Abstract
Human stem cell-derived β (SC-β) cells still exhibit limited glucose response required for insulin secretion due to glycolytic bottlenecks, yet how these metabolic abnormalities impact glucose response and functional maturation of SC-β cells remains unclear. In this study, we identify a metabolic checkpoint located at PEP accumulation that impedes the functional maturation, which is rescued by restoration of pyruvate kinase M1 (PKM1). Glucose-tracing metabolomics in human stem cell-derived islets reveal abnormal glycolytic PEP accumulation at resting condition, associated with impaired calcium response and insulin secretion upon high glucose or glycolytic metabolite stimulation. Mechanistically, elevated PEP significantly raises intracellular basal calcium levels and downregulates expression of genes involved in TCA cycle as elucidated by single cell transcriptomics. Furthermore, the activity of pyruvate kinase, which metabolizes PEP, is notably reduced due to low PKM1 expression. Overexpression of PKM1 alleviates PEP accumulation, restores TCA-related gene expression, and enhances glucose-stimulated calcium responses and insulin secretion. Together, our findings reveal a critical role of PKM1-regulated PEP metabolism in SC-β cell functional maturation and highlight the importance of metabolic reprogramming for advancing stem cell-based therapies for diabetes.
Similar content being viewed by others
Introduction
Functionally mature stem cell-derived β (SC-β) cells are essential for ensuring the effectiveness of SC-β cell transplantation therapy for diabetes treatment. One of the most critical characteristics of pancreatic β cell functional maturation is glucose-stimulated insulin secretion (GSIS). While recent protocols have shown improvements in GSIS in SC-β cells, these cells still exhibit significantly reduced GSIS compared to native human β cells, presenting a barrier to the effectiveness of β cell replacement therapies1,2.
In native β cells, glucose metabolism generates ATP through glycolysis and oxidative phosphorylation (OXPHOS), leading to the closure of KATP channels, Ca²⁺ influx, and subsequent insulin release3. While SC-β cells can secrete insulin in response to TCA cycle metabolites and depolarization, they are less responsive to high glucose levels, possibly due to a metabolic defect located at the glycolytic GAPDH and PGK1 proteins1,4,5. Additionally, inhibition of GAPDH leads to the accumulation of upstream metabolites, which impairs the OXPHOS necessary for insulin secretion in diabetic β-cells via mTORC1 signaling remodeling6, a pathway also involved SC-β cell maturation7. However, attempts to enhance GAPDH and PGK1 expression in SC-β cells have not restored GSIS4, suggesting the presence of unidentified downstream metabolic bottlenecks that may decouple glycolysis from OXPHOS8.
Impaired PEP metabolism is a metabolic obstacle hampering SC-β from achieving a proper glucose response1,4. Recently, PEP from glycolysis has gained attention for its role in generating ATP via pyruvate kinase (PK) to initiate KATP closure, Ca2+ influx, and triggering pathway9,10,11. Although this is still controversial12,13,14,15, abnormal PEP metabolism would inevitably impair downstream metabolism required for insulin release16. In SC-β cells, bypassing the PEP with extracellular PEP, oxaloacetic acid (OAA), and other permeable metabolites has significantly increased insulin secretion4,17. However, the role of PEP in SC-β function and maturation remain unclear.
The two main splicing isoforms of pyruvate kinase muscle (PKM), PKM1 and PKM2, catalyze the conversion of PEP to pyruvate. Beyond this, they exert distinct, isoform-specific effects on cell differentiation and function18. While PKM2 activity accounts for only 10% of PK activity in mature β cells, the constitutively active PKM1 accounts for 90%. Both PK isoforms are essential for PEP metabolism and insulin secretion11. Due to their sequence similarity, most transcriptomic and proteomic studies cannot distinguish between PKM1 and PKM2, leaving it unclear whether a deficiency in either isoform or PK activity could affect glucose metabolism and functional maturation via regulating PEP in SC-β cells.
In this study, we explore the metabolic bottlenecks that uncouple glucose sensing from functional maturation in SC-β cells. We found that PEP abnormally accumulates at basal glucose level, which interferes with basal glucose responsiveness and gene expression programs. Restoration of PKM1, which was markedly lower in SC-β cells compared to primary β cells, was able to partially restore GSIS, enhance Ca²⁺ responses, and normalize the expression of glycolytic and OXPHOS genes affected by PEP imbalance. Together, these findings highlight PEP accumulation as a metabolic checkpoint that impairs functional maturation of SC-β cells.
Results
Metabolomic profiling of SC-islets
We first generated SC-islets following established protocols (Supplementary Fig. 1A and Supplementary Data 1)2,19,20. In agreement with previous studies, SC-islets differentiated from human embryonic stem cell lines H1 and H9 showed increased β-cell-specific gene expression (INS, NKX6.1, MAFA) and relatively uniform size (Supplementary Fig. 1B, C). However, SC-islets displayed limited GSIS compared to mouse islets (Supplementary Fig. 1D).
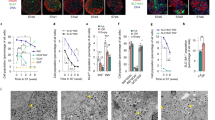
To investigate glucose metabolic defect in SC-islets, we compared glucose metabolic fate between mouse- and SC-islets by performing metabolite tracing experiment with labeled [U-¹³C₆]-glucose (Fig. 1A). Key glycolytic intermediates—glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), dihydroxyacetone phosphate (DHAP), and 3-phosphoglycerate (3-PG)/2-phosphoglycerate (2-PG)—showed similar glucose-dependent increases in both mouse and SC-islets, indicating that glucose metabolism from glucose through to the 3-PG/2-PG stage remains relatively intact (Fig. 1B–E). Interestingly, while high glucose-induced PEP levels were comparable between mouse and SC-islets, basal PEP levels at low glucose were significantly higher in SC-islets with lower stimulating index, as were pyruvate levels (Fig. 1F, G) and other TCA metabolites (Fig. 1H). In addition, glutamate was preferably metabolized to glutamine (Gln) rather than GSH, and ATP production was much lower in SC-islets (Fig. 1I), indicating that the disrupted glucose metabolism originated from dysregulated glycolytic metabolism downstream of PEP (Fig. 1J).
A Graphic overview of glucose tracing experimental procedure. Primary mouse islets and SC-islets were labelled with 2.8 mM (LG, low glucose) or 16.7 mM (HG, high glucose) [U13C6]-glucose for 60 min before LC-MS detection for metabolomics. B–G Labeled metabolites percentages (percentage of U-¹³C₆ labeling of total metabolites) of glycolytic metabolites. Stimulating index, the ratio of labeled metabolites between HG and LG. B G6P glucose-6-phosphate, n = 3 mice and 6 batches of SC-islets, p = 0.0258, 0.0423; C F6P fructose-6-phosphate, n = 4 mice and 6 batches of SC-islets, p = 0.0111, 0.0073; D DHAP dihydroxyacetone phosphate, n = 3 mice and 4 batches of SC-islets; E 3-PG/2-PG, 3-phosphoglycerate/2-phosphoglycerate, n = 5 mice and 6 batches of SC-islets; F PEP phosphoenolpyruvate, n = 5 mice and 6 batches of SC-islets, p = 0.0422, 0.0071 for the left graph, p = 0.0067 for the right graph; G pyruvate, n = 4 mice and 5 batches of SC-islets, p = 0.0144, 0.0432 for the left graph, p < 0.0001 for the right graph. H Labeled metabolites percentages of TCA cycle metabolites. ΟΑΑ, oxaloacetic acid, n = 4 mice and 5 batches of SC-islets, p = 0.0215; Malate, n = 5 mice and 6 batches of SC-islets, p = 0.0435; Citrate, n = 6 mice and 6 batches of SC-islets, p = 0.0024, 0.0417; Succinate, n = 4 mice and 3 batches of SC-islets, p = 0.0049, 0.0364. I Labeled metabolites percentages of glutamate-derived metabolites and ATP. Glutamate, n = 6 mice and 6 batches of SC-islets, p = 0.0015, 0.0344; Gln, glutamine, n = 6 mice and 6 batches of SC-islets, p = 0.0137, 0.0098; GSH, n = 5 mice and 6 batches of SC-islets, p = 0.018, 0.0195; ATP, n = 5 mice and 6 batches of SC-islets, p = 0.0351. (M + x) represents the count of carbon atoms that are labeled with 13C. J Overview of glucose metabolism with metabolic defect starting from PEP in SC-islets compared to mouse islets. All data are presented as mean ± SEM, with statistical significance determined using one-way ANOVA followed by Sidak’s multiple comparison test (B–I) and two-tailed unpaired Student’s t test (right graph from F, G). ns not significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Bottleneck downstream of PEP limits glucose-dependent function
To further investigate whether glycolytic bottlenecks downstream of PEP impedes glucose responsiveness, we assessed the effects of PEP and its upstream metabolites on SC-β cells. Cell-permeable metabolites upstream of PEP, including glyceraldehyde (GLA, a precursor to glyceraldehyde 3-phosphate) and dihydroxyacetone (DHA, a precursor to DHAP), elicited minimal insulin secretion (Fig. 2A, B) and calcium responses (Fig. 2C, D) in SC-β cells compared to mouse β cells. Glyceric acid (GA, a non-phosphorylated precursor to 3-PG and 2-PG4) also produced negligible effects in SC-β cells (Fig. 2E and Supplementary Fig. 2A), indicating a metabolic blockage downstream of 2-PG. Consistently, although 3-PG was not permeable to induce substantial calcium responses in mouse-β cells (Fig. 2F)4, the infusion (dialysis) of 50 \({{\rm{\mu }}}\)M 3-PG, 2-PG, or K+-PEP into the cell cytosol via whole-cell patch clamp significantly enhanced depolarization-induced exocytosis by ( ~ fivefold) compared to the control group in mouse-β cells. In contrast, SC-β cells, generated from CRISPR-Cas9 method and identified by EGFP (Supplementary Fig. 2B–D), showed no metabolite-dependent changes in exocytosis (Fig. 2G). Remarkably, this was because under control conditions, SC-β cells already exhibited a tenfold higher exocytosis compared to mouse β-cells, limiting further amplification by these metabolites (Fig. 2G). Additionally, SC-β cells displayed greater depolarization-induced calcium entry across various conditions compared to mouse β-cells (Fig. 2H). Together, these findings suggest that the saturating effect of metabolite accumulation downstream of PEP at basal glucose levels prevents additional metabolite- or glucose-dependent enhancement of exocytosis.
