyping was listed in Supplementary Table 3.
Animals
All mice were maintained in pathogen-free and humidity- and temperature-controlled (22 ± 2 °C) conditions with a 12-h light/dark cycle (lights on from 7:00 to 19:00), and 50–60% relative humidity at the National Institute of Biological Science. Standard chow and water were available without restriction. All animal studies were performed after being approved by the Institutional Laboratory Animal Care and Use Committee of Beijing Jiaotong University and conducted following the regulations of the Institutional Animal Care and Use Committee (IACUC) at the National Institute of Biological Sciences in Beijing. For animal studies, the mice were earmarked before grouping and randomly separated into groups by an independent person, and experimental/control animals were bred separately. All animal experiments involving tumor models were conducted in accordance with approved protocols. Mice were euthanized when tumor volume reached a maximum of 500 mm³ or upon exhibiting signs of distress, by exposure to a gradually increasing concentration of carbon dioxide followed by cervical dislocation to ensure death.
Induction of colitis and CAC
Eight- to ten-week-old male C57BL/6 mice were induced to colitis with 3.0% (w/v) Dextran sodium sulfate (DSS) (MW: 36,000–50,000 D; MP Biomedicals, Canada) dissolved in sterile, distilled water for 5 days, followed by 5 days of normal water (Number of induced mice, n = 5 for WT and n = 5 for Lypd6b−/−; n = 5 for Lypd6bfl/fl and n = 5 for CD8Lypd6b−/−).
For CAC, eight- to ten-week-old male C57BL/6 mice were injected intraperitoneally with 10 mg/kg azoxymethane (AOM) (Sigma, St Louis, MO). 5 days later, 2% DSS was given in drinking water for 5 days, followed by normal water for 14 days, repeated the cycle 3 times, and the mice were euthanized on the 120th day (Number of injected mice, n = 10 for WT and n = 10 for Lypd6b−/−).
CRC transplant tumor model
Six- to eight-week-old female C57BL/6 mice were selected, and 7 × 105 MC38 cells or 2 × 106 CMT-93 cells were injected subcutaneously into the flank of Lypd6b−/− and WT mice or Lypd6bfl/fl and CD8Lypd6b−/− mice. The tumor volumes of mice were measured every two days using calipers to measure tumor length (L) and width (W), and tumor volumes were estimated by the formula: V = (L × W2)/2. The mice were observed for 15–20 days and then euthanized to obtain the tumors (Number of injected mice, n = 5 for WT and n = 5 for Lypd6b−/−; n = 5 for Lypd6bfl/fl and n = 5 for CD8Lypd6b−/−).
In vivo CD8+ T cell depletion
100 µg anti-CD8a antibody (53–6.72; BioXCell) in phosphate-buffered saline (PBS) was injected intraperitoneally twice a week, starting from 1 day before DSS was given or tumor inoculation. PBS was injected following the same timetable as the control group. The depletion efficacy was confirmed by immunofluorescence staining or flow cytometry analysis.
OXP treatment for CRC
Six- to eight-week-old female C57BL/6 WT and Lypd6b−/− C57BL/6 mice were assigned to homogeneous groups of 5 animals. Therapy commenced when the MC38 transplanted tumor diameter reached 5 mm (day 4), with mice thereafter receiving intraperitoneal injections of oxaliplatin (Sigma-Aldrich, USA) at 20 mg/kg body weight three times a week. The tumors were measured with a digital caliper until the mice were euthanized (n = 5 mice per group).
Anti-PD1 antibody treatment for CRC
Tumor volume was monitored using a digital caliper and calculated according to the formula: V = (L × W2)/2. Therapy commenced when the MC38 transplanted tumor volume reached 50 mm3 (day 6). Female Lypd6bfl/fl and CD8Lypd6b−/− mice on a C57BL/6 background were selected, and intraperitoneally injected with a single dose of anti-PD1 (RMP1-14; BioXCell) at 100 μg twice a week (n = 5 mice per group). The tumors were measured with a digital caliper until the mice were euthanized.
