Abstract
Chimeric antigen receptor (CAR)-T cell therapy has transformed the treatment of B-cell malignancies, but its success in acute myeloid leukemia (AML) remains limited. Durable responses depend on the formation of long-lived memory T cells, whereas T cell exhaustion contributes to non-response and relapse. In patients with AML who achieved remission after cord blood transplantation, we here first observe enrichment of memory T cells with high expression of the chemokine receptor CXCR4. Next, we show that engineering CAR-T cells to co-express CXCR4 enhances their persistence and anti-leukemic activity in patient-derived xenograft models. Using single-cell profiling and metabolic analysis, we find that CXCR4 promotes memory-associated transcriptional programs, reduces exhaustion, and supports oxidative metabolism. These effects are observed with CAR-T cells targeting CD25 or CD96 as AML-associated targets. Our results indicate that CXCR4 strengthens CAR-T cell memory and durability, offering a strategy to improve immunotherapy outcomes in AML and beyond.
Similar content being viewed by others
Introduction
Targeted therapy using engineered immune cells, particularly chimeric antigen receptor (CAR)-T cells targeting CD19, has emerged as a transformative treatment for relapsed and refractory B cell acute lymphoblastic leukemia (B-ALL) and diffuse large B cell lymphoma (DLBCL)1,2,3,4. Much effort has been made in addressing two questions: (1) Can CAR-T cell therapy be successfully extended to non-B-lineage hematologic malignancies? (2) How can CAR-T cells be programmed to establish long-lived memory capable of sustaining long-term remission?
Acute myeloid leukemia (AML) is a genetically heterogenous hematologic malignancy characterized by a wide spectrum of somatic mutations, chromosomal abnormalities and cell surface protein expression5. Identifying optimal CAR-T cell targets in AML poses significant challenges6. First, many surface molecules are shared between AML cells and normal hematopoietic stem/progenitor cells (HSPCs) and essential myeloid lineages, making selective tumor targeting more difficult than in B cell malignancies. Second, while B-cell aplasia following CD19-targeted therapy can be managed by intravenous immunoglobulin supplementation, on-target, off-tumor toxicity against vital myeloid populations such as neutrophils may lead to serious complications, including febrile neutropenia and life-threatening infections. Third, unlike the near-universal expression of CD19 in B-ALL, AML surface antigen expression is highly heterogeneous across patients. Several AML-associated antigens such as CLL-17, CD338, CD1239, and tumor-associated antigen Lewis-Y (TAA-LeY)10 are currently under evaluation for clinical use in CAR-T therapy. Nevertheless, there remains a critical need to expand the antigenic repertoire for AML CAR-T therapy while minimizing toxicity to healthy hematopoietic cells.
A broader and persistent challenge in CAR-T therapy is disease relapse, frequently attributed to T cell exhaustion. Exhausted CAR-T cells exhibit diminished survival and impaired cytotoxic function, contributing to treatment failure. In contrast, acquisition of memory phenotypes is associated with sustained CAR-T cell persistence, long-term functionality, and durable antitumor immunity. In patients treated with CD19 CAR-T, T-cell exhaustion was more frequently observed in partial and non-responsive patients. Conversely, in patients achieving complete remission, CAR-T cells persisted and remained functional for over four years following infusion11,12,13,14.
The CXCR4 chemokine receptor is a rhodopsin-like GCPR that is widely expressed by most hematopoietic cells including T cells. It is best known for regulating cell trafficking and homing via interaction with its canonical ligand CXCL12/SDF-1 while CXCR4 expression in T cells is reported to facilitate their migration to the bone marrow (BM) and other disease-relevant tissue sites15,16,17,18.
To date, engineering CXCR4 into adoptive cell therapies has primarily focused on improving cell trafficking and tumor infiltration. For instance, CXCR4 co-expression on CAR-T cells enhanced tumor infiltration and antitumor activity in a murine model of pancreatic ductal adenocarcinoma19. Similarly, co-expression of CXCR4 on EGFRvIII-directed CAR-NK cells promoted glioblastoma infiltration and resulted in complete tumor regression and prolonged survival in vivo20 In the context of hematologic malignancies, CXCR4 expression has been leveraged to improve BM homing of engineered immune effector cells. CXCR4-enhanced BCMA-targeted CAR-NK cells exhibited improved BM migration and reduced tumor burden in a murine multiple myeloma model21. Likewise, CXCR4 co-expression on CD33-directed CAR-cytokine-induced killer (CIK) cells led to enhanced BM localization, improved anti-leukemic activity, and extended survival in an AML mouse model22.
Beyond its role in cell migration, CXCR4 is also recognized for its roles in T cell survival, persistence, and memory formation. CXCR4 plays a critical role in early thymic T cell development23 and continues to influence T cell function changes after maturation. CXCR4-deficient mice exhibit compromised T cell self-renewal and impaired memory generation18,24. Consistent with this, CXCR4 expression is enriched in naïve and memory T cell populations but is rapidly downregulated following TCR stimulatio25,26. Furthermore, CXCR4 is preferentially expressed in Th2 cells and IL-4-stimulated T cells, which are canonically long-lived cells associated with humoral immunity, whereas it is relatively low in Th1 cells, which are canonically short-lived cells that mediate cytotoxic effector functions26. Notably, recent findings suggest that effector and exhausted CAR-T cells can be epigenetically reprogrammed toward a memory-like state through exposure to CXCL12, highlighting a potential role for the CXCR4-CXCL12 axis in promoting long-term CAR-T functionality27.
Here, we show that profiling of T cells from patients with AML after cord blood (CB) transplantation reveals an enrichment of CD4+ memory populations in remission versus relapse. We find that expression of the chemokine receptor CXCR4 correlates positively with memory T cell formation and inversely with T cell exhaustion in patient T cells. In parallel, RNA sequencing of patient-derived leukemias and normal hematopoietic progenitors identifies CD25/IL2RA as a promising CAR-T target, with high expression in AML blasts and low risk of off-tumor toxicity. We demonstrate that CXCR4-expressing, CD25-targeted CAR T cells achieve superior elimination of AML in patient-derived xenograft (PDX) models driven by reduced exhaustion, enhanced memory differentiation, and durable protection against re-challenge. Furthermore, we observe similar benefits when targeting an independent AML antigen, CD96, highlighting the generalizability of CXCR4-mediated CAR-T cell enhancement. Together, these results establish CXCR4 expression as a strategy to strengthen CAR T cell persistence and efficacy, offering a path toward durable immunotherapy for high-risk AML.
Results
CXCR4 marks CD4+ memory T cells associated with AML remission
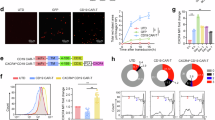
To identify features linked to durable anti-leukemic immunity, we profiled T cells from AML patients in remission or relapse following CB transplantation using Cytometry by Time-of-Flight (CyTOF) (Fig. 1a, Table S1-S2). Unsupervised clustering of T cell markers revealed subsets including CCR7+IL7RA+CD45RA+ T naïve/stem cell memory (Tn/scm), CCR7+IL7RA+CD45RA- T central memory (Tcm), and CCR7+CD45RA-GZMK+ T effector memory (Tem), amongst other T cell populations (Fig. 1b, c; Figure. S1a). Compared to remission, relapse samples showed marked depletion in the proportion of CD4+ Tn/scm cells (23.1-fold) and naïve T cells (10.6-fold), alongside a modest (3.7-fold) increase CD4+ effector cells (Fig. 1d; Figure. S1b). Consistent with observations in CD19 CAR-T clinical trials28, the majority (86.6%) of memory cells were CD4+.
a Schematic of experimental design. T cells were isolated from peripheral blood (PB) or bone marrow (BM) of primary AML patients in remission following ablative radiation therapy and cord blood transplant (CBT). CD33⁻CD3⁺ T cells were analyzed by CyTOF (see Table S1). b UMAP projection of patient-derived T cells showing expression of CD4/CD8, memory/stemness, and exhaustion markers. c Unsupervised clustering of T cell populations based on select CyTOF markers. Cell subtype designations are based on marker expression patterns. Number of cells in each T cell subset across all n = 14 samples from 11 patients is shown. d Comparison of T cell subsets in remission and relapse samples. Forest plot shows mean log₂ fold-enrichment of each subtype in n = 11 remission samples versus n = 3 relapse samples, with 95% confidence intervals. Statistical analysis was performed using a two-tailed T-test. The non-adjusted p-value is reported. e Violin and box plots show CXCR4 protein expression across T cell subsets. Dashed lines indicate median CXCR4 expression in remission (blue) and relapse (red). f UMAP projection of CD4⁺ tumor-infiltrating lymphocytes (TILs) from a previously published scRNA-seq dataset30, annotated by subtype. CXCR4 expression was imputed using ALRA. g Dot plot showing expression of canonical T cell marker genes across CD4⁺ TIL subtypes. h Violin and box plots showing CXCR4 expression across CD4⁺ TIL subtypes. i) Number of cells from each T-cell subtype is shown across different quartiles of CXCR4 expression (calculated across the entire dataset). For box and whisker plots in panels (d, h)central lines show median, lower and upper hinges show first and third quartiles (the 25th and 75th percentiles) and whiskers extend up to 1.5 * IQR. Source data are provided as a Source data file for (d). “Fig. 1a” created in BioRender. Liang, M. (https://BioRender.com/uexx577) licensed under CC BY 4.0.
Unsupervised clustering of CyTOF-derived protein expression identified CXCR4 as closely associated with memory markers CCR7, IL7RA, and CD45RA, despite not being used as a marker to define T cell subtypes (Fig. 1c). CXCR4 expression in remission samples correlated positively with memory markers CCR7 and IL7RA (r = 0.550 and 0.350, respectively), and negatively with exhaustion/effector markers GZMB and Tbet (r = −0.349 and −0.292) (p < 1.e-100 for all) (Fig. 1c). Consistent with the association between CXCR4 and central memory markers, CXCR4 expression was highest in CD4+ Tscm and CD8+ Tscm (Fig. 1e)29.
In a published CD4+ tumor infiltrating lymphocyte (TIL) scRNA dataset spanning 9 different solid tumor types30 (Fig. 1f), the cluster annotated as Tn/scm/cm was most similar in marker gene expression to CD4+ memory populations we defined by CyTOF (Fig. 1g). CXCR4 expression was elevated in Tn/scm/cm and EOMESHi populations and depleted in exhausted precursors and Th17 cells (Fig. 1g, h). High CXCR4-expressing cells were enriched for memory phenotypes and underrepresented in exhausted subsets (Fig. 1i). Together, our findings indicate a potential link between CXCR4 and CD4+ memory T cell populations in the acquisition of anti-tumoral immunity.
These data support a shared association between CXCR4 expression and CD4+ memory T cell identity, suggesting that CXCR4 may promote memory acquisition and persistence across tumor contexts. Based on these findings, we hypothesized that engineering of CXCR4 expression could be used to enhance CAR-T cell memory and antitumor function.
CD25 identified as a candidate CAR-T target with minimal off-tumor toxicity
To identify optimal CAR targets for CXCR4-engineered CAR-T cells, we screened for AML-specific surface antigens using bulk RNA-seq from AML blasts (n = 235 samples) and normal CD34⁺ HSPCs (n = 44 samples), selecting genes with high AML-specific expression (log₂FC > 2.5, padj < 0.001) and low expression in non-hematopoietic tissues (avg TPM < 5, max TPM < 10 across 31 tissues from GTEx) (Fig. 2b; Supplementary Data 1, 2). Four of six candidate targets identified in our cohort met the same criteria for expression level and DE significance in the BEAT AML cohort (Figure. S2a; Supplementary Data 3)31,32.
a Differential gene expression analysis of AML blasts (n = 235 samples from 155 patients) versus normal hematopoietic stem/progenitor cells (HSPCs; n = 44 samples from 14 donors). Genes encoding surface proteins are highlighted in pink. Validated AML CAR targets (black) and candidate targets identified in our screen (blue) are annotated. log₂ fold-changes and FDR-corrected p-values from DESeq2 (Wald-test) are reported. b Heatmap of surface molecule expression across 31 non-hematopoietic tissues (GTEx dataset; median TPM for n = 4 to 803 donors per tissue). Candidate targets with low off-tumor expression and canonical AML CAR-T targets are annotated. Novel targets discovered in our cohort and replicated in beatAML are labeled in blue. c Representative flow cytometry histogram of CD25 expression in CD25⁻ and CD25⁺ AML patient samples (pink = anti-CD25 antibody, gray = isotype control). CD25⁺ AML was defined as > 70% CD25⁺ blasts by flow cytometry (n = 12/66 patients). d AML cohort ranked by CD25 mRNA expression (black dots, n = 155 patient samples). For a subset, CD25 surface levels were validated by flow cytometry (pink bars; n = 55 patient samples). Middle: Expression log₂ fold-change relative to HSPCs for candidate and select validated AML CAR-T targets. Bottom: Mutational landscape for individual samples. For some samples, multiple individual measurements were made for different rounds of xenotransplantation. e In vitro cytotoxicity of CAR-T cells assessed by co-culture with primary AML blasts. Effector CAR constructs include: ACTT and CD19 (negative controls); CD33 (positive control); CD25; and CD96. Sorted PDX BM-derived AML blasts for a representative CD25/CD96 dual-positive patient sample (P12) were used as target cells. Co-culture experiments were setup for multiple effector:target ratios and %AML reduction was evaluated at 24 h and 72 h. Experiments were replicated using CAR-T cells derived from n = 3 CB donors (rep1-3). Each data point corresponds to average of three technical repicates. Y-axis reports the percentage of hCD33+ cells compared to AML blasts alone. Error bars correspond to mean ± SD. The non-adjusted p-value reported for each CAR construct versus ACTT (two-tailed T-test). Source data are provided as a Source data file for panel(s) (a, d, e).
