Abstract
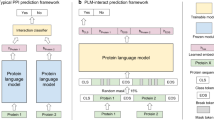
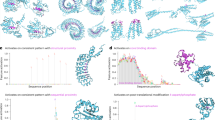
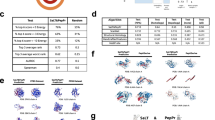
Protein Language Models (PLMs) trained on large databases of protein sequences have proven effective in modeling protein biology across a wide range of applications. However, while PLMs excel at capturing individual protein properties, they face challenges in natively representing protein-protein interactions (PPIs), which are crucial to understanding cellular processes and disease mechanisms. Here, we introduce MINT, a PLM specifically designed to model sets of interacting proteins in a contextual and scalable manner. Using unsupervised training on a large curated PPI dataset derived from the STRING database, MINT outperforms existing PLMs in diverse tasks relating to protein-protein interactions, including binding affinity prediction and estimation of mutational effects. Beyond these core capabilities, it excels at modeling interactions in complex protein assemblies and surpasses specialized models in antibody-antigen modeling and T cell receptor-epitope binding prediction. MINT’s predictions of mutational impacts on oncogenic PPIs align with experimental studies, and it provides reliable estimates for the potential for cross-neutralization of antibodies against SARS-CoV-2 variants of concern. These findings position MINT as a powerful tool for elucidating complex protein interactions, with significant implications for biomedical research and therapeutic discovery.
Similar content being viewed by others
Data availability
We retrieved physical PPI training data for MINT from STRING-DB20. We obtained the gold-standard PPI dataset from https://figshare.com/articles/dataset/PPI_prediction_from_sequence_gold_standard_dataset/21591618/322, the HumanPPI dataset from https://github.com/westlake-repl/SaProt67, and the YeastPPI dataset from PEER (https://miladeepgraphlearningproteindata.s3.us-east-2.amazonaws.com/ppidata/yeast_ppi.zip)12. The SKEMPI entries were downloaded from https://life.bsc.es/pid/skempi223 and the PDB-Bind dataset from https://www.pdbbind-plus.org.cn/28. The Mutational PPI data were obtained from https://github.com/jishnu-lab/SWING/tree/main/Data/MutInt_Model29. The FLAB antibody datasets are available at https://github.com/Graylab/FLAb/tree/main/data24, and the SARS-CoV-2 binding datasets at this link: https://www.biorxiv.org/content/10.1101/2020.04.03.024885v1.supplementary-material38. The TCR-epitope task from TDC-2 was downloaded from (https://tdcommons.ai/)44. The TCR-epitope-HLA data were retrieved from https://github.com/Armilius/PISTE/tree/main/data17, and the TCR-epitope interface prediction data were obtained from https://github.com/pengxingang/TEIM46. We obtained experimentally validated oncoPPI data from https://github.com/ChengF-Lab/oncoPPIs57. Finally, we obtained SARS-CoV-2 neutralization data from https://opig.stats.ox.ac.uk/webapps/covabdab/64. Source data for all figures is provided with this paper. Source data are provided with this paper.
Code availability
The code used to develop MINT, perform the analyzes, and generate results in this study is publicly available and has been deposited at https://github.com/VarunUllanat/mint under the MIT License. The publication release is deposited on Zenodo at https://doi.org/10.5281/zenodo.1717487574.
References
Bepler, T. & Berger, B. Learning protein sequence embeddings using information from structure. Int. Conf. Learn. Represent. (2019).
Bepler, T. & Berger, B. Learning the protein language: evolution, structure, and function. Cell Syst. 12, 654–669 (2021).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 118, e2016239118 (2021).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Adv. Neural Inf. Process. Syst. 34, 29287–29303 (2021).
Thadani, N. N. et al. Learning from prepandemic data to forecast viral escape. Nature 622, 818–825 (2023).
Hie, B., Zhong, E. D., Berger, B. & Bryson, B. Learning the language of viral evolution and escape. Science 371, 284–288 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. nature 596, 583–589 (2021).
Wu, R. et al. High-resolution de novo structure prediction from primary sequence. Preprint at https://www.biorxiv.org/content/10.1101/2022.07.21.500999v1 (2022).
Singh, R., Sledzieski, S., Bryson, B., Cowen, L. & Berger, B. Contrastive learning in protein language space predicts interactions between drugs and protein targets. Proc. Natl. Acad. Sci. USA 120, e2220778120 (2023).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Sledzieski, S., Singh, R., Cowen, L. & Berger, B. D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions. Cell Syst. 12, 969–982 (2021).
Xu, M. et al. Peer: a comprehensive and multi-task benchmark for protein sequence understanding. Adv. Neural Inf. Process. Syst. 35, 35156–35173 (2022).
Charih, F., Biggar, K. K. & Green, J. R. Assessing sequence-based protein–protein interaction predictors for use in therapeutic peptide engineering. Sci. Rep. 12, 9610 (2022).