Adoptive CD8+ T cell transfer experiment
Splenic CD8+ T cells from WT and Lypd6b−/− mice were sorted and activated for 48 h in the presence of CD3 and CD28 antibodies. 5 × 106 CD8+ T cells were injected into the tail vein of 8-week-old nude mice. On the second day, 7 × 105 MC38 cells were injected subcutaneously into the flank of nude mice.
Histopathological and immunofluorescence analysis
The obtained murine tissues were fixed in 4% paraformaldehyde (PFA) overnight, then embedded in paraffin, and cut into 3–5 μm sections. Paraffin sections of the tumor or colon were stained with hematoxylin and eosin (H&E) (Zhongshan Jinqiao Biotechnology). For IF staining, frozen sections were incubated with anti-Lypd6b (Abcam, USA), anti-CD8, anti-CD4 (BioLegend), anti-F4/80 (BioLegend), anti-CD11b, anti-Gr1, anti-B220 (BD Biosciences), and anti-E-cadherin (Santa Cruz Biotechnology) followed by incubation with Alexa Fluor 488-and 594-conjugated secondary antibodies (Jackson Immuno Research Labs). Sections were evaluated under the microscope (DP71; Olympus) using bright-field and fluorescence microscopy.
Isolation of intestinal lymphocytes from colitis mice
Colon tissues were dissected and obtained from mice with acute colitis. Using 40% and 80% percoll gradients (GE Healthcare Biosciences) isolated the lamina propria lymphocytes as previously described44.
Isolation of immune cells from transplanted tumors
After the tumor had grown to a certain size, dissected and mechanically sheared the tumor tissues, then they were digested in DMEM medium with DNase I (50 μg/mL) and Collagenase IV (1 mg/mL) (Millipore-Sigma) for 60 min at 37 °C accompanied by gentle shaking. The cells were collected after filtering cell suspension through a 40 μm cell strainer twice.
Flow cytometry analysis
We obtained the single-cell suspensions from the spleen, colon lamina propria, and transplanted tumors, and stained with conjugated mouse-specific mAbs as follows: Percp-anti-CD11b (clone M1/70), APC-anti-Gr1 (clone RB6-8C5), Percp-anti-CD8 (clone 53–6.7), APC-anti-CD8 (clone 53–6.7), APC-anti-CD4 (clone GK1.5), PE-anti-CD4 (clone GK1.5), FITC-anti-CD69 (clone IM7), FITC-anti-IFN-γ (clone XMG1.2), and APC-anti-IL-2 (clone JES6-5H4). Antibodies were purchased from BioLegend and applied at a 0.2 μg/ml concentration. All flow samples were read with Calibur (BD Biosciences), and data was analyzed with FlowJo software.
Isolation of CD8+ T cell
Naïve CD8+ T lymphocytes were isolated from mouse spleens by negative selection using the Naïve CD8+ T Cell Isolation Kit (BioLegend) according to the manufacturer’s instructions. Purified CD8+ T cells were stimulated with 2 μg/mL anti-CD3 and 1 μg/mL anti-CD28 in 6-well plates for the following RNA extraction, western blot analysis, and metabolism analysis.
T cell activation and staining
Primary T cells were isolated from the spleen of DSS-treated mice. Using the carboxyfluorescein diacetate succinimidyl ester (CFSE) dilution (Millipore-Sigma) to monitor the T cell proliferation. T Cells were incubated with CFSE (2 μM) for 10 min at 37 °C, and then washed and stimulated with 2 μg/mL anti-CD3 (BioLegend) and 1 μg/mL anti-CD28 (Multi Sciences) for 48 h. The dilution of CFSE intensity was detected by flow cytometry. For intracellular cytokine staining, isolated spleen cells were cultured with PMA/Ionomycin and BFA/Monensin mixtures (Multi Sciences) for 10 h. Then, the cells were stained with anti-IFN-γ, anti-IL-2, and anti-TNF (BioLegend) after fixation/permeabilization with Cell Permeabilization Kit (ThermoFisher). Stained cells were read with Calibur (BD Biosciences) and data was analyzed with FlowJo software.
