Abstract
Human organ structure and function are important endophenotypes for clinical outcomes. Genome-wide association studies (GWAS) have identified numerous common variants associated with phenotypes derived from magnetic resonance imaging (MRI) of the brain and body. However, the role of rare protein-coding variations affecting organ size and function is largely unknown. Here we present an exome-wide association study that evaluates 596 multi-organ MRI traits across over 50,000 individuals from the UK Biobank. We identified 107 variant-level associations and 224 gene-based burden associations (67 unique gene-trait pairs) across all MRI modalities, including PTEN with total brain volume, TTN with regional peak circumferential strain in the heart left ventricle, and TNFRSF13B with spleen volume. The singleton burden model and AlphaMissense annotations contributed 8 unique gene-trait pairs including the association between an approved drug target gene of KCNA5 and brain functional activity. The identified rare coding signals elucidate some shared genetic effects across organs, prioritize previously identified GWAS loci, and are enriched for drug targets. Overall, we demonstrate how rare variants enhance our understanding of genetic effects on human organ morphology and function and their connections to complex diseases.
Similar content being viewed by others
Introduction
Magnetic resonance imaging (MRI)-derived traits enable us to study the structure, function, and abnormalities of human organs in vivo. Many of these traits serve as established endophenotypes implicated in complex diseases and related traits. Therefore, it is of great interest to uncover genetic effects using imaging data to better understand the biology of human organs in health and disease. Recent genome-wide association studies (GWAS) have successfully identified common variants associated with multi-organ imaging traits, including brain structural, diffusion, and functional MRI1,2,3,4,5,6,7,8, as well as cardiovascular magnetic resonance imaging (CMR)9,10,11 and abdominal MRI12,13,14. However, a limitation of common variant signals identified by GWAS is that they often reside in non-coding regions and exhibit small effect sizes, which complicates the direct derivation of biological insights or the identification of causal genes15,16,17,18,19. By focusing on rare variants in protein-coding regions of the genome, whole exome sequencing (WES) studies aim to directly identify genes of interest.
Although existing exome-wide association studies16,17,20,21 (ExWAS) have identified associations for some imaging traits, our knowledge of the rare variant genetic architectures of human organs and their roles in various diseases remains substantially limited. Specifically, most previous ExWAS have focused on a single organ and/or a small set of imaging traits, lacking a multi-organ perspective that simultaneously explores the genetic effects of the human brain and body. For example, Park et al.20 analyzed CT imaging-derived hepatic fat, Haas et al.14 studied liver fat based on abdominal MRI using machine learning, Jurgen et al.21 studied several CMR traits, and Backman et al.16 and Karczewski et al.17 included brain MRI traits in their UK Biobank (UKB) WES studies of a wide range of phenotypes. In addition, these studies might be underpowered because of the relatively limited sample sizes, which are critical for rare variant association studies. Therefore, analyzing more and refined imaging traits, spanning multiple organs, in larger sample sizes will yield deeper insights into the genetic effects of rare variants across the whole body and shared genetic effects across organs10.
Here, we conducted ExWAS for 596 MRI traits derived from the brain, heart, liver, kidney, and lung (Supplementary Data S1) of over 50,000 participants from the UKB study. We used an internal discovery-replication design to make the best use of available data resources to identify rare variants and genes associated with human organ structure and function (Fig. 1). Specifically, all the primary findings and the corresponding interpretations are based on the joint sample for higher statistical power and more stable summary statistics. The joint sample was then split into the internal discovery sample and validation sample to partially verify the robustness of the findings within the UKB study (Fig. 1, Methods, and Supplementary Note). We evaluated various functional annotation approaches for missense variants in gene-level set-based association testing, including conventional methods such as SIFT22, PolyPhen2 HDIV23, PolyPhen2 HVAR23, LRT24, and MutationTaster25, as well as the deep learning-based method AlphaMissense26. We compared our results with previous GWAS on the same set of MRI traits in order to provide shared common and rare variant evidence for a gene’s involvement in a trait. We also used burden heritability regression (BHR)27 tests to characterize the genetic architecture for ultra-rare coding variants (minor allele frequency [MAF] <1 × 10−4) across functional classes. In summary, our study identified exome-wide associations for multi-organ structure and function, providing a valuable source of evidence that could be useful in drug discovery and clinical therapeutics. These findings may also enhance our understanding of the complex interrelations between the human organs, health, and disease.
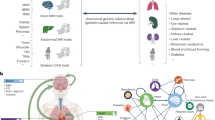
A An overview of the data included in our study. For imaging phenotypes, we made use of a broad range of multi-organ imaging phenotype data including brain imaging traits such as regional brain volume from structural MRI, diffusion tensor imaging (DTI) parameters from diffusion MRI, and functional connectivity and activity traits from resting and task fMRI; CMR traits such as measurements of the aorta (ascending/descending aorta) and heart chambers (left ventricle, right ventricle, left atrium, and right atrium); abdominal MRI traits such as abdominal organ traits (liver, kidney, lung, spleen, and pancreas) and abdominal composition measurements of fat and muscle. For genetic data, we included whole exome sequencing data and focused on rare coding variants (minor allele frequency [MAF] <0.01). Only European individuals with imaging traits were included in the present study (n = 54,365). Some elements in this figure were created in BioRender: Fan, Y. (2025) https://BioRender.com/oz6gd9i, Fan, Y. (2025) https://BioRender.com/f2ok1xo, Fan, Y. (2025) https://BioRender.com/x77lwka, Fan, Y. (2025) https://BioRender.com/29marqh, Fan, Y. (2025) https://BioRender.com/a8yoefm, and Fan, Y. (2025) https://BioRender.com/19zbtn5. B An overview of our study design. We adopted an internal discovery-replication procedure and finalized the summary statistics based on the joint sample (phases 1 to 5 sample). Specifically, we conducted association tests on phases 1 to 3 sample, phases 4 to 5 sample, and phases 1 to 5 sample respectively. We examined (i) whether the detected signals from phases 1 to 3 sample had concordant directions and remained significant in phases 4 to 5 sample; (ii) whether the detected signals from phases 1 to 3 sample had concordant directions and obtained stronger evidence (smaller P-values) in the joint sample. Based on this validated procedure, the significant results and other downstream analyses used the summary statistics generated from this joint sample. The average n indicates the average sample size used in the tests across all 596 phenotypes. Some elements in this figure were created in BioRender: Fan, Y. (2025) https://BioRender.com/n0j8gb2 and Fan, Y. (2025) https://BioRender.com/wawasqc.
Results
Overview of variant-level associations with multi-organ MRI traits
Based on European individuals from UKB phases 1 to 5 MRI data (released up to late 2023, average n = 40,038; Fig. 1), we identified 107 rare (MAF < 0.01) variant-level associations (Supplementary Data S2) using a conservative P-value threshold of 2.8 × 10−10 (Bonferroni adjusted for all variant-trait tests with minor allele count [MAC] > 5 as 0.05/178,280,016, Methods) (Fig. 2A). Only predicted loss-of-function variants (pLoF) and missense variants were included in our study. There were 75 gene-trait pairs between 24 unique genes and 62 MRI traits, including 3 abdominal MRI traits, 7 CMR traits of the heart and the aorta, 10 brain structural MRI traits (regional brain volumes), as well as 42 brain diffusion MRI traits (diffusion tensor imaging [DTI] parameters) (Fig. 2B). Additionally, 138 associations (92 unique genes and 107 MRI traits) showed suggestive evidence, surviving a more liberal P-value threshold of 1 × 10−8 (Supplementary Data S3, Methods). These associations spanned across all categories of MRI traits (Supplementary Data S2). As expected, larger effect sizes were linked with lower MAF, which revealed the process of negative selection17,28,29 (Fig. 2C).
A Manhattan plot for variant-level association tests across all 596 imaging phenotypes. Only coding variants of interest were included in the variant-level association tests and associations with P < 1 × 10−3 were plotted. P-values are derived from two-sided tests based on z-scores (or equivalently a χ2 test with one degree of freedom. The dashed lines indicate the variant-level significant P-value threshold at 2.8 × 10−10 from Bonferroni correction and the suggestive P-value threshold at 1 × 10−8 as rare variant genome-wide significance threshold (details are provided in Method). The x-axis shows all the categories of our imaging phenotypes, while we further separated the abdominal MRI traits into abdominal organs and body/fat composition, and separated CMR traits into heart chambers and aorta areas based on the specific traits. Significant coding-variant-level associations (P < 2.8 × 10−10) were labeled to the corresponding genes. For DTI parameters, the unlabeled signals all belong to VCAN gene. B Summary count for the number of signals, unique variants (unique genes in the parentheses), and unique traits within each imaging category that has significant variant-level associations. C The scatter plot for all the variant-level significant results (colored by imaging categories) and suggestive results (in grey color). The x-axis shows the minor allele frequency of each association, and the y-axis is the absolute value of the corresponding genetic effect size in s.d. units. D Graphical illustration of associations between two missense variants (rs72553883 and rs34557412) in TNFRSF13B and spleen volume. The rug plot at the first row demonstrates the locations of these two missense variants. The rug plot at the second row indicates the locations of rare variants in the ‘plof_alpha’ gene burden model, since gene-based burden test also revealed the association between TNFRSF13B and spleen volume. The summary statistics (P-values) of exome-wide associations between TNFRSF13B and blood traits based on burden tests were downloaded from Genebass17. Some elements in this figure were created in BioRender: Fan, Y. (2025) https://BioRender.com/f2ok1xo, Fan, Y. (2025) https://BioRender.com/rdhm596, and Fan, Y. (2025) https://BioRender.com/e81d7o9. E Visualization of the associations between two missense variants (rs2652098 and rs143368552) in VCAN and 12 mean diffusivity traits (β1 is the effect size in s.d. of rs2652098 and β2 is the effect size in s.d. of rs143368552).
Due to the lack of independent data sources and the inherent rarity of the variants, identifying rare variant associations typically requires large sample sizes, and findings are difficult to fully replicate. To assess the reliability of our findings derived from the joint sample and maximize the use of available data, we adopted an internal replication procedure30 (Fig. 1 and Methods). Specifically, we split our joint sample to the internal discovery dataset and validation dataset for internal replication. The discovery dataset only included individuals from phases 1 to 3 of MRI release (average n = 30,739), while the validation dataset is independent of it by design and contained individuals from phases 4 and 5 of the MRI release (Method). For the purpose of replication analysis, summary statistics from the discovery sample will be separately compared with results from the validation sample and the joint sample (Method and Supplementary Note). When we restricted our study to the discovery sample, which is the subset of the joint sample, 59 associations passed the Bonferroni threshold of P < 3.5 × 10−10 (Bonferroni adjusted for all variant-level tests with MAC > 5 in this subset, 0.05/142,935,657, Methods) (Supplementary Data S4) and were used for internal replication. Among these 59 associations, 48 had mutations with MAC > 5 suitable for association tests (Supplementary Data S4) in the independent validation sample from phases 4 and 5 of the MRI release. Of these, 45 (93.75% = 45/48) passed the P-value threshold of 2.9 × 10−2 (Benjamini–Hochberg false discovery rate [FDR] at the 0.05 level), and all had consistent effect size directions as those in the phases 1 to 3 data. We conducted the Pearson correlation test for the effect sizes of the identified associations in the discovery sample and the validation sample. Among these 48 associations with summary statistics available in both stages, we observed a substantial correlation (t = 17.825, df = 46, P = 2.8 × 10−22, rg = 0.935), which shows a high level of agreement between the associations identified in the discovery sample and validation sample. When we relaxed the P-value cutoff to 1 × 10−8, there was still a moderate to high correlation (t = 7.7585, df = 81, P = 2.2 × 10−11, rg = 0.653). Furthermore, 84.7% (= 50/59) of these associations had smaller P-values in the combined phases 1 to 5 sample, and all of them had consistent effect directions. Variants with decreased P-values in the joint analysis implied similar effects of the two sub-cohorts, and we found that the 50 signals that had smaller P-values in the combined sample indeed included all the 45 signals that were replicated in the independent phases 4 and 5 sub-cohort (Supplementary Data S4). A comparison between the observed summary statistics in the internal analysis and the expected improvements of these values in the enlarged joint sample is discussed in Supplementary Note. More details of the results from the internal replication analysis and summary statistics of variant-level associations across different stages can be found in Supplementary Data S4. Overall, this internal replication analysis showed the robustness and validity of our data results. Empirically, over 90% of the 107 variant-level associations identified in the UKB phases 1 to 5 sample could potentially be replicated at the 5% FDR level, should a replication dataset become available. In the following two sections, we highlight some interesting findings across different organs. The complete list of all these 107 variant-level associations is presented in Supplementary Data S2.