A, B Insulin secretion in response to 20 mM glyceraldehyde (GLA) (A, n = 3 mice, 5 batches of SC-islets, p < 0.001 on the left graph and p = 0.037 on the right graph) or 20 mM dihydroxyacetone (DHA) (B, n = 4 mice, 4 batches of SC-islets, p = 0.0286). C–F Average [Ca²⁺]ᵢ responses to 20 mM GLA (C, n = 19 mouse β cells, 14 SC- β cells respectively from 3 different experiments, p = 0.0202), 20 mM DHA (D, n = 8 mouse β cells, 8 SC- β cells respectively from 3 different experiments, p = 0.0072), 20 mM glyceric acid (GA) (E, n = 14 mouse β cells, 18 SC- β cells respectively from 3 different experiments, p = 0.0474), and 20 mM 3-PG (F, n = 30 mouse β cells, 11 SC- β cells respectively from 3 different experiments, p = 0.0282). G Representative traces (left), and average total normalized exocytotic responses of β-cell (right) with infusion of 50 \({{\rm{\mu }}}\)M pH-corrected 3-PG, 2-PG, and K+-PEP from mouse-β (n = 20, 15, 15, 17 cells) and SC-β (n = 11, 12, 13, 15 cells) at 2.8 mM glucose, p = 0.04, 0.002, <0.001, <0.001. H Representative traces (left), and average Ca2+ currents of β-cell (right) with infusion (dialysis) of 50 µM pH-corrected 3-PG, 2-PG, and K+-PEP from mouse-β (n = 20, 15, 15, 17 cells) and SC-β (n = 11, 12, 13, 15 cells), p < 0.001, 0.004, <0.001, <0.001. Glucose level was 2.8 mM unless indicated. For [Ca2+]i recordings, single mouse-β cell was loaded with Cal-500 AM and identified by immunostaining for insulin. Single SC-β cell was identified by using cells differentiated from H1-Ins-jGCaMP7f46. [Ca2+]i recordings were analyzed using smooth dynamics method59. To demonstrate the functional viability of β cells and response to KCl, 5 mM KCl and 20 mM K+-PEP (Phosphoenol-pyruvate PEP, monopotassium salt) were added at the end of [Ca2+]i recording. For patch clamp, single SC-β cell was identified by using cells differentiated from H1-Ins-EGFP. All data are presented as mean ± SEM, with statistical significance determined using unpaired two-tailed Mann–Whitney test (right graphs from A, B), unpaired two-tailed Student’s t test (right graphs from C–F), or two-way ANOVA followed by Sidak’s multiple comparison test (left graph from A, right graphs from G and H). *P < 0.05, **P < 0.01, ***P < 0.001.
Basal PEP metabolites elevates Ca²⁺ and hinders SC-β cell maturation
Since basal PEP and its downstream metabolites were upregulated in SC-β cell, we next examined their effects on cellular function. As expected, Na⁺-PEP (trisodium salt), which is expected to not freely permeable to the plasma membrane, failed to elicit a calcium response in mouse β cells (Fig. 3A). In contrast, Na⁺-PEP triggered calcium responses in CD49a-based magnetically sorted SC-β cells, which was blocked by KATP channel opener diazoxide (Dia), mitochondrial complex III inhibitor antimycin A and PKM2 inhibitor PKM2-IN-1 (PKi) (Fig. 3B–D and Supplementary Fig. 2E, F), suggesting that unlike mouse β cells, Na⁺-PEP can be transported into SC-β cells and metabolized to promote calcium influx via KATP channel. Similarly, although OAA, which was upregulated in SC-islets (Fig. 1H), did not evoke calcium responses in mouse β cells (Fig. 3E), but induced Dia- and antimycin A-sensitive calcium responses in sorted SC-β cells that were not inhibited by PKi (Fig. 3F–H). Permeable methyl-pyruvate (MP) can also induce a diazoxide- and antimycin A-dependent calcium response in SC-β cells (Supplementary Fig. 2G–J), albeit with lower sensitivity compared to mouse β cells (Supplementary Fig. 2J). Accordingly, Na+-PEP, OAA, and MP can trigger significant insulin secretion in SC-islets (Fig. 3I, J).
A Average [Ca2+]i recordings to Na+-PEP (5, 20 mM) followed by 30 mM KCl from mouse-β cells (n = 115 cells from 3 different experiments, p < 0.0001, 0.0001, 0.0001). B–D Average [Ca2+]i recordings to Na+-PEP (5, 20 mM) followed by treatment of diazoxide (Dia, 200 µM) (B, p < 0.0001, 0.0001, 0.0001), antimycin A (5 µM) (C, p < 0.0001, 0.0001, 0.0001) or PKM2-IN-1 (PKi, 10 µM) (D, p < 0.0001, 0.0001) from SC-β cells (n = 490, 324, 312 cells respectively from 3 different experiments). E Average [Ca2+]i recordings to OAA (20 mM) followed by 30 mM KCl from mouse-β cells (n = 82 cells from 3 different experiments, p < 0.0001). F–H Average [Ca2+]i recordings to OAA (20 mM) followed by treatment of diazoxide (Dia, 200 µM) (F, p < 0.0001, 0.0001), antimycin A (5 µM) (G, p < 0.0001, = 0.0393) or PKM2-IN-1 (PKi, 10 µM) (H, p < 0.0001, <0.0001) from SC-β cells (n = 59, 108, 161 cells respectively from 3 different experiments). [Ca2+]i recordings were analyzed using average fluorescence method59. Recordings were normalized to control condition. Glucose level was 5 mM unless indicated. I Insulin secretion to 20 mM Na+-PEP followed by KCl from mouse (n = 3) and SC-islets (n = 5), p = 0.0028. J Static insulin secretion to 2 mM glucose (LG), 20 mM glucose (HG), 20 mM MP, and 20 mM OAA (n = 5, 5 independent experiments in triplicate, p = 0.002, 0.002). K Single-cell RNA-seq transcriptomic profiling (DNBelab C Series) of SC-islets cultured with 50 or 500 µM K+-PEP throughout stage 6/7 and collected on day 14 of stage 7 (S7D14). A UMAP-based embedding projection of an integrated dataset comprising 77,623 SC-derived cells and adult human islet cells, separated by sample origin and clustered by cell type. L Cell number ratio for each cell type in the dataset. M Differentially expressed (DE) genes between control (50 µM KCl) and K+-PEP-treated (50 µM) β cell group, including adult and SC-β cells. Gene sets enriched analysis (GSEA) using Reactome pathways showed upregulated and downregulated pathways along with DE genes between control (50 µM KCl) and K+-PEP-treated (50 µM) β cell group (n = 6438 cells, 7360 cells). N Differentially expressed (DE) genes between control (500 µM KCl) and K+-PEP-treated (500 µM) β cell group, including adult and SC-β cells. Gene sets enriched analysis (GSEA) using Reactome pathways showed upregulated and downregulated pathways along with DE genes between control (500 µM KCl) and K+-PEP-treated (500 µM) β cell group (n = 7343 cells, 8263 cells). NES normalized enrichment score, DE differentially expressed. [Ca2+]i recordings were analyzed using average or smooth dynamics method59. All data are presented as mean ± SEM, with statistical significance determined using one-tailed unpaired Student’s t test (I), one-way ANOVA followed by Sidak’s multiple comparison test (A–H, J) or unpaired two-tailed Mann–Whitney test adjusted with multiple comparisons (left graphs from M, N). *P < 0.05, **P < 0.01, ***P < 0.001.
Due to the elevated basal PEP as the starting point of abnormal downstream TCA metabolism and its effects on calcium response, we further investigated the impact of PEP on SC-β cells differentiation. Since K+-PEP is the monopotassium salt of PEP, we used KCl as a control to eliminate the potential effect of K+. SC-islets were cultured with 50/500 \({{\rm{\mu }}}\)M KCl or K+-PEP throughout Stage 6/7 and collected for single-cell RNA sequencing at S7D14. Integration with human adult islet datasets yielded a combined atlas of 77,623 cells, including 19,126 adult islet cells21. Clustering by cell identity revealed distinct transcriptional profiles of SC-β cells compared with adult β cells.
Although most cells differentiated from our protocol were annotated as SC-β cells, a significant fraction showed high similarity to adult β and α cells (Fig. 3K). Notably, PEP treatment reduced the proportion of adult β, adult α, and endocrine progenitor cells, while increasing the proportion of SC-β, SC-α, SC-EC, and polyhormonal populations (Fig. 3L). We then focused on SC-β and adult β cell subpopulations to investigate the transcriptional changes associated with the PEP effect. Treatment with 50 \({{\rm{\mu }}}\)M PEP disrupted the expression of β-cell-specific genes (INS, INS-IGF2) and genes essential for insulin secretion and mitochondrial OXPHOS, including mitochondrial complex I (MT-ND1-5), complex III (MT-CYB), complex IV (MT-CO1-3), and Complex V (MT-ATP6, MT-ATP8)22 (Fig. 3M(left), Supplementary Data 2). Consistent with these findings, gene set enrichment analysis (GSEA) revealed downregulation of the TCA cycle, the respiratory transport chain, and mitochondrial complex I biogenesis, all critical for OXPHOS-driven insulin secretion23. ROBO signaling, which regulates islet morphology, cell identity, and insulin secretion24,25,26, was also downregulated following PEP treatment. Conversely, PEP treatment induced a neuronal-like transcriptional program involving calcium signaling (ERBB4, PRKCB, CACNA1E), a characteristic of immature β-cells, and the removal of this signature was essential for their maturation (Fig. 3M(right), Supplementary Data 2)27. Similarly, 500 \({{\rm{\mu }}}\)M PEP impaired genes related to β-cell identity and function (Fig. 3N and Supplementary Data 3). Together, these results demonstrate that while PEP enhances calcium responses and insulin secretion, it simultaneously drives transcriptional programs that hinder the further maturation of SC-β cells.