Amongst these, mRNA expression of IL-2 receptor alpha chain CD25/IL2RA was highly AML-specific and was associated with poor overall survival in the BEAT AML cohort (Figure. S2b). The minimal expression of CD25 other tissues and the absence of CD25 protein in hematopoietic populations other than CD4+ Tregs (Figure. S2c, d) supports a favorable safety profile. Furthermore, CD25 is a known marker of chemotherapy-resistant leukemia-initiating cells (LICs)33 and is frequently reported as a marker of poor prognosis in AML34,35. We therefore selected CD25 as a target based on its 1) AML-specific overexpression vs HSPCs; 2) low off-tumor toxicity risk; 3) enrichment in LICs and 4) potential to address unmet needs for high-risk, treatment-refractory disease.
CD25 expression varied across patients in our cohort, with high surface positivity ( > 70% of cells) detected in 18.2% (12/66) of AML cases (Fig. 2c). Among 155 samples with matched RNA-seq and mutational data, IL2RA mRNA expression was elevated in FLT3-ITD (6.7-fold, p = 1.6e-7) and NPM1 (3.4-fold, p = 0.0021) mutant AML (Fig. 2d, Figure. S2e). We selected four representative IL2RA-high samples for subsequent experiments (Figure. S3). Surface CD25 protein expression in these samples was highest in putative LIC-like CD34+CD38- fractions (Figure. S3).
As an additional target, we selected CD96, a previously reported LSC marker36 which also met the same criteria for high expression in AML blasts versus HSPCs and low expression in non-hematopoietic tissues. CD96 surface positivity was observed in 55.1% of AML samples and mRNA overexpression overlapped with 75% of IL2RA-high cases, offering broader coverage (Fig. 2d, Fig. 4a, b). We note that CLL1/CLEC12A, a previously validated AML CAR target, may also be a viable alternative based on overexpression in both our overall cohort (48.8%) and in IL2RA-high cases (43.8%) (Fig. 2d).
Generation of CAR-T from CB T cells
We generated CAR-T cells by lentiviral transduction of donor CD3+ T cells with second-generation CAR vectors, incorporating an extracellular CD8 leader, antigen-specific scFv, CD8 hinge, 4-1BB co-stimulatory, and CD3ζ signaling domains37 (Figure. S4a). CXCR4-expressing CAR-T cells were created by dual transduction with an additional vector encoding full-length CXCR4 derived from either mouse (mCXCR4) or human (hCXCR4) (Figure. S4b). Murine and human CXCR4 and CXCL12 share a high degree of homology, with 91% and 99% similarity, respectively. Furthermore, human CXCR4 knock-in mice exhibit normal hematopoiesis in vivo, suggesting functional conservation of the CXCR4-CXCL12 axis between humans and mice in hematopoietic cells38. Therefore, in our initial CD25-CAR experiments, we used mCXCR4 to facilitate monitoring of transduced CXCR4 expression in human CAR-T cells. In subsequent experiments, we transitioned to hCXCR4 to align with clinical translation goals, as humanized receptors are associated with a lower risk of immune rejection39,40. See “Methods” and Data Availability for full vector sequences.
We considered several donor T cell sources for CAR-T cell generation, including autologous patient-derived peripheral blood (PB), healthy adult PB, and CB. We initially attempted to generate autologous CAR-T cells using patient PB samples isolated during active disease. However, patient T cells exhibited poor viability and minimal expansion in vitro. Viable T cell numbers never reached the threshold ( ~ 1 × 10⁵) required for lentiviral transduction, even after extended activation (Figure. S4b). Phenotypic analysis of the starting CD3⁺ populations revealed higher proportions of exhausted and lower proportions of naïve and memory subsets in patient samples (Figure. S4b), suggesting that poor quality of T cells severely limits the feasibility of generating functional autologous CAR-T cells from AML patients with relapse or refractory disease.
Given the poor expansion and exhaustion-prone phenotype of autologous T cells from AML patients, we explored alternative sources for CAR-T generation. While healthy adult PB is traditionally the most commonly used source of allogeneic immune cells, CB is increasingly recognized as a promising alternative, offering several advantages for adoptive immune cell transfer, including less differentiated T cell phenotype, an abundance of naïve T cells, reduced risk of graft-versus-host disease (GVHD), less stringent HLA matching requirements, and the potential for development of off-the-shelf CAR-T products41,42. CB- and healthy donor PB-derived T cells both showed robust expansion in vitro and high transduction efficiency for CAR constructs and dual CAR/CXCR4 (Figure. S4c-f). In consideration of the superior T cell phenotype, expansion potential, and translational advantages, we selected CB as the primary donor source for CAR-T cell generation.
In vitro assessment of CD25-targeted CAR-T cytotoxicity and on-target off-tumor toxicity
We generated CD25-targeted CAR-T cells as above, substituting the scFv domain with an anti-CD25 scFv, which we developed in-house as previously described (sequence in Data Availability)43 (Figure. S4c). We evaluated cytotoxicity and specificity of CD25-targeted CAR-T cells against patient-derived CD25+ AML blasts in vitro. Controls included non-specific activated T cells (ACTT), CD19-targeted CAR-T (negative), and CD33-targeted CAR-T (positive). CD25-targeted CAR-T cells showed potent killing of CD25-expressing primary AML cells, slightly weaker than CD33-targeted CAR-T (90.0 ± 3.6 % versus 99.9 ± 0.06 % AML reduction, respectively; 72 h; E:T = 3:1) but significantly greater than ACTT or CD19-CAR-T controls (Fig. 2e). Cytotoxicity against AML blasts was more pronounced at later time points and at higher effector:target ratio for all AML-specific CARs, confirming cytotoxicity is mediated by CAR-T cells. CD25-targeted CAR-T were ineffective against primary AML cells for which CD25 was knocked out, confirming antigen specificity (Figure. S2f). In vitro cytotoxicity data for additional patient samples are provided in (Figure. S2f).
Co-culture of CAR-T cells with autologous CB-derived CD34+ HSPCs resulted in comparable decrease in HSPC number between ACTT, CD19- and CD25-targeting CAR-T cells (Figure. S2g), suggesting that that off-tumor toxicity against HSPCs may be mediated by CAR-T-derived inflammatory cytokines rather than CD25-directed killing44. Activated T-cells and regulatory T cells (Tregs) normally express CD25, posing a risk for on-target off-tumor toxicity (Figure. S2d). During CAR-T generation, no significant difference in T cell viability or number was observed between CD25- and CD19-targeted CAR-T or ACTT within the first 2–3 days post-transduction (Figure. S5a). In contrast, co-culture with autologous Tregs showed reduction in Treg counts after 3 days with CD25-targeted CAR-T (72.2 ± 11.6%) compared to CD19 CAR-T (0.1 ± 30.9%) (Figure. S5b). These results support the specificity of CD25-targeted CAR-T cells, with minimal on-target off-tumor toxicity against normal HSPCs and modest cytotoxicity against normal Tregs. The long-term in vivo impact of CD25-targeted CAR-T treatment on Tregs and other hematopoietic cells remains to be determined.
Structural assessment of CD25 CAR targeting specificity
Given difference in the extent of CD25-targeted CAR-T toxicity against AML and T cells, we investigated potential structural differences in CD25 presentation on AML blasts versus T cells. CD25/IL2RA forms part of the IL-2 receptor complex with CD122/IL2RB and CD132/IL2RG. IL2RB mRNA was ~11-fold lower in CD25+ AML blasts than in T cells (Figure. S5c), suggesting that AML cells predominantly express single-chain IL2RA (scIL2RA). In vitro reconstitution confirmed stable complex formation between our anti-CD25 scFv (S417 Fab) and both scIL2RA and the full IL-2R heterotrimer (Figure. S5d), but not with IL2RA and IL2RG alone in the absence of IL2RB (Figure. S5e). Single-particle cryo-EM analysis of the S417 Fab and IL-2R heterotrimer suggests that antibody binding does not interfere with IL-2 binding or complex assembly (Figure S5f).
CXCR4 improves therapeutic efficacy of CD25-targeted CAR-T cells in vivo
Having identified CD25 as a viable AML target, we next investigated whether CXCR4 overexpression enhances CD25-targeted CAR-T cell persistence and antitumor efficacy in vivo. We generated PDX models of CD25+ AML using four primary AML samples with high surface CD25 expression (75.7-85.5% of blasts, Figure. S3; and Table S1; see Supplementary Data 4 for full cohort details) via xenotransplantation of sorted AML-initiating cells into newborn NSG mice. After confirming successful AML engraftment (hCD45⁺CD33⁺) in PB, mice were given one intravenous dose (5 × 10⁵ − 4.6 × 10⁶ cells per mouse, see Supplementary Data 4) of CAR-T cells, followed by weekly monitoring by phlebotomy (Fig. 3a). mCXCR4(+) CD25-targeted CAR-T cells exhibited significantly improved clearance of AML from PB (mean %AML chimerism change = −6.2 ± 21.7% in mCXCR4(+) CD25-targeted CAR-T, versus +18.3 ± 34.3%, p = 0.002 and versus +63.1% ± 25.6%, p = 2.9e-5 in CXCR4(-) CD25-targeted CAR-T and untreated PDX, respectively) (Fig. 3b). AML clearance was also improved in BM, spleen, and liver (Fig. 3c).
a Schematic of study design. b Peripheral blood %AML chimerism pre-CAR-T treatment and at endpoint in untreated (n = 22 animals), CD25-targeted CAR-T-treated (n = 32 animals), and mCXCR4(⁺) CD25 CAR-T-treated (n = 27 animals). c AML chimerism in PB, spleen, bone marrow, and liver of PDX mice, stratified by treatment. Group sizes by treatment: untreated (n = 4, 9, 8, 15 animals), CXCR4(⁻)CD25-targeted CAR-T (n = 10, 10, 8, 7 animals), and mCXCR4(⁺) CD25-targeted CAR-T (n = 12, 8, 7, 8 animals), PDX mice of Patients 10-13, respectively. Statistical comparisons made using unpaired two-sided Mann-Whitney U tests. The non-adjusted p-value is reported. For b, c Box and whisker plots: central lines show median, lower and upper hinges show first and third quartiles (the 25th and 75th percentiles) and whiskers extend up to 1.5 * IQR. d Longitudinal kinetics of AML cells (hCD45⁺CD33⁺), CAR-T cells (hCD45⁺CD3⁺), and murine leukocytes (mCD45⁺) in nucleated cell fraction from PB (red cells were excluded by lysis pre-FACS). Lines and shaded regions indicate mean ± SD. The number of mice included at each time point is shown. e Kaplan-Meier survival curves for PDX animals stratified by treatment arm including CXCR4(-) CD25-targeted CAR-T (n = 27 animals), mCXCR4(+) CD25-targeted CAR-T (n = 32 animals), and mock/ACTT-treated (n = 10 animals) groups. Individuals removed for paired analysis or time-controlled sampling were censored (see Table S4). Shaded regions show 95% confidence intervals. f H&E and CD33 IHC staining of femur sections from PDX (P11). Representative images selected from sections prepared from n = 5 mice per treatment group (scale: 20 µm) g) Laser scanning microscopy (LSM) imaging and 3D reconstruction of liver sections acquired from PDX (P10), harvested 10 days after injection with CXCR4(-) or mCXCR4(+) CD25-targeted CAR-T. IF staining was performed for CXCL12 (green), CD3 (red) and DAPI (blue). Images were acquired on a Zeiss Axio Observer 7. Z-stack 3D reconstruction was performed using ImarisViewer. Representative images selected from sections prepared from n = 3 mice per treatment group (scale: 30 µm). Source data are provided as a Source data file for (b, c, d, e). “Fig. 3a” created in BioRender. Liang, M. (https://BioRender.com/5hfwv50) licensed under CC BY 4.0.