Variant-level tests identified associations for hepatic and spleen MRI traits
To our knowledge, associations between rare variants and abdominal MRI-derived traits were only studied in Hass et al.14 with the focus restricted to liver fat and only 18,103 subjects were included. Another study20, with a similar focus on hepatic fat trait, used CT imaging data from 9594 individuals to study the exome-wide associations. Thus, the roles of rare variants in a wider range of abdominal MRI traits remain largely unexplored. In our variant-level analysis, we identified associations between spleen volume and two missense variants in TNFRSF13B (rs72553883, MAF = 0.55%, χ2 = 79.20, 1 df, effectorg = 0.025 L, effect = 0.37 s.d. units, 95% CI = [0.29, 0.45], P = 5.6 × 10−19, and rs34557412, MAF = 0.66%, χ2 = 122.25, 1 df, effectorg = 0.028 L, effect = 0.42 s.d. units, 95% CI = [0.34, 0.49], P = 2.0 × 10−28), while the latter was the top hit among all variant-level associations. TNFRSF13B is a well-known risk gene of common variable immunodeficiency and mutations in TNFRSF13B typically occur in patients who developed splenomegaly31,32,33. In addition, the missense variant rs72553883 was associated with a series of blood-related traits including platelet, myeloid white cell, and lymphoid white cell indices in a previous GWAS34 using genotyping array data while similar associations between rare variants in TNFRSF13B and multiple blood biomarkers were also observed in Karczewski et al.17 using exome data (Fig. 2D). Consistent with these results on spleen abnormalities derived from immunodeficiency and the central role of the spleen in blood filtration and blood cell turnover, our finding points out a direct association between spleen volume and missense mutations in TNFRSF13B. Another missense variant in the manganese transporter SLC30A10 (rs188273166, MAF = 0.09%, χ2 = 94.79, 1 df, effectorg = 63.72 ms35,36 [ms], effect = 1.18 s.d. units, 95% CI = [0.94, 1.42], P = 2.1 × 10−22) was associated with higher corrected T1 liver iron. SLC30A10 is known to be involved in maintaining the manganese level37. This association aligned with recent GWAS that reported the role of rs188273166 in hypermagnesemia symptoms and a SLC30A10-targeted study found its positive effect on corrected T1 liver iron38,39. Interestingly, we also identified a missense variant in DDX51 associated with the fat-free muscle volume of right posterior thigh (rs200735214, MAF = 0.01%, χ2 = 41.06, 1 df, effectorg = −0.87 L, effect = −1.09 s.d. units, 95% CI = [−1.42, −0.76], P = 1.5 × 10−10), which constituted the only signal that passed our stringent P value threshold for human muscle/fat composition traits. Notably, at a more permissive P-value threshold at 1 × 10−8 as our suggestive evidence, we observed that rs200735214 in DDX51 was also associated with the fat-free muscle volume for total thigh (MAF = 0.01%, χ2 = 36.02, 1 df, effectorg = −2.26 L, effect = −0.90 s.d. units, 95% CI = [−1.19, −0.60], P = 1.95 × 10−9) among other signals for human muscle/fat composition traits such as rs534069616 in PLIN4 and posterior thigh muscle fat infiltration (MAF = 0.2%, χ2 = 36.94, 1 df, effectorg = −0.95%, effect = −0.40 s.d. units, 95% CI = [−0.53, −0.27], P = 1.2 × 10−9 for the left; MAF = 0.2%, χ2 = 34.52, 1 df, effectorg = −0.91%, effect = −0.38 s.d. units, 95% CI = [−0.51, −0.26], P = 4.2 × 10−9 for the right). DDX51, a member of the DEAD-box helicase family, plays a crucial role in ribosomal RNA processing and is essential for ribosome biogenesi40,41. Although no direct link between DDX51 and muscle function has been reported previously, the strong association identified in our exome data highlights its potential involvement in muscle composition traits. Meanwhile, PLIN4 encodes a protein known as Perilipin 4, which belongs to the perilipin family of proteins that coat lipid droplets and involve in regulating lipid metabolism42. Perilipins are essential for the proper storage and release of lipids within cells. In particular, PLIN4 has been shown to be highly expressed in skeletal muscle and was found at periphery of skeletal muscle fibers42, which is consistent with our findings.
Variant-level tests identified associations for brain and heart MRI traits
We highlight some rare variants associations with regional brain volumes, brain DTI parameters, and CMR traits of the heart and aorta. A missense variant in ADRA1A was associated with the volume of the left ventral diencephalon (rs771722367, MAF = 0.01%, χ2 = 42.67, 1 df, effectorg = 683.56 mm3, effect = 1.28 s.d. units, 95% CI = [0.90, 1.67], P = 6.5 × 10−11), though the P value for this variant and right ventral diencephalon was P = 3.6 × 10−8 (χ2 = 30.37, 1 df), which did not pass our liberal threshold at 1 × 10−8. ADRA1A was previously reported to be associated with schizophrenia43,44. The ventral diencephalon area was also associated with a missense variant in PKD1 (rs1181041827, MAF = 0.01%, χ2 = 45.71, 1 df, effectorg = 923.88 mm3, effect = 1.73 s.d. units, 95% CI = [1.21, 2.24], P = 5.2 × 10−11 for right ventral diencephalon; MAF = 0.01%, χ2 = 43.11, 1 df, effectorg = 945.24 mm3, effect = 1.77 s.d. units, 95% CI = [1.26, 2.29], P = 1.4 × 10−11 for left ventral diencephalon). For DTI parameters, two missense variants (rs2652098 and rs143368552) in Versican (VCAN) contributed the largest number of associations (63 in total, effect range = [−0.48, 0.42] s.d. units, P < 2.8 × 10−10). Figure 2E visualizes the associations between these two missense variants and 12 mean diffusivity traits. Common variants in VCAN have been extensively associated with white matter traits in previous GWAS2,5,8,45,46,47. Notably, Elliot et al.5 identified an intronic variant rs67827860 with strong associations with DTI traits. Thus, we conducted conditional analysis for all the 63 associations between these two variants in VCAN and DTI traits by conditioning on this nearby common variant as a covariate (Methods). We found that 85.7% of these associations still survived our stringent threshold at 2.8 × 10−10 while all the associations passed our suggestive threshold at 1 × 10−8, which supports the identified missense variants in VCAN as independent signals in addition to the previously identified common variant (Supplementary Data S5). VCAN plays a pivotal role in various neural processes, which may influence the pathophysiology of neurological disorders such as multiple sclerosis48,49,50. Another missense variant rs201680145 in NOTCH3 was also associated with multiple DTI parameters (effect range = [−1.50, 1.70] s.d. units, P < 2.5 × 10−10). Previous studies have revealed the role of NOTCH3 in white matter hyperintensities and several neurodegenerative diseases51,52,53.
We found associations between a missense variant rs189569984 in RBM20 and three CMR traits, including left ventricular end-systolic volume (LVESV), right ventricular end-systolic volume (RVESV) (MAF = 0.87%, χ2 = 64.60, 1 df, effectorg = −3.77 mL, effect = −0.20 s.d. units, 95% CI = [−0.25, −0.15], P = 9.1 × 10−16 for LVESV; MAF = 0.87%, χ2 = 44.88, 1 df, effectorg = −3.37 mL, effect = −0.16 s.d. units, 95% CI = [−0.21, −0.11], P = 2.1 × 10−11 for RVESV), and left ventricular ejection fraction (LVEF) (MAF = 0.87%, χ2 = 56.37, 1 df, effectorg = 1.49 %, effect = 0.24 s.d. units, 95% CI = [0.18, 0.30], P = 6.0 × 10−14). Mutations in RBM20 were previously known to be related to cardiovascular diseases including heart failure and dilated cardiomyopathy54,55,56, and were associated with LVEF in a previous GWAS on CMR traits9. Our rare variant analysis prioritized RBM20 and provided additional evidence for its impact on heart structure and function. Furthermore, we found that ascending aorta maximum/minimum areas were associated with a missense variant in ANO1 (rs201870990, MAF = 0.61%, χ2 = 42.35, 1 df, effectorg = 41.95 mm2, effect = 0.22 s.d. units, 95% CI = [0.15, 0.29], P = 7.6 × 10−11 for maximum; MAF = 0.61%, χ2 = 45.61, 1 df, effectorg = 42.05 mm2, effect = 0.23 s.d. units, 95% CI = [0.16, 0.29], P = 1.4 × 10−11 for minimum). ANO1 was among the loci identified by a recent GWAS57 on ascending aorta diameter (see Fig. 1 in their study57) but was not pointed out or discussed explicitly. Indeed, ANO1 was also reported to be effective in preventing cardiac fibrosis and may be a potential target for therapy58,59. However, future research might elucidate its role in aortic development and/or geometric remodeling. In summary, our analysis of rare variants directly prioritized and implied a small set of genes related to human organs. Due to the rarity of these variants and their lack of linkage disequilibrium (LD) with common variants, they could not be effectively studied in previous GWAS that focused on common variants.
Gene-based burden tests identified complementary signals
Gene-based burden tests enable us to study the collective effects of rare variants within a gene, thus boosting power. In burden tests, the variants of interest defined by annotation and MAF (Methods) are collapsed into a burden mask. Then, the burden score is calculated as the maximum genotype dosage across the single variant sites in the mask, and this score is tested for association (Methods). However, the involved burden models pose challenges to the adjustment for multiple testing since they have unknown correlated structure. Thus, we prioritized a P-value threshold at 1 × 10−9 based on two empirical null distributions60,61 (Methods). A total number of 17,474 genes are available for burden tests in the joint sample and discovery sample. Using the same joint sample as in variant-level tests, we identified 224 significant associations in gene-based tests (P < 1 × 10−9, Methods; Supplementary Data S6 for all the significant results). As different burden models or MAF cutoffs may implicate the same gene-trait associations (Methods), we further summarize the nonredundant results of 67 unique gene-trait pairs60 (involving 26 genes and 57 MRI traits) in Table 1 (a more comprehensive version is provided in Supplementary Data S7). For suggestive evidence, we additionally put all the associations that passed a more relaxed P-value threshold at 1 × 10−8 in Supplementary Data S8. Manhattan plot for all the gene-level associations were presented in Fig. 3A, across all MRI phenotype categories. Three gene-trait pairs overlapped across variant-level and gene-level tests (two associations between ANO1 and max/min aorta area, and an association between TNFRSF13B and spleen volume), indicating the robustness of these gene-trait pairs as both a single rare variant in this region and the joint effects of variants across the region are strong enough to drive an association. Consistent with observations in previous ExWAS17,61, we identified 21 additional genes, highlighting the power of gene-based burden tests in rare variant association studies. In particular, we found 3 genes and 18 gene-trait associations for brain functional MRI (fMRI) traits, which did not have any signals in variant-level tests. To evaluate the reportability of our findings, we wanted to follow a similar procedure in variant-level tests to perform an internal replication. However, many rare mutations, particularly those with a MAF of less than 0.01%, accounted for a large proportion of our discoveries but were not observed in the smaller independent dataset from phases 4 to 5 (average n = 8989). Therefore, we only made use of the internal discovery sample and examined whether the associations in the combined phases 1 to 5 sample had smaller P-values than those in the phases 1 to 3 data, similar to internal-replication analysis for variant-level tests (Methods and Supplementary Note). When we restricted the discovery sample to individuals in phases 1 to 3, there were 96 significant associations for 25 unique gene-trait pairs (P < 1 × 10−9, Supplementary Data S9). Within these 96 associations, 79.2% (76/96, 19 gene-trait pairs) had smaller P-values in the combined phases 1 to 5 sample, all of which had concordant directions of effects (Supplementary Data S9). The detailed summary statistics and results of internal replication analysis for gene-based associations have been provided in Supplementary Data S9. We discuss some of the 67 gene-trait pairs in the following two sections.