SC-β cells show absence of PKM1 expression
To assess whether PEP accumulation results from altered production or consumption, we examined glycolytic enzymes in SC-islets. ALDOA, PGK1, and PGAM1 showed a trend of reduction in SC-islets, while enolase (ENO1 and ENO2), which directly produced PEP from upstream metabolites, were comparable between mouse- and SC-islets. Remarkably, PKM1 gene expression, which metabolized PEP to pyruvate, was ~100-fold lower in SC-islets compared to mouse islets (Fig. 4A) and nearly absent as demonstrated by western blotting (Fig. 4B). Interestingly, PKM2 expression was higher in SC-islets (Fig. 4A) and seemed to be prone to complete denaturation compared to mouse islets (Fig. 4B). Immunostaining confirmed that mouse β cells expressed both PKM1 and PKM2, whereas SC-β cells expressed much less PKM1 compared to mouse islets (Fig. 4C, D) and human islets (Supplementary Fig. 3A–C). Notably, PKM1 was highly expressed in human islets relative to surrounding exocrine tissues (Supplementary Fig. 3A). Accordingly, PK activity, rather than PGK1 and GAPDH, was markedly reduced in SC-islets but restored by PKM1 overexpression (Fig. 4E, F). These findings suggest that loss of PKM1 might limit PEP consumption and drive its accumulation in SC-β cells.
A Glycolytic gene expression of mouse- and SC-islets from S7D14 (n = 3 different experiments in duplicate, p = 0.000101, 0.00012, 0.011199, 0.000002). B Western blotting of PKM1 with normalized band intensity calculation (top) and PKM2 with normalized band intensity calculation (bottom) of mouse- and SC-islets from S7D14 (n = 3, p = 0.000111, 0.002916, 0.022479, 0.000002). C Representative image of dissociated mouse islet cells immunostained for insulin, PKM1, and PKM2 (n = 3 independent experiments). Scale bar = 20 µm. D Representative image of dissociated SC-islet cells immunostained for insulin, PKM1, and PKM2 (n = 3 independent experiments). Scale bar = 20 µm. E Enzymatic activity of pyruvate kinase (n = 3, 8, 3), PGK1 (n = 3, 5), and GAPDH3,6 of mouse- and SC-islets or overexpressed with PKM1-mcherry (p < 0.0001, <0.0001). F Representative image of dissociated SC-islet cells immunostained for insulin, PKM1, and PKM2 after PKM1-mcherry overexpression (n = 3 independent experiments). Scale bar = 20 µm. (Additional images are provided for C, D, F in source data file). All data are presented as mean ± SEM, with statistical significance determined using multiple unpaired two-tailed t test adjusted with Holm–Sidak method (A, B) or one-way followed by Sidak’s multiple comparison test (E). *P < 0.05, **P < 0.01, ***P < 0.001.
PKM1 reverses PEP-induced changes and promoted maturation
Given that PEP impaired SC-β cell maturation at the transcriptomic level, we asked whether PKM1 overexpression could reverse these effects. PKM1-overexpressing SC-islets were cultured with 50 \({{\rm{\mu }}}\)M K+-PEP throughout Stage 6/7 and profiled by single-cell RNA-seq at S7D14. Integration with datasets from KCl- or K⁺-PEP-treated SC-islets and human islets (59,833 cells) revealed that PKM1 overexpression restored the proportions of “Adult β” and “Adult α” cells while reducing “SC-β” cells (Fig. 5A and Supplementary Fig. 4A–C). Since PKM1 cannot be distinguished from PKM2 in sequencing data, we used mCherry expression as an indicator of successful PKM1 overexpression. PKM1 promoted genes related to β-cell-specific genes (INS, INS-IGF2), and mitochondrial OXPHOS, including mitochondrial complex I (MT-ND1-5), complex III (MT-CYB), complex IV (MT-CO1-3), and Complex V (MT-ATP6, MT-ATP8) (Fig. 5B and Supplementary Data 4). Consistently, genes in TCA cycle and ROBO signaling pathway were upregulated while genes related to neuronal system was downregulated (Fig. 5C and Supplementary Data 4). Similarly, in SC-islets exposed to higher K+-PEP (5 mM) for a shorter period (Stage 6 and collected at S7D1), PKM1-overexpression restored PEP-induced genes and pathways linked to β-cell identity, insulin secretion, glucose metabolism, and TCA cycle (Supplementary Fig. 5A–E and Supplementary Data 5).
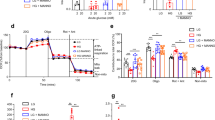
A Cell number ratio for each cell type in the dataset (Adult islets, 50 µM KCl, 50 µM K+-PEP, and K+-PEP with PKM1-overexpression). B, C Differentially expressed (DE) genes (B) and gene sets enriched analysis (GSEA) using Reactome pathways (C) between K+-PEP-treated (50 µM) and K+-PEP-treated (50 µM) β cell group with PKM1-overexpression, including adult and SC-β cells. NES normalized enrichment score (n = 7360 cells for 50 µM PEP and 4659cells for 50 µM PKM1-PEP). D Static insulin secretion to 1, 11, 20 mM glucose and 30 mM KCl from CTR and PKM1-overexpressing SC-islets (n = 3, 3 experiments, p = 0.0073, 0.0212, 0.0014, 0.0054). E Oxygen consumption rate (OCR) measured by Seahorse assay (n = 8 CTR, 8 PKM1 in at least triplicate), with relevant respiration parameters calculated at right as shown by italicized numbers. P = 0.018, 0.0128, 0.0083, 0.0069, 0.0053, 0.0038, 0.0087, 0.0189, 0.0277, 0.033, 0.0309, 0.0288, 0.0282, 0.0277 in the left graph; P = 0.042, 0.0223 in the right graph. F Average [Ca2+]i recordings to different concentration of glucose level and 20 mM K+-PEP from CTR and PKM1-overexpressing SC-β (n = 30, 28 cells from 5 different experiments, p = 0.049, 0.031, 0.014, 0.001). G Static insulin secretion to 1, 11, 20 mM glucose and 30 mM KCl from sorted CTR and PKM1-overexpressing SC- β cells (n = 4, 4 experiments, p = 0.0013, 0.0144). H Average total normalized exocytotic responses of CTR and PKM1-overexpressing SC-β cell (n = 21, 17, 18, 18 cells, p = 0.002, 0.02). I Average Ca2+ currents of CTR and PKM1-overexpressing SC-β cell (n = 21, 17, 18, 18 cells). LG, 2.8 mM glucose. HG, 20 mM glucose. 2 G, 2 mM glucose. 2.8 G, 2.8 mM glucose. 11 G, 11 mM glucose. 20 G, 20 mM glucose. FBP fructose 1,6-bisphosphate. SC-β differentiated from H1-Ins-EGFP cells or mouse-β were loaded with Cal-500 AM for calcium imaging and was used for patch clamp measurement. PKM1-overexpressing SC-β was identified by mCherry overexpression. All data are presented as mean ± SEM, with statistical significance determined using unpaired two-tailed Mann–Whitney test adjusted with multiple comparisons (B, C), one-way (H) or two-way ANOVA (D, left graph from E, right graph from F, G) followed by Sidak’s multiple comparison test, or unpaired two-tailed Student’s t test (right graph from E). *P < 0.05, **P < 0.01, ***P < 0.001.
Functionally, PKM1-overexpressing SC-islets displayed enhanced GSIS (Fig. 5D), oxygen consumption rate (Fig. 5E), and calcium responses (Fig. 5F). To further examine if this effect was due to the metabolism of SC-β cell, SC-β cells were magnetically sorted based on CD49a and reaggregated. Sorted CD49a⁺ SC-β cells with PKM1 overexpression showed reduced basal insulin release, leading to a greater glucose-dependent fold increase (Fig. 5G), and restored glucose-dependent exocytosis (Fig. 5H) with preserved calcium entry (Fig. 5I). Metabolically, CD49a⁺-sorted PKM1-overexpressing SC-β cells showed a glucose-dependent increase in F6P, DHAP, and 3-PG/2-PG comparable to controls (Supplementary Fig. 6A–C), but with lower basal PEP and pyruvate levels, resulting in greater glucose-dependent changes (Supplementary Fig. 6D, E). Downstream metabolites, including citrate, malate, and succinate, also displayed modest improvements in glucose dependency (Supplementary Fig. 6F–H). However, ATP increased only at basal glucose (Supplementary Fig. 6I), and no further improvements were observed in other metabolites (Supplementary Fig. 6J–L). FBP, an upstream glycolytic metabolite and activator of PKM2, could induce higher insulin secretion and calcium response in PKM1-overexpressing SC-islets (Supplementary Fig. 7A, B), but these effects cannot be inhibited by PKi, antimycin A, or diazoxide (Supplementary Fig. 7C–E). Acute treatment of PKM2 agonist TEPP-46 (PKa), can only modestly but not significantly enhance insulin (Supplementary Fig. 7F) or calcium responses (Supplementary Fig. 7G) in both control and PKM1-overexpressing SC-islets. Overall, PKM1 overexpression improved glucose-stimulated function in SC-β cells.