We tracked temporal changes in PB cell chimerism and observed transient reduction of murine CD45⁺ cells during peak AML and CAR-T cell chimerism, followed by recovery as AML burden declined (Fig. 3d). AML clearance correlated with a rise in CAR-T chimerism in both groups between weeks 2-4 post-injection. However, CXCR4(-) CD25-targeted CAR-T cells failed to persist, coinciding with AML relapse, while mCXCR4(+) CAR-T cells expanded until weeks 6-8 and were associated with durable AML suppression (Fig. 3d). We assessed percent survival in PDX post CAR-T injection, counting mice that either died or had to be sacrificed prior to a designated experiment endpoint (Fig. 3e). Although survival time could not be precisely determined due to our time-controlled experiment design, CD25-targeted CAR-T treated mice exhibited significantly better survival at 8 weeks post-treatment for both CXCR4(-) (n = 27; OS = 65%; 95% CI: 46-92%) and mCXCR4(+) (n = 32; OS = 92%; 95% CI: 82-100%) when compared to mock (ACTT-treated) mice (n = 10; OS = 20%; 95% CI: 4.1-97.1%). Furthermore, treatment with mCXCR4(+) versus CXCR4(-) CD25-targeted CAR-T resulted in an 83% reduction in mortality (Cox hazard ratio 0.17; 95% CI: 0.03-0.89).
Immunohistochemistry for human CD33 revealed near-complete clearance of AML and robust recovery of leukocytes, erythroid cells, and megakaryocytes in the BM of mCXCR4(+) CD25-targeted CAR-T-treated mice (Figure. S6a, b). In contrast, CXCR4(-) CAR-T-treated mice showed persistent AML and limited recovery of hematopoietic elements (Fig. 3f; and S6b). Notably, mCXCR4(+) CAR-T cells localized near CXCL12⁺ stromal cells in BM at day 10 post-infusion (Fig. 3g; and S6c). Mirroring our findings in post-CBT patients, T cells isolated from BM and spleen of animals treated with mCXCR4(+) CAR-T were predominantly CD4+ with a significantly higher proportion of CD4+ cells versus in CXCR4(-) CAR-T -treated animals (Figure. S6d).
mCXCR4(+) CD19-targeted CAR-T failed to eliminate CD25⁺CD19⁻ AML cells (Figure. S6e), showing that the improved anti-AML activity of mCXCR4(+) CAR-T was target-specific. In cases with ≥4/8 HLA matches between CB donors and AML, CD25-targeted CAR-T cells cleared AML while ACTT could not, indicating the effect is not due to alloreactivity (Figure. S6f). Prior studies suggest CXCR4 enhances effector cell trafficking to tumor sites via CXCL1219,22,45, which is also expressed by BM stromal cells. We tested whether CXCR4 improved CAR-T trafficking by comparing animals receiving mCXCR4(+) CD25-targeted CAR-T with matched PDX littermates receiving CXCR4(-) CD25-targeted CAR-T. CAR-T chimerism was significantly higher in BM of mCXCR4(+) versus CXCR4(-) CAR-T-treated animals. However, these differences disappeared upon normalization for circulating (PB) CAR-T chimerism, indicating increased CAR-T presence in BM was due to overall survival and persistence, not selective BM trafficking/retention (Figure. S6g). Moreover, mCXCR4(+) CAR-T were not sequestered in BM but circulated systemically. Signs of graft versus host disease such as liver pathology, dermal thickening, elevated AST/ALT, and abnormal CD3⁺ infiltration were absent in both HSC- and AML-engrafted mice up to 8+ weeks post-CAR-T infusion (Figure. S7a, b).
Together, our findings demonstrate that CXCR4 expression enhances the anti-AML activity of CD25-targeted CAR-T cells in vivo, promoting leukemia clearance in PB, BM, and peripheral tissues, facilitating hematopoietic recovery, and increased survival.
CXCR4 improvement of CAR-T cell therapeutic efficacy in vivo is replicated against an independent CAR target, CD96
We constructed CD96-targeted CAR-T cells from CB-derived donor T cells as was done for CD25-targeted CAR-T cells, switching the anti-CD25 scFv sequence with a previously published anti-CD96 scFv sequence46. For CD96 CAR-T cells, we substituted full-length murine CXCR4 with full-length human CXCR4 (hCXCR4).
In vitro co-culture revealed potent killing of CD96+ AML blasts by CD96-targeted CAR-T cells (97.2 ± 2.2 % AML reduction; 72 h; E:T = 3:1) as compared to ACTT and CD19-targeted CAR-T controls, with efficacy comparable to that of CD33- and CD25-targeted CAR-T cells (Fig. 2g). In PDX mice engrafted with CD96+ AML, treatment with hCXCR4(+) CD96-targeted CAR-T cells led to improved AML clearance in PB (%AML chimerism change = +7.7 ± 30.5% in hCXCR4(+) CD96-targeted CAR-T, versus +47.5 ± 40.2%, p = 0.01 in CXCR4(-) CD96-targeted CAR-T and versus +59.8 ± 21.3%, p = 2.7e-5 in untreated PDX, respectively), plus improved AML clearance in BM, spleen, and liver (Fig. 4c, d). IHC staining of BM revealed recovery of murine hematopoietic elements in mice treated with hCXCR4(+) CD96 CAR-T, while CXCR4(-) treated animals showed persistence of AML and impaired hematopoiesis (Fig. 4e).
a AML samples ranked by normalized CD96 mRNA expression (black dots, n = 155 patients). CD96 surface expression (flow cytometry) shown for a subset (blue bars, n = 95 patients). Bottom heatmap shows mutational profiles per sample, as in Fig. 2d.b Representative flow cytometry histogram showing CD96 surface expression in CD96⁻ and CD96⁺ patient-derived AML samples, as in Fig. 2c. Anti-CD96 antibody staining is shown in blue, isotype control in gray. 98 AML samples were analyzed; cases with >70% CD96⁺ leukemic blasts were defined as CD96⁺ (blue, n = 54), others as CD96⁻ (gray, n = 44). c Peripheral blood %AML chimerism pre-CAR-T injection and at endpoint in untreated (n = 21), CXCR4(-) CD96 CAR-T-treated (n = 23), and hCXCR4(+) CD96 CAR-T-treated (n = 18) animals. Red dots indicate animals that achieved complete remission (0% AML chimerism in PB). d AML chimerism in peripheral blood, spleen, bone marrow, and liver of PDX mice as in (c), stratified by treatment. Group sizes by treatment: untreated (n = 3, 9, 4, 5), CXCR4(-) CD96 CAR-T (n = 6, 9, 5, 3), and hCXCR4(+) CD96 CAR-T (n = 4, 6, 4, 4), PDX mice of Patients 12–15, respectively (See Table S5). Statistical comparisons were made using unpaired two-sided Mann-Whitney U tests. The non-adjusted p-value is reported. For (c, d) Box and whisker plots: central lines show median, lower and upper hinges show first and third quartiles (the 25th and 75th percentiles) and whiskers extend up to 1.5 * IQR. e) H&E and CD33 immunohistochemistry of bone marrow from Patient 13-derived PDX (untreated, CXCR4(-), or hCXCR4(+) CD96 CAR-T treated) (scale: 20 µm). Representative images selected from sections prepared from n = 3 animals per treatment group. Source data are provided as a Source data file for (a, c, d).
These findings demonstrate that CXCR4-driven enhancement of in vivo CAR-T cell efficacy extends beyond CD25 targeting, supporting a broader role for CXCR4 in improving CAR-T cell function against AML.
CXCR4 expression enhances memory CAR-T cell development in vitro and in vivo
To investigate how CXCR4 influences the differentiation of CAR-T cell subsets, we profiled memory and exhaustion markers on CD25-targeted CAR-T cells after extended in vitro co-culture (1 week) under normoxic (21% pO₂, mimicking peripheral circulation) and hypoxic (2.5% pO₂, mimicking the bone marrow microenvironment) conditions, with or without hCXCR4 overexpression (Fig. 5a). To assess whether the beneficial effects of CXCR4 are specific to CB-derived T cells, we also included CAR-T cells generated from healthy donor PB.
a Timeline of extended co-culture experiments. b Proportion of CD4+ Tcm cells (as % of CD3 + ) and (c) PD-1 surface expression (MFI) were measured by flow cytometry on day 6 of co-culture. Data shown for n = 3 donors for both cord blood (CB) and peripheral blood (PB). The average of 3 technical replicates per sample is plotted. Statistical analysis was performed using a paired two-tailed T-test. The non-adjusted p-value is reported. d (Left) Unsupervised clustering of CD25-targeted CAR-T cells based on CyTOF protein expression. Cells from bone marrow and spleen of PDX treated with CXCR4(-) or mCXCR4(+) CD25-targeted CAR-T (n = 5 mice per group). (Right) Expression of marker proteins defining each UMAP cluster. e Relative abundance of each cluster in CD3⁺ CAR-T population. Forest plot shows log₂ fold-enrichment of each subtype in CXCR4(-) or mCXCR4(+) treatment groups, with 95% confidence intervals. Diamonds indicate mean values; whiskers represent 95% confidence intervals. Statistical analysis was performed using a two-tailed T test. The non-adjusted p-value is reported. f Proportion of CD4⁺ naïve/T stem cell memory (Tscm) and central memory (Tcm) CAR-T cells in bone marrow and spleen of PDX treated with CXCR4(-) (n = 14 animals) or mCXCR4(+) (n = 30 animals) CD25-targeted CAR-T cells. g Frequencies of immune-surveillant (CD226⁺TIGIT⁻), memory (IL7RA⁺, TCF1⁺), and checkpoint-expressing (TIM3⁺TIGIT⁺) CD4⁺ CAR-T cells in bone marrow and spleen. mCXCR4(+) CD25-targeted CAR-T cells exhibited enhanced memory and immune-surveillant phenotypes, while checkpoint markers were enriched in CXCR4(-) CD25-targeted CAR-T cells. Data shown for n = 14 and n = 30 (n = 4 and n = 14 remission) animals for CXCR4(-) and mCXCR4(+) CD25-targeted groups, respectively. h Proportion of CD4⁺ naïve/Tscm and Tcm CAR-T cells in PDX treated with CXCR4(-) (n = 15) or hCXCR4(+) (n = 16) CD96-targeted CAR-T cells. i Frequency of immune-surveillant and memory phenotype CD4⁺ CAR-T cells in CD96 CAR-T-treated PDX, as in (g). Data shown for n = 15 (n = 2 remission) and n = 16 (n = 9 remission) animals for CXCR4(-) and hCXCR4(+) CD96-targeted groups, respectively. Note: TCF1 was not included in CD96-targeted CAR-T profiling. Graphs in panels (f–i) show mean ± SD. Statistical comparisons were performed using unpaired two-tailed t-test. Source data are provided as a Source data file for panel(s) (b, c, f–i).
Although in vitro T cell differentiation demonstrates large inter-donor variability, hCXCR4(+) CAR-T cells exhibited a trend of higher Tcm percentage compared to CXCR4(-) CAR-T cells under normoxia for both CB- and PB-derived cells (Fig. 5b). Additionally, we observed a reduction in PD-1 surface expression in hCXCR4(+) cells under normoxia (Fig. 5c). These effects were also seen in PB-derived hCXCR4(+) CD25-targeted CAR-T cells, suggesting that the effects of CXCR4 on memory acquisition and exhaustion resistance is may not limited to CB-derived T cells. The beneficial effects of CXCR4 co-expression were abrogated under hypoxic conditions, potentially suggesting that these cells require the specific bone marrow niche for memory acquisition in vivo.
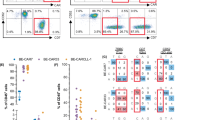
We next assessed memory and exhaustion of CAR-T cells in vivo by performing high-dimensional proteomic profiling of functional T cell memory and exhaustion-associated markers on CAR-T cells isolated from AML PDX mice treated with CD25-targeted CAR-T cells (Figure. S8). Unsupervised clustering of CAR-T cells from bone marrow and spleen at 4-6 weeks post-infusion revealed distinct populations: naïve/stem-like T cells (CD4⁺ and CD8⁺ N/Tscm), central memory T cells (CD4⁺ Tcm), effector T cells (CD4⁺ and CD8⁺ Teff), and checkpoint-expressing/exhausted T cells (CD4⁺ and CD8⁺ Tex) (Fig. 5d).
mCXCR4(+) CAR-T cells exhibited higher proportions of CD4⁺ naïve/stem-like (3.3-fold increase) and central memory populations (3.9-fold increase), marked by elevated expression of canonical memory markers TCF1, IL7RA, and the immune-surveillance receptor CD226 (Fig. 5e). Additionally, BCL-2 expression was enriched in Tcm subsets, suggesting that CXCR4 signaling may enhance memory development through BCL-2-mediated survival pathways (Fig. 5d). Conversely, T cell populations expressing immune checkpoint and exhaustion markers, including CD4⁺ cells expressing TIM3, TIGIT, and PD-1, and CD8⁺ cells co-expressing LAG3 and TIM3, were diminished in mCXCR4(+) CAR-T cells.
Across both bone marrow and spleen, mCXCR4(+) CAR-T cells showed a consistently higher frequency of CD4⁺ cells with memory and immune surveillance phenotypes, and a lower frequency of cells expressing exhaustion and checkpoint molecules (Fig. 5f, g). In the small number of CXCR4(-) CD25-targeted CAR-T-treated PDX that successfully eliminated AML (n = 4), we found lower frequencies of central memory T cells and higher frequencies of exhausted T cells compared with mCXCR4(+) CD25-targeted CAR-T recipients, indicating that CXCR4-induced memory and evasion of exhaustion is not solely due to reduced AML burden (Fig. 5g, red labels).