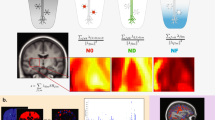
A Manhattan plot for gene-level burden tests across all 596 imaging phenotypes. For significant gene-trait pairs appearing in multiple burden models, we plotted the association with the smallest P-value. P-values are derived from two-sided tests based on z-scores (or equivalently a χ2 test with one degree of freedom. The dashed lines indicate the gene-level significant P-value threshold at 1 × 10−9 as well as the suggestive P-value threshold at 1 × 10−8. The multiple testing thresholds were derived from empirical null distribution with details provided in Method. The y-axis is capped at 1 × 10−30 and only gene-trait pairs with P < 1 × 10−3 were included. The color legend and x-axis are same as the Manhattan plot for variant-level associations in Fig. 2A. Significant genes were labeled. B Effect sizes for all non-redundant significant gene-trait pairs (n = 67, corresponding to Table 1) within each imaging category. The dashed line indicates an effect size of zero. We labelled all the genes that had the largest positive and negative effects within each imaging category and genes that had an absolute effect size greater than 1.5. C Distribution of total brain volume (in cm3) of mutation carriers (N = 7 individuals) versus non-carriers (N = 40,739 individuals) of PTEN among all the individuals (N = 40,746) included in the exome-wide association test for total brain volume. To illustrate, we select the burden model with the smallest P-value, that is “plof_int5”, which means the aggregation of pLoF variants and missense variants with a minor allele frequency (MAF) cutoff at 1 × 10−4. In the violin and boxplots shown, the violin plot illustrates the full distribution of total brain volume across carriers and non-carriers of PTEN burden variants using kernel density estimation, with the width of the violin representing data density at each value. Overlaid boxplots summarize key statistics: the center line denotes the median; the bounds of each box represent the first (Q1) and third (Q3) quartiles; and the whiskers extend to the most extreme data points within 1.5 times the interquartile range (IQR) from the box. The black diamonds indicate group medians. D The effect size estimates (circles, triangles, and diamonds at the centers of bars for distinct MAF cutoffs) and the corresponding 95% confidence intervals (bars) across different burden models of PTEN (N = 40,476 individuals). The color indicates distinct MAF cutoffs. The y-axis includes all the burden models (Methods). Only burden models with minor allele count >5 (after aggregation) were tested (Methods) and plotted.
Genes associated with abdominal, brain, and heart MRI traits in burden tests
As shown in Table 1, we identified 5 genes for abdominal MRI traits, 7 for CMR traits, 7 for regional brain volumes, 8 for brain DTI parameters, and 3 for brain fMRI traits. Figure 3B highlights the genes with large effect sizes. Below, we highlight several interesting findings for each trait category.
We found that 4 out of 5 signals of abdominal MRI traits from the combined phases 1 to 5 sample also present in the phases 1 to 3 data (Supplementary Data S4). The highest hit among all the gene-level signals was the association between spleen volume and TNFRSF13B when aggregating the effects of pLoF and Alpha damaging (Methods) missense variants (MAF = 1.4%, χ2 = 246.24, 1 df, effectorg = 0.027 L, effect = 0.40 s.d. units, 95% CI = [0.35, 0.45], P = 1.7 × 10−55) (Fig. 2D). In addition to TNFRSF13B, we observed that SH2B3 was also associated with spleen volume (MAF = 0.4%, χ2 = 63.40, 1 df, effectorg = 0.025 L, effect = 0.38 s.d. units, 95% CI = [0.29, 0.48], P = 1.7 × 10−15), where spleen abnormalities (e.g. splenomegaly) and associated blood traits have been previously reported62,63,64,65. For muscle measurements, we found an association between fat-free muscle volume of the posterior thigh and TANC1 (MAF = 0.01%, χ2 = 37.63, 1 df, effectorg = −1.06 L, effect = −1.32 s.d. units, 95% CI = [−1.74, −0.90], P = 8.5 × 10−10). TANC1 had genetic overlaps with the identified loci in previous GWAS on heel bone mineral density66 and its role in muscle development and rhabdomyosarcoma has been discussed in previous studies67,68.
As shown in Fig. 4A, TTN was associated with 8 CMR traits, including LVESV (MAF = 0.39%, χ2 = 109.60, 1 df, effectorg = 7.34 mL, effect = 0.40 s.d. units, 95% CI = [0.32, 0.47], P = 1.2 × 10−25), LVEF (MAF = 0.39%, χ2 = 140.48, 1 df, effectorg = −3.48 %, effect = −0.56 s.d. units, 95% CI = [−0.66, −0.47], P = 2.1 × 10−32), as well as global and regional peak circumferential strain measurements (effect range = [0.32, 0.60] s.d. units, P range = [1.2 × 10−21, 2.4 × 10−11]). Figure 4C visualizes the original effect sizes (in percentage) of TTN on different regional peak circumferential strain measurements and a global peak circumferential strain measurement. We observed that they generally produced similar and consistent results. These results make sense as TTN is a well-known gene associated with cardiac structure9 and cardiovascular diseases such as heart failure69, dilated cardiomyopathy70,71, atrial fibrillation72, supraventricular tachycardia, and mitral valve disease21. In addition to the previously known exome-wide association between TTN and LVESV21, our findings suggest a broader influence of this gene on cardiac structure and function, using a larger sample size and an expanded set of CMR traits.
A Graphical illustration of 8 CMR traits of left ventricle (LV) associated with TTN. LVESV, left ventricular end-systolic volume; LVEF, left ventricular ejection fraction; and Ecc, peak circumferential strain, including both global (“Global”) and regional traits (such as “AHA_9”). B Five genes (ANO1, COL21A1, GEM, PLCE1, and TAGLN) associated with ascending aorta and descending aorta areas. PLCE1 was also associated with brain fMRI traits. Three ICA-based functional activity traits (Net25_Node3, Net25_Node5, and Net25_Node9) associated with PLCE1 are illustrated, with their major brain regions and networks labeled. The color represents the weight profile of the ICA node. In addition, COL21A1 was associated with cerebrospinal fluid volume. C [Top]: original effect sizes (in percentage; circles) and the corresponding 95% confidence intervals (bars) of TTN on different regional peak circumferential strain measurements (N = 48,948 individuals for AHA_9; 48,948 individuals for AHA_13; 48,940 individuals for AHA_14; 48,943 individuals for AHA_15; 48,947 individuals for AHA_16) and a global peak circumferential strain measurement (N = 48,934 individuals). [Bottom left]: absolute values of original effect sizes (in mm2; circles) and the corresponding 95% confidence intervals (bars) of ANO1, COL21A1 and GEM on ascending aorta maximum and minimum area (N = 48,132 for ascending aorta maximum area and N = 48,135 for ascending aorta minimum area); we use ‘+’ to indicate the positive effects and ‘-’ to indicate the negative effects. [Bottom right]: absolute values of original effect sizes (in mm2; circles) and the corresponding 95% confidence intervals (bars) of PLCE1 and TAGLN on descending aorta maximum and minimum area (N = 48,143 for ascending aorta maximum area and N = 48,150 for ascending aorta minimum area). Association between TAGLN and descending aorta maximum area was among the suggestive results (Supplementary Data S8).
We would like to highlight our exome-wide associations with CMR traits of aorta. Notably, three genes (ANO1, COL21A1, GEM) were associated with both maximum and minimum area of ascending aorta, while PLCE1 was associated with maximum and minimum area of descending aorta (Fig. 4B, C). In particular, both COL21A1 and GEM lower the size of the ascending aorta (effectorg range = [−61.16, −54.95] mm2, P range = [1.7 × 10−13, 6.4 × 10−13] for COL21A1 and effectorg range = [−87.68, −87.06] mm2, P range = [1.3 × 10−12, 1.06 × 10−11] for GEM, respectively). Rare variants in COL21A1 and GEM were reported to be linked to higher pulse blood pressure in a previous rare variant association study of blood pressure73. A smaller aorta leads to a wider pulse pressure because it increases aortic characteristic impedance, which is highly dependent on aortic diameter. Our results on these aorta area-lowering genes further provide the evidence for the underlying mechanisms of previous observations. Moreover, GEM is involved in cardiac activities74 and is related to heart failure74,75. Another study76 pointed out that such effects of GEM could potentially be used as a gene therapy target in treating ventricular arrhythmias and heart failure. Our discovery indicated that GEM may also impact the structural integrity of the ascending aorta, thus improving our understanding of the role of GEM in cardiovascular health. In contrast, missense variants in ANO1 have positive effect. The role of ANO1 has been discussed in our variant-level analysis and we recaptured these associations again by aggregating the missense variants in ANO1 using burden test (effectorg range = [32.43, 32.79] mm2, P range = [1.7 × 10−13, 6.4 × 10−13]), suggesting the similar effects and aligned directions across a group of missense variants within ANO1. In addition, TAGLN was also associated with descending aorta minimum area (MAF = 0.05%, χ2 = 38.71, 1 df, effectorg = 62.28 mm2, effect = 0.67 s.d. units, 95% CI = [0.46, 0.88], P = 4.91 × 10−11) while its association with descending aorta maximum area was observed at a more liberal P value threshold at 1 × 10−8 (Supplementary Data S8). At the bottom of Fig. 4C, we found that the effects (in mm2) of different genes on the aorta area could largely vary in terms of both directions and the sizes. GOLM1 was found to be associated with descending aorta distensibility (MAF = 0.01%, χ2 = 44.23, 1 df, effectorg = 2.85 mmHg, effect = 2.5 s.d. units, 95% CI = [1.77, 3.24], P = 2.93 × 10−11). A recent study demonstrated that GOLM1 may present a potential therapeutic target for treat sepsis-induced cardiac dysfunction in animal models77. GOLM1 was also related to Alzheimer’s disease and the implied cognitive deficits78. However, its role in aortic wall remodeling requires further study. Using GTEx data resources79, we found that four (ANO1, COL21A1, GEM, TAGLN) out of five genes (ANO1, COL21A1, GEM, TAGLN, PLCE1) associated with ascending/descending aorta areas had moderate to high expression and overexpressed in aorta while PLCE1 had high expression in artery tissues in general, which further supports their roles in affecting the structure of aorta.
For brain MRI traits, we highlight in Fig. 3C, D the strongest association between PTEN and total brain volume with a large effect size when aggregating the pLoF and missense variants (MAF = 0.01%, effectorg = 386,703.40 mm3, effect = 2.77 s.d. units, 95% CI = [2.22, 3.32], P = 5.8 × 10−23). The distribution of total brain volume in cm3 for PTEN mutation carriers versus non-carriers is provided in Fig. 3C. Notably, as illustrated in Fig. 3D, different burden masks would yield associations with various strengths, and we observed that the signal became stronger if we aggregate ultrarare (MAF < 1 × 10−4) pLoF variants and damaging missense variants in PTEN, which may not be observed and testable in the previous ExWAS16,17 with a sample size smaller than ours due to the low MAF. In a recent ExWAS80, PTEN was associated with autism spectrum disorder. Indeed, PTEN is a well-known gene that has been consistently linked to autism spectrum disorder81,82,83,84, brain development85,86,87, and brain clinical phenotypes including brain overgrowth88,89,90, with an enlarged brain volume representing an associated phenotype in these conditions. To our knowledge, no previous GWAS or ExWAS has linked PTEN to any brain MRI measurements, thus our analysis provided direct evidence connecting rare variants in PTEN with total brain volume. Another gene OMA1 was observed to be associated with 7 regional brain volume traits including superior frontal gyrus, lateral orbitofrontal cortex, and precentral gyrus for both sides of the brain and right superior parietal lobule (effectorg range = [−524.83, −171.02] mm3 units, P range = [1.77 × 10−14, 6.35 × 10−10]). Notably, PTEN and OMA1 regulate the PTEN-induced kinase 1 (PINK1) and thus may be useful in preventing epileptogenesis91 while a protective role of OMA1 in neurodegeneration92,93,94 was previously reported. The remarkably strong association PTEN demonstrate that even within a general healthy cohort (as opposed to disease-focused or case-control studies), we can identify biological links between genes associated with neurodevelopmental disorders using MRI data.
For DTI parameters, in contrast to variant-level analysis where missense variants in VCAN accounted for the most associations, our gene-based burden tests revealed a more diverse range of effects on white matter, with 8 distinct genes accounting for 10 gene-trait associations. For brain fMRI traits, PLCE1, GLUL, and KCNA5 were identified to be associated with 18 phenotypes, where PLCE1 contributed to 16 of them. We found that many gene-trait pairs would not have been identified if singleton burden models or the recent deep learning-based AlphaMissense26 burden models were not included. We will provide more detailed discussions about these interesting associations in the following sections.
Overall, gene-based burden tests examined the aggregated effects of rare variations within genes and advanced our understanding of the underlying genetic dispositions across the abdomen, heart, and brain organs. Our findings prioritized a set of rare genes previously unknown to be associated with human organs. We further discussed examples of genetic overlaps between these MRI signals and other health-related phenotypes, which may help to understand complex diseases by providing exome-wide insights into the genetic mechanisms underlying multiple human organs including abdominal, cardiac, and neurological pathologies.