Sledzieski, S., Devkota, K., Singh, R., Cowen, L. & Berger, B. TT3D: leveraging precomputed protein 3d sequence models to predict protein–protein interactions. Bioinformatics 39, btad663 (2023).
Singh, R., Devkota, K., Sledzieski, S., Berger, B. & Cowen, L. Topsy-turvy: integrating a global view into sequence-based PPI prediction. Bioinformatics 38, i264–i272 (2022).
Sledzieski, S. et al. Democratizing protein language models with parameter-efficient fine-tuning. Proc. Natl. Acad. Sci. USA 121, e2405840121 (2024).
Feng, Z. et al. Sliding-attention transformer neural architecture for predicting T cell receptor–antigen–human leucocyte antigen binding. Nat. Mach. Intell. 6, 1216–1230 (2024).
Kenlay, H. et al. Large scale paired antibody language models. Preprint at https://arxiv.org/abs/2403.17889 (2024).
Singh, R. et al. Learning the language of antibody hypervariability. Proc. Natl. Acad. Sci. USA 122, e2418918121 (2025).
Szklarczyk, D. et al. The string database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic acids Res. 51, D638–D646 (2023).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30 (2017).
Bernett, J., Blumenthal, D. B. & List, M. Cracking the black box of deep sequence-based protein–protein interaction prediction. Brief. Bioinforma. 25, bbae076 (2024).
Jankauskaitė, J., Jiménez-García, B., Dapkūnas, J., Fernández-Recio, J. & Moal, I. H. Skempi 2.0: an updated benchmark of changes in protein–protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics 35, 462–469 (2019).
Chungyoun, M., Ruffolo, J. A. & Gray, J. J. Flab: benchmarking deep learning methods for antibody fitness prediction. Preprint at https://www.biorxiv.org/content/10.1101/2024.01.13.575504v1 (2024).
Grazioli, F. et al. Attentive variational information bottleneck for TCR–peptide interaction prediction. Bioinformatics 39, btac820 (2023).
Devlin, J. et al. BERT: Pre-training of deep bidirectional transformers for language understanding. Proc. NAACL-HLT. 1, 4171–4186 (2019).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold3. Nature 630, 493–500 (2024).
Liu, Z. et al. Forging the basis for developing protein–ligand interaction scoring functions. Acc. Chem. Res. 50, 302–309 (2017).
Siwek, J. C. et al. Sliding Window Interaction Grammar (SWING): a generalized interaction language model for peptide and protein interactions. Nat. Methods 22, 1707–1719 (2025).
Rao, R. M. et al. MSA Transformer. In Proceedings of the 38th International Conference on Machine Learning. (eds Meila, M. & Zhang, T.) 8844–8856 (PMLR, 2021).
Madani, A. et al. Progen: language modeling for protein generation. Preprint at https://arxiv.org/abs/2004.03497 (2020).
Elnaggar, A. et al. Prottrans: towards cracking the language of life’s code through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
Greenfield, E. A. Antibodies: A Laboratory Manual, Second Edition (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar], 2014).
Gabrielli, E. et al. Antibody complementarity-determining regions (cdrs): a bridge between adaptive and innate immunity. PLoS ONE 4, e8187 (2009).
Shanehsazzadeh, A. et al. Unlocking de novo antibody design with generative artificial intelligence. Preprint at https://www.biorxiv.org/content/10.1101/2023.01.08.523187v4 (2023).
Warszawski, S. et al. Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLoS Comput. Biol. 15, e1007207 (2019).
Koenig, P. et al. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc. Natl. Acad. Sci. USA 114, E486–E495 (2017).
Desautels, T., Zemla, A., Lau, E., Franco, M. & Faissol, D. Rapid in silico design of antibodies targeting SARS-CoV-2 using machine learning and supercomputing. Preprint at https://www.biorxiv.org/content/10.1101/2020.04.03.024885v1 (2020).
Zhu, Z. et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA 104, 12123–12128 (2007).
Delgado, J., Radusky, L. G., Cianferoni, D. & Serrano, L. Foldx 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics 35, 4168–4169 (2019).
Leaver-Fay, A. et al. Scientific benchmarks for guiding macromolecular energy function improvement. In Methods in Enzymology, Vol. 523, 109–143 (Elsevier, 2013).
Barlow, K. A. et al. Flex ddg: Rosetta ensemble-based estimation of changes in protein–protein binding affinity upon mutation. J. Phys. Chem. B 122, 5389–5399 (2018).
Peters, B., Nielsen, M. & Sette, A. T cell epitope predictions. Annu. Rev. Immunol. 38, 123–145 (2020).
Velez-Arce, A. et al. Signals in the cells: multimodal and contextualized machine learning foundations for therapeutics. NeurIPS Workshop on AI for New Drug Modalities (2024).
Yoo, S., Jeong, M., Seomun, S., Kim, K. & Han, Y. Interpretable prediction of SARS-CoV-2 epitope-specific TCR recognition using a pre-trained protein language model. IEEE/ACM Trans. Comput. Biol. Bioinforma. 21, 428–438 (2024).