To further explore the transcriptional changes induced by PKM1, single-cell sequencing datasets generated from control islets and PKM1-overexpressing islets collected at S7D1 and S7D14 were integrated with adult islets dataset21 (43,866 cells). The cell dataset was clustered according to cell identity. A total of twelve major cell-type classes with marker genes were identified (Supplementary Fig. 8A–D), including around 25% SC-β cells across SC-islets dataset (Supplementary Fig. 8B). To maintain condition across PKM1-non-expressing and -expressing cells more consistent, only PKM1-overexpressing islets collected at S7D1 and S7D14 were integrated with human adult islets for downstream analysis and categorized into four groups based on time of origin and PKM1-overexpression (6,264 cells): S7D1PKM1−, S7D1PKM1+, S7D14PKM1−, and S7D14PKM1+ (Fig. 6A). Compared to S7D1PKM1-, as expected, extended culture of SC-islets to S7D14PKM1− showed a more mature β-cell gene profile (INS, IAPP, HOPX) accompanied by higher gene expression related to insulin secretion (PCSK1, FXYD2, CPE, RBP4, FFAR1, PCSK1N, TTR)1 and OXPHOS pathway (Aerobic respiration and respiratory electron transport) (Fig. 6B and Supplementary Data 6), most of which were recapitulated by the comparison between S7D1PKM1+ and S7D14PKM1+ (Fig. 6C and Supplementary Data 6). Notably, S7D1PKM1+, compared to S7D1PKM1-, induced similar transcriptional changes and enriched pathways, including aerobic respiration and respiratory electron transport pathway (Fig. 6D and Supplementary Data 6), which were further demonstrated in the comparison between S7D14PKM1− and S7D14PKM1+ (Fig. 6E and Supplementary Data 6). As shown in (Fig. 6F), genes related to β cell identity, insulin secretion, glycolysis, OXPHOS, and exocytosis, such as INS, PCSK1N, PKM, MT-CO1, and VAMP2 showed a trend of culture time- and PKM1-dependent increase. Consistently, PKM expression in adult β-cells, including PKM1 and PKM2, was significantly higher than in non-PKM1-overexpressing cells and comparable to the PKM1-overexpressing group. Taken together, PKM1 overexpression triggered a transcriptional program that supports both β-cell functional maturation.
A Single-cell RNA-seq transcriptomic profiling (10× Genomics) of SC-islets overexpressing PKM1 collected on day 1 of stage 7 (S7D1) and day 14 of stage 7 (S7D14). A UMAP-based embedding projection of an integrated dataset comprising 25,380 SC-derived cells and adult human islet cells, separated by sample origin and clustered by cell type. B–E Differentially expressed (DE) genes between B S7D1PKM− and S7D14PKM− (n = 692, 674cells) C S7D1PKM+ and S7D14PKM+ (n = 39, 129 cells) D S7D1PKM− and S7D1PKM+ (n = 692, 39 cells) E S7D14PKM− and S7D14PKM+ (n = 674, 129 cells) β cell group, including adult and SC-β cells (left). DE genes were submitted to Metascape for enrichment analysis. F DE signature genes related to cell identity, insulin secretion, glycolysis, TCA cycle and OXPHOS, exocytosis, and those downregulated genes (n = 692, 39, 674, 129, 7370 cells). P = 0.00059, <2.22 × 10−16, = 0.00012, = 0.0002, <2.22 × 10−16, <2.22 × 10−16 (INS); P = 1.8 × 10−6, <2.22 × 10−16, = 2.6 × 10−13, = 3 × 10−6, <2.22 × 10−16, <2.22 × 10−16 (PCSK1N); P = 0.00039, = 9.3 × 10−9, = 0.00037, = 7.2 × 10−11 (PKM); P = 0.0074, <2.22 × 10−16, = 4.4 × 10−9, = 0.0044, <2.22 × 10−16, <2.22 × 10−16 (MT-CO1); P = 3.5 × 10−6, <2.22 × 10−16, = 0.043, = 0.0021, <2.22 × 10−16, <2.22 × 10−16 (VAMP2). All data are presented as mean ± SEM, with statistical significance determined using two-tailed Mann–Whitney test with Bonferroni correction for multiple comparisons (B–F). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
Aberrant glucose metabolism has been observed in SC-β cells, but key metabolic bottlenecks that decouples the glucose response from GSIS remain unknown8. Our studies demonstrated that elevated basal PEP, largely due to the lack of PKM1, represents one important metabolic block that contributing to immature β-cell function. While PEP is critical for coupling glycolysis to insulin secretion, excessive basal PEP disrupts glucose responsiveness and impairs SC-β cell maturation. Restoring PKM1 normalizes PEP metabolism and promotes the functional maturation of SC-β cells.
PEP metabolism is essential for GSIS10,28. In mature islets, glucose stimulation elevates PEP levels in a controlled manner29. In contrast, SC-β cells exhibit abnormally high basal PEP, which contributes to elevated basal exocytosis and calcium currents. These findings indicate that SC-β cells are highly exocytosis-competent, but remain unable to properly trigger glucose-stimulated secretion. Previous studies have reported that, at basal glucose, immature SC-β cells exhibit spontaneous action potentials and elevated exocytosis30. Elevated PEP may not only amplify basal exocytosis but also locally increase ATP10, thereby triggering calcium influx even at low glucose. This pre-activation of the triggering machinery can limit the dynamic range of glucose-stimulated responses and even impede maturation. Similarly, PKM1 overexpression, which should enhance glycolytic flux into ATP, actually reduce basal insulin secretion. This can be explained by a difference between global versus local ATP pools9,10,11,12. In SC-β cells lacking PKM1, residual PKM2 activity can metabolize the abnormally high basal PEP to generate localized ATP in microdomains near KATP channels. This local ATP is sufficient to close KATP channels, elevate basal Ca²⁺ influx, and drive inappropriate basal exocytosis. By contrast, PKM1 efficiently consumes PEP, preventing its abnormal accumulation and thereby suppressing the local ATP generation for triggering insulin secretion at low glucose. While PKM1 also increases pyruvate flux into mitochondria and elevates basal mitochondrial ATP, this global ATP pool may be less effective at triggering secretion than PEP-driven local ATP at low glucose.
Despite promoting pyruvate flux into TCA, enhancing OXPHOS-related gene expression, and lowering basal PEP, PKM1 overexpression in sorted SC-β cells does not fully increase ATP or insulin secretion at high glucose. This reflects a persistent mitochondrial bottleneck: TCA cycle flux and OXPHOS remain limited, so extra pyruvate cannot be fully converted into ATP. As a result, PKM1 primarily expands the dynamic range of GSIS by lowering basal insulin secretion rather than increasing insulin secretion at high glucose. Thus, PEP accumulation represents one, but not the only, block in the metabolic network. A previous study identified a metabolic bottleneck at the GAPDH and PGK1 steps4. However, in our study, GAPDH and PGK activity showed only a slight decrease. Additionally, the glucose-dependent increase in 3-PG/2-PG was still maintained. Similar to PEP metabolism, pyruvate metabolism also exhibited elevated basal pyruvate level and loss of glucose-stimulated increase. Pyruvate cycling is essential in insulin secretion23,31,32. Recent study reports that pyruvate metabolism is also mediated by LDHB, which is required for inhibiting basal insulin secretion33. However, LDHB level is much lower in SC-islets (Fig. 6F), indicative of a pyruvate metabolic bottleneck. Therefore, functional immaturity of SC-β cells is likely a consequence of multiple metabolic bottlenecks at both glycolytic and mitochondrial levels5.
Unlike mouse islets, SC-β cells appear permeable to several metabolites, including PEP and OAA, consistent with previous studies17. High concentrations of these metabolites elicit significant insulin secretion and calcium responses, which are blocked by inhibitors of PK, mitochondrial metabolism, and KATP channels, indicating that the metabolites enter SC-β cells and are metabolized intracellularly. Although the specific transporter for PEP and OAA remains unknown, several reports indicate that PEP can cross the plasma membrane in specific cell types, including red blood cells34 via an anion-transport protein, as well as human mesenchymal stem cells35 and Th17 cells36, where it can influence differentiation. This suggests there might be a transporter particularly active in immature SC-islets but not in mature islets, which could potentially be exploited for metabolic programming. It is worth noting that significant depolarization was observed only at 5 mM and 20 mM PEP, concentrations that exceed physiological levels but can serve as a metabolic substrate to probe β cell function. However, the actual amount of PEP entering the cell under these concentrations remains unknown, and it is possible that only a fraction is internalized and contributes to depolarization. Notably, extracellular 50 \({{\rm{\mu }}}{{\rm{M}}}\) PEP, which cannot trigger obvious depolarization, was sufficient to induce transcriptional changes that are unfavorable for β-cell maturation. Although this concentration cannot elicit overt depolarization, its effects may still be mediated through disrupted Ca²⁺ handling and signaling37. PEP may increase basal Ca²⁺ levels, pushing cells closer to the threshold for depolarization. Consistently, PEP upregulated calcium- and potassium-channel genes (CACNA1E, CACNA2D1, CACNA2D3) while downregulating OxPhos and glucose metabolism pathways. Thus, PEP may engage both metabolic/transcriptional mechanisms and a feedforward Ca²⁺-dependent pathway to influence β cell maturation. Glucose-stimulated calcium influx is not only a hallmark of functional maturation in β cells but also a crucial event in regulating β cell maturation16,38. During pancreatic β cell maturation, the calcium influx induced by glucose stimulation precedes the acquisition of insulin secretion ability39. As glucose metabolism matures, proper glucose metabolism triggers calcium influx, activating the calcium-dependent phosphatase Calcineurin (CaN). This activation promotes the dephosphorylation and nuclear translocation of nuclear factor of activated T cells (NFAT), which binds to β cell-specific DNA and regulates transcription of specific genes, promoting insulin vesicle formation and functional maturation in β cells40,41,42. However, excessive and sustained calcium influx can delink NFAT transcriptional activity, leading to β cell dedifferentiation43. Therefore, calcium mishandling by PEP in SC-β cells may be a key factor impeding functional maturation.