Pleiotropic effects of PLCE1 and COL21A1 on brain and heart MRI traits
In our gene-based burden tests, PLCE1 was associated with the largest number of phenotypes including 16 brain fMRI traits (including 5 traits from parcellation-based approach95 and 11 traits from whole brain spatial independent component analysis [ICA]96,97,98) and 2 CMR traits (descending aorta maximum/minimum areas) (Fig. 4B). In addition, COL21A1 was associated with both brain cerebrospinal fluid (CSF) volume and ascending aorta maximum/minimum areas (Fig. 4B). We found that a recent GWAS99 discovered the associations of descending and ascending aorta areas separately with PLCE1 and COL21A1 while PLCE1 was also found in previous GWAS3,5,100 for brain fMRI. PLCE1 encodes a phospholipase (PLCε) that catalyzes the hydrolysis of phosphatidylinositol-4,5-bisphosphate to generate inositol 1,4,5-triphosphate and diacylglycerol. They are common second messengers regulating multiple cellular processes, including cell activation, growth, differentiation, and gene expression101. Interestingly, prior mechanistic study identified a causal role for PLCε on the development of thoracic dilation102 and dissection103. Similarly, COL21A1 encodes the alpha-1 chain of collagen 21, a known extracellular matrix component in the arterial wall, secreted by vascular smooth muscle cells. While the previous GWAS99 suggested distinct genetic bases between the ascending and descending aorta and linked aortic traits to brain small vessel disease, it did not establish direct associations of these genes with other brain MRI traits. Our results provide exome-wide evidence highlighting the associations of PLCE1 and COL21A1 with both brain and heart MRI traits. Additionally, a previous study suggested that the observed GWAS effects of PLCE1 on fMRI traits may be blood-derived100. Our exome-wide evidence supporting PLCE1’s role in both aortic and fMRI traits aligns with this hypothesis. Therefore, it might be helpful to integrate cardiac features and cardiac-related genetic information when analyzing fMRI data, given that fMRI phenotypes are blood oxygen level-dependent and are influenced by blood flow.
The connection between the heart and brain has increasingly garnered attention10,104. Moreover, the role of hemodynamic role of the aorta involved the cushioning of pressure and flow pulpability caused by the intermittent ventricular ejection, and an impaired cushioning function for changes in aortic stiffness and/or diameter has been identified as a key mechanism for target organ damage, including the brain and the heart105. The associations we identified, involving rare coding mutations in the same genes (PLCE1 and COL21A1) linked to heart, aortic and brain structure and function, suggest that similar biological mechanisms may underlie the shared pathways of these organs that influence both cardiac and neurological health.
Associations uniquely identified by singletons and AlphaMissense
Singleton variants are those observed only once in the study cohort. AlphaMissense26 is a recent deep learning-based method that offers a novel approach to annotating damaging missense variants. The Singleton burden model captured the rarest category in our association tests and may be of special interest16,106, and the incorporation of AlphaMissense offered additional genetic findings in our study. Therefore, we would like to highlight the contributions of these two groups of burden models in this section. In our gene-based burden tests, 10 gene-trait pairs would not be detected without the singleton burden models or including AlphaMissense as part of our annotation resources. Among these signals, 8 of them did not even have any counterparts that passed the relaxed P < 1 × 10−8 threshold (Supplementary Data S10).
Singleton damaging missense variants in glutamate-ammonia ligase GLUL, which encodes the glutamine synthetase protein, were associated with functional connectivity of the somatomotor and secondary visual networks (MAF = 0.01%, χ2 = 37.54, 1 df, effect = 2.48 s.d. units, 95% CI = [1.68, 3.27], P = 8.9 × 10−10). GLUL knockout mice were reported to be neuronally affected in multiple regions including somatosensory and visual cortices107. Our singleton analysis provides consistent evidence in human genetics. In addition, a recent study suggested that visual-somatosensory integration may be a biomarker for preclinical Alzheimer’s disease108. Together, rare mutations in GLUL may play a potential role in human neurodegenerative diseases109,110,111,112. Another gene, KCNA5 related to potassium voltage-gated channel, was associated with functional activity in the subcortical-cerebellum network when we combined the rare pLoF variants and Alpha damaging missense variants (MAF = 0.1%, χ2 = 44.21, 1 df, effect = 0.59 s.d. units, 95% CI = [0.41, 0.76], P = 2.9 × 10−11). Notably, voltage-gated potassium channels are essential for neurons and cardiac activities and previous GWAS have discovered the common variants mapped to KCNA5 associated with cortical surface7 and thickness113. In addition, a previous study on the rat cerebellum model revealed strong KCNA5 immunoreactivities in the cerebellar nuclei114. Consistent with these findings, we provided additional evidence for KCNA5’s role in brain function. Moreover, some studies also found close relationships between KCNA5 and heart diseases including atrial fibrillation115 and pulmonary arterial hypertension116. Indeed, KCNA5 is an approved drug target for the treatment of cardiac arrhythmias. Inspired by this additional context, we investigated whether KCNA5 was associated with any CMR traits. Interestingly, we found that when aggregating the singleton pLoF and damaging missense variants, KCNA5 was associated with a regional peak circumferential strain measurement (MAF = 0.02%, χ2 = 21.69, 1 df, effectorg = 5.11%, effect = 1.08 s.d. units, 95% CI = [0.63, 1.54], P = 3.2 × 10−6; “Ecc_AHA_4”, Supplementary Data S1) at a P-value threshold of 1.74 × 10−5 (0.05/2870, Bonferroni adjusted for 82 CMR phenotypes across all the 7 variant function classes and 5 MAF classes). These discussions further suggested that multi-organ imaging genetic studies would bring about insights to the complex interplay across the brain-heart system. In summary, our results underscore the efficacy of our gene-based burden test approach and highlight the importance of using multiple MAF cutoffs and innovative annotation tools.
Comparison with previous ExWAS for brain MRI and CMR traits
In this section, we discuss the links between our gene-based rare variant signals and existing ExWAS results on MRI traits. To our knowledge, there were no rare variant associations reported for brain fMRI or abdominal MRI traits. The studies most similar to ours are those by Backman et al.16, which included regional brain volumes and DTI parameters among a broad range of phenotypes, and Jurgen et al.21, which focused on CMR traits, while Pirruccello et al.117 declared no findings of rare variant associations for aorta traits. Both studies used data from the UKB cohort but with smaller sample sizes compared to our current study. Importantly, we analyzed different brain MRI and DTI parameters, extracted from raw images using our own pipeline, compared to those investigated by Backman et al.16. Additionally, we incorporated a broader set of CMR traits, including strain and thickness metrics, expanding upon the traits analyzed by Jurgen et al.21.
For regional brain volumes and DTI parameters (corresponding to STR and dMRI traits in Backman et al.16), 5 genes (AMPD3, HTRA1, MYCBP2, RBL1, SCUBE2) whose associations passed our stringent P-value threshold (P < 1 × 10−9) were also significantly associated with their brain MRI traits. For example, associations between AMPD3 and mean MD of splenium of corpus callosum passed the stringent P value threshold both in our study and in Backman et al.16. SCUBE2 was significantly associated with the regional volume of the left cerebellum exterior in our analysis, while its association with the volume of the cerebellum cortex in the left hemisphere was reported in their result as suggestive evidence. MYCBP2 was associated with the external capsule in both our analysis and Backman et al.16. Additionally, several associations between PLEKHG3 and multiple dMRI traits were identified in Backman et al.16 and two gene-trait pairs within these associations passed our relaxed threshold (P < 1 × 10−8). For CMR traits, we replicated the only exome-wide significant association between TTN and LVEF in Jurgen et al.21, while their suggestive association between TTN and LVESV also passed our stringent threshold. In addition, we found 7 associations between TTN and 7 peak circumferential strain metrics. We summarized the overlapping associations in these previous ExWAS studies in Supplementary Data S11.
Concordant evidence with GWAS signals
Associations with rare coding variants could prioritize genes among the numerous loci identified in GWAS for polygenic complex traits by providing concordant evidence16,21. In this section, we leveraged the previous GWAS summary statistics1,2,3,10,118 of all the same 596 imaging traits to compare the identified signals between common variants and rare coding variants (Methods). As expected, we observed convergent evidence for all categories of MRI traits. Briefly, more than half of the 174 rare coding signals (53.4% = 93/174, that is, 64/107 variant-trait associations and 29/67 gene-trait pairs) were within the 1 Mb range of the independent GWAS signals, which is consistent with the observation in a previous large-scale UKB phenotype screening16. Based on our results, we found that variant-level associations with regional brain volumes and CMR traits were not within the neighborhood of GWAS signals, while all categories of MRI traits had at least one rare coding association within the 1 Mb of GWAS signal for signals from gene-level tests.
We zoomed into these shared signals between GWAS and ExWAS to provide more detailed insights. The association between SH2B3 and spleen volume identified in our gene-level burden test was further supported by an independent GWAS signal rs2239194, which is an expression quantitative trait locus (eQTL) in spleen tissue79. For DTI parameters, two missense variants (rs2652098 and rs143368552) in VCAN contributed the most to the variant-level associations and all these associations were within 1 Mb of the GWAS signals. However, similar to findings from a previous GWAS study47, we did not identify any brain tissue-related eQTLs. Associations between AMPD3 and DTI parameters of splenium of corpus callosum were also accompanied by signals from GWAS. These common variants are eQTLs of multiple brain tissues including cerebellum and spinal cord where brain white matter also presents. For fMRI traits, all the associations between PLCE1 and 18 MRI traits (16 brain fMRI traits and 2 CMR traits) were consistent with previous GWAS results, though there were not eQTLs in brain cortex tissues. On the other hand, we found that the only signal for brain volumes that fell into the GWAS loci was the association between COL21A1 and CSF volume. The corresponding common variants rs3857615 and rs9475654 were eQTLs in cerebellum and cerebellum hemisphere.
In addition, we identified gene-trait associations that were not reported in our previous GWAS of the same MRI traits but appeared in other studies (e.g., RBM20 and LVEF in Pirruccello et al.9). We also observed that some rare coding associations in our study were linked to GWAS loci of related traits within the same phenotype cluster. For example, though both ExWAS and GWAS revealed the associations between TTN and multiple regional peak circumferential strain measurements, the phenotype for the signals did not exactly match. In summary, rare variant associations with MRI traits not only identified novel signals but also prioritized genes among thousands of GWAS loci. These associations provide new insights and context for understanding the genetic basis of structure and function in the human brain and body. We provided these exome-wide signals that showed convergent evidence with previous GWAS signals in Supplementary Data S12 and S13.
Multi-organ imaging genetics and drug targets
We used the Therapeutic Target Database (TTD)119 to query potential drug target genes (Methods). Among the 26 unique genes identified by gene-based burden tests, 6 genes were approved drug targets (KCNA5 and ANO1) or in clinical trials (TTK, GEM, LPAR3, and TNFRSF13B). The identified genes in gene-based burden tests were significantly enriched for potential drug targets (6 out of 26, compared with 1735 out of 17,448 genes, OR = 2.7, P = 0.0391, Supplementary Data S14). Moreover, when including a broader set of potential drug genes that were documented in TTD to be reported in the literature, the enrichment became stronger (13 out of 26, compared with 3468 out of 17,448 genes, OR = 4.0, P = 5.737 × 10−4, Supplementary Data S14). Consistent with the procedure in a previous study61 and observations in other ExWAS16,61,120, our results suggest that rare coding variants identified in imaging genetics may facilitate the discovery of promising drug target genes121,122. Given the complicated nature of human organ interplay and the corresponding biological pathways involved, we would emphasize that such enrichment should be interpreted carefully as they might not directly point to the corresponding diseases and related traits for existing drugs. However, these associations with human organs could provide novel drug targets if they are supported by further evidence, and in turn these MRI traits themselves might be useful to validate the new drugs if the underlying mechanisms are well established123. Moreover, As endophenotypes for complex diseases, MRI traits can offer additional information on known drug targets, such as potential off-targets and/or side effects121,124, and also aid in drug repurposing (e.g., ANO1 and cardiac disease as discussed in the previous section58,59).
Burden heritability and genetic correlation
To investigate the rare coding genetic architecture for multi-organ MRI traits, we applied burden heritability regression (BHR)27 to estimate the heritability for pLoF and damaging missense variants (i.e., “int1 missense variants”, Methods), while the heritability for synonymous variants is also calculated, and it serves as negative control. Designed for rare variants with MAF < 1 × 10−3, BHR stratifies variants based on functional annotation and MAF to allow for different variant classes to have different mean effect sizes. Given our relatively small sample size (average n = 40,038), we restricted our analysis to the ultra-rare MAF bin (MAF < 1 × 10−4) which contained most of the coding variants in our analysis (Supplementary Data S15). As expected, ultra-rare pLoF variants consistently demonstrated higher heritability than damaging missense variants across all MRI categories (Fig. 5A). Supplementary Data S16 and S17 show the complete list of burden heritability for ultra-rare pLoF variants and damaging missense variants. In contrast, there is no evidence that synonymous variants have significant heritability, showing that the estimates from our results for rare variants are well calibrated (Supplementary Data S18). Furthermore, the highest average heritability of ultra-rare pLoF variants were observed in the DTI parameters category. For ultra-rare pLoF variants, the trait had the highest heritability was the caudal anterior cingulate (h² = 0.012, SE = 0.0035) among regional brain volumes, the mean axial diffusivity of the body of the corpus callosum (h² = 0.020, SE = 0.0048) among DTI parameters, functional connectivity between the second visual and auditory networks (h² = 0.020, SE = 0.0047) among brain fMRI traits, regional peak circumferential strain (h² = 0.016, SE = 0.0053) among CMR traits, and pancreas iron (h² = 0.019, SE = 0.0053) among abdominal MRI traits. We note that these burden h² estimates are relatively large, which is similar to the high common variant heritability for many imaging traits. This in general, supports an endophenotype model where genes have more direct connections with imaging phenotypes than downstream health outcomes.