Western blot analysis
Colon tissue and sorted CD8+ T cells were analyzed using the following primary antibodies: anti-Lypd6b (Abcam, USA), anti-PI3K, anti-p-PI3K (Affinity), anti-mTOR, anti-p-mTOR (Cell signaling), LDHA (Proteintech), and β-actin (Lab lead). The secondary antibodies were HRP-conjugated goat anti-mouse IgG (Lab lead) and goat anti-rabbit IgG (BioLegend). Blots were scanned using a Clinx Science Instrument (Shanghai Qinxiang Scientific Instrument).
RT-PCR
Total RNA was isolated with TRIzol, and reverse transcription (RT) was performed using PrimeScript RT Master Mix (TIANGEN). Then, qPCR was performed using a SYBR Premix Ex TaqTM Kit (TIANGEN). Primers for qPCR were listed in Supplementary Table 4.
Enzyme-linked immunosorbent assay (ELISA)
The IFN-γ and interleukin-2 (IL-2) in CD8+ T cell culture was detected by the ELISA kit (Cloud-Clone Corp) according to the manufacturer’s instructions.
Seahorse assays
Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured with an XFe96 Extracellular Flux Analyzer (Agilent Technologies) with the manufacturer’s recommended protocol. CD8+ T cells were isolated from the spleens and sorted by magnetic beads. Then they were activated with 2 μg/mL anti-CD3 and 1 μg/mL anti-CD28 antibodies for 48 h. Activated CD8+ T cells were seeded on XFe96 microplates (3 × 105 cells/well) pre-coated with Cell-Tak adhesive (BD Biosciences). Rapid centrifugation causes the cells to adhere to the bottom plate. Cells were rested in a non-buffered assay medium in a non-CO2 incubator for 25 min. Glycolysis or OXPHOS associated parameters were measured by Seahorse XFe Glycolysis Stress test kit. In a glycolysis assay, three compounds are injected separately: 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxyglucose (2-DG). In an OXPHOS assay, three compounds are injected separately: 2 μM oligomycin, 2 μM carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP), and a combination of 1 μM antimycin A and 1 μM rotenone.
Measurement of glucose consumption and lactate production
Extracellular glucose in the CD8+ T cell culture medium was determined using a glucose assay kit (Nanjing Jiancheng Bioengineering Institute) recommended by the manufacturer’s instructions. Glucose consumption was calculated by subtracting the concentration in the culture media from the original glucose concentration. Extracellular lactate in CD8+ T cell culture medium was determined with a lactate assay kit (Nanjing Jiancheng Bioengineering Institute) recommended by the manufacturer’s instructions. The values were normalized to protein concentration.
CD8+ T cellular glucose uptake analysis
CD8+ T cells from the spleen of WT and Lypd6b−/− mice were sorted and activated with 2 μg/mL CD3 and 1 μg/mL CD28 antibodies for 48 h. Then incubated CD8+ T cells were treated with 100 μM 2-NBDG (Beyotime) for 30 min before measuring fluorescence by flow cytometry.
Tetramethylrhodamine ethyl ester (TMRE measurement)
Mitochondrial membrane potential (MMP) of CD8+ T cells was detected by TMRE staining kit (Beyotime). Briefly, the activated CD8+ T cells were cultivated in a 24-well plate at 2 × 106 cells. Then, the media were discarded, and the cells were cultured in media containing 10 μM TMRE in the 37 °C incubator for 20 min, washed with PBS. Fluorescence intensity was detected by Calibur (BD Biosciences), λex = 550 nm, λem = 575 nm.
Mito-tracker deep red FM staining
Activated CD8+ T cells from the spleen of WT and Lypd6b−/− mice were collected, and centrifuged at 1000 × g for 5 min, and gently re-suspend the cells with Mito Tracker Deep Red FM preheated at 37 °C, incubated at 37 °C for 20 min. After incubation, centrifuge at 1000 × g for 5 min, discard the supernatant, and analyze by Calibur (BD Biosciences) with FlowJo software.