Although PKM1 overexpression in SC-β cells enhanced glucose-stimulated responses—calcium entry, insulin secretion, oxygen consumption, and exocytosis, this effect should not be solely due to PKM1’s enzymatic role in restoring glycolysis-anaplerosis coupling. Given that the transcriptional profile and glucose-stimulus coupling in SC-β cells are still far from those of primary pancreatic β cells, attempts to restore glucose response by acutely fixing one or two metabolic blocks are unlikely to succeed4. We also failed to elicit significant changes of insulin secretion with acute treatment of PKM2 agonist, consistent with previous study17. Additionally, although FBP seemed to enhance insulin secretion in PKM1-overexpressing islets, this effect was likely due to enhanced functional maturation rather than PKM2 activation, as acute exposure to TEPP-46 did not produce a significant effect and the precise mechanism by which FBP modulated insulin secretion remains unknown. Unlike short-term exposure, chronic TEPP-46 treatment has been shown to enhance functional maturation of SC-β cells17. In fact, long-term PKM1 overexpression reversed the functional impairment induced by PEP and improved the functional state of SC-β cells by upregulating genetic programs related to cell identity, glycolysis, and OXPHOS. Therefore, rather than by just reconnecting glycolysis-anaplerosis coupling, constituent activation of PK activity, either through long-term PKM2 activation or PKM1 restoration, primarily enhances glucose response by promoting functional maturation via regulating PEP metabolism. Previous studies have individually deleted PKM1 or PKM2 to assess their effects on β cell function11. However, these effects appeared to be modest. It would be intriguing to investigate the dual deletion of both PKM1 and PKM2 to determine how this impacts PEP levels as well as the functional maturation of SC-β cells.
Interestingly, SC-islets with PKM1-overexpression showed a higher proportion of SC-α cells. During extended differentiation, some progenitor-like populations, including SC-ECs, may progressively adopt an α cell fate (Supplementary Fig. 8B)44. Our data further suggest that altered PEP metabolism and PKM1 activity may influence α cell differentiation (Fig. 5A) and function9. Sorted β cells overexpressing PKM1 primarily showed reduced basal insulin secretion without a notable increase in glucose-stimulated response. Therefore, enhanced insulin secretion in whole SC-islets at high glucose may reflect indirect effects of PKM1 on other cell types, including α cells, where altered glucagon secretion could further modulate β cell function45. PKM1 appears to regulate insulin secretion in SC-islets not only through direct modulation of β cell glycolytic and metabolic fluxes, but also indirectly via α cell–mediated mechanisms. Further investigation is needed to elucidate the specific role of PKM1 in α cell function.
In summary, PEP accumulation disrupts β-cell function by elevating basal calcium levels, thereby suppressing GSIS, decoupling glycolysis from OXPHOS, and inhibiting β-cell maturation. PKM1 overexpression enhances GSIS through a dual mechanism: restoring the glycolysis-OXPHOS linkage and promoting β-cell functional maturation (Supplementary Fig. 9). This suggests that PKM1 plays a crucial role in facilitating β-cell maturation and insulin secretion through metabolic regulation. Importantly, this highlights the potential of targeting PKM1 as a strategy to improve the functional maturation of stem cell-derived β-cells for cell replacement therapy in diabetes, and to optimize β-cell function in disease contexts. This study has several limitations. First, due to the scarcity of human islets, not all experiments could include human islets as controls. Although species-specific differences remain an important consideration, mouse islets offer a reproducible and controlled model system and exhibit many structural and functional similarities to human islets. Second, while PKM1 overexpression enhances the functional maturation of SC-β cells, the exact mechanisms by which PKM1 and PEP influence SC-β functional maturation remain unknown. Identifying these mechanisms is essential for advancing our understanding.
Methods
Cell culture and differentiation of SC-β cells
In vitro differentiation of H1 and H9. H1 or H9 human embryonic stem cells (hESC) were cultured on 1:100 diluted Matrigel (Corning, 354277) in mTeSR medium (Stemcell, 85850). After rinsing with Mg2+/Ca2+-free DPBS (Thermo Fisher, 14190250) and digestion with TrypLE Express Enzyme (1×) (Thermo Fisher, 12605028) for 3 min at 37 °C, the digestion was stopped with mTeSR. Single cells were collected and spun at 300 × g for 5 min. The cell pellet was resuspended in mTeSR medium supplemented with Y-27632 (10 μM; MCE, HY-10583), and single cells were seeded at approximately 1 × 105 cells/well on Matrigel-coated surfaces. Cells were fed with mTeSR medium daily and differentiated after 48 h at approximately 90% confluency. The differentiation protocol was modified based on previous protocols2,19 (see also Supplementary Data 1). The Institutional Review Board of Guangzhou National Laboratory and People’s Hospital of Xinjiang Uygur Autonomous Region reviewed and approved all work involving human pluripotent stem cells carried out in this manuscript.
S1: Definitive endoderm (3d). Undifferentiated pluripotent stem cells plated on 1:100 Matrigel-coated surfaces were cultured in MCDB 131 medium (Reprocell, PM151210) with 0.5% BSA (fatty acid-free BSA, Proliant, 69760), 1.7 × g/l sodium bicarbonate (SIAL, S6014), 10 mM final glucose concentration (Sigma, G7528), 0.25 mM Vitamin C (Sigma, A1300000), 1% Glutamax (Thermo Fisher Scientific, 35050061), 1% Penicillin-Streptomycin (Thermo Fisher Scientific, 15140122), 100 ng/ml Activin A (MCE, HY-P70311), 3 μM ChIR99021 (MCE, HY-10182), 50 nM PI-103 (Selleck, S1038), and 10 μM Y-27632 for day 1 only. For day 2, cells were cultured in MCDB131 with 0.5% BSA, 1.7 × g/l sodium bicarbonate, 10 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), 50 ng/ml Activin A, and 0.1 μM ChiR99021. On day three, cells were cultured in MCDB with 0.5% BSA, 1.7 × g/l sodium bicarbonate, 10 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), and 50 ng/ml Activin A.
S2: Primitive gut tube (2d). S1 cells were transferred to MCDB 131 medium containing 0.5% BSA, 1.7 × g/l sodium bicarbonate, 10 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), and 50 ng/ml FGF7 (Stemcell, 78046.2) for 2 days.
S3: Posterior foregut (2d). S2 cells were cultured for an additional 2 days in MCDB 131 medium supplemented with 2% BSA, 2.5 × g/l sodium bicarbonate, 10 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), 1:200 ITS-X (Reprocell, pb180431), 50 ng/ml FGF7, 2 μM retinoic acid (Sigma, R2625), 0.25 μM SANT-1 (MCE, HY-100224), 100 nM LDN193189 (BMP receptor inhibitor, MCE, HY-12071), 500 nM PdBu (Millipore, 524390), and 10 μM Y-27632.
S4: Pancreatic endoderm (4d). S3 cells were further differentiated in MCDB 131 medium supplemented with 2% BSA, 2.5 × g/l sodium bicarbonate, 10 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), 1:200 ITS-X, 2 ng/ml FGF7, 2 μM RA, 0.25 μM SANT-1, 100 nM LDN193189, 100 nM PdBu, 10 μM Y27632, 10 μM ALK5 inhibitor II (Selleck, s7223), 100 ng/ml EGF (MCE, HY-P7109), and 10 mM Nicotinamide (Sigma-Aldrich, N0636) for 4 days.
S5: pancreatic endocrine precursors (3d). On the last day of S4 culture, cells were treated with accutase (Stemcell, 7920) for 8 min at 37 °C, followed by gentle pipetting to break into single cell suspension. Next, cell suspension were transferred into low-attached 6-well culture plate in suspension at 300 × g with S5 medium, which consisted of MCDB 131 medium with 2% BSA, 2 × g/l sodium bicarbonate, 20.6 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), 1:200 ITS-X, 0.05 μM RA, 0.25 μM SANT-1, 100 nM LDN193189, 10 μM zinc sulfate (ZnSO4, Sigma, Z0251), 10 μM ALK5 inhibitor II, 1 μM T3 (MCE, HY-A0070), 1 μM compound E(MCE, HY-14176), 20 ng/ml Betacellulin (Stemcell, 78105), 10 μg/ml Heparin (Sigma, H3149), 10 μM Isoxazole 9 (ISX9, MCE, HY-12323) and 100 nM Wnt-C59 (MCE, 1243243-89-1) for 3 dads. For lentivirus infection, mCherry or PKM1-mCherry lentivirus were added at S5D1, achieving stable PKM1 expression at S5D3. After 48 h, lentivirus media was replaced with new S5 media.
S6: S6 cells, characterized by NKX6.1 and insulin expression, were cultured for 5 days. The culture medium consisted of MCDB 131 supplemented with 2% BSA, 1.8 × g/l sodium bicarbonate, 20.6 mM glucose, 0.25 mM Vitamin C, 1% Glutamax, 1% Pen-Strep (Gibco), 1:200 ITS-X, 100 nM LDN193189, 10 μM ZnSO4, 10 μM ALK5 inhibitor II, 1 μM T3, 0.1 μM compound E, 10 μg/ml Heparin, 100 nM Wnt-C59, and 1 mM N-acetylcysteine (Sigma, A9165) for 5 days. For PEP treatment, 5 mM PEP was added to media and media pH was corrected with NaHCO3 as that of S6 media.
S7: S7 cells, characterized by NKX6.1, insulin, and MAFA expression, were cultured for 14-21 days. The culture medium consisted of MCDB 131 supplemented with 2% BSA, 1.5 × g/l sodium bicarbonate, 5.6 mM glucose, 1% Glutamax, 1% Pen-Strep (Gibco), 1:200 ITS-X, 10 μM ZnSO4, 10 μM ALK5 inhibitor II, 1 μM T3, 10 μg/ml Heparin, 1 mM N-ace, 10 μM Trolox (Sigma, T3251), 1:2000 Trace element A (Corning, 25-021-CI), 1:2000 Trace element B (Corning, 25-022-CI), 2 μM R428 (SelleckChem, S2841), 1:2000 chemically defined lipid concentrate (lipid, Gibco, 11905031), and 10 μM Forskolin (Aladdin, 66575-29-9).
Generation of reporter cell line
H1-Ins-EGFP wase generated as previously reported for H1-Ins-jGCaMP7f46. We utilized low off-target CRISPR/Cas9n technology for in situ insertion of the EGFP gene in hESC (H1). By employing the Cas9 D10A nickase (PX335, Addgene #42335) with dual sgRNAs targeting each side of the insulin gene stop codon, we inserted P2A-EGFP-PGK-Puro-pA. This insertion enables simultaneous insulin protein expression for identifying insulin-producing cells in differentiated pancreatic endocrine cells. Cells were transfected at 70% confluence using Lipofectamine™ Stem Reagent (STEM00008), with puromycin added after 24 h at 1 µg/ml to select for successful insertions, ensuring elimination of non-transfected cells.