These findings were corroborated in PDX models treated with CD96-targeted CAR-T cells, where hCXCR4 overexpression similarly promoted expansion of Tcm populations and CD4⁺ memory subsets, along with acquisition of a CD226⁺TIGIT⁻ immune-surveillant phenotype (Fig. 5h, i). Together, these results demonstrate that CAR-T cells can establish durable memory phenotypes through interaction with human AML cells in a xenogeneic context. Importantly, CXCR4 expression enables CAR-T cells to evade exhaustion and acquire memory, immune surveillance, and longevity features that are not restricted to a specific target antigen.
Single-cell RNA sequencing reveals T cell and AML transcriptional programs associated with CXCR4-driven therapeutic efficacy
To investigate the impact of CXCR4 on CAR-T cell fate at single-cell resolution, we performed single-cell RNA sequencing (scRNA-seq) on CD25-targeted CAR-T cells isolated from the bone marrow of PDX mice. We profiled CXCR4(-) CAR-T cells at 4 weeks (n = 3) and mCXCR4(+) CAR-T cells at both 4 weeks (n = 3) and 5 months (n = 2) post-injection (Fig. 6a; and Figure. S9). Unsupervised clustering revealed 13 T cell clusters, which we annotated using established lineage and state-specific marker genes (Fig. 6b, c; and Figure. S10a). Three CD4+ clusters expressed canonical memory-associated markers (e.g., TCF1/TCF7, CCR7, IL7R) and scored highly for memory gene modules (Figure. S10b), which we designated Tscm-cm1-3. These represent stem-like memory (Tscm) and central memory (Tcm) populations, which share transcriptional profiles29. Notably, Tscm-cm1 and Tscm-cm2 clusters expressed high levels of CD226, a costimulatory receptor involved in tumor immune surveillance whereas Tscm-cm3 cells expressed EOMES, a transcriptional regulator of tissue-resident memory fate. Expression of CAR transgene (CD25) and mCXCR4 transgene persisted in CAR-T cells up to 5 months post-injection (Figure. S10c), indicating durable transgene expression in vivo.
a Schematic of experimental design detailing CAR-T cell collection for single-cell RNA sequencing. Cells were obtained from AML PDX BM, spleen, and liver at 2.5 weeks, 4 weeks, and 5 months post-injection. b UMAP projection of BM-derived CD25-targeted CAR-T cells collected at 4 weeks and 5 months post-injection. Cells are colored by CD4/CD8 identity, cell cycle score, and select memory/exhaustion markers. Cell type annotations were made based on canonical T-cell subtype markers (see Fig. S7c). c Relative abundance of each cluster across treatment conditions. Forest plot shows log₂ fold-enrichment of each subtype in CXCR4(-) or mCXCR4(+)treatment groups (n = 3 animals per group) at 4 weeks, with 95% confidence intervals. Diamonds indicate mean; whiskers represent 95% confidence intervals. Statistical comparisons were made using two-tailed t-tests. Non-adjusted p-values are reported. See also Figure. S7e. d Dot plot showing expression of representative marker genes across T-cell subtypes and treatment conditions. Additional markers and full subtype profiles are shown in Figure. S7f. e Differential expression (DE) analysis comparing CXCR4(-) versus mCXCR4(+) CD25-targeted CAR-T cells at 4 weeks and 5 months. Genes with base mean >15, FDR < 0.05, and log₂ fold-change ≥ 0.58 in at least one comparison are shown. Genes with early (4-week) or late (5-month) CXCR4-associated upregulation are highlighted in red and blue, respectively. f Gene set enrichment analysis (GSEA) of DE genes ranked by log₂ fold-change. GSEA p-values were calculated using the fgsea package in R. FDR-adjusted p-values are reported. Only pathways with FDR < 0.05 in at least one contrast and enrichment FDR < 0.01 are displayed. g UMAP visualization of liver- and spleen-derived CAR-T cells harvested at 2.5 weeks post-injection. Cells are colored by CXCR4 expression status and tissue origin. ALRA-imputed expression of select memory and exhaustion markers is shown. Source data are provided as a Source data file for (c, f). “Fig. 6a” created in BioRender. Liang, M. (https://BioRender.com/j4jhcy1) licensed under CC BY 4.0.
Quantitative analysis of T cell subtype populations revealed substantial enrichment of memory-associated CD4+ CAR-T cells in mCXCR4(+) samples at both 4 weeks and 5 months compared to CXCR4(-) counterparts (52.4% and 36.2% vs. 18.7% Tscm-cm cells, respectively; (Fig. 6c; and Figure. S10d, e) In contrast, proliferative and exhausted subsets were markedly overrepresented in CXCR4(-) samples (29.2% vs. 3.4% and 1.1% at 4 weeks and 5 months, respectively). Consistently, memory- and resting-state gene modules were more strongly expressed in mCXCR4(+) CAR-T cells, while expression of exhaustion-associated markers was reduced, particularly at the 5-month time point even within the same T cell subtypes (Fig. 6d; and Figure. S10f). CD8⁺ and proliferative/exhausted subsets declined over time in mCXCR4(+) CAR-T cells, whereas CD4⁺ Tscm-cm2 cells persisted, supporting a model in which long-lived CD4⁺ memory cells mediate durable immune surveillance following AML clearance.
Differential gene expression and gene set enrichment analysis comparing mCXCR4(+) and CXCR4(-) CAR-T cells at both 4 weeks and 5 months, as well as memory-enriched (Tscm-cm) versus non-memory subsets (Fig. 6e), revealed significant enrichment of T cell activation, proliferation, mTOR signaling, and cytotoxicity pathways in CXCR4(-) and non-memory populations. In contrast, mCXCR4(+) and Tscm-cm cells were enriched for TNFα signaling (Fig. 6f), a pathway implicated in T cell survival, memory maintenance, and anti-leukemic activity47. Notably, TNFα target genes JUNB, FOS, MKP-1/DUSP1, and CD69, all key regulators of T cell self-renewal and long-term persistence, were prominently expressed and further upregulated at 5 months (Figure. S10g). Additionally differentially expressed genes included BCL2, ZFP36 (a negative regulator of T cell activation), and GADD34/PPP1R15A (an inhibitor of mTOR signaling), collectively suggesting a transcriptional program that favors memory formation by mitigating exhaustion and senescence.
To investigate the timing of memory and exhaustion switch, we performed scRNA-seq at 2.5 weeks post-injection (Fig. 6a,g; Figure. S9b), an early phase when CAR-T cells begin to engage AML and cytotoxic activity starts to decline. At this point, both CXCR4(-) and mCXCR4(+) CAR-T cells exhibited similar proportions of proliferating cells. However, mCXCR4(+) CAR-T cells were enriched for CD4⁺ T cells (65.5% vs. 5.8%) and expressed elevated levels of TCF7, IL7R, CCR7, and CD226, markers indicative of early memory commitment. While PD-1 expression was observed across all conditions, CXCR4(-) CAR-T cells showed greater expression of exhaustion markers, including LAG3, TIM3/HAVCR2, and 4-1BB/TNFRSF9, the latter associated with regulatory T cell activity in tumors48,49 (Fig. 6g). These findings indicate that CXCR4 expression in CAR-T cells promotes CD4+ fate and memory gene expression as early as 2.5 weeks post-injection.
Collectively, these data demonstrate that mCXCR4 expression programs CAR-T cells toward a memory-enriched CD4⁺ phenotype characterized by reduced exhaustion and enhanced expression of survival and longevity-associated genes. This transcriptional reprogramming is evident as early as 2.5 weeks post-infusion and persists for at least 5 months, suggesting durable remodeling of T cell fate.
Stemness- and longevity-associated gene expression in mCXCR4(+) CAR-T cells
Quiescence, self-renewal, and extended longevity are defining features of both memory T cells and hematopoietic stem cells (HSCs)50. To determine whether these lineages share common transcriptional programs, we analyzed a published scRNA-seq dataset of purified CD34⁺/CD45RA⁻ populations from cord blood. Long-term HSCs were identified by low expression of lineage/maturation markers (CD33, CD38) and high expression of canonical stem cell markers (CD90/THY1, CD45b/ITGA2, CD133/PROM1) (Figure. S11a).
Differential gene expression analysis comparing HSCs with non-HSC progenitors revealed substantial transcriptional overlap with the signatures observed in mCXCR4(+) versus CXCR4(-) CAR-T cells, as well as in Tscm-cm versus non-memory CAR-T subsets (Figure. S11b). Unsupervised clustering of shared DEGs showed that genes downregulated across both comparisons were enriched for cell cycle and mitotic processes, whereas upregulated genes were associated with oxidative stress response (Figure. S11c). Notably, these included regulators of stress tolerance and survival (FOS, FOSB, TXNIP), quiescence-promoting transcription factors (NURR2/NR4A2, HLF, KLF2), and stemness-associated factors (CITED2, SESN1). Together, these findings suggest that memory T cells and HSCs achieve persistence through shared transcriptional programs that suppress proliferation and enhance resistance to oxidative stress.
Memory-favoring metabolic programs in mCXCR4(+) CAR-T cells reflect adaptation to hypoxic bone marrow conditions
T cell fate is tightly regulated by metabolic state: while effector and exhausted T cells (Teff, Texh) predominantly rely on glycolysis51, while memory populations such as Tscm and preferentially utilize oxidative phosphorylation (OXPHOS) and fatty acid oxidation52. The BM, a physiologically hypoxic environment, imposes unique metabolic constraints on infiltrating T cells 53,54,55,56. Consistent with their quiescent phenotype, Tscm-cm subsets displayed enriched expression of OXPHOS-related genes, alongside upregulation of oxidative stress response pathways shared with hematopoietic stem cells (Fig. 6f; and Figure. S11c).
To investigate this further, we assessed metabolite and metabolism-related gene expression changes in mCXCR4(+) versus CXCR4(-) CAR-T cells. Unsupervised clustering of CAR-T cell subpopulations based on glycolytic, TCA cycle, and OXPHOS gene expression clearly separated exhausted T cells (Texh) from other subsets, characterized by high expression of glycolytic enzymes (HK, PGM) and lactate transporters (SLC16A1, SLC16A4) and low expression of OXPHOS/TCA cycle genes (Figure. S12a). Similarly, CXCR4(-) CAR-T cells showed elevated expression of glycolytic genes (GAPDH, ALDOC, TPI1) and reduced expression of TCA cycle enzymes (OGDH, SDHD, ACO1, ACO2, CS), (Figure. S12a–c), together supporting a link between metabolic programming and memory differentiation.
We measured oxygen saturation and lactate levels in non-engrafted NSG mice and in AML PDX mice with or without CAR-T treatment. As expected, untreated PDX BM was more hypoxic than that of non-engrafted NSG mice; however, lactate levels were comparable, suggesting that AML alone does not significantly elevate BM lactate production (Figure. S12d, e). In contrast, BM from CXCR4(-) CAR-T-treated PDX mice showed markedly elevated lactate compared to all other groups, including non-engrafted, untreated PDX, and mCXCR4(+) CAR-T-treated mice whereas lactate levels in mCXCR4(+) CAR-T-treated mice remained similar to baseline (Figure. S12e). These results suggest that CXCR4(-) CAR-T cells rely more heavily on anaerobic glycolysis, possibly due to impaired TCA cycle activity, leading to increased lactate production, metabolic exhaustion, and reduced therapeutic efficacy. Notably, CXCR4 overexpression enhanced memory formation most prominently under hypoxic conditions (Fig. 5b), further supporting a model in which CXCR4 facilitates memory differentiation by promoting metabolic adaptation to the bone marrow microenvironment.
CXCR4 expression promotes long-term functional immune memory and durable protection against AML rechallenge in vivo
Lastly, we investigated the formation of functional immune memory in vivo. In PDX models treated with mCXCR4(+) CD25-targeted CAR-T cells, TCF1⁺ memory T cells were observed in close proximity to CXCL12⁺ stromal cells in the spleen, while CCR7⁺ memory T cells localized near CXCL12⁺ stromal cells in the liver beyond one month post-transfusion, suggesting persistent maintenance of central memory T cells within peripheral tissue niches (Fig. 7a, b).
a HE (left) and TCF1 IHC (middle) staining in Patient 13-derived PDX spleens harvested 30 days post-treatment with mCXCR4(+) CD25-targeted CAR-T cells. CXCL12 IHC staining (right) in spleens of untransplanted NSG mice reveals stromal expression. Inset panels show 20x magnification. Scale: 20 µm. Representative images selected from sections prepared from n = 3 animals (b) CXCL12, CCR7 and CD3 IHC staining in Patient 11-derived PDX liver harvested 40 days post-treatment with mCXCR4(+) CD25-targeted CAR-T cells. CCR7-expressing naïve/Tscm and Tcm cells are observed to co-localized CXCL12-expressing stromal cells in the liver. Representative images selected from sections prepared from n = 5 animals. Scale: 50 µm c) Frequencies of immune-surveillant (CD226⁺TIGIT⁻), memory (IL7RA⁺, TCF1⁺), and checkpoint-expressing (TIM3⁺TIGIT⁺) CD4⁺ CAR-T cells in bone marrow and spleen of PDX mice treated with mCXCR4(+) CD25-targeted CAR-T cells, stratified by time post-injection. Short-term (ST) samples (n = 8 animals) were collected <30 days post-treatment; long-term (LT) samples (n = 22 animals) were collected >90 days post-treatment. Graphs show mean ± SD. Statistical analysis performed using an unpaired two-tailed t-test. d In vivo re-challenge of AML PDX previously cleared of peripheral blood AML following mCXCR4(+) CD25-targeted CAR-T treatment. Patient-derived CD25⁺ AML cells failed to engraft or expand in previously treated animals (pink lines and dots) across four independent experiments, whereas robust AML proliferation occurred in control littermates lacking CAR-T therapy. PDX IDs correspond to those in Supplementary Data 4. Source data are provided as a Source data file for (c, d).