A The distribution of burden heritability point estimates for ultra-rare (minor allele frequency <1 × 10−4) pLoF variants and damaging missense variants (i.e., the int1 damaging missense variants) across 591 multi-organ imaging traits (including 36 abdominal MRI traits, 82 CMR traits, 101 Regional Brain Volumes, 110 DTI traits, 82 resting-state ICA-based fMRI traits, 90 parcellation-based fMRI traits, and 90 parcellation-based fMRI traits) that had a sample size larger than 10,000 (5 abdominal imaging traits were excluded due to insufficient sample size). Burden heritability was estimated based on the summary statistics of each variant-level tests and all the individuals (N = 54,365) were covered. In the boxplots shown, each box represents the distribution of burden heritability estimates across a certain imaging phenotype category for pLoF or damaging missense variant groups. The central line within each box denotes the median value; the upper and lower bounds of the box correspond to the third (Q3) and first (Q1) quartiles, respectively. Whiskers extend to the most extreme data points within 1.5 times the interquartile range (IQR) from the box, and points beyond the whiskers indicate outliers. The dashed horizontal line marks the null expectation. B Burden genetic correlations of ultra-rare (minor allele frequency <1 × 10−4) pLoF variants for selected imaging phenotypes and other complex traits. P-values are derived from two-sided tests based on z-scores. Only top hits are presented here. Specifically, we first selected seven trait pairs from pairs that had ten smallest P-values among all the trait pairs and additionally included three pairs that had smallest or second smallest P-values for regional brain volumes and abdominal MRI traits, which resulted in nine imaging traits across all MRI categories and seven Genebass traits. Filled circles indicate the selected top hits (P < 0.0032) while filled triangles indicate other pairs that passed a nominal significance threshold at P < 0.05. “Phenotype_ID” was used to define the selected imaging traits. Specifically, abdominal fat ratio from abdominal MRI traits, left atrium maximum volume (LAV_max), regional radial strain (Err_AHA_13) and peak circumferential strain (Err_AHA_4) traits from CMR, left and right insular volumes, functional activity trait from resting fMRI (ICA-based, Net100_Node55) traits, Visual2-Auditory network functional activity trait from task fMRI (parcellation-based), and Somatomotor-Dorsal-Attention network functional activity trait from resting fMRI (parcellation-based) traits were shown here (bottom-to-top order). Supplementary Data S1 includes more details of these phenotypes.
We further explored the burden genetic correlation pattern for ultra-rare pLoF variants between MRI traits (MAF < 1 × 10−4) and 186 complex traits and diseases (MAF < 1 × 10−5) downloaded from Genebass17 (Supplementary Data S19). We focused on 67 heritable MRI traits that passed the Bonferroni adjusted threshold at P < 8.4 × 10−5 (=0.05/596). Consequently, we estimated 12,462 (67 × 186) pairs of burden genetic correlations. We found 3 pairs that passed the MRI-multiple testing threshold at P < 7.5 × 10−4 (0.05/67) and 591 pairs passed a nominal P-value threshold at P < 0.05. Although no pairs survived the most stringent P-value threshold adjusted for both the imaging traits and Genebass traits at P < 4.0 × 10−6 (0.05/12,462), we observed that there were interesting pairs in the top-ranking subset for continuous traits from Genebass (Supplementary Data S20). The results for binary traits can be found in Supplementary Data S21. Figure 5B shows the selected top hits of burden genetic correlation pairs, including insular volume and reaction time125, regional radial strain and systolic blood pressure, left atrium maximum volume and triglycerides to high-density lipoprotein cholesterol ratio, and abdominal fat ratio and birth weight. These burden genetic correlations between MRI traits and health-related traits may reveal a shared genetic basis in ultra-rare pLoF variants. While the current MRI sample size was slightly underpowered, future larger samples may enable a more robust quantification and confirmation of these genetic links.
Discussion
We conducted a large-scale ExWAS for 596 multi-organ imaging traits including brain, heart, liver, kidney, and lung. By using both WES and MRI data from over 50,000 participants from the UKB study, we uncovered how rare coding variants contribute to human organ structure and function. The consequences of these rare coding variants to human organs were largely understudied. For example, in abdominal organs, a prior ExWAS focused solely on hepatic fat measurement from CT imaging20, while another GWAS included a broader set of traits13. However, with a relatively limited sample size of over 10,000, it failed to identify rare variant associations. For CMR traits, our study incorporated refined measurements of the left ventricle, right ventricle, ascending/descending aorta, and left/right atrium. These were not examined in previous ExWAS21 but resulted in additional discoveries in our analysis. Additionally, our results helped prioritize genes among previously identified GWAS loci9,10. For brain MRI traits, we analyzed a different set of regional brain volumes and DTI parameters compared to previous ExWAS16,17. Additionally, we discovered associations between rare variants and fMRI traits, which had not been previously reported.
Analyzes at the variant level and gene-based burden level may yield complementary insights into gene associations. For example, gene-based burden testing is more powerful for identifying signals driven by pLoF variants as these variants are rare individually but have consistent effect directions17, while the pattern for missense variants signals in variant-level tests and gene-level tests is less obvious16,17,61. In our study, we found that pLoF signals were exclusively detected by gene-based tests, whereas three gene–trait pairs were observed across both variant- and gene-level analyzes. This pattern aligns with previous findings and underscores the importance of incorporating both approaches. Moreover, although gene-based burden tests can enhance signal detection16,17, annotating the deleterious variants is still an active research area. In group-based tests, variants are collapsed into functional-frequency groups, but different approaches may lead to inconsistent and/or incomplete results126. Therefore, it is helpful to leverage the annotation information from diverse resources to better empower the set-based association test. We incorporated AlphaMissense26 as part of our annotations, resulting in the discovery of an additional set of genes (GOLM1, KCNA5, LPAR3, PIGX, TAGLN, and WIPF3). AlphaMissense is a novel deep learning model for pathogenic missense variant prediction and achieved good performance in predicting the unknown clinical significance of many missense variants.
There is also ongoing research regarding the best practice of performing rare variant association tests. For example, Auer et al.127 pointed out that rare variant associations for quantitative traits might be susceptible to outliers and non-normality and ignoring such issues could result in false discoveries. Although outliers were removed in our pre-processing steps for phenotype data (Methods), there is still potential concern about non-normality. We verified all the identified associations by applying rank-based inverse normal transformation (RINT) (Methods). The results largely supported our original findings (Supplementary Data S22 and S23 and Supplementary Note). Briefly, all associations except one (LPAR3 with the mean MD of uncinate fasciculus) passed the nominal threshold adjusted for the number of signals undergoing RINT analysis (4.7 × 10−4 = 0.05/107 for variant-level tests and 2.2 × 10−4 = 0.05/224 for gene-based burden tests, respectively). The majority of these associations also remained significant under our primary stringent threshold. In addition, Greer et al.128 suggested retaining all the variants before conducting association tests and examining the Hardy–Weinberg equilibrium afterward to avoid the over-removal of variants included in the study. We examined the variants filtered out by Hardy–Weinberg equilibrium step in our quality control step and no new discoveries were identified, suggesting that this step had minimal impact on our study (Supplementary Note).
Over half of the identified rare variant signals were within 1 Mb of GWAS common variants. These rare variant associations could prioritize the causal genes among many identified GWAS loci and uncover novel signals. Human organ MRI traits are widely used as endophenotypes for diseases and health-related traits; our rare variant results deepen our understanding from two aspects. First, we showed that many of the identified gene-trait pairs were consistent with existing literature on the associated genes and their links to organ-related diseases or complex traits. This alignment includes findings from previous GWAS and ExWAS, observed clinical outcomes related to these genes, and animal models that elucidate the biological mechanisms connecting the gene to the phenotype. Second, we observed a significant enrichment of potential drug targets among our rare variant signals. This suggests that rare variant imaging genetics can play a role in identifying and repurposing drug targets, as well as understanding potential side effects during drug development. These findings have practical implications for both real-world clinical applications and future scientific research.
Along with these significant new insights, the present study has a few limitations. A primary limitation is the relatively modest sample size for identifying rare variant associations with MRI traits. For example, when the sample size increased from an average of n = 30,739 in the phases 1 to 3 analysis to an average of n = 40,038 in the phases 1 to 5 analysis, we observed a substantial increase in identified signals (84 vs. 174, that is 59 variant-level associations and 25 unique gene-trait pairs in the phases 1 to 3 analysis, and 107 variant-level associations and 67 unique gene-trait pairs in the phases 1 to 5 analysis). It is reasonable to hypothesize that a large number of rare variant associations have not been discovered for many MRI traits. Compared with recent ExWAS studies for other phenotypes encompassing nearly or more than 500,000 participants16,17,21,61,129,130, there remains substantial room for improvement in imaging studies. This is especially important for MRI traits that have a highly polygenic architecture, such as regional brain volumes, and is also critical for simultaneously comparing multiple traits. As the UKB imaging project completes data collection from 100,000 subjects131, we anticipate detecting additional signals and obtaining more robust results. This will not only help identify associated rare coding variants and genes but also elucidate the genetic architecture, such as burden heritability and burden genetic correlation, facilitating downstream analyzes for rare coding variants.
Another limitation is the lack of diversity in imaging ExWAS data resources. First, no independent non-UKB database currently exists that combines both MRI traits and WES data in a sample size comparable to the UKB study. As discussed above, rare variants typically require a large sample size for detection and replication. Although our study leveraged the largest available dataset and had enhanced statistical power, we were unable to perform independent non-UKB replications because many rare variants may not be observed in smaller samples. Consequently, this led us to use joint analysis with a stringent P-value cutoff, and an internal validation procedure to assess the robustness of our findings. Nonetheless, future independent studies with more diverse data resources are essential to replicate the signals we identified. Second, our analysis primarily focused on individuals of European ancestry. It is critical to extend these studies to underrepresented ancestry groups in genetic research as data become available. Different ethnicities may exhibit heterogeneous genetic architectures, and including a diverse range of ancestries could provide a more comprehensive understanding of genetic influences on organ structure and function. When future data resources become available, it will be valuable to replicate our findings and extend the scope of WES studies across multiple cohorts. Meta-analysis may be a robust and efficient approach for identifying and replicating associations using only summary statistics.
In conclusion, we have identified associations between rare coding variants and imaging traits across human organs. These findings enhance our understanding of the genomic mechanisms underlying human organ structure and function from the perspective of protein-coding variants, potentially contributing to the identification and prioritization of novel targets for pre-clinical and clinical drug development. Compared with GWAS of similar sample size (at the scale of 50,000) to our study, the identified associations are much fewer partially due to the limited statistical power induced by the low frequency of the protein-coding variants17,18,61. However, rare coding variants offer more direct and stronger effects with interpretable biological implications, which would contribute to complementary genetic information with common variants, prioritize the effector genes, and elucidate the underlying mechanisms between gene-trait pairs. Looking ahead, it is important to increase sample sizes, integrate data from diverse populations, and expand ExWAS to include a broader spectrum of imaging phenotypes. We anticipate that future collaborative efforts will further elucidate the genetic landscape of human organs, advancing our knowledge of human biology and health.
Methods
Ethics and data resources
This study made use of the data from UK Biobank (UKB) involving ~500,000 participants aged from 40 to 69 when recruited between 2006 and 2010 (https://www.ukbiobank.ac.uk/). The UKB study received the ethics approval from the North West Multi-centre Research Ethics Committee (reference number: 11/NW/0382) with informed consent obtained by all the participants. The present study was under UKB application number 76139.
Whole-exome sequencing (WES) data were obtained from the UK Biobank and the detailed sequencing protocols can be found elsewhere18. By following a unified OQFE protocol121, raw sequencing reads were aligned to the GRCh38 reference genome, and small variants were called on a per-sample basis using DeepVariant. Per-sample gVCFs were then joint-genotyped using GLnexus to produce multi-sample project-level VCFs (pVCF) suitable for association analysis. We made use of the OQFE WES files in BGEN format provided by UK Biobank after these centrally pre-processing steps.