RNA sequencing analysis
RNA-sequencing analyses were performed in CD8+ T cells sorted from MC38 tumors of WT and Lypd6−/− mice. Total RNA was extracted with TransZol Routine RNA Extraction Kit (TransGen Biotech, Beijing). Using the principle of component analysis for the Stats package and plots with the ggplot2 package in R (version 3.5). After removing adaptor sequences, the raw transcriptomic reads were mapped to the C57BL/6 genome using HISAT40/Bowtie241 tools, including low-quality and polyN sequence reads. Normalization was performed by using RESM software. DEGs were identified with the threshold of padj < 0.05, and the absolute value of log2 Ratio ≥ 0. The Kyoto Encyclopedia of KEGG enrichment analysis was performed using phyper in R. All data mining and Fig. presentations were conducted on the Dr. Tom network platform of BGI.
Human CRC patient specimens
Human CRC tissues and paracancerous tissues were gathered from CRC patients of different ages at Dazhou Central Hospital with ethical approval for human IRB00000003-17003. Informed consent was obtained from all patients. Relevant information on the patients can be found in Supplementary Table 2.
Tissues including 30 human CRC cancer tissue samples and 30 paracancerous tissue samples for LYPD6B expression were determined by IHC staining using a rabbit anti-LYPD6B antibody (Abcam, USA). The evaluation of LYPD6B staining was carried out according to a method described in a previous study45. The intensity of LYPD6B expression was scored as follows: 0, negative; 1, weak; 2, moderate; 3, strong. The extent of staining was scored as follows: 1, 0 to<25%; 2, 25 to<50%; 3, 50 to<75%; or 4, 75 to 100%. Five randomly selected fields were observed under a light microscope. The final score was determined by multiplying the intensity scores by the extent of staining. Sums from 0 were defined as negative for LYPD6B; sums from>0 to 4 were defined as weak expression of LYPD6B; sums from>4 to 8 were defined as moderate expression of LYPD6B, sums from>8 to 12 were defined as strong expression of LYPD6B.
Transcriptome sequencing (n = 154) from patients with CRC tumor tissues and paracancerous tissues was collected at the Dazhou Central Hospital with informed consent46. The Ethics Committee of Beijing Jiaotong University approved the use of human specimens in accordance with the Declaration of Helsinki.
Public database analysis
The pan-cancer expression data of LYPD6B was obtained from the Tumor Immune Estimation Resource (TIMER) 2.0 website (https://cistrome.shinyapps.io/timer/). Using the bioGPS database (http://biogps.org/#goto=welcome) to explore the expression profile of Lypd6b in mouse cell lines. In addition, the Kaplan–Meier plotter (http://kmplot.com) is an online database used to estimate the colon adenocarcinoma (COAD) patient’s survival with LYPD6B expression. Moreover, the processed sequencing data were available in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24555 for sing-cell RNA-seq, GSE184093 for RNA-seq. UALCAN database (http://ualcan.path.uab.edu) is applied to explore LYPD6B expression between tumor and normal tissues. Additionally, the correlation analysis between LYPD6B expression and CD8+ T cells or immunostimulators was obtained from the TISIDB.
Ethics committee approval and patient consent
All animal experiments were performed in accordance with the Beijing Jiaotong University, Institutional Animal Care and Use Committee Guideline (BJTUBS202003). All CRC patient samples were approved by the Dazhou Central Hospital Ethics Committee (IRB00000003-17003).
Statistical analysis
All data were expressed as the mean ± SEM, they were analyzed by the GraphPad Prism V8.0 software. The difference between the two groups was compared by two-tailed unpaired Student’s t test. Multiple comparisons were made using two-way ANOVA analysis. The data are representative of at least three independent experiments. P < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The publicly sequencing data were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE24555 for single-cell RNA-seq, GSE184093 for RNA-seq. The raw transcriptome of CRC patients has been deposited in the National Genomics Data Center database with the accession number of HRA000235, https://ngdc.cncb.ac.cn/search/specific?db=hra&q=HRA000235. The raw transcriptome of CD8+ T cells from mice has been deposited in the ArrayExpress database with the accession number of E-MTAB-16060, https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-16060?key=c517ce45-af5d-4728-90ad-14453d2c0864. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. Source data are provided with this paper.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Eng, C. et al. A comprehensive framework for early-onset colorectal cancer research. Lancet Oncol. 23, e116–e128 (2022).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Shah, S. C. & Itzkowitz, S. H. Colorectal cancer in inflammatory bowel disease: mechanisms and management. Gastroenterology 162, 715–730.e713 (2022).
