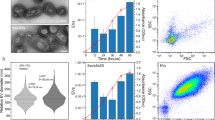
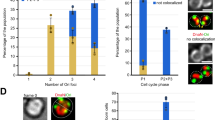
Abstract
Despite its fundamental importance for the propagation of all cellular life, very little is known about the molecular basis of chromosome segregation in archaea. This is a particularly intriguing issue in members of the genus Sulfolobus which encode homologs of a “bacterial-like” candidate segregation machinery, termed SegAB, related to the bacterial ParA-based systems, yet rely on a “eukaryotic-like” ESCRT machinery for cell division. While investigating the temporal dynamics of Sulfolobus chromosome architecture, we uncovered conformational changes at the candidate segregation locus. Loss of the Seg system results in altered nucleoid morphology and induction of a DNA damage response. Significantly, loss of the Seg system also leads to precocious assembly of the key cell division protein CdvA. Finally, we observe interactions between SegA and CdvA, indicating dual roles for the Seg system in effecting chromosome segregation and defining the positioning of the cell division apparatus.
Similar content being viewed by others
Data availability
The sequence reads for the 3C-Seq, RNA-Seq and ChIP-Seq data have been deposited in the Sequence Read Archive, BioProject PRJNA1272008; submissions SUB15364289 and SUB15720780. Source data will be provided with this paper. Strains are available on request from RYS. Source data are provided with this paper.
References
Jalal, A. S. B. & Le, T. B. K. Bacterial chromosome segregation by the ParABS system. Open Biol. 10, 200097 (2020).
Reyes-Lamothe, R. & Sherratt, D. J. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 17, 467–478 (2019).
Bernander, R. & Poplawski, A. Cell cycle characteristics of thermophilic Archaea. J. Bacteriol. 179, 4963–4969 (1997).
Samson, R. Y. & Bell, S. D. Cell cycles and cell division in the Archaea. Curr. Opin. Microbiol. 14, 350–356 (2011).
Lindas, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J. & Bernander, R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA 105, 18942–18946 (2008).
Samson, R. Y., Obita, T., Freund, S. M., Williams, R. L. & Bell, S. D. A role for the ESCRT system in cell division in Archaea. Science 322, 1710–1713 (2008).
Samson, R. Y. et al. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol. Cell 41, 186–196 (2011).
Risa, G. T. et al. The proteasome controls ESCRT-III-mediated cell division in an archaeon. Science 369, 642-+ (2020).
Kalliomaa-Sanford, A. K. et al. Chromosome segregation in Archaea mediated by a hybrid DNA partition machine. Proc. Natl. Acad. Sci. USA 109, 3754–3759 (2012).
Yen, C. Y. et al. Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly. Nucleic Acids Res. 49, 13150–13164 (2021).
Pulianmackal, L. T. et al. Multiple ParA/MinD ATPases coordinate the positioning of disparate cargos in a bacterial cell. Nat. Commun. 14, 3255 (2023).
Lutkenhaus, J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev. Biochem. 76, 539–562 (2007).
Gruber, S. & Errington, J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009).
Sullivan, N. L., Marquis, K. A. & Rudner, D. Z. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137, 697–707 (2009).
Tran, N. T., Laub, M. T. & Le, T. B. K. SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 20, 2057–2071 (2017).
Wang, X., Brandao, H. B., Le, T. B., Laub, M. T. & Rudner, D. Z. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017).
Wang, X. et al. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675 (2015).
Hirano, T. Condensin-based chromosome organization from bacteria to vertebrates. Cell 164, 847–857 (2016).
Takemata, N., Samson, R. Y. & Bell, S. D. Physical and functional compartmentalization of archaeal chromosomes. Cell 179, 165–179 e118 (2019).
Duggin, I. G., McCallum, S. A. & Bell, S. D. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. USA 105, 16737–16742 (2008).
Takemata, N. & Bell, S. D. Multi-scale architecture of archaeal chromosomes. Mol. Cell 81, 473–487.e476 (2021).
Lundgren, M. & Bernander, R. Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. USA 104, 2939–2944 (2007).
Gómez-Raya-Vilanova, M. V. et al. Transcriptional landscape of the cell cycle in a model thermoacidophilic archaeon reveals similarities to eukaryotes. Nat. Commun. 16, 5697 (2025).