Multi-organ imaging phenotypes
The information of imaging phenotypes can be found in previous studies1,2,3,4,10. Briefly, we studied 596 imaging traits encompassing brain MRI traits1,2,3,4, heart CMR traits10, and abdominal MRI traits. Three major modalities of brain MRI were included. First, we included 101 regional brain volume traits1 derived from structural MRI. Second, we used 110 tract-averaged parameters2 from DTI capturing the microstructure of brain white matter. Third, for fMRI traits, we involved 76 node amplitude traits and six global functional connectivity traits based on ICA3 as part of resting fMRI traits; we also included 180 (90 + 90) parcellation-based4,95 resting fMRI traits and task fMRI traits. Regarding CMR traits, we made use of 82 traits10 including regional and local measurements from cardiac chambers and the aorta. For abdominal MRI traits, we contained 41 imaging traits based on MRI data of the liver, kidneys, lungs, pancreas, spleen, and body muscle/fat composition. For each phenotype, values greater than five times the median absolute deviation from the median value were removed as in our previous work1,2,3,4,10. A full list of these 596 traits can be found in Supplementary Data S1.
Internal replication and joint analysis design
Given the limited sample size (average n = 40,038 across imaging traits with non-missing data) and lack of independent data sources, we prioritized our signals through joint analysis30 and adopted an internal validation procedure to evaluate the robustness of our discoveries. Specifically, we first restricted our sample to include only participants from phases 1 to 3 of MRI release (average n = 30,739) and then excluded any individuals related to phases 1 to 3 participants in the rest of the sample from phases 4 to 5 of MRI release (average n = 8,989, relatives of the phases 1 to 3 were removed). The joint sample included all the individuals from phases 1 to 5 and was used to report significant results. To investigate the robustness of variant-level associations, we tested the associations on phases 1 to 3 individuals and phases 4 to 5 individuals separately. Then, focusing on the significant level and effect sign, we investigated (i) whether signals identified from the phases 1 to 3 sample could be replicated in the independent phases 4 to 5 sample and (ii) whether the signals identified from phases 1 to 3 sample had stronger evidence (i.e., smaller P values and concordant effect directions) in the joint sample. For gene-based burden tests, we noted that many rare mutations, especially those variants with MAF < 1 × 10−4, accounted for a large proportion of our discoveries but were not observed in the smaller independent dataset from phases 4 to 5 (average n = 8989; even within a burden model with MAF cutoff at 0.01, the lack of those rare variants in the sample from phase 4 to phase 5 would make the burden associations incomparable). Thus, we only investigated whether the identified associations from phases 1 to 3 dataset had stronger evidence in the joint sample. Consequently, based on this internal replication procedure, we were able to expect the level to which our discoveries from the joint analysis could be replicated.
Quality control for UK Biobank exome data
We used Plink v.2.0132 to conduct the quality control steps, restricting the sample to individuals with imaging data (n = 54,365). For exome sequencing data, we included all the variants that had a MAF below 0.01. Variant-level quality control excluded all the variants that had missing rate larger than 10% or had Hardy–Weinberg equilibrium P < 1 × 10−15. Sample-level quality control excluded any sample with missing rate over 10%. Consequently, no individual was excluded from the quality control process, while 8,127,841 variants remained for the downstream analyzes before annotation.
Functional annotation for protein-coding variants
We used Variant Effect Predictor133 (VEP v.108) for variant annotation. Each variant was mapped to the most severe consequence across the canonical transcripts. We defined the loss-of-function variants using the Loss-of-Function Transcript Effect Estimator134 (LOFTEE) plugin, which further collapsed stop-gained, essential splice, and frameshift variants into high-confidence predicted loss-of-function variants (hcpLoF or pLoF hereafter) or low-confidence predicted loss-of-function variants (lcpLoF). Missense variants were prioritized by dbNSFP135,136 (v.4.5a) plugin using five prediction algorithms16: SIFT22, PolyPhen2 HDIV23, PolyPhen2 HVAR23, LRT24, and MutationTaster25. Missense variants were defined as “int5 damaging missense variants” if predicted damaging or possible damaging by the intersection of all the five algorithms and “int1 damaging missense variants” if predicted damaging or possible damaging by any one of the five algorithms. Parallelly, we also used AlphaMissense26 to assign the missense variants to be “Alpha damaging” if predicted as “pathogenic”. Synonymous variants served as empirical null control to support the study-wise P-value threshold for burden tests and the method would be detailed below in the section for determining study-wise significance level. Predicted loss-of-function variants (including both hcpLoF and lcpLoF) and missense variants were included in our downstream association tests. They were referred to as non-synonymous variants or coding variants of interest (n = 2,143,707).
Exome-wide association tests
The covariate adjustment for brain MRI, DTI parameters, brain fMRI, and heart CMR traits was consistent with our previous GWAS1,2,3,10,118. In short, we adjusted a set of basic covariates for all the imaging traits including age (at imaging), age-squared, sex, age-sex interaction, age-squared-sex interaction, imaging site, and the top 40 genetic PCs (for the phases 4 to 5 sample replication analysis, we only adjusted for top 10 genetic PCs). For brain structural MRI traits, we additionally adjusted for total brain volume (for traits other than itself). For brain fMRI traits, we additionally adjusted for the effects of volumetric scaling, head motion, head motion-squared, brain position, and brain position-squared. For heart CMR traits, we additionally adjusted for the effects of standing height and weight. For abdominal MRI traits137, we additionally adjusted for the effects of standing height and body mass index.
We used REGENIE (v. 3.1.3)138 to conduct the exome-wide association tests. Common variants from genotyping array data (MAF > 0.01, genotype missing rate <10%, Hardy–Weinberg equilibrium P > 1 × 10−15, LD pruning with r2 < 0.9) were included in the step 1 of REGENIE to capture genome-wide polygenic effects. Then, the predictors obtained from step 1 were used in step 2 for both variant-level tests and gene-based burden tests. For all the variant-level tests and gene-based burden tests, we followed the default setting of REGENIE that only variants/burden masks with MAC > 5 were tested. For variant-level tests, we tested all the coding variants of interest as defined above. For gene-based burden tests, we included three general types of burden masks: (i) hcpLoF variants only, (ii) damaging missense variants only, and (iii) the combinations hcpLoF, lcpLoF, and damaging missense variants. For damaging missense variants, we further defined three categories, that is “int5 damaging”, “int1 damaging”, and “Alpha damaging” missense variants as described above, which resulted in seven finer variant sets for each gene: (i) hcpLoF variants only, (ii) “Alpha damaging” missense variants only, (iii) “int5 damaging” missense variants only, (iv) “int1 damaging” missense variants only, (v) hcpLoF variants and “Alpha damaging” missense variants, (vi) hcpLoF variants and “int5 damaging” missense variants, and (vii) hcpLoF variants, lcpLoF variants, and “int1 missense” variants. Additionally, for each gene set, we considered four levels of variants based on the alternative allele frequency (REGENIE used alternative allele frequency for separation, but we would keep the notation MAF as they were mostly concordant in our analysis): singletons only, MAF ≤ 1 × 10−4, MAF ≤ 1 × 10−3, and MAF ≤ 1 × 10−2. Then for each given gene set, the score is calculated as the maximum of the genotype dosage across all the variants in this set (the “max rule” as the default implementation in REGENIE), and the score is tested for association. We noted that some sets may not be testable due to lack of qualified variants (for example, considering a gene that only had variants with MAF > 1 × 10−3) while some sets would produce repetitive results (for example, considering a gene that only had variants with MAF ≤ 1 × 10−4). To account for these issues, we adopted an empirical null based P-value threshold for multiple testing adjustment as described below.
Study-wise significance level for association tests
For variant-level associations, we applied the Bonferroni correction at 0.05 level, which yields a conservative P-value cutoff at 2.8 × 10−10 (=0.05/178,280,016, that is 0.05 adjusted for all the variant-level tests for coding variants of interest, see Supplementary Data S24). The variant-level tests include all the coding variants of interest (n = 2,143,707) across all 596 phenotypes, but only those with sufficient MAC (MAC > 5) were tested and the MAC was calculated for each phenotype respectively by REGENIE, so a large number of tests of variants with MAC < 5 were ignored (Supplementary Note). The summary count and more details are available in Supplementary Data S24. Briefly, 2,143,707 coding variants of interest were included for variant-level association tests, and an average of 299,128 tests were conducted for each phenotype in joint sample and similarly an average of 239,825 tests per phenotype were conducted for the discovery sample (Supplementary Data S24 and Supplementary Note). We note that our primary Bonferroni-type P-value threshold is quite stringent as the phenotypes are not entirely independent, while we also provided the results for the variant-level associations that passed the rare variant genome-wide significance threshold at 1 × 10−8 as suggestive evidence139,140. The P-value cutoff for phases 1 to 3 sample was 3.5 × 10−10 (=0.05/142,935,657, Supplementary Data S24).
For gene-based burden tests, Bonferroni correction for all the tests conducted may be too strict and inappropriate here since distinct burden models and MAF cutoffs were highly correlated and may produce repetitive results. Moreover, the dependence structure made it unclear whether the Benjamini and Hochberg procedure could provide valid false discovery rate control. Alternatively, we used resampling-based methods to derive the empirical distributions, which suggested setting 1 × 10−9 as our conservative P-value threshold for burden tests. Specifically, we investigated the empirical null distributions of P-values from both permutation test60,61 and synonymous burden test61,141 separately. We conducted the permutation for the imaging traits once for every phenotype while preserving the genetic structure and the burden models60,61. At the tail of 243,691,511 P-values derived from the permutation-based null test, we only observed 5 results that had P-values smaller than 1 × 10−9 (Supplementary Data S25). Therefore, based on this permutation-derived threshold at 1 × 10−9, the expected false discoveries would be 5 out of 224 significant associations (3 out of 67 significant gene-trait pairs) across all burden models and imaging phenotypes.
Furthermore, we found that under this permutation-based P-value cutoff, only synonymous variants in NOSTRIN and HIGD1B were observed to be associated with the mean diffusivity of the cingulum (cingulate gyrus) tract in brain white matter and the volume of left rostral middle frontal region, respectively, at the tail of the distribution of 40,545,911 P-values from synonymous burden tests (Supplementary Data S26). These two genes were previously reported to be associated with white matter142,143 and other brain-related traits144,145, although the biological significance of the roles of synonymous variants remains unclear. Nevertheless, the very few significant associations further supported the validity of our choice of this P-value cutoff through a complementary perspective as synonymous variants generally would not contribute to the gene-trait associations61,141. We observed similar patterns of the tail distributions of P-values from permutation test and synonymous burden test from the phases 1 to 3 sample (Supplementary Data S27 and S28). Thus, the P-value cutoff for burden tests of this sample (used in our internal replication) was set to be 1 × 10−9. We also provided a more liberal P-value cutoff at 1 × 10−8 as suggestive evidence (approximately empirical false discovery proportion 0.1, Supplementary Data S25 and S26) for gene-based burden test.
Cross-reference with previous GWAS
For the comparison with GWAS signals, we leveraged the summary statistics derived from previous studies1,2,3,4,10 on the same study cohort. The P-value cutoff for GWAS signals was set to be the Bonferroni adjustment for all the traits (P < 5 × 10−8 / trait numbers for each category) as described previously. Moreover, we focused on reported independent variants146 for GWAS signals and examined whether our exome-wide signals fell within the 1 Mb range of such independent GWAS signals to prioritize the effector genes among thousands of identified GWAS loci. The genetic build for previous GWAS was GRCH37, so we did liftover147 for our exome-wide results back from GRCH38 to GRCH37 and made comparisons based on the same genetic coordinates.
Imaging genetics and drug detection
We downloaded Therapeutic Target Database (TTD, last updated by January 10th, 2024)119 and evaluated all the entries documented in TTD that were claimed to be approved drug targets, in clinical trials, or supported by literature. We performed enrichment tests to see whether the identified genes in our gene-based burden tests were enriched in TTD drug target genes. We conducted Fisher’s exact tests for two classes of drug target genes in TTD. Specifically, we first included all drug targets that are either approved drug targets or in clinical trials (documented as “Successful” or “Clinical trial” in TTD), and then additionally included drug targets that were supported by literature (documented as “Literature-reported” in TTD).
Ultra-rare burden heritability and genetic correlation with health-related traits
We used the recently proposed BHR27 method to quantify the burden heritability and the burden genetic correlation for rare variants. BHR required one to divide the rare variants into multiple subgroups based on MAF cutoff and annotation outcome. Following the practical guide and previous examples, we investigated the genetic architecture of ultra-rare variants (MAF < 1 × 10−4) in our sample across 596 imaging traits. We used “univariate” mode and ran BHR to estimate the burden heritability for ultra-rare pLoF variants and “int1 damaging” missense variants. Synonymous variants were used as negative control. The corresponding P-values were manually calculated using one-sided test. Moreover, we downloaded 186 health-related traits or diseases from Genebass17 to estimate the burden genetic correlation with imaging traits in terms of the ultra-rare pLoF variants. Among the 596 imaging traits examined, those that showed significant heritability after multiple testing adjustment (P = 8.4 × 10−5 = 0.05/596, that is Bonferroni adjusted for 596 imaging traits) were included in burden correlation analysis. Then, we ran “bivariate” mode of BHR to calculate the burden genetic correlation for the selected imaging traits and downloaded phenotypes.