Generation of lentivirus
All overexpression lentiviral plasmids were constructed via homologous recombination. During the construction process, the following steps were undertaken: 1. puc57 was consistently used as the plasmid backbone; 2. The lentiviral integration site from the lentiCRISPR v2 plasmid (addgene: #52961) was utilized; 3. The EF1a promoter was employed to enhance gene expression, followed by the target gene’s ORF (with the stop codon removed), linked to P2A and mCherry for monitoring plasmid transfection efficiency and viral overexpression in 293 T cells; 4. All plasmids were recombined using the EZ-HiFi Seamless Cloning Kit (Genstar #T196), and the accuracy of the sequences was verified by DNA sequencing. Pspax2, VSVG, and PLV were mixed in a ratio of 1:0.5:1 for transfection of 293 T cells using PEI. 12 h post-transfection, the medium was replaced with DMEM containing 5% FBS and antibiotics. At 48- and 72-h post-transfection, the supernatants were collected, centrifuged at 2000 rcf for 10 min to remove cell debris, sterile filtered through a Millex 0.22 µm filter, and the viruses were finally concentrated using an Amicon® Ultra filter (#UFC9100).
13C-glucose labeling metabolomics
All animal studies were conducted with approval from Institutional Animal Care and Use Committee of Guangzhou Medical University. ~400 primary mouse islets or SC-islets were used for each replicate with at least 3 biological replicates and preincubated at KRB solution with 2.8 mM glucose for 60 min, followed by incubation at 2.8 (low) or 16.7 mM (high) [U-¹³C₆] glucose (Cambridge Isotope Laboratories, CLM 1396) for 60 min at 37 °C and 5% CO2. Afterwards, islets were washed with PBS 3 times and added 50 μL “extraction solution” (Acetonitrile/Methanol/Aqueous = 4:4:2 kept at −20 °C overnight. The mixture was kept in ice to sonicate for 5 min followed by future centrifugation at 12,000 × g for 5 min. Supernatant was transferred to vials and run with 4 μL injection volume by LC-MSMS system. Chromatographic separation was performed using an Agilent 1290II ultra-high-pressure liquid chromatography system equipped with 6546 quadrupole time-of-flight (QTOF) mass spectrometry. A Waters ACQUITY UPLC BEH Amide column (2.1 mm × 100 mm × 1.7 μm) and guard column (2.1 mm × 5 mm × 1.7 μm) at 35 °C was used to separate metabolites with mobile phase A: 100% aqueous containing 15 mM ammonium acetate and 0.3% ammonium hydroxy and mobile phase B: 90% acetonitrile (v/v) aqueous containing 15 mM ammonium acetate and 0.3% ammonium hydroxy. The linear gradient was set as follows: 10% A (0.0–8.0 min), 50% A (8.0–10.0 min), 50% A (10.0–11.0 min), and 10% A (11.0–20.0 min). The total run time was 20 min and flow rate was 0.3 mL/min. The mass spectrometer was equipped with Agilent Jet-stream source operating in negative and positive ion mode with source parameters set as follow: Nebulizer gas, 45 psi; Sheath gas temperature, 325 °C; Sheath gas flow, 10 L/min; Dry gas temperature, 280 °C; Dry gas flow, 8 L/min; Capillary voltage, 3500 V for two ion modes and nozzle voltage, 500 V for positive and 1000 V for negative mode. The QTOF scan parameters were set as follows: Scan speed, 1.5 scan/s; scan range, 50–1700 m/z and ion fragmentor voltage, 140 v. The acquired data quality was monitored by Amino Acid standards (Merck, Sigma-Aldrich Production GmbH, Switzerland), mix-samples quality controls, and blanks. Peak integration and metabolite isotopologue identification was accomplished using Profinder 10.0 (Agilent). Nature abundance was assayed using non-labelled samples and removed to avoid any possible confounding effect.
Patch clamp analysis
Patch Clamp experiment was performed as previously reported47,48. Briefly, dispersed single cells cultured on 35 mm dishes overnight were preincubated at 2.8 mM glucose for 1 hour and patched in bath solution containing (in mM): 118 NaCl, 5.6 KCl, 20 TEA, 1.2 MgCl2, 2.6 CaCl2, 5 HEPES at indicated glucose level with a pH of 7.4 (adjusted by NaOH) at 32–35 °C. Whole-cell capacitance was recorded with the sine+DC lock-in function of an EPC10 amplifier and PatchMaster software (HEKA Electronics). Exocytotic responses and inward Ca2+ currents were measured 1–2 min after obtaining the whole-cell configuration in response to ten 500 ms depolarizations to 0 mV from a holding potential of −70 mV. Changes in capacitance and integrated Ca2+ charge entry were normalized to cell size (fF/pF and pC/pF, respectively). The intracellular solution contained (in mM): 125 Cs-Glutamate, 10 CsCl, 10 NaCl, 1 MgCl2, 5 HEPES, 0.05 EGTA, 3 MgATP, and 0.1 cAMP with pH = 7.15 (pH adjusted with CsOH). For dialysis/infusion experiment, metabolites were added to pipette solution (pH-corrected) as indicated. Measurement was performed 3 min after whole cell mode, which achieved homogenous distribution of metabolites. SC-β cells were identified by EGFP fluorescent signal. Mouse β-cells were identified by size ( > 4 pF) and a Na+ channel half-maximal inactivation at around −90 mV.
Enzymatic activity measurement
PK activity was measured using the CheKine™ Micro PK Assay Kit (KTB1120) according to the manual. PK catalyzes phosphoenolpyruvate and ADP reaction into ATP and pyruvate, and lactate dehydrogenase further catalyzes the production of lactic acid and NAD + by NADH and pyruvate. The rate of NADH decline at 340 nm can reflect PK activity. Cell lysates from 400 islets were sonicated on ice using the provided Extraction Buffer and quantified by BCA assay. 10 μL sample, 10 μL Working Reagent and 180 μL Substrate Mix Working Reagent were mixed and added to the 96-well UV plate. Measure the absorbance value at 340 nm with a microplate reader. The data shown represent the first ten data points of NADH consumption in this reaction.
PGK1 activity was measured using the Grace Biotech Micro PGK Assay Kit (G0885W) according to the manual. Phosphoglycerate kinase (PGK) catalyzes the reaction between 3-PG and ATP to produce 1,3-bisphosphoglycerate and ADP. In a subsequent reaction, 1,3-bisphosphoglycerate is catalyzed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reacts with NADH to generate 3-phosphoglyceraldehyde, NAD⁺, and inorganic phosphate. Since NADH absorbs light at 340 nm, the decrease in NADH concentration during the reaction results in a reduction in absorbance at this wavelength. This absorbance change at 340 nm can be used to monitor PGK activity. Cell lysates from 400 islets were sonicated on ice using the provided Extraction Buffer and quantified by BCA assay. 20 μL sample was mixed with the Working Reagent added to the 96-well UV plate. After 10 minutes, measure the absorbance value at 340 nm with a microplate reader. The data shown represent the first ten data points of NADH consumption in this reaction.
GAPDH activity was determined using the Abcam GAPDH colorimetric assay kit (ab204732) as previously reported4. This assay tracks the conversion of the substrate glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate along with NADH production. NADH, generated by GAPDH activity present in cell lysates, is linked to a proprietary developer included in the kit, which utilizes the NADH from GAPDH to initiate a secondary colorimetric reaction in each well. Cell lysates from 400 islets were sonicated on ice using the provided Extraction Buffer and quantified by BCA assay. After adding 50 µL sample into the reaction mix, the plate was incubated at 37 °C for the assay duration. NADH accumulation was indirectly measured at 450 nm by monitoring the developer reaction product in kinetic mode over a 10-min period. The data presented display the raw conversion values for the first 10 time points of the reaction.
Oxygen consumption
The Agilent Seahorse XFE24 Analyzer with islet capture microplates was used, following a protocol previously outlined49. Specifically, 70 islets were loaded per well and treated sequentially with 2 mM glucose, 20 mM glucose, 5 µM oligomycin, 5 µM FCCP, and 5 µM rotenone/antimycin A. For each data point, the experiment included a 3-min mixing phase, a 2-min waiting phase, and a 3-min measurement phase. Each biological sample had 3 ~ 4 technical replicates, and the oxygen consumption rate was normalized islet protein content.
scRNA sequencing analysis
Briefly, approximately 10,000 cells were captured, reverse-transcribed, barcoded, and amplified50. cDNA libraries were prepared using 10× Genomics Chromium Single Cell Reagent Kits v3 or DNBelab C Series High-throughput Single-Cell RNA Library Preparation Set V3.0 (MGI, #940-001818-00), quantified with Qubit 3.0, analyzed on Agilent 2100, and sequenced on the Illumina NovaSeq 6000 (PE150) by Annoroad Gene Technology or high-throughput DNBSEQ-T7RS sequencer at the Advanced Cell Technology Core Facility, Guangzhou National Laboratory (Guangzhou, China) with read length 30 bp for read1, 100 bp for read2, and 10 bp for sample index. Raw FASTQ files were processed with Cell Ranger (v4.0.0) (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines /latest/advanced/references) using default settings, aligning reads to the GRCh38 human genome. We included 12 samples of primary adult human pancreatic islets as reference scRNAseq dataset for further analysis21. High quality cells ( > 300 genes, <10% mitochondrial genes, <3% red blood cell gene expression) were selected for data normalization, scaling, and principal component analysis with default methods. Top 3000 variable genes were identified from each sample. The Harmony method was applied on the first 30 principal components, using sample as a covariate to integrate different samples51. The integrated principal components were used to construct the UMAP, identify neighboring cells (via shared nearest neighbor), and define cell clusters using default Seurat methods. The clustering resolution was adjusted to 0.5. Differentially expressed genes among clusters were identified using Seurat’s FindMarkers function. The annotation of the clusters was performed using the GPTCelltype package according to the known marker genes52. Adult and SC- β cells were used for identifying differentially expressed gene with an adjusted P value < 0.005 and log2 fold change >0.15. We performed gene set enrichment analysis using Reactome53 and Metascape v.3.554 with an adjusted P value cutoff of <0.05. Detailed analytical result related to differentially genes and enrichment can be seen in Supplementary Data 2–6.