Zhang, C., Phillips, A. P. R., Wipfler, R. L., Olsen, G. J. & Whitaker, R. J. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun. 9, 4908 (2018).
Gotz, D. et al. Responses of hyperthermophilic crenarchaea to UV irradiation. Genom. Biol. 8, R220 (2007).
Frols, S. et al. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J. Bacteriol. 189, 8708–8718 (2007).
Ajon, M. et al. UV-inducible DNA exchange in hyperthermophilic Archaea mediated by type IV pili. Mol. Microbiol 82, 807–817 (2011).
Recalde, A. et al. New components of the community-based DNA-repair mechanism in Sulfolobales. Microlife 6, uqaf002 (2025).
van Wolferen, M., Pulschen, A. A., Baum, B., Gribaldo, S. & Albers, S. V. The cell biology of Archaea. Nat. Microbiol 7, 1744–1755 (2022).
van Wolferen, M. et al. Species-specific recognition of Sulfolobales mediated by UV-inducible Pili and S-layer Glycosylation patterns. mBio 11, https://doi.org/10.1128/mBio.03014-19 (2020).
van Wolferen, M., Wagner, A., van der Does, C. & Albers, S. V. The archaeal Ced system imports DNA. Proc. Natl. Acad. Sci. USA 113, 2496–2501 (2016).
Kabli, A. F. et al. Coupling chromosome organization to genome segregation in Archaea. Nat. Commun. 16, 6759 (2025).
Yang, Y. et al. A novel RHH family transcription factor aCcr1 and its viral homologs dictate cell cycle progression in Archaea. Nucleic Acids Res. 51, 1707–1723 (2023).
Xuyang, L., Cristina, L. M., Laura, M. A. & Xu, P. A clade of RHH proteins ubiquitous in Sulfolobales and their viruses regulates cell cycle progression. Nucleic Acids Res. 51, 1724–1739 (2023).
Laursen, S. P., Bowerman, S. & Luger, K. Archaea: the Final Frontier of Chromatin. J. Mol. Biol. 433, 166791 (2021).
Samson, R. Y., Dobro, M. J., Jensen, G. J. & Bell, S. D. The Structure, function and roles of the Archaeal ESCRT apparatus. Subcell. Biochem. 84, 357–377 (2017).
Takemata, N. & Bell, S. D. Chromosome conformation capture assay combined with biotin enrichment for hyperthermophilic Archaea. STAR Protoc. 2, 100576 (2021).
Takemata, N. & Bell, S. D. High-resolution analysis of chromosome conformation in hyperthermophilic Archaea. STAR Protoc. 2, 100562 (2021).
Samson, R. Y., Duggin, I. G. & Bell, S. D. Analysis of the Archaeal ESCRT apparatus. Methods Mol. Biol. 1998, 1–11 (2019).
Cezanne, A., Hoogenberg, B. & Baum, B. Probing archaeal cell biology: exploring the use of dyes in the imaging of Sulfolobus cells. Front. Microbiol. 14, 1233032 (2023).
Samson, R. Y. et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496 (2013).
Acknowledgements
S.D.B.’s lab is funded by NIH R35GM152171. This work was additionally funded by NIH 1R01GM135178, the College of Arts and Sciences, Indiana University and the College of Arts and Sciences, The Ohio State University. We are indebted to Veronica A. Bell for invaluable assistance with microscopy. We thank Buzz Baum for discussions prior to publication. We are also grateful to Mart Krupovic and Miguel Gomez Raya Vilanova for sharing protocols and advice for acetic acid-mediated arrest and release of S. islandicus.
Author information
Authors and Affiliations
Contributions
R.Y.S. and N.T. performed the baby machine synchronization, and N.T. performed the Hi-C experiments on this material. All other experiments were performed and analyzed by R.Y.S. S.D.B. assisted in experimental design, obtained funding and wrote the initial draft of the manuscript. All three authors contributed to the final text of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Nature Communications thanks Qihong Huang and the other anonymous reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Samson, R.Y., Takemata, N. & Bell, S.D. Coordination of chromosome segregation and cell division in the archaeon Sulfolobus acidocaldarius. Nat Commun (2025). https://doi.org/10.1038/s41467-025-67934-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67934-8