Rank-based inverse normal transformation and conditional analysis
To verify the robustness of the identified associations, rank-based inverse normal transformation (RINT) was applied by calling “--apply-rint” in both step 1 and step 2 of REGENIE for all identified associations in our study, while we followed the same testing procedure. The summary statistics produced after applying RINT were compared with the original ones. Conditional analysis was performed by specifying the nearby common variants as covariates to assess the independence of the identified signals. The conditional analysis is restricted to step 2 in REGENIE, while the genome-wide predictors in step 1 were reused.
Statistics and reproducibility
No statistical method was used to predetermine sample size. We made full use of the sample with both imaging phenotypes and exome sequencing data available. No data were excluded from the analyzes after quality control step as described previously. Specific details on study design and association analyzes have been provided in the previous sections. The experiments were not randomized, and the investigators were not blinded to allocation during analysis, because this is an observational association analysis of existing cohort data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The individual-level data used in this study can be obtained from UK Biobank (https://www.ukbiobank.ac.uk/). Other datasets in this paper can be obtained from Genebass (https://app.genebass.org/), GWAS summary statistics for imaging traits in BIG-KP (https://bigkp.org/), dbNSFP v.4.5a (https://sites.google.com/site/jpopgen/dbNSFP), the Therapeutic Target Database (https://idrblab.net/ttd/), and the GTEx dataset v8 (https://gtexportal.org/home/). The ExWAS summary statistics generated in this study have been deposited in Zenodo (https://doi.org/10.5281/zenodo.17496486)148.
Code availability
We made use of publicly available software and tools. The code used in this study has been deposited in Zenodo (https://doi.org/10.5281/zenodo.17496486)148.
References
Zhao, B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 51, 1637–1644 (2019).
Zhao, B. et al. Common genetic variation influencing human white matter microstructure. Science 372, eabf3736 (2021).
Zhao, B. et al. Common variants contribute to intrinsic human brain functional networks. Nat. Genet. 54, 508–517 (2022).
Zhao, B. et al. Genetic influences on the intrinsic and extrinsic functional organizations of the cerebral cortex. Preprint at https://www.medrxiv.org/content/10.1101/2021.07.27.21261187v1 (2023).
Elliott, L. T. et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018).
Smith, S. M. et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat. Neurosci. 24, 737–745 (2021).
Grasby, K. L. et al. The genetic architecture of the human cerebral cortex. Science 367, eaay6690 (2020).
Zhao, B. et al. Large-scale GWAS reveals genetic architecture of brain white matter microstructure and genetic overlap with cognitive and mental health traits (n = 17,706). Mol. Psychiatry 26, 3943–3955 (2021).
Pirruccello, J. P. et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat. Commun. 11, 2254 (2020).
Zhao, B. et al. Heart-brain connections: phenotypic and genetic insights from magnetic resonance images. Science 380, abn6598 (2023).
Khurshid, S. et al. Clinical and genetic associations of deep learning-derived cardiac magnetic resonance-based left ventricular mass. Nat. Commun. 14, 1558 (2023).
Streicher, S. A. et al. Genome-wide association study of abdominal MRI-measured visceral fat: the multiethnic cohort adiposity phenotype study. PLoS ONE 18, e0279932 (2023).
Liu, Y. et al. Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. eLife 10, e65554 (2021).
Haas, M. E. et al. Machine learning enables new insights into genetic contributions to liver fat accumulation. Cell Genom. 1, 100066 (2021).
Edwards, S. L., Beesley, J., French, J. D. & Dunning, A. M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93, 779–797 (2013).
Backman, J. D. et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 599, 628–634 (2021).
Karczewski, K. J. et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genom. 2, 100168 (2022).
Van Hout, C. V. et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020).
Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim. 1, 59 (2021).
Park, J. et al. Exome-wide association analysis of CT imaging-derived hepatic fat in a medical biobank. Cell Rep. Med. 3, 100855 (2022).
Jurgens, S. J. et al. Analysis of rare genetic variation underlying cardiometabolic diseases and traits among 200,000 individuals in the UK Biobank. Nat. Genet. 54, 240–250 (2022).
Vaser, R., Adusumalli, S., Leng, S. N., Sikic, M. & Ng, P. C. SIFT missense predictions for genomes. Nat. Protoc. 11, 1–9 (2016).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet Chapter 7, Unit7 20 (2013).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Schwarz, J. M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7, 575–576 (2010).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Weiner, D. J. et al. Polygenic architecture of rare coding variation across 394,783 exomes. Nature 614, 492–499 (2023).
Park, J. H. et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc. Natl. Acad. Sci. USA 108, 18026–18031 (2011).
Zeng, J. et al. Widespread signatures of natural selection across human complex traits and functional genomic categories. Nat. Commun. 12, 1164 (2021).
Skol, A. D., Scott, L. J., Abecasis, G. R. & Boehnke, M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 38, 209–213 (2006).
Salzer, U. et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat. Genet. 37, 820–828 (2005).
Artac, H., Bozkurt, B., Talim, B. & Reisli, I. Sarcoid-like granulomas in common variable immunodeficiency. Rheumatol. Int. 30, 109–112 (2009).
Pulvirenti, F. et al. Clinical associations of biallelic and monoallelic TNFRSF13B variants in italian primary antibody deficiency syndromes. J. Immunol. Res. 2016, 8390356 (2016).
Astle, W. J. et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167, 1415–1429 e19 (2016).
Parisinos, C. A. et al. Genome-wide and Mendelian randomisation studies of liver MRI yield insights into the pathogenesis of steatohepatitis. J. Hepatol. 73, 241–251 (2020).
Mojtahed, A. et al. Reference range of liver corrected T1 values in a population at low risk for fatty liver disease-a UK Biobank sub-study, with an appendix of interesting cases. Abdom. Radiol. 44, 72–84 (2019).
Mercadante, C. J. et al. Manganese transporter Slc30a10 controls physiological manganese excretion and toxicity. J. Clin. Invest. 129, 5442–5461 (2019).
Ward, L. D. et al. GWAS of serum ALT and AST reveals an association of SLC30A10 Thr95Ile with hypermanganesemia symptoms. Nat. Commun. 12, 4571 (2021).
Seidelin, A. S., Nordestgaard, B. G., Tybjaerg-Hansen, A., Yaghootkar, H. & Stender, S. A rare genetic variant in the manganese transporter SLC30A10 and elevated liver enzymes in the general population. Hepatol. Int. 16, 702–711 (2022).
Martin, R., Straub, A. U., Doebele, C. & Bohnsack, M. T. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 10, 4–18 (2013).
Hussain, A. DEAD Box RNA helicases: biochemical properties, role in RNA processing and ribosome biogenesis. Cell Biochem. Biophys. 82, 427–434 (2024).
Pourteymour, S. et al. Perilipin 4 in human skeletal muscle: localization and effect of physical activity. Physiol. Rep. 3, e12481 (2015).
Cheng, C. et al. Association of the ADRA1A gene and the severity of metabolic abnormalities in patients with schizophrenia. Prog. Neuropsychopharmacol. Biol. Psychiatry 36, 205–210 (2012).
Clark, D. A. et al. Polymorphisms in the promoter region of the alpha1A-adrenoceptor gene are associated with schizophrenia/schizoaffective disorder in a Spanish isolate population. Biol. Psychiatry 58, 435–439 (2005).
Persyn, E. et al. Genome-wide association study of MRI markers of cerebral small vessel disease in 42,310 participants. Nat. Commun. 11, 2175 (2020).
Sargurupremraj, M. et al. Cerebral small vessel disease genomics and its implications across the lifespan. Nat. Commun. 11, 6285 (2020).
Rutten-Jacobs, L. C. A. et al. Genetic study of white matter integrity in UK Biobank (N=8448) and the overlap with stroke, depression, and dementia. Stroke 49, 1340–1347 (2018).
Ghorbani, S. et al. Versican promotes T helper 17 cytotoxic inflammation and impedes oligodendrocyte precursor cell remyelination. Nat. Commun. 13, 2445 (2022).
Gu, W. L. et al. Expression and regulation of versican in neural precursor cells and their lineages. Acta Pharm. Sin. 28, 1519–1530 (2007).
Dours-Zimmermann, M. T. et al. Versican V2 assembles the extracellular matrix surrounding the nodes of ranvier in the CNS. J. Neurosci. 29, 7731–7742 (2009).
Ramirez, J. et al. Parkinson’s disease, NOTCH3 genetic variants, and white matter hyperintensities. Mov. Disord. 35, 2090–2095 (2020).
Ceroni, M. et al. Migraine with aura and white matter abnormalities: Notch3 mutation. Neurology 54, 1869–1871 (2000).
Masoli, J. A. H., Pilling, L. C., Kuchel, G. A. & Melzer, D. Clinical outcomes of CADASIL-associated NOTCH3 mutations in 451,424 European Ancestry Community Volunteers. Transl. Stroke Res. 10, 339–341 (2019).
Larson, E. J. et al. Rbm20 ablation is associated with changes in the expression of titin-interacting and metabolic proteins. Mol. Omics 18, 627–634 (2022).
Li, N., Hang, W., Shu, H. & Zhou, N. RBM20, a therapeutic target to alleviate myocardial stiffness via titin isoforms switching in HFpEF. Front. Cardiovasc. Med. 9, 928244 (2022).
Radke, M. H. et al. Therapeutic inhibition of RBM20 improves diastolic function in a murine heart failure model and human engineered heart tissue. Sci. Transl. Med. 13, eabe8952 (2021).
Tcheandjieu, C. et al. High heritability of ascending aortic diameter and trans-ancestry prediction of thoracic aortic disease. Nat. Genet. 54, 772–782 (2022).
Gao, Y. et al. ANO1 inhibits cardiac fibrosis after myocardial infraction via TGF-beta/smad3 pathway. Sci. Rep. 7, 2355 (2017).
Tian, X. et al. ANO1 regulates cardiac fibrosis via ATI-mediated MAPK pathway. Cell Calcium 92, 102306 (2020).
Dhindsa, R. S. et al. Rare variant associations with plasma protein levels in the UK Biobank. Nature 622, 339–347 (2023).
Wang, Q. et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature 597, 527–532 (2021).
Blombery, P. et al. Biallelic deleterious germline SH2B3 variants cause a novel syndrome of myeloproliferation and multi-organ autoimmunity. eJHaem 4, 463–469 (2023).
Morales, C. E., Stieglitz, E., Kogan, S. C., Loh, M. L. & Braun, B. S. Nf1 and Sh2b3 mutations cooperate in vivo in a mouse model of juvenile myelomonocytic leukemia. Blood Adv. 5, 3587–3591 (2021).
Perez-Garcia, A. et al. Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood 122, 2425–2432 (2013).
Arfeuille, C. et al. Germline bi-allelic SH2B3/LNK alteration predisposes to a neonatal juvenile myelomonocytic leukemia-like disorder. Haematologica 109, 2542–2554 (2023).
Kim, S. K. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE 13, e0200785 (2018).
Avirneni-Vadlamudi, U. et al. Drosophila and mammalian models uncover a role for the myoblast fusion gene TANC1 in rhabdomyosarcoma. J. Clin. Invest. 122, 403–407 (2012).
Granados, V. A. et al. Selective targeting of myoblast fusogenic signaling and differentiation-arrest antagonizes rhabdomyosarcoma cells. Cancer Res. 79, 4585–4591 (2019).
Povysil, G. et al. Assessing the role of rare genetic variation in patients with heart failure. JAMA Cardiol. 6, 379–386 (2021).
Micaglio, E. et al. Clinical considerations for a family with dilated cardiomyopathy, sudden cardiac death, and a novel TTN frameshift mutation. Int. J. Mol. Sci. 22, 670 (2021).
Liu, J. S. et al. Whole-exome sequencing identifies two novel TTN mutations in Chinese families with dilated cardiomyopathy. Cardiology 136, 10–14 (2017).
Choi, S. H. et al. Monogenic and polygenic contributions to atrial fibrillation risk: results from a National Biobank. Circ. Res. 126, 200–209 (2020).
Surendran, P. et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat. Genet. 52, 1314–1332 (2020).
Murata, M., Cingolani, E., McDonald, A. D., Donahue, J. K. & Marban, E. Creation of a genetic calcium channel blocker by targeted gem gene transfer in the heart. Circ. Res. 95, 398–405 (2004).
Tan, F. L. et al. The gene expression fingerprint of human heart failure. Proc. Natl. Acad. Sci. USA 99, 11387–11392 (2002).
Paar, V. et al. Pathophysiology of calcium mediated ventricular arrhythmias and novel therapeutic options with focus on gene therapy. Int. J. Mol. Sci. 20, 5304 (2019).