Immunophenotypic analysis of CAR-T cells isolated at short-term ( < 30 days) and long-term ( > 90 days) time points post-treatment revealed sustained and increasingly pronounced CXCR4-associated advantages. These included elevated frequencies of CD226⁺TIGIT⁻ immune-surveillant T cells and IL7RA⁺ Tscm-cm populations, alongside reduced proportions of TIGIT⁺TIM3⁺ exhausted CD4⁺ T cells and Ki67⁺ proliferating cells (Fig. 7c).
To directly assess functional memory, we performed an AML rechallenge experiment. PDX mice that achieved complete remission following mCXCR4(+) CD25-targeted CAR-T treatment were re-injected with CD25-expressing AML cells 4-5 weeks post-remission, a stage when CAR-T cell numbers typically contract (Fig. 3f). AML cells failed to engraft or initiate disease (Fig. 7d), demonstrating functional memory and long-lasting anti-leukemic activity of mCXCR4(+) CAR-T cells.
Together, these findings show that CXCR4 expression supports the formation and maintenance of long-lived, immune-surveillant memory CAR-T cells capable of preventing relapse through sustained functional persistence. CXCR4 signaling thus plays a critical role in overcoming T cell exhaustion and enabling long-term disease suppression in vivo.
Discussion
CAR-T cell therapies have revolutionized the treatment of several B cell malignancies, including B-cell acute lymphoblastic leukemia (B-ALL), B-cell non-Hodgkin lymphoma, mantle cell lymphoma, and multiple myeloma. Extending their application to myeloid malignancies such as acute myeloid leukemia (AML) remains a significant challenge. A major barrier is the onset of CAR-T cell exhaustion, driven by chronic antigen stimulation and excessive CAR signaling, which compromises long-term persistence and leads to treatment failure and disease relapse57.
Several strategies have been developed to address CAR-T exhaustion, including optimization of CAR architecture to reduce tonic signaling, incorporation of immune checkpoint blockade, and engineering of cytokine and chemokine circuits13,58,59. Additional strategies under investigation include HLA genome editing, cytokine/chemokine engineering, and epigenetic reprogramming to reverse T cell exhaustion57,60,61,62. Enhancing CAR-T memory formation has emerged as a promising approach to prolong antitumor immunity. This has been achieved through the selection of naïve or stem-like T cell subsets, culture methods that preserve early memory phenotypes, and use of 4-1BB co-stimulatory domains to upregulate memory-associated gene programs63. Beyond these approaches, directly programming of memory-inducing signals into CAR-T cells offers an attractive strategy to enhance therapeutic durability.
To improve CAR-T cell efficacy and persistence against AML, we employed two complementary strategies: selection of CB as a T cell source and engineering of the chemokine receptor CXCR4. While CD19 CAR-T therapies for B-cell malignancies typically use autologous PB from patients, our attempts to generate CAR-T cells from AML patient-derived PB were unsuccessful due to poor T cell viability and limited expansion in vitro. These challenges likely reflect the immunosuppressive environment and low T cell counts commonly seen in AML, where the malignancy arises in early hematopoietic progenitors. These limitations prompted us to explore alternative T cell sources, particularly CB-derived T cells, which offer several advantages in adoptive cell therapy. CB-derived T cells are enriched for naïve T cells with superior proliferative capacity, longevity, and memory potential59. Their early differentiation state also contributes to reduced risk of GvHD and more permissive HLA matching, expanding the donor pool64,65. Clinically, CB transplantation has been associated with reduced relapse risk in acute leukemia, and established infrastructure for CB processing and banking supports scalable, off-the-shelf CAR-T manufacturing31,60,66,.
Building on this platform, we identified CXCR4 as a memory-enhancing surface receptor through analysis of T cell populations from AML patients in remission after CB transplantation. Enforced CXCR4 expression in CAR-T cells promoted a shift toward stem and central memory phenotypes while reducing exhaustion. Our in vitro experiments using healthy donor PB-derived CAR-T cells also demonstrated enhanced acquisition of memory phenotypes upon CXCR4 overexpression, suggesting that the benefits of CXCR4-mediated reprogramming may extend to more widely used PB-derived CAR-T products, broadening clinical applicability.
Transcriptomic and proteomic profiling revealed upregulation of genes associated with anti-apoptotic, anti-inflammatory, and oxidative stress resistance pathways, consistent with the role of the CXCR4/CXCL12 axis in maintaining hematopoietic stem cell quiescence67,68. To further understand the basis for this memory advantage, we profiled metabolic gene expression and found that CXCR4( + ) CAR-T cells may preferentially utilize oxidative phosphorylation over glycolysis. Metabolic profiling revealed that CXCR4( + ) CAR-T cells maintained lower lactate production in vivo, consistent with metabolic gene expression changes reflecting adaptation to the hypoxic bone marrow niche. These findings support a model in which CXCR4 enhances memory acquisition not only by modulating transcriptional and phenotypic states, but also by promoting metabolic fitness within the tumor microenvironment.
In vivo memory-associated gene expression emerged as early as 2.5 weeks in CXCR4( + ) CAR-T cells and persisted for at least 5 months. CXCR4( + ) CAR-T cells demonstrated superior AML killing and enhanced persistence in peripheral tissues, with memory T cells maintained in close proximity to CXCL12⁺ stromal niches in the spleen and liver. CXCR4( + ) CAR-T cells also exhibited elevated CD4:CD8 ratios, a hallmark of effective immune surveillance seen in clinical CAR-T therapy28. Finally, CXCR4( + ) CAR-T cells provided durable protection in rechallenge models, confirming the formation of functional long-term memory.
Target selection remains a fundamental challenge in CAR-T design, requiring careful consideration of both tumor specificity and safety. We selected CD25 based on its enriched expression in AML relative to hematopoietic stem cells, lack of expression in non-hematopoietic tissues, and its association with chemotherapy resistance, poor survival outcomes, and leukemia-initiating potential. While CD25 is also expressed on activated T cells and Tregs, fratricide was modest and CAR-T cells exhibited greater cytotoxicity against AML blasts over Tregs in vitro. We observed potential differences in CD25 surface presentation between AML blasts and T cells: AML cells predominantly expressed single-chain CD25, whereas T cells expressed the heterotrimeric high-affinity IL-2 receptor. Although the CD25 epitope recognized by our S417 Fab remains accessible in both configurations, it is possible that these structural differences affect immune synapse formation and quality—key determinants of CAR-T antigen sensitivity and cytotoxicity69,70. Whether structural differences in CD25 presentation contribute to differences in CAR-T antigen recognition and how these differences can be leveraged to enhance specificity is a topic for future research.
Nevertheless, the potential for on-target off-tumor effects on Tregs warrants careful consideration. Treg depletion in mice results in the spontaneous development of autoimmune diseases71, while human genetic disorders that involve defects in Treg function are characterized by a variety of autoimmune phenomena72. On the other hand, CD25-targeted antibodies have reported minimal adverse autoimmune outcomes in both pre-clinical nonhuman primate models73,74, and in clinical trials in patients (e.g., daclizumab)75. Clinical trials of CD7-targeted CAR-T cells, which also target normal T cells and Tregs, similarly report low to manageable autoimmune effects, often treatable with standard immunosuppression76. Overall, we believe autoimmune risks posed by our CD25-targeted CAR-T cells will be comparable to those reported for existing therapies that deplete Tregs. In the context of aggressive CD25⁺ AML, transient Treg depletion may be an acceptable risk, especially if followed by supportive care or hematopoietic stem cell transplant. In addition, engineering approaches such as bi-specific CAR circuits or optimized antibody design can potentially reduce toxicity against Tregs.
Given that CD25 is expressed in only a subset of AML cases, we also investigated CD96, a second target identified through our antigen screen, which is expressed in over 55% of AML patient samples in our cohort. CD96 is an immune checkpoint molecule found on AML blasts as well as certain solid tumors, making it a promising target for broader therapeutic applications77. We demonstrated that CXCR4 expression enhances the efficacy of CD96-targeted CAR-T cells in vivo, mirroring the improvements in anti-AML activity and memory formation observed with CD25-targeted CAR-T cells. These results underscore the versatility of CXCR4 as a modular enhancement strategy that can be applied across CAR constructs to improve T cell durability, regardless of the antigen target.
In conclusion, our study identifies CXCR4 as a powerful engineering target to enhance CAR-T cell efficacy, persistence, and immune memory in the treatment of AML. By leveraging a developmental insight from patient T-cells in remission post-CBT and integrating CXCR4 into CAR-T constructs, we improved both the functional quality and durability of anti-leukemic responses in preclinical models. CXCR4-mediated memory enhancement was antigen-independent and sustained over time, offering protection against relapse through durable immune surveillance. These findings support the development of CXCR4-engineered CAR-T therapies not only for AML but potentially for other malignancies where long-term T cell persistence is critical to therapeutic success. Further investigation into the safety, timing, and modulation of CXCR4 expression will be important to guide clinical translation and maximize its therapeutic benefit. Collectively, our findings lay the foundation for clinical trials of CXCR4-enhanced CAR-T therapies in AML and beyond.
Methods
Human samples
Experiments were performed with authorization from the Institutional Review Board for Human Research at RIKEN Center for Integrative Medical Sciences. Leukemia samples were from patients at Toranomon Hospital (Tokyo, Japan). CB samples were obtained from Central Japan Cord Blood Bank (Aichi, Japan). Healthy donor PB and CB samples for extended in vitro co-culture experiments were obtained at Nippon Medical School (Tokyo, Japan). Written informed consent was obtained from all patients and healthy donors for their participation in the study, including on the publication of indirectly identifying information (age, sex, and diagnosis). Written informed consent was also obtained for the collection of cord blood and peripheral blood from healthy donors. No bias or consideration was made on the sex of human patient or donor samples, as our study primarily focuses on cells either in vitro or in a xenogeneic in vivo environment. No compensation, financial or otherwise, was provided for participation in the study.
Cell isolation from primary samples
AML BM MNCs and CB MNCs were isolated with density gradient centrifugation. CD34+ and CD34- cells were separated by autoMACS (Miltenyi Biotec) with anti-human CD34 immunomagnetic beads. T cells were further enriched with autoMACS using Pan T Cell Isolation Kit (human) (Miltenyi Biotec) from CD34- population.
CyTOF experiments
Information on antibodies (clone and manufacturer) are summarized in Table S2. When available, antibodies pre-conjugated to metals were used for detection of target proteins. For detection of other proteins except LAG3 and TIM3, purified antibodies were conjugated with metals using MaxPAR X8 or X9 Antibody Labeling Kit as per manufacturer recommendation (Standard Biotools). Antibody for Granzyme K was a kind gift of Dr. Inagaki and Dr. Hirata, Nippon Medical School, Tokyo, Japan. CD25, LAG3 and TIM3, binding of biotin-, FITC- and APC-conjugated primary antibodies, respectively, were detected by metal-conjugated anti-biotin, -APC and -FITC secondary antibodies. Cells were first incubated with biotin-conjugated CD25, FITC-conjugated LAG3 and APC-conjugated TIM3 antibodies for 30 minutes on ice, with 1 µl Cell-ID intercalator-Rh (500 µM) added for detection of dead cells during the last 15 minutes. After washing with Maxpar Cell Staining Buffer (CSB) (Standard Biotools), cells were incubated with metal-conjugated primary antibodies and anti-biotin, -FITC and -APC secondary antibodies for 30 minutes at room temperature (RT). Cells were washed with CSB, fixed and permeabilized using Transcription Factor (TF) Buffer Set (BD Biosciences) as per manufacturer recommendation and stained with metal-conjugated intracellular antibodies for 30 minutes at RT. Cells were then washed twice with TF Perm/Wash Buffer and incubated for 1 h or overnight in 1 ml Maxpar Fix and Perm Buffer containing 1 µl Cell-ID Intercalator-Ir (Standard Biotools). Directly before analysis cells were re-suspended in ddH2O(Maxpar Cell Acquisition Solution; FLUIDIGM #201240), filtered (20 µm Celltrix, Sysmex (35um Cell strainer snap cap 5 mL tube; BD #352235)), counted and adjusted to 5 − 10 × 105 cells ml−1. EQ four-element calibration beads were added at a final concentration of 1:10 of the sample volume to be able to normalize the data to compensate for signal drift and day-to-day changes in instrument sensitivity. Stained cells were analyzed on a Helios mass cytometer (Standard Biotools) using the CyTOF Helios MS software v7.0.8493 operated with default settings.
