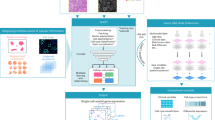
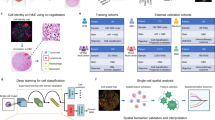
Abstract
Understanding the spatial organization of individual cell types within tissue and how this organization is disrupted in disease, is a central question in biology and medicine. Hematoxylin and eosin-stained slides are widely available and provide detailed morphological context, while spatial gene expression profiling offers complementary molecular insights, though it remains costly and limited in accessibility. Predicting gene expression directly from histological images is therefore an attractive goal. However, existing approaches typically rely on small image patches, limiting resolution and the ability to capture fine-grained morphological variation. Here, we introduce a deep learning approach that predicts single-cell gene expression from morphology, matching patch-based methods on spot level prediction tasks. The model recovers biologically meaningful expression patterns across two cancer datasets and distinguishes fine cell populations. This approach enables molecular-level interpretation of standard histological slides at scale, offering new opportunities to study tissue organization and cellular diversity in health and disease.
Similar content being viewed by others
Data availability
We accessed the spatial transcriptomic data used in this study within the HEST database33 hosted at https://huggingface.co/datasets/MahmoodLab/hest with the provided cell segmentation. For each cancer type, we include the slide ID from HEST along with links to the original publication or source website. We excluded slides which were not preserved with FFPE and where the H&E staining quality was insufficient for the cell segmentation algorithm to perform effectively. Visium slides HEST ids: • Prostate: INT25, INT26, INT27, INT28, INT35 • Kidney32: INT13, INT14, INT15, INT17, INT18, INT19, INT21, INT24 • Breast: TENX39• Ovary: TENX65. Xenium slides HEST ids (with publication of raw dataset links): • NCBI783, NCBI784, NCBI7854• TENX94, TENX95 https://www.10xgenomics.com/datasets/ffpe-human-breast-with-pre-designed-panel-1-standard, • TENX96, TENX97 https://www.10xgenomics.com/datasets/ffpe-human-breast-with-custom-add-on-panel-1-standard, • TENX98, TENX99 https://www.10xgenomics.com/datasets/ffpe-human-breast-using-the-entire-sample-area-1-standard, single cell RNA dataset: • Ovary41: https://datasets.cellxgene.cziscience.com/73fbcec3-f602-4e13-a400-a76ff91c7488.h5ad• Breast43: https://datasets.cellxgene.cziscience.com/fabd4946-3f41-459c-ba79-188749a8baa4.h5ad. Source data are provided with this paper.
Code availability
The code used to develop the model is publicly available and has been deposited in sCellST at (https://github.com/loicchadoutaud/sCellST58)under CC-BY 4.0 license. The code used to perform the analyses and generate results in this study is publicly available and has been deposited in sCellST_reproducibility at (https://github.com/loicchadoutaud/sCellST_reproducibility.git).
References
Song, A. H. et al. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng. 1, 930–949 (2023).
Rao, A., Barkley, D., França, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14, 8353 (2023).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Wan, X. et al. Integrating spatial and single-cell transcriptomics data using deep generative models with SpatialScope. Nat Commun. 14, 7848 (2023).
Jiang, X. et al. Reconstructing spatial transcriptomics at the single-cell resolution with BayesDeep https://www.biorxiv.org/content/10.1101/2023.12.07.570715v1 (2023).
Schmauch, B. et al. A deep learning model to predict RNA-Seq expression of tumours from whole slide images. Nat. Commun. 11, 1–15 (2020).
Hu, J. et al. Deciphering tumor ecosystems at super resolution from spatial transcriptomics with TESLA. Cell Systems https://www.sciencedirect.com/science/article/pii/S2405471223000844 (2023).
He, B. et al. Integrating spatial gene expression and breast tumour morphology via deep learning. Nat. Biomed. Eng. 4, 827–834 (2020).
Monjo, T., Koido, M., Nagasawa, S., Suzuki, Y. & Kamatani, Y. Efficient prediction of a spatial transcriptomics profile better characterizes breast cancer tissue sections without costly experimentation. Sci. Rep. 12, 1–12 (2022).
Rahaman, M. M., Millar, E. K. A. & Meijering, E. Breast cancer histopathology image-based gene expression prediction using spatial transcriptomics data and deep learning. Sci. Rep. 13, 13604 (2023).