Xing, Y. et al. Golgi protein 73 promotes LPS-induced cardiac dysfunction via mediating myocardial apoptosis and autophagy. J. Cardiovasc. Pharm. 83, 116–125 (2024).
Inkster, B. et al. Genetic variation in GOLM1 and prefrontal cortical volume in Alzheimer’s disease. Neurobiol. Aging 33, 457–465 (2012).
Consortium, G. T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584 e23 (2020).
Frazier, T. W. et al. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol. Psychiatry 20, 1132–1138 (2015).
Rademacher, S. & Eickholt, B. J. PTEN in autism and neurodevelopmental disorders. Cold Spring Harb. Perspect. Med. 9, a036780 (2019).
Busch, R. M. et al. Neurobehavioral phenotype of autism spectrum disorder associated with germline heterozygous mutations in PTEN. Transl. Psychiatry 9, 253 (2019).
Cummings, K., Watkins, A., Jones, C., Dias, R. & Welham, A. Behavioural and psychological features of PTEN mutations: a systematic review of the literature and meta-analysis of the prevalence of autism spectrum disorder characteristics. J. Neurodev. Disord. 14, 1 (2022).
Clipperton-Allen, A. E. et al. Pten haploinsufficiency disrupts scaling across brain areas during development in mice. Transl. Psychiatry 9, 329 (2019).
Clipperton-Allen, A. E. et al. Pten haploinsufficiency causes desynchronized growth of brain areas involved in sensory processing. iScience 25, 103796 (2022).
Chen, Y., Huang, W. C., Sejourne, J., Clipperton-Allen, A. E. & Page, D. T. Pten mutations alter brain growth trajectory and allocation of cell types through elevated beta-catenin signaling. J. Neurosci. 35, 10252–10267 (2015).
Koboldt, D. C. et al. PTEN somatic mutations contribute to spectrum of cerebral overgrowth. Brain 144, 2971–2978 (2021).
Vanderver, A. et al. Characteristic brain magnetic resonance imaging pattern in patients with macrocephaly and PTEN mutations. Am. J. Med. Genet. A 164A, 627–633 (2014).
Dhamija, R. & Hoxworth, J. M. Imaging of PTEN-related abnormalities in the central nervous system. Clin. Imaging 60, 180–185 (2020).
Cui, Y. et al. Inhibition of PTEN-induced kinase 1 autophosphorylation may assist in preventing epileptogenesis induced by pentylenetetrazol. Neurochem. Int. 172, 105644 (2024).
Korwitz, A. et al. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J. Cell Biol. 212, 157–166 (2016).
Franchino, C. A. et al. Sustained OMA1-mediated integrated stress response is beneficial for spastic ataxia type 5. Brain 147, 1043–1056 (2024).
Anderson, C. J. et al. Prohibitin levels regulate OMA1 activity and turnover in neurons. Cell Death Differ. 27, 1896–1906 (2020).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Beckmann, C. F. & Smith, S. M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. imaging 23, 137–152 (2004).
Hyvarinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 10, 626–634 (1999).
Alfaro-Almagro, F. et al. Image processing and quality control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage 166, 400–424 (2018).
Francis, C. M. et al. Genome-wide associations of aortic distensibility suggest causality for aortic aneurysms and brain white matter hyperintensities. Nat. Commun. 13, 4505 (2022).
Sprooten, E., Beckmann, C. F., Franke, B. & Bralten, J. Shared genetic influences on resting-state functional networks of the brain. Hum. Brain Mapp. 43, 1787–1803 (2022).
Smrcka, A. V., Brown, J. H. & Holz, G. G. Role of phospholipase Cepsilon in physiological phosphoinositide signaling networks. Cell Signal 24, 1333–1343 (2012).
Hodgkinson, C. P. & Gomez, J. A. Loss of PLCepsilon activity as a culprit of ascending aortic dilation and aortic aneurysm. Am. J. Physiol. Heart Circ. Physiol. 324, H659–H661 (2023).
Atchison, D. K. et al. Phospholipase Cepsilon insufficiency causes ascending aortic aneurysm and dissection. Am. J. Physiol. Heart Circ. Physiol. 323, H1376–H1387 (2022).
Testai, F. D. et al. Cardiac contributions to brain health: a scientific statement from the American Heart Association. Stroke 55, e425–e438 (2024).
Chirinos, J. A., Segers, P., Hughes, T. & Townsend, R. Large-artery stiffness in health and disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 74, 1237–1263 (2019).
Zuk, O. et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA 111, E455–E464 (2014).
Zhou, Y. et al. Selective deletion of glutamine synthetase in the mouse cerebral cortex induces glial dysfunction and vascular impairment that precede epilepsy and neurodegeneration. Neurochem. Int. 123, 22–33 (2019).
Mahoney, J. R. & Verghese, J. Visual-somatosensory integration and quantitative gait performance in aging. Front. Aging Neurosci. 10, 377 (2018).
Chen, J. & Herrup, K. Chapter 70 - glutamine as a potential neuroprotectant in Alzheimer’s disease. in Diet and Nutrition in Dementia and Cognitive Decline (eds Martin, C. R. & Preedy, V. R.) 761–771 (Academic Press, San Diego, 2015).
Andersen, J. V. et al. Deficient astrocyte metabolism impairs glutamine synthesis and neurotransmitter homeostasis in a mouse model of Alzheimer’s disease. Neurobiol. Dis. 148, 105198 (2021).
Robinson, S. R. Neuronal expression of glutamine synthetase in Alzheimer’s disease indicates a profound impairment of metabolic interactions with astrocytes. Neurochem. Int. 36, 471–482 (2000).
Jayakumar, A. R. & Norenberg, M. D. Glutamine synthetase: role in neurological disorders. Adv. Neurobiol. 13, 327–350 (2016).
Shadrin, A. A. et al. Vertex-wise multivariate genome-wide association study identifies 780 unique genetic loci associated with cortical morphology. NeuroImage 244, 118603 (2021).
Chung, Y. H., Shin, C., Kim, M. J., Lee, B. K. & Cha, C. I. Immunohistochemical study on the distribution of six members of the Kv1 channel subunits in the rat cerebellum. Brain Res. 895, 173–177 (2001).
Yang, Y. et al. Novel KCNA5 loss-of-function mutations responsible for atrial fibrillation. J. Hum. Genet. 54, 277–283 (2009).
Vera-Zambrano, A. et al. Novel loss-of-function KCNA5 variants in pulmonary arterial hypertension. Am. J. Respir. Cell Mol. Biol. 69, 147–158 (2023).
Pirruccello, J. P. et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat. Genet. 54, 40–51 (2022).
Zhao, B. et al. An atlas of trait associations with resting-state and task-evoked human brain functional organizations in the UK Biobank. Imaging Neurosci. 1, 1–23 (2023).
Zhou, Y. et al. TTD: therapeutic target database describing target druggability information. Nucleic Acids Res. 52, D1465–D1477 (2024).
Sun, B. B. et al. Genetic associations of protein-coding variants in human disease. Nature 603, 95–102 (2022).
Szustakowski, J. D. et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 53, 942–948 (2021).
Trajanoska, K. et al. From target discovery to clinical drug development with human genetics. Nature 620, 737–745 (2023).
Steckler, T. & Salvadore, G. Chapter 7 - neuroimaging as a translational tool in animal and human models of schizophrenia. in Translational Neuroimaging (ed McArthur, R. A.) 195–220 (Academic Press, 2013).
Nguyen, P. A., Born, D. A., Deaton, A. M., Nioi, P. & Ward, L. D. Phenotypes associated with genes encoding drug targets are predictive of clinical trial side effects. Nat. Commun. 10, 1579 (2019).
Vicario, C. M., Nitsche, M. A., Salehinejad, M. A., Avanzino, L. & Martino, G. Time processing, interoception, and insula activation: a mini-review on clinical disorders. Front Psychol. 11, 1893 (2020).
McCarthy, D. J. et al. Choice of transcripts and software has a large effect on variant annotation. Genome Med. 6, 26 (2014).
Auer, P. L., Reiner, A. P. & Leal, S. M. The effect of phenotypic outliers and non-normality on rare-variant association testing. Eur. J. Hum. Genet 24, 1188–1194 (2016).
Greer, P. J. et al. A reassessment of Hardy-Weinberg equilibrium filtering in large sample genomic studies. Preprint at https://www.medrxiv.org/content/10.1101/2024.02.07.24301951v1 (2024).
Akbari, P. et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science 373, eabf8683 (2021).
Tian, R. et al. Whole-exome sequencing in UK Biobank reveals rare genetic architecture for depression. Nat. Commun. 15, 1755 (2024).
Littlejohns, T. J. et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat. Commun. 11, 2624 (2020).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, s13742-015–0047-8 (2015).
McLaren, W. et al. The ensembl variant effect predictor. Genome Biol. 17, 122 (2016).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Liu, X., Li, C., Mou, C., Dong, Y. & Tu, Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 12, 103 (2020).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 32, 894–899 (2011).
Agrawal, S. et al. Inherited basis of visceral, abdominal subcutaneous and gluteofemoral fat depots. Nat. Commun. 13, 3771 (2022).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Fadista, J., Manning, A. K., Florez, J. C. & Groop, L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur. J. Hum. Genet. 24, 1202–1205 (2016).
Gao, X. R., Chiariglione, M. & Arch, A. J. Whole-exome sequencing study identifies rare variants and genes associated with intraocular pressure and glaucoma. Nat. Commun. 13, 7376 (2022).
Dhindsa, R. S. et al. A minimal role for synonymous variation in human disease. Am. J. Hum. Genet. 109, 2105–2109 (2022).
Cui, X. et al. Endothelial nitric oxide synthase regulates white matter changes via the BDNF/TrkB pathway after stroke in mice. PLoS ONE 8, e80358 (2013).
Elahi, F. M. et al. Liquid biopsy” of white matter hyperintensity in functionally normal elders. Front. Aging Neurosci. 10, 343 (2018).
Winkler, E. A. et al. A single-cell atlas of the normal and malformed human brain vasculature. Science 375, eabi7377 (2022).
Crouch, E. E., Joseph, T., Marsan, E. & Huang, E. J. Disentangling brain vasculature in neurogenesis and neurodegeneration using single-cell transcriptomics. Trends Neurosci. 46, 551–565 (2023).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Raney, B. J. et al. The UCSC Genome Browser database: 2024 update. Nucleic Acids Res. 52, D1082–D1088 (2024).
Fan, Y. Summary statistics for mapping rare protein-coding variants on multi-organ imaging traits. https://doi.org/10.5281/zenodo.17496459 (2025).
Acknowledgements
Research reported in this publication was supported by National Institute of Mental Health under Award Number R01MH136055 and National Institute on Aging under Award Numbers RF1AG082938 and R01AG085581. Assistance for this project was provided by the UNC Intellectual and Developmental Disabilities Research Center (NICHD; P50 HD103573; H.Z.), and by grants RF1AG098697 (H.Z.), R01AR082684 (H.Z.), OT2OD038045-01 (H.Z.), R21HD120911 (Tengfei L.), and K01AG095286 (Tengfei L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study has also been partially supported by funding from the Purdue University Statistics Department, Department of Statistics and Data Science at the University of Pennsylvania, Wharton Dean’s Research Fund, Analytics at Wharton, Wharton AI & Analytics Initiative, Perelman School of Medicine CCEB Innovation Center Grant, and the University Research Foundation Grant (B.Z.). This research has been conducted using the UK Biobank resource (application number 76139), subject to a data transfer agreement. We would like to thank the individuals who represented themselves in the UK Biobank for their participation and the research teams for their efforts in collecting, processing, and disseminating these datasets. We would like to thank the research computing groups at the University of North Carolina at Chapel Hill, Purdue University, and the Wharton School of the University of Pennsylvania for providing computational resources and support that have contributed to these research results.
Author information
Authors and Affiliations
Contributions
Y.F. and B.Z. designed the study. Y.F., Jie C., Z.F., D.Y.Z., and Z.S. analyzed the data and generated the results. T.L., S.H., Z.J., P.Y.G., X.Q., T.L., and H.L processed the MRI data. J.C., J.L.S., P.F.S., R.W., A.N., M.D.R., J.M.O., W.W., D.J.R., and H.Z. provided feedback on study design and helped results interpretations. Y.F. and B.Z. wrote the manuscript with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
R.W. is a current employee and/or stockholder of Regeneron Genetics Center or Regeneron Pharmaceuticals. Other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Andre Altmann and Akl Fahed for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fan, Y., Chen, J., Fan, Z. et al. Mapping rare protein-coding variants on multi-organ imaging traits. Nat Commun 17, 739 (2026). https://doi.org/10.1038/s41467-025-67431-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67431-y