CyTOF data analysis
FlowJo (BD Biosciences) was used for initial manual gating on live single cells and selection of T cells (for patient samples) and human cells (for PDX samples). Gated FCS files were exported for downstream exploratory and statistical analysis using the CATALYST package in R78 following a recommended workflow79. Samples were downsampled to include a maximum of 3000 cells. Data were transformed with the CATALYST default arcsinh transformation (scale factor 5). Self-organizing map lustering and UMAP embedding were performed using default hyperparameter settings. The following 12 markers were used for clustering and UMAP embedding of AML patient T cells: CD4, CD8, CCR7, IL7RA, CD45RA, TCF7, CD45RO, Tbet, GZMK, GZMB, TIGIT. The following 21 markers were used for clustering and UMAP embedding of PDX-derived AML and CAR-T cells: CD3, CD4, CD8, CD33, TIGIT, Tbet, GZMK, CCR7, CTLA4, Ki67, CD62L, PD1, IL7RA, CD45RA, CD45RO, TCF1, CD226, CASP3, GZMB, LAG3, TIM3. Cell types were designated by manual inspection of marker intensities. We found that results were robust to multiple iterations of downsampling, seeds, and inclusion/exclusion of some markers.
Mice and xenogeneic transplantation
NOD.Cg-PrkdcscidIl2rgtmlWjl/Sz (NSG) mice were bred and maintained under defined flora at the animal facility of RIKEN and at The Jackson Laboratory conditioned at 23 + /−2 °C in 50 + /−10% humidity, on 12h-dark/light cycle. All experiments were performed with authorization from and according to guidelines established by the Institutional Animal Committees at RIKEN (AEY2024-004(2)) and The Jackson Laboratory. No specifications for maximal hematological tumor burden were specified; instead, mice were regularly monitored and euthanized in accordance with humane endpoint criteria outlined by the respective institutional review boards, including rapid weight loss, prolonged abnormal appearance, and prolonged abnormal lethargy. Both female and male newborn NSG mice received 1.5 Gy total body irradiation followed by intravenous injection of 103 to 105 sorted human cells within 72 h of birth as described80. The extent of engraftment of human cells in NSG recipients was assessed by retro-orbital phlebotomy and flow cytometry. Sex of PDX host animals was not considered as part of the study design, which focuses on xenotransplanted human cells.
Patient cohort bulk RNA-seq and bioinformatic analysis
AML cells used for bulk RNA-seq included sorted blast populations from original patient samples (taken from BM or PB) and PDX engrafted samples (taken from recipient BM or spleen). Sorting strategies were chosen case-by-case according to each sample’s immunophenotypic characteristics. HSPC samples consisted of sorted cord blood cells. The specific source and sorting strategy used per library are detailed in Supplementary Data 5. RNA was extracted using TRIzol reagent (Catalog No. 15596018; Invitrogen). NEB Next Ultra RNA Library Prep Kit for Illumina (Catalog No. E7530; New England Biolabs) was used for RNA library preparation. Final library size distribution was validated using Bioanalyzer (Agilent) and quantified using quantitative PCR. The DNA libraries were sequenced on a HiSeq 2500 (Illumina, using 50-base single-end read mode). Reads were mapped to the human genome (NCBI version 19) using TopHat2 version 2.0.8 and botwie2 version 2.1.0 with default parameters, and gene annotation was provided by NCBI RefSeq. The data of the non-hematopoietic tissues used for the analyses described in this manuscript were obtained from GTEx Analysis V8 in the GTEx Portal on 4/22/22 (https://gtexportal.org/home/). Differential expression analysis was performed using DESeq2 v1.46.0. The programming framework R version 4.0.3 was used in these analyses.
Construction of lentiviral vectors
Antigen-specific CAR and CXCR4 sequences (mouse, NM_009911.4; human, NM_003467.3) were synthesized as gBlocks gene fragments (IDT) and were transduced into pHR_SFFV (gift from Wendell Lim (Addgene plasmid #79121)) using the In-Fusion HD Cloning Kit (TAKARA). Replication-incomplete lentivirus was packaged via transfection of 293 T cells with pHR_SFFV vector containing human or mouse CXCR4 cDNA, and/or pHR_SFFV vector containing the CAR construct and packaging plasmids (pCMVR, pL2, pMD2.G-VSV-G, pAdV) (Promega) using JetPEI transfection reagent (Polyplus). Lentiviral particals for CXCR4( + ) CAR-T cells were created by dual transfection of 293 T cells with pHR_SFFV_[m/h]CXCR4 and pHR_SFFV_CDxx-CAR (CD25, CD96, CD33, and CD19-targeted CAR-T cells) in a ratio of 1:3. Viral supernatant was harvested four days after transfection and concentrated using Vivaspin200 (Merck). Concentrated viral supernatant was centrifuged overnight at 4 C and the pellet resuspended in culture medium for T cell infection. We used an in-house anti-human CD25 scFv sequence, generated as previously described43. For CARs targeting other antigens, we used the following published scFv sequences: CD1981, CD3382, CD9646. Sequences for all viral vectors have been deposited on GenBank (accession numbers in the Data Availability section).
CB-derived CAR-T cell generation, Treg differentiation, in vitro cytotoxicity assays and in vivo treatment in AML PDX mice
T cells were prepared from CB mononuclear cells as described above and cultured in X-Vivo20 (Lonza) supplemented with 5% fetal bovine serum, 10mM N-acetyl-cysteine (Sigma-Aldrich #A9165) and 55 µm 2-mercaptoethanol (Thermo Scientific). 106 T cells were stimulated with 25 µl Human T-activator CD3/CD28 DynaBeades (Thermo Scientific). Next day, T cells were transduced with CAR lentivirus with vectofusin-1 (10 µ g/ml) (Miltenyi Biotec). At 48 h post-transduction, T cells were analyzed for CAR and CXCR4 transgene expression by flow cytometry using anti-G4S linker and anti-(mouse/human)CXCR4, respectively (antibody information in Table S3). After confirmation of surface protein expression, CD25-targeted CAR-T cells were retro-orbitally injected into AML PDX mice. CB samples used for PDX were HLA genotyped and matched to an appropriate patient sample where possible. Degree of matching was from 0-2/8 loci for most cases. A detailed analysis of PDX treated with CAR-T HLA-match meeting the clinical standard for allogeneic CBT ( > = 4/8 loci) is provided in Figure. S3c. Details of AML PDX mice and CAR-T treatment are summarized in Supplementary Data 4.
In vitro cytotoxicity assays were performed by co-culture of ~105 CAR-T cells with either AML, autologous CD34 + HSC, or autologous Tregs in a ratio of 1:1 for 72 h. Tregs were differentiated from CB-derived naïve T cells using the CellXVivo Human Treg Cell Differentiation Kit (R&D Systems; #CDK0006). In all cases, surviving target cells were assessed as a percentage of the respective viable cell count after 72 h of solo culture.
Long-term co-culture experiments CB- or PB-derived T cells were performed as above with the addition of hCXCR4 (100 ng/mL) at the start of co-culture on d3. Co-cultures were performed under normoxic (21% pO₂) and hypoxic (2.5% pO₂) conditions, with or without hCXCR4. Co-culture was performed using CD25+ TL1 as target cells and maintained for 6 days as primary AML blasts typically lose viability during prolonged culture. Memory and exhaustion markers were assessed by flow cytometry on d8. All long-term co-culture samples and experiments were performed at Nippon Medical School (Tokyo Japan).
Flow cytometry
Information on antibodies (clone, label, manufacturer) are summarized in Table S3. Analysis was performed using FACSAriaIII or FACSCanto II (BD Biosciences) using FACSDiva v6.1.3 or FACSDiva v8.0.1, respectively.
Histopathological analysis
Tissue sections (3μm) were cut from 4% paraformaldehyde-fixed paraffin-embedded recipient organs. Sections were deparaffinized using xylene and ethanol, and antigen retrieval was performed (Histofine pH9.0 NICHIREI BIOSCIENCE INC.). Non-specific background was reduced by incubating the slides in methanol and H2O2 (Wako). After blocking with horse serum, slides were incubated with mouse anti-human TCF1 antibody (R&D systems, #2203) (1:200), anti-human CD33 (R&D systems, MAB11371) (1:100), anti-human and mouse CXCL12 (R&D systems, MAB350) (1:20), or anti-human CCR7 (Abcam, ab253187) (1:100) and then HRP-conjugated horse anti-rabbit or anti-mouse IgG antibody (ImmPRESS, MP-7500). Slides were then stained with 3,3′-diaminobenzine or hematoxylin and eosin, dehydrated, mounted using VectaMount (Vector Laboratories) and analyzed with the Axiovert 200 microscope (Zeiss). Photos were taken using the AxioCam MRc 5 and LSM980 (Zeiss), S6 E(Leica) and AxioVision release 4.6 software.
Statistical analysis
Error bars and in-text ranges report mean + SD unless specified otherwise. Difference testing was performed using unpaired two-tailed student’s t-test unless specified otherwise. P-value less than 0.05 was considered significant. For box and whisker plots: Central lines show median, lower and upper hinges show first and third quartiles (the 25th and 75th percentiles) and whiskers extend up to 1.5 * IQR, unless specified otherwise. Pearson’s r values reported in-text were calculated using the cor.test() function in R. Survival analysis was performed using the ‘survival’ package in R.
scRNA-seq
Approximately 13,000 cells from each condition were loaded onto the 10x Genomics Chromium Controller using Chromium Single Cell 5’ Library and Gel Bead Kit v1.1. The RT reaction proceeded for 45 minutes at 53 °C, and then the temperature was raised to 85 °C for 5 minutes to inactivate the reverse transcriptase. PCR amplification: The cDNA molecules were amplified using PCR according to the manufacturer’s instructions. The reaction mixture contained a PCR master mix from the Chromium Single Cell 3’ Library and Gel Bead Kit v2, along with the following cycling conditions: 98 °C for 45 seconds, followed by 12 cycles of 98 °C for 20 seconds, 67 °C for 30 seconds, and 72 °C for 1 minute, and then a final extension step of 72 °C for 1 minute. The PCR product was purified using SPRI beads according to the manufacturer’s instructions. The purified PCR product was fragmented, end-repaired, and ligated to adapters according to the manufacturer’s instructions for the Illumina sequencing platform. The final library was then size-selected, PCR amplified, and sequenced on an Illumina instrument according to standard protocols. Fastq were aligned to hg38 (refdata-gex-GRCh38-2020-A) using cellranger v5.0.0 (cellranger multi with gex and vdj libraries). For each library cells were processed using Seurat v4.0.183 filtering excluding those with more than 6 median absolute deviations from the median for UMI count, gene count, and mitochondrial UMI percentage (up to a max of 25% mitochondrial UMI). Counts normalized to log(1 + UMI per 10000), 2000 variable features were selected, scaled to mean 0 unit variance and reduced to 30 principal components. Clustering was performed with FindClusters using the leiden84 algorithm and resolution 1. RunTSNE and RunUMAP were run with default parameters.
Libraries were merged as follows: SelectIntegrationFeatures was used to select common variable features. Libraries were combined and these features were scaled to mean 0 unit variance for 20 principal components. Harmony v0.185 was applied to batch correct the principal components with default parameters. Clustering was performed with FindClusters using the leiden algorithm, resolution 1 and method = “igraph”. RunTSNE and RunUMAP were run with default parameters. T cell subtype designations: Graph-based clusters were designated identities based on the inspection of expressed T cell subtype markers (see Figure. S7a). Expression of CXCR4 and other HSC marker genes was performed using Adaptively thresholded Low-Rank Approximation (ALRA)86. Differential Expression Analysis: DE analysis was performed using DESeq2 using recommended settings for scRNA analysis (minReplicatesForReplace = Inf, minmu = 1e-6, useT = TRUE). For comparisons with biological replicates (ie, between CXCR4(-) and CXCR4( + ) 4 weeks and 5 months), pseudobulk counts were calculated per cluster per sample and used to construct the DESeq object. Hypothesis testing was performed using the Wald test with the design ~ treatment + cellType. For comparisons where only one biological replicate was available, the likelihood ratio test was used, as per recommendations for scRNA-seq. Expression of CXCR4 transgene and CAR construct was determined by re-mapping unmapped scRNA-seq reads to a reference FASTA containing the CAR, mouse CXCR4, and human CXCR4 cDNA sequences. Only re-mapped reads that mapped uniquely to the CAR and mouse CXCR4 cDNA sequences were counted.