Waldner, M., Schimanski, C. C. & Neurath, M. F. Colon cancer and the immune system: the role of tumor invading T cells. World J. Gastroenterol. 12, 7233–7238 (2006).
Li, X., Shao, C., Shi, Y. & Han, W. Lessons learned from the blockade of immune checkpoints in cancer immunotherapy. J. Hematol. Oncol. 11, 31 (2018).
Li, Y. et al. A mini-review for cancer immunotherapy: molecular understanding of PD-1/PD-L1 pathway & translational blockade of immune checkpoints. Int. J. Mol. Sci. 17, 1151 (2016).
Lines, J. L., Sempere, L. F., Broughton, T., Wang, L. & Noelle, R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer Immunol. Res. 2, 510–517 (2014).
Bidgood, G. M., Keating, N., Doggett, K. & Nicholson, S. E. SOCS1 is a critical checkpoint in immune homeostasis, inflammation and tumor immunity. Front. Immunol. 15, 1419951 (2024).
Chung, J. S. et al. DC-HIL/Gpnmb is a negative regulator of tumor response to immune checkpoint inhibitors. Clin. Cancer Res. 26, 1449–1459 (2020).
Makaremi, S. et al. Immune checkpoint inhibitors in colorectal cancer: challenges and future prospects. Biomedicines 9, 1075 (2021).
Miwa, J. M., Anderson, K. R. & Hoffman, K. M. Lynx prototoxins: roles of endogenous mammalian neurotoxin-like proteins in modulating nicotinic acetylcholine receptor function to influence complex biological processes. Front. Pharm. 10, 343 (2019).
Demars, M. P. & Morishita, H. Cortical parvalbumin and somatostatin GABA neurons express distinct endogenous modulators of nicotinic acetylcholine receptors. Mol. Brain 7, 75 (2014).
Ni, J. et al. Cloning and characterization of a human LYPD7, a new member of the Ly-6 superfamily. Mol. Biol. Rep. 36, 697–703 (2009).
Venkatesan, S. et al. Chrna5 and lynx prototoxins identify acetylcholine super-responder subplate neurons. iScience 26, 105992 (2023).
Ochoa, V., George, A. A., Nishi, R. & Whiteaker, P. The prototoxin LYPD6B modulates heteromeric α3β4-containing nicotinic acetylcholine receptors, but not α7 homomers. Faseb J. 30, 1109–1119 (2016).
Chung, B. H. et al. Severe intellectual disability and autistic features associated with microduplication 2q23.1. Eur. J. Hum. Genet. 20, 398–403 (2012).
Koroknai, V. et al. DNA hypermethylation is associated with invasive phenotype of malignant melanoma. Exp. Dermatol 29, 39–50 (2020).
Liu, Z., Liu, H., Wang, Y. & Li, Z. A 9‑gene expression signature to predict stage development in resectable stomach adenocarcinoma. BMC Gastroenterol. 22, 435 (2022).
Koebel, C. M. et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 450, 903–907 (2007).
Cao, J. et al. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis. 14, 407 (2023).
Boudreau, A. et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 12, 779–786 (2016).
Chapman, N. M. & Chi, H. Hallmarks of T-cell exit from quiescence. Cancer Immunol. Res. 6, 502–508 (2018).
Chi, H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12, 325–338 (2012).
Xu, K. et al. Glycolysis fuels phosphoinositide 3-kinase signaling to bolster T cell immunity. Science 371, 405–410 (2021).
Kelly, C. J. et al. Regulation of O2 consumption by the PI3K and mTOR pathways contributes to tumor hypoxia. Radiother. Oncol. 111, 72–80 (2014).
Hashemi, M. et al. Biological landscape and nanostructural view in development and reversal of oxaliplatin resistance in colorectal cancer. Transl. Oncol. 40, 101846 (2024).
Ma, W., Gilligan, B. M., Yuan, J. & Li, T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J. Hematol. Oncol. 9, 47 (2016).
Maimela, N. R., Liu, S. & Zhang, Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput. Struct. Biotechnol. J. 17, 1–13 (2019).
Okazaki, T. et al. PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J. Exp. Med. 208, 395–407 (2011).