Jiang, Y., Xie, J., Tan, X., Ye, N. & Nguyen, Q. Generalization of deep learning models for predicting spatial gene expression profiles using histology images: a breast cancer case study. preprint, Preprint at http://biorxiv.org/lookup/doi/10.1101/2023.09.20.558624 (2023).
Gao, R. et al. Harnessing TME depicted by histological images to improve cancer prognosis through a deep learning system. Cell Rep. Med. https://www.cell.com/cell-reports-medicine/abstract/S2666-3791(24)00205-2 (2024).
Zeng, Y. et al. Spatial transcriptomics prediction from histology jointly through Transformer and graph neural networks. Brief. Bioinforma. 23, bbac297 (2022).
Xie, R. et al. Spatially resolved gene expression prediction from histology images via bi-modal contrastive learning. In Advances in Neural Information Processing Systems 36. 70626–70637 (NeuIPS, 2023).
Pang, M., Su, K. & Li, M. Leveraging information in spatial transcriptomics to predict super-resolution gene expression from histology images in tumors. Preprint at https://www.biorxiv.org/content/10.1101/2021.11.28.470212v1 (2021).
Bergenstråhle, L. et al. Super-resolved spatial transcriptomics by deep data fusion. Nat. Biotechnol. 40, 476–479 (2021).
Huang, C.-H., Park, Y., Pang, J. & Bienkowska, J. R. Single-cell Gene Expression Prediction Using H&E Images Based On Spatial Transcriptomics. 12471, 1247105 (2023).
Zhang, D. et al. Inferring super-resolution tissue architecture by integrating spatial transcriptomics with histology. Nat. Biotechnol. https://www.nature.com/articles/s41587-023-02019-9 (2024).
Hörst, F. et al. CellViT: vision transformers for precise cell segmentation and classification. Med. Image Anal. 94, 103143 (2024).
Graham, S. et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563 (2019).
Balestriero, R. et al. A cookbook of self-supervised learning. Preprint at http://arxiv.org/abs/2304.12210 (2023).
Zimmermann, E. et al. Virchow2: scaling self-supervised mixed magnification models in pathology. Preprint at http://arxiv.org/abs/2408.00738 (2024).
Nakhli, R. et al. VOLTA: an environment-aware contrastive cell representation learning for histopathology. Nat. Commun. 15, 3942 (2024).
Koch, V. et al. DinoBloom: a foundation model for generalizable cell embeddings in hematology. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2024 (eds. Linguraru, M. G.) LNCS, Vol. 15012, 520–530 (Springer Nature Switzerland, 2024).
Chen, X., Xie, S. & He, K. An empirical study of training self-supervised vision transformers. In Proc. IEEE/CVF Int. Conf. Comput. Vis. (ICCV). 9620–9629 (2021). https://doi.org/10.1109/ICCV48922.2021.00950.
Dietterich, T. G., Lathrop, R. H. & Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 89, 31–71 (1997).
Jia, Y., Liu, J., Chen, L., Zhao, T. & Wang, Y. THItoGene: a deep learning method for predicting spatial transcriptomics from histological images. Brief. Bioinforma. 25, bbad464 (2023).
Min, W., Shi, Z., Zhang, J., Wan, J. & Wang, C. Multimodal contrastive learning for spatial gene expression prediction using histology images. Brief. Bioinforma. 25, bbae551 (2024).
Radford, A. et al. Learning transferable visual models from natural language supervision. In Proceedings of the 38th International Conference on Machine Learning. Vol. 139, 8748–8763 (PMLR, 2021).
Meylan, M. et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity 55, 527–541.e5 (2022).
Jaume, G. et al. HEST-1k: a dataset for spatial transcriptomics and histology image analysis. In Advances in Neural Information Processing Systems 37. 53798–53833 (NeurIPS, 2024). https://doi.org/10.52202/079017-1704.
Tirosh, I. & Suva, M. L. Cancer cell states: Lessons from ten years of single-cell RNA-sequencing of human tumors. Cancer Cell 42, 1497–1506 (2024).
Gough, M. et al. Receptor CDCP1 is a potential target for personalized imaging and treatment of poor outcome HER2+, triple negative and metastatic ER+/HER2- breast cancers. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-24-2865 (2025).
Bao, H., Wu, W., Li, Y., Zong, Z. & Chen, S. WNT6 participates in the occurrence and development of ovarian cancer by upregulating/activating the typical Wnt pathway and Notch1 signaling pathway. Gene 846, 146871 (2022).