CD4 TIL analysis
A reprocessed high-quality subset of the CD4 TIL dataset by Zheng et al. 30 was obtained from (https://doi.org/10.6084/m9.figshare.21981536.v1). Previously defined clusters were used as-is, but cell type labels were re-evaluated and assigned by inspection of marker genes. Raw counts were used to infer expression levels with ALRA.
HSPC scRNA-seq analysis
Raw gene x count matrix for a previously published CD34+ cord blood dataset87 was downloaded from GEO under accession GSE162950 / GSM4968836. Cells were filtered for detected features ≥1000 and nCount RNA≤ 25000 prior to dimensionality reduction and clustering as described above. CD34+ HSPCs were annotated based on the expression of known HSPC marker genes. DE analysis comparing HSPCs to non-HSPCs was performed on single cells as described above. GO enrichment analysis of shared CXCR4( + ) CAR-T, T central memory, and HSPCs was performed using ActivePathways88.
in vivo hypoxia detection
Hypoxia detection was performed using the EF5 Hypoxia detection kit, Alexa Fluor 488 (Merck). Following the manufacturer’s protocol, EF5 was diluted in 100% ETOH and subsequently adjusted to a 10 mM EF5 solution with 5% glucose. The 10 mM EF5 solution was administered via the retro-orbital sinus at a dose of 30 mg/kg. Three hours after the injection, tissue samples were prepared by freezing them in OCT compound and fixing them with 4% PFA and EtOH. For flow cytometry analysis, they were fixed with 4% PFA and EtOH after being dissociated into single cells. In both methods, detection was carried out using the Alexa Fluor 488 anti EF5 antibody included in the kit.
Tissue Lactate concentration
The tissues were weighed and submerged in 500 µL of ice-cold PBS (-). Tissues were then dissected with scissors and homogenized using a glass homogenizer. The supernatant was collected by centrifugation at 500 × g for 5 minutes and subsequently filtered through a 10 kDa filtration membrane (AMICON ULTRA 0.5 ML 10 K, Merck Millipore) at 14000 × g. The lactate concentration in the supernatant was measured using the Lactate assay Kit (DOJINDO) following the manufacturer’s instructions. The concentration in µmol/mL was calculated and converted to nmol/mg based on the weight per tissue.
Cryo-EM expression, and purification of IL-2 receptor subunits and Fabs
The baculovirus transfer vector for extracellular domain of human IL-2Rb (residues 27-240) and human IL-2Rg (residues 23-255), N-terminally fused with a gp67 signal peptide and C-terminally fused with 8 x His tag, were respectively subcloned into the pFastBac1 by In-Fusion cloning technology (Clontech). IL-2Rg was further mutated to remove glycosylation sites at Asn-75 by replacement with Gln. The final plasmid was transformed into DH10MultiBac E. Coli cells (ATG:biosynthetics GmbH, Germany) competent cells to generate the recombinant bacmid DNA. Bacmid preparation and P0, P1, P2 and P3 virus production were performed as described previously89. The generated bacmid was transfected into sf9 cells at a cell density of 1.0 × 106 cells/ml using FuGENE HD (Promega) according to the manufacturer’s instructions. P0 viral stock was collected after 72 h post-transfection and isolated into individual plaque. Isolated clones were continually amplified for production of higher-titer P1 to P3 viral stock. High Five cells were selected for IL-2Rb and IL-2Rg overexpression. High Five cells cultured at a cell density of 2 × 106 cells/ml in Express Five™ SFM (Thermo Fisher Scientific) were infected with the P3 virus at Moi of 1.0. After 96 h post-infection at 19 °C under constant shaking, cell culture supernatant was harvested by centrifugation (2200 × g for 10 min at 4 °C followed by 15000 × g for 30 min at 4 °C) to remove the cell pellet and debris and was then filtrated with the 0.45-mm pore sized membrane filter.
The gene encoding extracellular domain of human IL-2Ra (residues 22-240), N-terminally fused with a signal peptide derived from an immunoglobulin kappa light chain and C-terminally fused with the linker sequence 3x(GGGGS) and 6xHis tag was subcloned into the pcDNA3.4 vector by In-Fusion cloning technology (Clontech). The protein was expressed using the Expi293 Expression System according to the manufacturer’s instructions. On day 3. Cell culture supernatant was harvested by centrifugation (4 °C, 11000 × g, 20 min) to remove the cell pellet and was filtrated with the 0.45-mm and 0.22-mm pore sized membrane filter.
Each protein secreted in the culture medium was purified by a HisTrap HP column (cytiva, 17524801). followed by gel-filtration chromatography on a Superdex 200 Increase10/300 GL column (cytiva, 2899044) pre-equilibrated with a wash buffer containing 20 mM Tris-HCl (pH8.0),150 mM NaCl.
Chimeric Fab heavy chains (VH-Rituximab CH1) attached with the C-terminally TEV protease cleavage site and 6 x His tag and chimeric light chains (VL-Rituximab CL) were, respectively, subcloned into the pcDNA3.4 vector. Each Fab protein was expressed, with plasmids for both chimeric Fab heavy chain and chimeric light chain with an amount ratio of 1:3, using the Expi293 Expression System. On day 7, cell culture supernatant was harvested by centrifugation (4 °C, 11000 × g, 20 min) to remove the cell pellet and was filtrated with the 0.45-mm and 0.22-mm pore sized membrane filters. Fab protein secreted in the culture medium was purified by a HisTrap HP column and then the affinity tags were cleaved by TEV protease at 4 °C overnight. The cleaved tags and protease were removed by a HisTrap HP column, followed by gel-filtration chromatography on a Superdex 200 Increase10/300 GL column pre-equilibrated with the wash buffer.
To prepare complexes of recombinant proteins for cryo-EM, the purified IL-2R subunits (each 0.7 mg/ml) and IL-2 (PROSPEC-TANY, CYT-209) were mixed with a molar ratio of 1:1:1 and then incubated at 4 °C overnight. The ligand-receptor complex was purified by gel-filtration chromatography on a Superdex 200 Increase10/300 GL column equilibrated in 20 mM Tris-HCl (pH8.0), 150 mM NaCl, followed by incubation with each Fab in two-fold molar ratio to the complex at 4 °C for 30 min. Heteropentamers were purified by gel-filtration chromatography. The fraction containing the heteropentamers (0.15 mg/ml, 3 μl) was applied onto UV/ozone freshly treated graphene grid (Quantifoil 300 mesh Au R1.2/1.3) and subsequently vitrified for 3 s at 4 °C in 100% humidity and plunge-frozen in liquid ethane using the Vitrobot Mark IV.
Cryo-EM data acquisition and image processing
Cryo-EM imaging of the complex was performed on a 300 kV Titan Krios G4 equipped with a BioQuantum K3 direct electron detector in the electron counting mode. The imaging was performed at a nominal magnification of ×105,000, corresponding to a calibrated pixel size of 0.829 Å per pixel. Each micrographic movie was recorded for a total of 14.5 electrons per pixel per second for 2.4 s, resulting in an accumulated exposure of 50.4 e-/A2. A total of ∼6,006 movies were collected. All the data were automatically acquired using the EPU software, with a defocus range of −0.8 to −2.0 μm.
All image processing was performed with cryoSPARC90. All of the dose-fractionated movies were subjected to beam-induced motion correction using the patch motion correction and the patch contrast transfer function (CTF) values were estimated with a default setting. 2,309,717 particles were picked using blob picker from the micrographs and extracted with a box size 448 pixels and binning to 128 pixels. Initial 3D maps were obtained from ab-initio 3D reconstruction with 805,113 particles rigorously selected by 2D classification followed by heterogenous refinement was performed using the 3D maps and 1,994,470 particles roughly selected by 2D classification. The 528,825 selected particles were extracted with a box size of 380 pixels without binning, subjected to homogenous refinement, 2D and 3D classifications yielded 353,002 particles for the final refinement. The particles were polished with reference-based motion correction and subjected to non-uniform refinements with per-particle CTF refinements. This resulted in a final map with an overall resolution of 3.0 Å according to Fourier shell correlation (FSC) = 0.143 criterion.
Model building and validation
The model of the protein complex was constructed by AlphaFold391 as an initial model, and fitted into density maps using ChimeraX92 and ISOLDE93 in UCSF ChimeraX. The model of the complex of the variable domains was iteratively refined by using phenix.real_space_refine of the Phenix package94. Molecular graphics and density map were prepared with UCSF ChimeraX.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw sequence level datasets are deposited under controlled access at the Japanese Genotype-phenotype Archive (JGA) in accordance to privacy and data protection terms stipulated in the research ethics agreement. Approval for raw data access can be obtained through a formal submission process via the National Bioscience Database Center Human Database. Sequence-level bulk RNA-seq data are deposited in the JGA under accession code JGAD000356 (https://ddbj.nig.ac.jp/search/entry/jga-dataset/JGAD000356); for data access request, refer to (https://humandbs.dbcls.jp/en/hum0243-v1#rna). Sequence-level scRNA-seq data are deposited in the JGA under accession code JGAD000990 (https://ddbj.nig.ac.jp/search/entry/jga-dataset/JGAD000990); for data access request, refer to (https://humandbs.dbcls.jp/en/hum0116-v2#scrna). De-identified count level bulk RNA-seq data for AML patient cohort are deposited on Zenodo under accession code 15606803. De-identified processed scRNA are deposited on Zenodo under accession code 17150046. CyTOF data are deposited on Zenodo under accession code 17247607. Cryo-EM models are deposited on the PDB under accession number 9KMC. Publicly available datasets used in this study can be accessed as below: Pre-processed CD4 + TIL scRNA-seq are available on Figshare under accession code 21981536.v1. scRNA-seq counts for CB-derived CD34+ HSPCs are available on NCBI GEO under accession code GSE162950 / GSM4968836. BeatAML2 cohort gene expression and metadata are available through the public portal (https://biodev.github.io/BeatAML2/]. GTEx V8 bulk tissue expression data is available through the official GTEx website (https://gtexportal.org/home/downloads/adult-gtex#bulk_tissue_expression) (Filename: GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_median_tpm.gct.gz; accessed 4/22/22) Full sequences of lentiviral constructs used in this study are available on Genbank: PX372844 (https://www.ncbi.nlm.nih.gov/nuccore/PX372844.1/) pHR_SFFV_hCXCR4. PX372845 (https://www.ncbi.nlm.nih.gov/nuccore/PX372845.1/) pHR_SFFV_mCXCR4. PX372846 (https://www.ncbi.nlm.nih.gov/nuccore/PX372846.1/) pHR_SFFV_CD25CAR. PX372847 (https://www.ncbi.nlm.nih.gov/nuccore/PX372847.1/) pHR_SFFV_CD96CAR. PX372848 (https://www.ncbi.nlm.nih.gov/nuccore/PX372848.1/) pHR_SFFV_CD19CAR. PX372849 (https://www.ncbi.nlm.nih.gov/nuccore/PX372849.1/) pHR_SFFV_CD33CAR. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Code availability
An R markdown file of all code used to analyze scRNA-seq and patient CyTOF datasets and minimal datasets are available on Zenodo under accession code 15460722 (https://doi.org/10.5281/zenodo.15460722).
References
Zhao, Y. et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res 70, 9053–9061 (2010).
Grupp, S. A. et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med 368, 1509–1518 (2013).
Lee, D. W. et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet 385, 517–528 (2015).
Park, J. H. et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med 378, 449–459 (2018).
Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med 374, 2209–2221 (2016).
Sterner, R. C. & Sterner, R. M. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 11, 69 (2021).
Jin, X. et al. First-in-human phase I study of CLL-1 CAR-T cells in adults with relapsed/refractory acute myeloid leukemia. J. Hematol. Oncol. 15, 88 (2022).
Tambaro, F. P. et al. Autologous CD33-CAR-T cells for treatment of relapsed/refractory acute myelogenous leukemia. Leukemia 35, 3282–3286 (2021).
Mardiros, A., Forman, S. J. & Budde, L. E. T cells expressing CD123 chimeric antigen receptors for treatment of acute myeloid leukemia. Curr. Opin. Hematol. 22, 484–488 (2015).
Ritchie, D. S. et al. Persistence and efficacy of second generation CAR T cell against the LeY antigen in acute myeloid leukemia. Mol. Ther. 21, 2122–2129 (2013).
Fraietta, J. A. et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med 24, 563–571 (2018).
Deng, Q. et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med 26, 1878–1887 (2020).
Kouro, T., Himuro, H. & Sasada, T. Exhaustion of CAR T cells: potential causes and solutions. J. Transl. Med 20, 239 (2022).
Porter, D. L. et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl. Med 7, ra139 (2015).
Kobayashi, N., Takata, H., Yokota, S. & Takiguchi, M. Down-regulation of CXCR4 expression on human CD8+ T cells during peripheral differentiation. Eur. J. Immunol. 34, 3370–3378 (2004).
Contento, R. L. et al. CXCR4-CCR5: a couple modulating T cell functions. Proc. Natl. Acad. Sci. USA 105, 10101–10106 (2008).
Kohli, K., Pillarisetty, V. G. & Kim, T. S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 29, 10–21 (2022).