Fluorescence activated cell sorting
For SC-β (H1-Ins-EGFP) cell sorting, cell aggregates were dissociated into a single-cell suspension using Accutase and resuspended in S7 media containing 10 μM Y27632. EGFP-positive cells were sorted into 15 ml Falcon tubes containing S7 medium and 10 μM Y27632 using a tube sorting mode (MA900, Sony).
Magnetic sorting of SC-β cells (CD49a⁺) cells
SC-β cells were sorted as previously described55. Briefly, organoid spheres were dissociated into single cells by Accutase (37 °C, 20–30 min) with gentle pipetting. Cells were collected by centrifugation (300 g, 5 min). The pellet was resuspended in 10 mL sorting buffer (DPBS supplemented with 1% BSA and 2 mM EDTA) and passed through a 37-µm cell strainer to remove residual aggregates. Approximately 1 × 107 cells were obtained, counted, and pelleted for downstream sorting. Cells were resuspended in sorting buffer at 3.33 × 107 cells/mL and incubated with anti-human CD49a-PE antibody (1:100 dilution) for 20 min at room temperature in the dark, with gentle mixing. After three washes, cells were incubated with anti-PE UltraPure magnetic beads (40 µL per 1 × 107 cells) for 15 min at 4 °C with intermittent mixing. Cells were washed three additional times with sorting buffer, counted, and resuspended at 5–6 × 107 cells/mL. An MS column (Miltenyi Biotec) was pre-equilibrated with 0.5 mL sorting buffer and placed in a magnetic separator prior to cell loading. The labeled cell suspension was applied to the column, followed by three washes with 1 mL sorting buffer, allowing the liquid to flow through completely each time. The column was then transferred to a clean 15-mL tube, and CD49a⁺ cells were eluted with 1 mL sorting buffer using the supplied plunger. Eluted cells were counted, centrifuged, and resuspended in S7 medium. For reaggregation, 1.5 × 106 cells per well were seeded into AggreWell™ 400 plates (24-well format; STEMCELL Technologies), yielding ~1200 reaggregated spheres per well.
Quantitative RT-PCR
RNA was extracted with the RNeasy Mini Kit (Qiagen) and treated with DNase. cDNA was synthesized using the RT reagent Kit with gDNA Eraser (TAKARA). Real-time qPCR was performed with SYBR Premix Ex TaqII (TAKARA) and analyzed using the ΔΔCt method, normalizing expression to GAPDH. Primer details are in Supplementary Data 7.
Western blotting
SC-islets from S7D14 and mouse islets were collected in RIPA lysis buffer with protease inhibitor (Beyotime). Lysates were centrifuged, and supernatants were separated by SDS-PAGE, transferred to PVDF membranes, blocked, and incubated with primary and HRP-conjugated secondary antibodies. Detection was performed using ECL reagent (Millipore) and captured on a G-Box. Protein quantification was done with ImageJ, normalizing target protein intensity to housekeeping proteins (beta-actin). Antibodies are detailed in Supplementary Data 8.
Immunohistochemistry
For cryo-sectioning, S7 organoids were fixed in 4% PFA (Sangon Biotech) at room temperature for 1 h, rinsed with PBS, and incubated overnight at 4 °C in 30% sucrose. They were then embedded in OCT, snap-frozen in liquid nitrogen, and stored at −80 °C. Sections were cut at 10 μm thickness and mounted on Superfrost Plus slides. For immunostaining of re-plated cells, SC-islets were dissociated, plated on Matrigel-coated confocal dishes and fixed with 4% PFA at 4 °C overnight. Cells were then permeabilized with Quickblock™ Blocking Buffer (BEYOTIME) at room temperature for 1 h. Primary antibodies were incubated overnight at 4 °C, followed by PBS washes, and staining with secondary antibodies and Hoechst 33342 (Thermo Fisher Scientific) for 2 h at room temperature. Imaging was done on a NIKON A1 confocal microscope. Antibody information is listed in Supplementary Data 8. For Paraffin-embedded islet sections immunostaining, the procedure were performed as previously described47. Donor information is listed in Supplementary Data 9.
Insulin secretion
For dynamic insulin secretion, 25 (mouse islets) or (SC-islets) were handpicked into home-made organoid chip for dynamic insulin secretion as previously described56. Islets were pre-perifused for 1 h at KRB solution with 2 mM or 2.8 mM glucose followed by stimulation of different levels of glucose, permeable metabolites and drugs at a sample collection rate of 10 ml/min every 3–6 min. For static insulin secretion, 15 islets per triplicates were preincubated with 2.8 mM for 1 h followed by stimulation of indicated conditions for 1 h. The pH of all solutions containing metabolites or drugs was adjusted to 7.4 using NaOH. D-mannitol (Aladdin Scientific, Shanghai, China) was used to balance the osmolarity of KRB solutions as previously reported57. The samples and lysates of islets were stored at −80 °C until further assayed by Insulin Detection Kit (STELLUX® Chemi Rodent Insulin ELISA kit; STELLUX® Chemi Human Insulin ELISA kit, Alpco). The total Na⁺ concentration in the KRB solution was adjusted to its original value by including the sodium contributed by sodium salts of metabolites (e.g., Na⁺-PEP) and NaOH used for correcting pH at 7.4.
Information of metabolites and drug is listed in Supplementary Data 10.
Intracellular Ca2+ and membrane potential imaging
For Ca2+ imaging, islets or dissociated cells cultured on 35 mm glass bottom were loaded with Cal-500 AM (2.5 μM, AAT Bioquest, USA) or Fluo-8AM (2.5 μM, AAT Bioquest, USA) for 30 min and perifused with pH-corrected (NaOH) and osmolarity-balanced KRB solution at indicated conditions. D-mannitol (Aladdin Scientific, Shanghai, China) was used to balance the osmolarity of KRB solutions as previously reported57. The fluorescence signal was excited and detected at an excitation/emission wavelength of 405 nm/500 nm for Cal-500 AM, and at 490 nm/514 nm for both Fluo-8AM cells. Ca2+ were imaged at 1 Hz on an inverted confocal microscope (Carl Zeiss LSM 980) for single cells and an upright confocal microscope (Carl Zeiss LSM 980 NLO) for whole islets58. Mouse β-cells were marked and identified by immunostaining and SC-β cells were identified by using H1-Ins-EGFP for experiment. Fluorescence ratios were calculated using a tool box from Matlab with indicated analytical method (average fluorescence and smooth dynamics)59. The total Na⁺ concentration in the KRB solution was adjusted to its original value by including the sodium contributed by sodium salts of metabolites (e.g., Na⁺-PEP, Na+-FBP) and NaOH used for correcting pH at 7.4. Information of metabolites and drug is listed in Supplementary Data 10.
Statistics and reproducibility
Data analysis was conducted using FitMaster (HEKA Electronik) and GraphPad Prism (v10.0c). Statistical outliers were identified and removed using the unbiased robust regression and outlier identification test. For normally distributed data, statistical comparisons were made using a 2-tailed Student’s t-test (for two groups) or ANOVA followed by Sidak’s multiple comparison test, as specified in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw data and count matrices of scRNA-seq data are available under the accession number GSE281603. Metabolomics data are available under the accession number PRJCA051828. All other data are available from the corresponding authors upon request. Source data are provided with this paper.
Code availability
The code for analyzing 10× Genomics sequencing data is available at Seurat (https://satijalab.org/seurat/). Additional detailed code for specific data analysis steps can be provided upon request.
References
Balboa, D. et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 40, 1042–1055 (2022).
Du, Y. et al. Human pluripotent stem-cell-derived islets ameliorate diabetes in non-human primates. Nat. Med. 28, 272–282 (2022).
Merrins, M. J., Corkey, B. E., Kibbey, R. G. & Prentki, M. Metabolic cycles and signals for insulin secretion. Cell Metab. 34, 947–968 (2022).
Davis, J. C. et al. Glucose response by stem cell-derived β cells in vitro is inhibited by a bottleneck in glycolysis. Cell Rep. 31, 107623 (2020).
Barsby, T. et al. Aberrant metabolite trafficking and fuel sensitivity in human pluripotent stem cell-derived islets. Cell Rep. 42, 112970 (2023).
Haythorne, E. et al. Altered glycolysis triggers impaired mitochondrial metabolism and mTORC1 activation in diabetic β-cells. Nat. Commun. 13, 6754 (2022).
Helman, A. et al. A Nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metab. 31, 1004–1016.e5 (2020).
Lin, H. et al. Glucose metabolism and regulation in establishing human stem cell-derived β cell maturation. Cell Rep. 44, 115892 (2025).
Ho, T., Potapenko, E., Davis, D. B. & Merrins, M. J. A plasma membrane-associated glycolytic metabolon is functionally coupled to KATP channels in pancreatic α and β cells from humans and mice. Cell Rep. 42, 112394 (2023).
Lewandowski, S. L. et al. Pyruvate kinase controls signal strength in the insulin secretory pathway. Cell Metab. 32, 736–750.e5 (2020).
Foster, H. R. et al. β-cell deletion of the PKm1 and PKm2 isoforms of pyruvate kinase in mice reveals their essential role as nutrient sensors for the KATP channel. eLife 11, e79422 (2022).
Merrins, M. J. & Kibbey, R. G. Glucose regulation of β-Cell KATP channels: it is time for a new model! Diabetes. 73, 856–863 (2024).
Rutter, G. A. & Sweet, I. R. Glucose regulation of β-cell KATP channels: is a new model needed? Diabetes. 73, 849–855 (2024).
Corradi, J. et al. KATP channel activity and slow oscillations in pancreatic beta cells are regulated by mitochondrial ATP production. J. Physiol. 601, 5655–5667 (2023).