Peng, X. et al. Characterizing the interaction conformation between T-cell receptors and epitopes with deep learning. Nat. Mach. Intell. 5, 395–407 (2023).
Vita, R. et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Shugay, M. et al. Vdjdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res. 46, D419–D427 (2018).
Tickotsky, N., Sagiv, T., Prilusky, J., Shifrut, E. & Friedman, N. Mcpas-tcr: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017).
Yang, M. et al. Mix-tpi: a flexible prediction framework for TCR–PMHC interactions based on multimodal representations. Bioinformatics 39, btad475 (2023).
Montemurro, A. et al. Nettcr-2.0 enables accurate prediction of TCR-peptide binding by using paired tcrα and β sequence data. Commun. Biol. 4, 1060 (2021).
Gao, Y. et al. Pan-peptide meta learning for T-cell receptor–antigen binding recognition. Nat. Mach. Intell. 5, 236–249 (2023).
Jiang, Y., Huo, M. & Cheng Li, S. Teinet: a deep learning framework for prediction of TCR–epitope binding specificity. Brief. Bioinforma. 24, bbad086 (2023).
Weber, A., Born, J. & Rodriguez Martínez, M. Titan: T-cell receptor specificity prediction with bimodal attention networks. Bioinformatics 37, i237–i244 (2021).
Lu, T. et al. Deep learning-based prediction of the T cell receptor–antigen binding specificity. Nat. Mach. Intell. 3, 864–875 (2021).
Leem, J., de Oliveira, S. H. P., Krawczyk, K. & Deane, C. M. Stcrdab: the structural T-cell receptor database. Nucleic Acids Res. 46, D406–D412 (2018).
Cheng, F. et al. Comprehensive characterization of protein–protein interactions perturbed by disease mutations. Nat. Genet. 53, 342–353 (2021).
Frazer, J. et al. Disease variant prediction with deep generative models of evolutionary data. Nature 599, 91–95 (2021).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Sahni, N. et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161, 647–660 (2015).
Fragoza, R. et al. Extensive disruption of protein interactions by genetic variants across the allele frequency spectrum in human populations. Nat. Commun. 10, 4141 (2019).
Wang, Y. et al. Alox5 exhibits anti-tumor and drug-sensitizing effects in MLL-rearranged leukemia. Sci. Rep. 7, 1853 (2017).
Fan, Y. et al. SARS-CoV-2 omicron variant: recent progress and future perspectives. Signal Transduct. Target. Ther. 7, 1–11 (2022).
Raybould, M. I., Kovaltsuk, A., Marks, C. & Deane, C. M. Cov-abdab: the coronavirus antibody database. Bioinformatics 37, 734–735 (2021).
Cho, A. et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature 600, 517–522 (2021).
Liu, Y. et al. Inactivated vaccine-elicited potent antibodies can broadly neutralize SARS-CoV-2 circulating variants. Nat. Commun. 14, 2179 (2023).
Su, J. et al. Saprot: protein language modeling with structure-aware vocabulary. Preprint at https://www.biorxiv.org/content/10.1101/2023.10.01.560349v1 (2023).
Hayes, T. et al. Simulating 500 million years of evolution with a language model. Science eads0018 (2025).
Steinegger, M. & Söding, J. MMSEqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Chen, J.-Y. et al. Evaluating the advancements in protein language models for encoding strategies in protein function prediction: a comprehensive review. Front. Bioeng. Biotechnol. 13, 1506508 (2025).
Guo, Y., Yu, L., Wen, Z. & Li, M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic acids Res. 36, 3025–3030 (2008).
Pan, X.-Y., Zhang, Y.-N. & Shen, H.-B. Large-scale prediction of human protein-protein interactions from amino acid sequence based on latent topic features. J. Proteome Res. 9, 4992–5001 (2010).
Luo, S. et al. Rotamer Density Estimator is an unsupervised learner of the effect of mutations on protein–protein interaction. Proc. ICLR (2023).
Ullanat, V., Jing, B., Sledzieski, S. & Berger, B. Learning the language of protein-protein interactions. varunullanat/mint: Publication release https://doi.org/10.5281/zenodo.17174876 (2025).
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number 1R35GM141861 and by a research gift from Quanta Computer. B.J. was partially supported by the Department of Energy Computational Science Graduate Fellowship under Award Number DESC0022158. S.S. was partially supported by the NSF Graduate Research Fellowship under Grant No. 2141064. We would also like to acknowledge Aditya Parekh and Anish Mudide for their helpful discussions and comments.
Author information
Authors and Affiliations
Contributions
B.J., S.S., and B.B. conceptualized the project. V.U. and B.J. constructed the training pipeline for MINT. V.U. and B.J. ran the training. V.U. performed downstream computational analysis, including model benchmarking and case studies. B.B. designed and led the study. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Nimisha Ghosh, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ullanat, V., Jing, B., Sledzieski, S. et al. Learning the language of protein-protein interactions. Nat Commun (2026). https://doi.org/10.1038/s41467-025-67971-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67971-3