Goedhart, M. et al. CXCR4, but not CXCR3, drives CD8(+) T-cell entry into and migration through the murine bone marrow. Eur. J. Immunol. 49, 576–589 (2019).
Sun, R. et al. CXCR4-modified CAR-T cells suppresses MDSCs recruitment via STAT3/NF-kappaB/SDF-1alpha axis to enhance efficacy against pancreatic cancer. Mol. Ther. 31, 3193–3209 (2023).
Muller, N. et al. Engineering NK Cells Modified With an EGFRvIII-specific Chimeric Antigen Receptor to Overexpress CXCR4 Improves Immunotherapy of CXCL12/SDF-1alpha-secreting Glioblastoma. J. Immunother. 38, 197–210 (2015).
Moles, M. W. et al. CXCR4 has a dual role in improving the efficacy of BCMA-redirected CAR-NK cells in multiple myeloma. Front Immunol. 15, 1383136 (2024).
Biondi, M. et al. Selective homing of CAR-CIK cells to the bone marrow niche enhances control of the acute myeloid leukemia burden. Blood 141, 2587–2598 (2023).
Ara, T. et al. A role of CXC chemokine ligand 12/stromal cell-derived factor-1/pre-B cell growth stimulating factor and its receptor CXCR4 in fetal and adult T cell development in vivo. J. Immunol. 170, 4649–4655 (2003).
Chaix, J. et al. Cutting edge: CXCR4 is critical for CD8+ memory T cell homeostatic self-renewal but not rechallenge self-renewal. J. Immunol. 193, 1013–1016 (2014).
Bleul, C. C., Wu, L., Hoxie, J. A., Springer, T. A. & Mackay, C. R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94, 1925–1930 (1997).
Jourdan, P. et al. IL-4 induces functional cell-surface expression of CXCR4 on human T cells. J. Immunol. 160, 4153–4157 (1998).
Ando, M. et al. Rejuvenating effector/exhausted CAR T cells to stem cell memory-like CAR T cells by resting them in the presence of CXCL12 and the NOTCH ligand. Cancer Res Commun. 1, 41–55 (2021).
Melenhorst, J. J. et al. Decade-long leukaemia remissions with persistence of CD4(+) CAR T cells. Nature 602, 503–509 (2022).
Gattinoni, L., Speiser, D. E., Lichterfeld, M. & Bonini, C. T memory stem cells in health and disease. Nat. Med 23, 18–27 (2017).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Bottomly, D. et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 40, 850–864 e859 (2022).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018).
Saito, Y. et al. Identification of therapeutic targets for quiescent, chemotherapy-resistant human leukemia stem cells. Sci Transl Med 2, 17ra19 (2010).
Li, J. et al. Role of CD25 expression on prognosis of acute myeloid leukemia: A literature review and meta-analysis. PLoS One 15, e0236124 (2020).
Gonen, M. et al. CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900. Blood 120, 2297–2306 (2012).
Hosen, N. et al. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 104, 11008–11013 (2007).
Milone, M. C. et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 17, 1453–1464 (2009).
Costa, M. J. et al. A mouse model for evaluation of efficacy and concomitant toxicity of anti-human CXCR4 therapeutics. PLoS One 13, e0194688 (2018).
Sommermeyer, D. et al. Fully human CD19-specific chimeric antigen receptors for T-cell therapy. Leukemia 31, 2191–2199 (2017).
Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020).
Liu, D. D. et al. Umbilical cord blood: a promising source for allogeneic CAR-T cells. Front Oncol. 12, 944248 (2022).
Rassek, K. et al. New player in CAR-T manufacture field: comparison of umbilical cord to peripheral blood strategies. Front Immunol. 16, 1561174 (2025).
Tanaka, M. et al. Development of a simple new flow cytometric antibody-dependent cellular cytotoxicity (ADCC) assay with excellent sensitivity. J. Immunol. Methods 464, 74–86 (2019).
Read, J. A., Rouce, R. H., Mo, F., Mamonkin, M. & King, K. Y. Apoptosis of hematopoietic stem cells contributes to bone marrow suppression following chimeric antigen receptor T cell therapy. Transpl. Cell Ther. 29, 165 e161–165 e167 (2023).
Ng, Y. Y., Du, Z., Zhang, X., Chng, W. J. & Wang, S. CXCR4 and anti-BCMA CAR co-modified natural killer cells suppress multiple myeloma progression in a xenograft mouse model. Cancer Gene Ther. 29, 475–483 (2022).
Renart-Depontieu, F. et al., Florence RENART-DEPONTIEUSachiko TAKAMIGermain MARGALL-DUCOSNicola BeltraminelliPierre GarroneAnne ROGELAymen Al-ShamkhaniXavier Preville. inventors; Cd96-binding agents as immunomodulators (WO2019030377A1). (2018).
Mehta, A. K., Gracias, D. T. & Croft, M. TNF activity and T cells. Cytokine 101, 14–18 (2018).
Freeman, Z. T. et al. A conserved intratumoral regulatory T cell signature identifies 4-1BB as a pan-cancer target. J. Clin. Invest 130, 1405–1416 (2020).
Swatler, J. et al. 4-1BBL-containing leukemic extracellular vesicles promote immunosuppressive effector regulatory T cells. Blood Adv. 6, 1879–1894 (2022).
Luckey, C. J. et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 103, 3304–3309 (2006).
Schurich, A. et al. Distinct metabolic requirements of exhausted and functional virus-specific CD8 T cells in the same host. Cell Rep. 16, 1243–1252 (2016).
Rangel Rivera, G. O. et al. Fundamentals of T cell metabolism and strategies to enhance cancer immunotherapy. Front Immunol. 12, 645242 (2021).
Chen, Y. & Gaber, T. Hypoxia/HIF modulates immune responses. Biomedicines 9, 260 (2021).
Cascone, T. et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987 e974 (2018).
Singer, K. et al. Warburg phenotype in renal cell carcinoma: high expression of glucose-transporter 1 (GLUT-1) correlates with low CD8(+) T-cell infiltration in the tumor. Int J. Cancer 128, 2085–2095 (2011).
Renner, K. et al. Metabolic hallmarks of tumor and immune cells in the tumor microenvironment. Front Immunol. 8, 248 (2017).
Weber, E. W. et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science 372, eaba1786 (2021).
Zhu, X., Li, Q. & Zhu, X. Mechanisms of CAR T cell exhaustion and current counteraction strategies. Front Cell Dev. Biol. 10, 1034257 (2022).
Lopez-Cantillo, G., Uruena, C., Camacho, B. A. & Ramirez-Segura, C. CAR-T Cell Performance: How to Improve Their Persistence? Front Immunol. 13, 878209 (2022).
Qasim, W. Genome-edited allogeneic donor “universal” chimeric antigen receptor T cells. Blood 141, 835–845 (2023).
Allen, G. M. et al. Synthetic cytokine circuits that drive T cells into immune-excluded tumors. Science 378, eaba1624 (2022).
Adachi, K. et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat. Biotechnol. 36, 346–351 (2018).
Guedan, S. et al. Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 3, e96976 (2018).
Milano, F. et al. Cord-blood transplantation in patients with minimal residual disease. N. Engl. J. Med 375, 944–953 (2016).
Sun, Y., Zhao, J., Zhang, L., Li, Z. & Lei, S. Effectiveness and safety of stem cell therapy for diabetic foot: a meta-analysis update. Stem Cell Res Ther. 13, 416 (2022).
Hiwarkar, P. et al. Cord blood CD8+ T-cell expansion following granulocyte transfusions eradicates refractory leukemia. Blood Adv. 4, 4165–4174 (2020).
Zhang, Y. et al. CXCR4/CXCL12 axis counteracts hematopoietic stem cell exhaustion through selective protection against oxidative stress. Sci. Rep. 6, 37827 (2016).
Tzeng, Y. S. et al. Loss of Cxcl12/Sdf-1 in adult mice decreases the quiescent state of hematopoietic stem/progenitor cells and alters the pattern of hematopoietic regeneration after myelosuppression. Blood 117, 429–439 (2011).
Enbar, T. & Tang, L. Mimicking the TCR immune synapse for improved CAR-T cell function. Cell Res. 35, 549–550 (2025).
Xiong, W. et al. Immunological synapse predicts effectiveness of chimeric antigen receptor cells. Mol. Ther. 26, 963–975 (2018).
Ellis, J. S., Wan, X. & Braley-Mullen, H. Transient depletion of CD4+ CD25+ regulatory T cells results in multiple autoimmune diseases in wild-type and B-cell-deficient NOD mice. Immunology 139, 179–186 (2013).
Scheinecker, C., Goschl, L. & Bonelli, M. Treg cells in health and autoimmune diseases: New insights from single cell analysis. J. Autoimmun. 110, 102376 (2020).
Solomon, I. et al. CD25-T(reg)-depleting antibodies preserving IL-2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat. Cancer 1, 1153–1166 (2020).
Song, D. et al. Two novel human anti-CD25 antibodies with antitumor activity inversely related to their affinity and in vitro activity. Sci. Rep. 11, 22966 (2021).
Giovannoni, G. et al. Safety and tolerability profile of daclizumab in patients with relapsing-remitting multiple sclerosis: An integrated analysis of clinical studies. Mult. Scler. Relat. Disord. 9, 36–46 (2016).
Zhang, Y. et al. Allogenic and autologous anti-CD7 CAR-T cell therapies in relapsed or refractory T-cell malignancies. Blood Cancer J. 13, 61 (2023).
Liu, F. et al. CD96, a new immune checkpoint, correlates with immune profile and clinical outcome of glioma. Sci. Rep. 10, 10768 (2020).
Crowell, H. L. et al. An R-based reproducible and user-friendly preprocessing pipeline for CyTOF data. F1000Res 9, 1263 (2020).
Nowicka, M. et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res 6, 748 (2017).
Ishikawa, F. et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat. Biotechnol. 25, 1315–1321 (2007).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic notch receptors. Cell 164, 780–791 (2016).
Cameron, J. T., Roland, B. W., George, S. & Laszlo, S. F. Chimeric antigen receptors targeting CD33. https://patents.google.com/patent/US20230220103A1/en?oq=US20230220103Al+CD33 (2023).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Linderman, G. C. et al. Zero-preserving imputation of single-cell RNA-seq data. Nat. Commun. 13, 192 (2022).
Calvanese, V. et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature 604, 534–540 (2022).
Paczkowska, M. et al. Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 11, 735 (2020).
Fitzgerald, D. J. et al. Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021–1032 (2006).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D. Struct. Biol. 74, 519–530 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Acknowledgements
F.I. is supported by the RIKEN President’s Discretionary Fund, Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 22H04997 and 22H04925(PAGES), and SECOM Science and Technology Foundation Grant Number SJ0446100Y. A.I-N was supported by the JSPS KAKENHI Grant Numbers 22K07243 and 25K10572. M.L. was supported by Grant-in-Aid for JSPS Fellows, 22KF0405. L.D.S is supported by NIH Grant Number P30 CA034196. Support from colleagues at Toranomon Hospital is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
A.I.-N. performed in vitro, in vivo, and metabolomics experiments, data analysis, and manuscript writing; M.L. performed data analysis, bulk and single-cell genomics analysis, figure preparation, and manuscript writing; M.S. collected patient samples following cord blood transplant; C.B. designed and tested CD96 CAR constructs; M.S. and C.B. contributed equally to this project. M.T.-M., S.F., K.S., A.K., H.K., N.S., M.I., R.N. performed in vitro and in vivo experiments involving AML samples; SF performed IHC and IF imaging; M.T.-M. performed CyTOF experiments; T.F. contributed to the choice of treatment targets for translational aspects. A.K., E.K., M.H., and S.A. performed analysis and interpretation of patient RNA-seq data. Y.A., H.A.-I., and M.K. coordinated preparation and sequencing of scRNA libraries. J.M. performed initial processing of scRNA datasets; K.H., S.M., and K.K. performed experiments, imaging, and structural analysis of cryoEM samples; T.M. and S.M. developed scFvs against CD25; S.T., Y.S., Y.N., K.T., and N.U. provided clinical samples; Y.S. and K.T. performed and supervised metabolism experiments. Y.N. contributed discussion on CAR-T cell application to clinical medicine. M.K., M.Kato, and Y.N. provided CB samples and performed experiments for the paper revision. TM and MS supervised cryoEM experiments. P.C., C.C.H., Y.M., and J.W.S. supervised the collection, sequencing, and analysis of scRNA-seq samples. M.deHoon supervised analysis of genomics data. LDS supervised animal experiments and preparation of the manuscript. Y.S. supervised experiments, analysis, and preparation of the manuscript. FI conceptualized and supervised the entire project. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sébastien Wälchli, Mark Leick and the other anonymous reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Itoh-Nakadai, A., Liang, M., Shindo, M. et al. CXCR4 induces memory formation over exhaustion in CAR-T cells to achieve durable leukemia targeting. Nat Commun 17, 101 (2026). https://doi.org/10.1038/s41467-025-67745-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67745-x