Cai, L., Li, Y., Tan, J., Xu, L. & Li, Y. Targeting LAG-3, TIM-3, and TIGIT for cancer immunotherapy. J. Hematol. Oncol. 16, 101 (2023).
Lasry, A., Zinger, A. & Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17, 230–240 (2016).
Yamazaki, T. et al. Expression of programmed death 1 ligands by murine T cells and APC. J. Immunol. 169, 5538–5545 (2002).
Seliger, B. & Quandt, D. The expression, function, and clinical relevance of B7 family members in cancer. Cancer Immunol. Immunother. 61, 1327–1341 (2012).
Kirchmair, A. et al. (13)C tracer analysis reveals the landscape of metabolic checkpoints in human CD8(+) T cell differentiation and exhaustion. Front. Immunol. 14, 1267816 (2023).
Reina-Campos, M., Scharping, N. E. & Goldrath, A. W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 21, 718–738 (2021).
Liu, S., Liao, S., Liang, L., Deng, J. & Zhou, Y. The relationship between CD4(+) T cell glycolysis and their functions. Trends Endocrinol. Metab. 34, 345–360 (2023).
Dáňová, K. et al. NF-κB, p38 MAPK, ERK1/2, mTOR, STAT3 and increased glycolysis regulate stability of paricalcitol/dexamethasone-generated tolerogenic dendritic cells in the inflammatory environment. Oncotarget 6, 14123–14138 (2015).
Xu, X., Ye, L., Araki, K. & Ahmed, R. mTOR, linking metabolism and immunity. Semin Immunol. 24, 429–435 (2012).
Peng, M. et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484 (2016).
Ganesh, K. et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375 (2019).
Tison, A., Garaud, S., Chiche, L., Cornec, D. & Kostine, M. Immune-checkpoint inhibitor use in patients with cancer and pre-existing autoimmune diseases. Nat. Rev. Rheumatol. 18, 641–656 (2022).
Yang, J. & Hu, L. Immunomodulators targeting the PD-1/PD-L1 protein-protein interaction: from antibodies to small molecules. Med. Res. Rev. 39, 265–301 (2019).
Zhang, J. et al. S100A4 promotes colon inflammation and colitis-associated colon tumorigenesis. Oncoimmunology 7, e1461301 (2018).
Yuan, Q. et al. MyD88 in myofibroblasts regulates aerobic glycolysis-driven hepatocarcinogenesis via ERK-dependent PKM2 nuclear relocalization and activation. J. Pathol. 256, 414–426 (2022).
Chu, Y. et al. A novel model based on CXCL8-derived radiomics for prognosis prediction in colorectal cancer. Front. Oncol. 10, 575422 (2020).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32571089, U24A20812) (to J.Z.), the Natural Science Foundation of Beijing (7232102) (to J.Z.) and National Science and Technology Major Project (2024ZD0520505) (to F.Z.).
Author information
Authors and Affiliations
Contributions
J.Z. designed the study and supervised the experiments. T.L., L.C., X. Y., and H.C. performed experiments and analyzed the data. F.Z., Q.L., and X.L. provided the clinical sample and analyzed the clinical data. J.Y., Z.L., and Z.L. performed single-cell sequencing data analysis. F.W. constructed Lypd6b−/− and Lypd6bfl/fl mice, D.X. bred Lypd6b−/− and Lypd6bfl/fl mice. T.L., J.Z., L.C., and X.Y. wrote and revised the manuscript. Z.C. and J.W. took part in the discussion of the research project.
Corresponding authors
Ethics declarations
Competing interests
The author declares the following competing interests: J.W. has received sponsored research funding from Remunix Inc. within the past 12 months, serves as a consultant for LAV, Henlius, and Hanmi, and is the founder, advisor, and a shareholder of Remunix. All of these efforts are not relevant to this study. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Vassiliki Boussiotis, Anastasios Giannou and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, T., Zeng, F., Li, Z. et al. Lypd6b depletion promotes CD8+ T cell-mediated anti-tumor immunity via metabolic reprogramming in colorectal cancer. Nat Commun 17, 675 (2026). https://doi.org/10.1038/s41467-025-67344-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67344-w