Croizer, H. et al. Deciphering the spatial landscape and plasticity of immunosuppressive fibroblasts in breast cancer. Nat. Commun. 15, 1–28 (2024).
Hu, Y. et al. INHBA(+) cancer-associated fibroblasts generate an immunosuppressive tumor microenvironment in ovarian cancer. npj Precis. Oncol. 8, 1–13 (2024).
Schumacher, T. N. & Thommen, D. S. Tertiary lymphoid structures in cancer. Sci. (N. Y., N. Y.) 375, eabf9419 (2022).
Hoque, M. Z., Keskinarkaus, A., Nyberg, P. & Seppänen, T. Stain normalization methods for histopathology image analysis: A comprehensive review and experimental comparison. Inf. Fusion 102, 101997 (2024).
Vázquez-García, I. et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature 612, 778–786 (2022).
CZI Cell Science Program et al. CZ CELLxGENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. Nucleic Acids Res. 53, D886–D900 (2025).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021).
Diao, J. A. et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 12, 1613 (2021).
Kömen, J., Marienwald, H., Dippel, J. & Hense, J. Do histopathological foundation models eliminate batch effects? a comparative study. Preprint at http://arxiv.org/abs/2411.05489 (2024).
Chen, J. et al. STimage-1K4M: a histopathology image-gene expression dataset for spatial transcriptomics. In 2009 Advances in Neural Information Processing Systems 37. 35796–35823 (NeurIPS, 2024). https://doi.org/10.52202/079017-1129.
Oliveira, M.F.d. et al. High-definition spatial transcriptomic profiling of immune cell populations in colorectal cancer. Nat. Genet. 57, 1512–1523 (2025).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Pan, S. J. & Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010).
Deng, J. et al. ImageNet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 https://ieeexplore.ieee.org/document/5206848/ (2009).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (2016). https://doi.org/10.1109/CVPR.2016.90.
Bendidi, I. et al. No free lunch in self supervised representation learning. Preprint at http://arxiv.org/abs/2304.11718 (2023).
Ilse, M., Tomczak, J. M. & Welling, M. Attention-based deep multiple instance learning. In Proceedings of the 35th International Conference on Machine Learning (Proceedings of Machine Learning Research 80), 2127–2136 (PMLR, 2018).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems 32 (NeurIPS 2019) (Curran Associates, 2019).
Loshchilov, I. & Hutter, F. Decoupled weight decay regularization. In Proceedings of the International Conference on Learning Representations (ICLR) (2019).
Chadoutaud, L. sCellST: code for sCellST predicts single-cell gene expression from H&E Images https://doi.org/10.5281/zenodo.17602576 (2025).
Acknowledgements
We would like to thank Raphaël Bourgade, Lucie Gaspard-Boulinc, Nicolas Captier, Nicolas Servant and Loredana Martignetti for helpful discussions. This work was funded by the French government under the management of Agence Nationale de la Recherche as part of the “Investissements d’avenir” program, reference ANR-19-P3IA-0001 (PRAIRIE 3IA Institute; grants to TW and EB) and ANR-23-IACL-0008 (PRAIRIE-PSAI; grants to TW and EB). Furthermore, this work was supported by ITMO Cancer (20CM107-00; grant to TW). Furthermore, this work was supported by a government grant managed by the Agence Nationale de la Recherche under the France 2030 program, with the reference numbers ANR-24-EXCI-0001, ANR-24-EXCI-0002, ANR-24-EXCI-0003, ANR-24-EXCI-0004, ANR-24-EXCI-0005 (grants to TW and EB).
Author information
Authors and Affiliations
Contributions
L.C., M.L., E.B., and T.W. designed and planned the study. L.C., M.L., and J.O. developed the tool. L.C., D.H., and J.F. performed the analysis. E.B. and T.W. supervised the study. L.C., D.H., J.F., E.B., and T.W. wrote the manuscript. All authors reviewed and/or edited the manuscript prior to submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Chao-Hui Huang, Tingying Peng, Qianqian Song and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chadoutaud, L., Lerousseau, M., Herrero-Saboya, D. et al. sCellST predicts single-cell gene expression from H& E images. Nat Commun (2026). https://doi.org/10.1038/s41467-025-67965-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67965-1