Satin, L. S., Corradi, J. & Sherman, A. S. Do we need a new hypothesis for KATP closure in β-cells? distinguishing the baby from the bathwater. Diabetes 73, 844–848 (2024).
Mu-u-min, R. B. A., Diane, A., Allouch, A. & Al-Siddiqi, H. H. Ca2+-Mediated signaling pathways: a promising target for the successful generation of mature and functional stem cell-derived pancreatic beta cells in vitro. Biomedicines 11, 1577 (2023).
Wang, C. et al. Cell identity dynamics and insight into insulin secretagogues when employing stem cell-derived islets for disease modeling. Front. Bioeng. Biotechnol. 12, 1392575 (2024).
Zhang, Z. et al. PKM2, function and expression and regulation. Cell Biosci. 9, 52 (2019).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Pagliuca, F. W. et al. Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014).
Xin, Y. et al. Pseudotime ordering of single human β-cells reveals states of insulin production and unfolded protein response. Diabetes 67, 1783–1794 (2018).
Lang, A. L. et al. A defect in mitochondrial complex III but not in complexes I or IV causes early β-cell dysfunction and hyperglycemia in mice. Diabetes 72, 1262–1276 (2023).
Campbell, J. E. & Newgard, C. B. Mechanisms controlling pancreatic islet cell function in insulin secretion. Nat. Rev. Mol. Cell Biol. 22, 142–158 (2021).
Adams, M. T., Gilbert, J. M., Hinojosa Paiz, J., Bowman, F. M. & Blum, B. Endocrine cell type sorting and mature architecture in the islets of Langerhans require expression of Roundabout receptors in β cells. Sci. Rep. 8, 10876 (2018).
Escot, S., Willnow, D., Naumann, H., Di Francescantonio, S. & Spagnoli, F. M. Robo signalling controls pancreatic progenitor identity by regulating Tead transcription factors. Nat. Commun. 9, 5082 (2018).
Yang, Y. H. C., Manning Fox, J. E., Zhang, K. L., MacDonald, P. E. & Johnson, J. D. Intraislet SLIT–ROBO signaling is required for beta-cell survival and potentiates insulin secretion. Proc. Natl. Acad. Sci. USA 110, 16480–16485 (2013).
Schmidt, M. D., Ishahak, M., Augsornworawat, P. & Millman, J. R. Comparative and integrative single cell analysis reveals new insights into the transcriptional immaturity of stem cell-derived β cells. BMC Genomics 25, 105 (2024).
Stark, R. et al. Phosphoenolpyruvate cycling via mitochondrial phosphoenolpyruvate carboxykinase links anaplerosis and mitochondrial GTP with insulin secretion*. J. Biol. Chem. 284, 26578–26590 (2009).
Sugden, M. C. & Ashcroft, S. J. H. Phosphoenolpyruvate in rat pancreatic islets: a possible intracellular trigger of insulin release?. Diabetologia 13, 481–486 (1977).
Bayly, C. L. et al. An INSULIN and IAPP dual reporter enables tracking of functional maturation of stem cell-derived insulin producing cells. Mol. Metab. 89, 102017 (2024).
Lu, D. et al. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS). Proc. Natl. Acad. Sci. USA 99, 2708–2713 (2002).
Ronnebaum, S. M. et al. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J. Biol. Chem. 281, 30593–30602 (2006).
Cuozzo, F. et al. LDHB contributes to the regulation of lactate levels and basal insulin secretion in human pancreatic β cells. Cell Rep. 43, 114047 (2024).
Hamasaki, N., Hardjono, I. S. & Minakami, S. Transport of phosphoenolpyruvate through the erythrocyte membrane. Biochem. J. 170, 39 (1978).
Jeong, G.-J. et al. Metabolites can regulate stem cell behavior through the STAT3/AKT pathway in a similar trend to that under hypoxic conditions. Sci. Rep. 9, 6112 (2019).
Huang, T.-Y. et al. Phosphoenolpyruvate regulates the Th17 transcriptional program and inhibits autoimmunity. Cell Rep. 42, 112205 (2023).
Wang, S.-Q. Ca2+ sparks and beyond. Biophys. Rep. 10, 257–258 (2024).
Wortham, M. & Sander, M. Transcriptional mechanisms of pancreatic β-cell maturation and functional adaptation. Trends Endocrinol. Metab. 32, 474–487 (2021).
Sanavia, T. et al. Temporal transcriptome analysis reveals dynamic gene expression patterns driving β-cell maturation. Front. Cell Dev. Biol. 9, 648791 (2021).
Goodyer, W. R. et al. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev. Cell 23, 21–34 (2012).
Zhao, J. et al. In vivo imaging of β-cell function reveals glucose-mediated heterogeneity of β-cell functional development. eLife https://doi.org/10.7554/eLife.41540 (2019).
Miranda, J. G. et al. Dynamic changes in β-cell [Ca2+] regulate NFAT activation, gene transcription, and islet gap junction communication. Mol. Metab. 57, 101430 (2022).
Darden, C. M. et al. Calcineurin/NFATc2 and PI3K/AKT signaling maintains β-cell identity and function during metabolic and inflammatory stress. iScience 25, 104125 (2022).
Zhu, H. et al. Understanding cell fate acquisition in stem-cell-derived pancreatic islets using single-cell multiome-inferred regulomes. Dev. Cell 58, 727–743.e11 (2023).
Capozzi, M. E. et al. β Cell tone is defined by proglucagon peptides through cAMP signaling. JCI Insight 4, 126742 (2019).
Liu, X. et al. Generation of INS-jGCaMP7f knock-in Ca2+ reporter human embryonic stem cell line, GZLe001-C, using CRISPR/Cas9-based gene targeting. Stem Cell Res. 82, 103633 (2025).
Lin, H. et al. β-Cell knockout of SENP1 reduces responses to incretins and worsens oral glucose tolerance in high-fat diet-fed mice. Diabetes 70, 2626–2638 (2021).
Huo, J. & Yang, H.-Q. Electrophysiological analysis of cardiac KATP channel. Biophys. Rep. 11, 77–86 (2025).
Lin, H. et al. A role and mechanism for redox sensing by SENP1 in β-cell responses to high fat feeding. Nat. Commun. 15, 334 (2024).
Chen, L. et al. Directed differentiation of pancreatic δ cells from human pluripotent stem cells. Nat. Commun. 15, 6344 (2024).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Hou, W. & Ji, Z. Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis. Nat. Methods 21, 1462–1465 (2024).
Milacic, M. et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 52, D672–D678 (2024).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Kelley, A. B. et al. Controlling human stem cell-derived islet composition using magnetic sorting. Biotechnol. Bioeng. 122, 2206–2217 (2025).
Yin, J. et al. Heterogenous glucose-stimulated insulin secretion at single islet level. Eng. Regen. 4, 387–395 (2023).
Gerber, K. M. et al. The capacity to secrete insulin is dose-dependent to extremely high glucose concentrations: a key role for adenylyl cyclase. Metabolites 11, 401 (2021).
Wang, H., Yang, B., Mo, L. & Yang, H.-Q. Intracellular calcium imaging for agonist screening. Biophys. Rep. 11, 164–171 (2025).
Romano, S. A. et al. An integrated calcium imaging processing toolbox for the analysis of neuronal population dynamics. PLoS Comput. Biol. 13, e1005526 (2017).
Acknowledgements
We thank the Proteomics and Metabolomics Platform, Guangzhou Laboratory for liquid chromatography-time-of-flight mass spectrometry (Q-TOF), and Nannan Wang for assistance with data collection and analysis. We also thank the Advanced Cell Technology Core Facility, Guangzhou National Laboratory for our Single Cell Preparation and Sequencing, and Ping Sun for assistance with data collection. We are grateful to Suli Li from People’s Hospital of Xinjiang Uygur Autonomous Region for procuring human donor pancreatic sections for research approved by People’s Hospital of Xinjiang Uygur Autonomous Region Ethics Board (KY2025101501). This work was supported by grants from the National Key Research and Development Program of China (2021YFA1101300 to Huisheng Liu), National Natural Science Foundation of China (32400682 to Haopeng Lin, 82200884 to Zonghong Li and 32570979 to Huisheng Liu), the High-Level Talent Program of Guangzhou Medical University (06-445-1247 to Haopeng Lin), Guangdong Province High-level Talent Youth Project (2021QN02Y939 to Zonghong Li), R&D Program of Guangzhou Laboratory (SRPG22-021 to Huisheng Liu), Guangdong Basic and Applied Basic Research Foundation (2023A1515010483 to Lihua Chen), the Young Scientists Program of Guangzhou National Laboratory (QNPG23-02 to Lihua Chen and QNPG23-10 Jiaxiang Yin), the National Key Research and Development Program of China (2020YFA0908200 to Huisheng Liu), Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01D51 to Yanying Guo) and Special Funds of the Government to Guide Local Science and Technology Development (ZYYD2024CG14 to Yanying Guo).
Author information
Authors and Affiliations
Contributions
H.S.L., Y.Y.G., Z.H.L., and T.X. conceived and supervised the study. H.P.L., X.L., D.Q.C., F.Z., W.P., and L.H.C. designed and performed experiments, data analysis, and interpretation. Z.H.D. performed the paraffin sectioning and immunostaining. Y.L.J. performed the cell differentiation and CD49a magnetic sorting. K.J. and P.X. performed single cell transcriptomics analyses and interpretation. Q.F.D. and J.W.Y. participated in experiments related to stem differentiation, gene expression measurement, and calcium imaging. J.X.Y. and Z.R.B. designed the perifusion apparatus for dynamic insulin secretion and calcium imaging. F.Z., D.Q., X.L., and T.R.Z. established the reporter cell lines. D.Q.C., X.L., and X.X.X. participated in mouse management and islet isolation. H.S.L. and H.P.L. wrote the paper, with contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Matthew Merrin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, H., Liu, X., Zhang, F. et al. Restoration of PKM1 improves functional maturation of human stem-cell derived-β cell by regulating PEP metabolism. Nat Commun 17, 914 (2026). https://doi.org/10.1038/s41467-025-67635-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67635-2