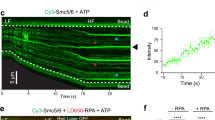
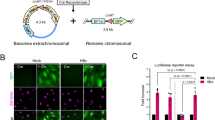
Abstract
Smc5/6 is an essential genome maintenance complex that interacts with double-stranded (ds) DNA, single-stranded (ss) DNA, and ss-dsDNA junctions. DNA association underlies Smc5/6’s functions in managing intermediates generated during genome replication and repair. However, the mechanisms of this activity are not fully understood. Here, we report a single-molecule study examining Smc5/6 association with a dsDNA substrate containing a ssDNA gap with defined 3’ and 5’ junctions. We found that Smc5/6 associates with both 3’ and 5’ junctions but prefers the 3’ junction in the presence of the ssDNA-binding complex RPA. Further, Smc5/6’s junction association frequency and dwell time are regulated by two non-SMC subcomplexes and DNA binding residues of Smc6. Moreover, Smc5/6 prefers binding to junction sites free of the sliding clamp PCNA over those occupied with it. These results suggest that Smc5/6 utilizes its multiple structural modules to associate with junction sites in coordination with other genome maintenance factors.
Similar content being viewed by others
Data availability
Kymographs used for analysis are available via Zenodo at https://zenodo.org/records/1779265868. Source data are provided with this paper.
Code availability
All specified scripts used to process and analyze C-trap data can be accessed on the LUMICKS Harbor site (“CTrapVis”, https://github.com/lumicks/harbor/tree/main/Visualization/C-Trap%20.h5%20File%20Visualization%20GUI)69.
References
Uhlmann, F. SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016).
Burmann, F. & Lowe, J. Structural biology of SMC complexes across the tree of life. Curr. Opin. Struct. Biol. 80, 102598 (2023).
Roy, S., Adhikary, H. & D’Amours, D. The SMC5/6 complex: folding chromosomes back into shape when genomes take a break. Nucleic Acids Res. 52, 2112–2129 (2024).
Peng, X. P. & Zhao, X. The multi-functional Smc5/6 complex in genome protection and disease. Nat. Struct. Mol. Biol. 30, 724–734 (2023).
Palecek, J. J. SMC5/6: multifunctional player in replication. Genes 10, 7 (2019).
Aragón, L. The Smc5/6 complex: new and old functions of the enigmatic long-distance relative. Annu. Rev. Genet. 52, 89–107 (2018).
Grange, L. J. et al. Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy. Nat. Commun. 13, 6664 (2022).
van der Crabben, S. N. et al. Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J. Clin. Investig. 126, 2881–92 (2016).
Payne, F. et al. Hypomorphism in human NSMCE2 linked to primordial dwarfism and insulin resistance. J. Clin. Investig. 124, 4028–38 (2014).
Yu, Y. et al. Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes. Proc. Natl. Acad. Sci. USA 119, e2202799119 (2022).
Alt, A. et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat. Commun. 8, 14011 (2017).
Zabrady, K. et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 44, 1064–1079 (2016).
Kanno, T., Berta, D. G. & Sjogren, C. The Smc5/6 complex Is an ATP-dependent intermolecular DNA linker. Cell Rep. 12, 1471–82 (2015).
Pradhan, B. et al. The Smc5/6 complex is a DNA loop-extruding motor. Nature 616, 843–848 (2023).
Chang, J. T. H. et al. Smc5/6’s multifaceted DNA binding capacities stabilize branched DNA structures. Nat. Commun. 13, 7179 (2022).
Tanasie, N. L., Gutiérrez-Escribano, P., Jaklin, S., Aragon, L. & Stigler, J. Stabilization of DNA fork junctions by Smc5/6 complexes revealed by single-molecule imaging. Cell Rep. 41, 111778 (2022).
Ampatzidou, E., Irmisch, A., O’Connell, M. J. & Murray, J. M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell Biol. 26, 9387–9401 (2006).
Menolfi, D., Delamarre, A., Lengronne, A., Pasero, P. & Branzei, D. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance. Mol. Cell 60, 835–846 (2015).
Branzei, D. et al. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 (2006).
Bermúdez-López, M. et al. Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 30, 1339–56 (2016).
Bonner, J. N. et al. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 16, 368–378 (2016).
Pond, K. W., de Renty, C., Yagle, M. K. & Ellis, N. A. Rescue of collapsed replication forks is dependent on NSMCE2 to prevent mitotic DNA damage. PLoS Genet. 15, e1007942 (2019).
Duan, X. et al. Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5-6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J. Biol. Chem. 284, 8507–15 (2009).
Yu, Y. et al. Integrative analysis reveals unique structural and functional features of the Smc5/6 complex. Proc. Natl. Acad. Sci. USA 118, e2026844118 (2021).
Taschner, M. et al. Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding. EMBO J. 40, e107807 (2021).
Hallett, S. T. et al. Nse5/6 is a negative regulator of the ATPase activity of the Smc5/6 complex. Nucleic Acids Res. 49, 4534–4549 (2021).
Hallett, S. T. et al. Cryo-EM structure of the Smc5/6 holo-complex. Nucleic Acids Res. 50, 9505–9520 (2022).
Gutierrez-Escribano, P. et al. Purified Smc5/6 complex exhibits DNA substrate recognition and compaction. Mol. Cell 80, 1039–1054 e6 (2020).
Serrano, D. et al. The Smc5/6 core complex is a structure-specific DNA binding and compacting machine. Mol. Cell 80, 1025–1038 e5 (2020).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102, 4777–82 (2005).
Duan, X. et al. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell 35, 657–68 (2009).
Taschner, M. & Gruber, S. DNA segment capture by Smc5/6 holocomplexes. Nat. Struct. Mol. Biol. 30, 619–628 (2023).
Johansson, E. & Dixon, N. Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 5, a012799 (2013).
Waterman, D. P., Haber, J. E. & Smolka, M. B. Checkpoint responses to DNA double-strand breaks. Annu. Rev. Biochem. 89, 103–133 (2020).
Choe, K. N. & Moldovan, G. L. Forging Ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell 65, 380–392 (2017).
Caldwell, C. C. & Spies, M. Dynamic elements of replication protein A at the crossroads of DNA replication, recombination, and repair. Crit. Rev. Biochem. Mol. Biol. 55, 482–507 (2020).
Barth, R. et al. SMC motor proteins extrude DNA asymmetrically and can switch directions. Cell 188, 749–763.e21 (2025).
Wasserman, M. R. & Liu, S. A tour de force on the double helix: exploiting DNA mechanics to study DNA-based molecular machines. Biochemistry 58, 4667–4676 (2019).
Chua, G. N. L. et al. A non-catalytic role for RFC in PCNA-mediated processive DNA synthesis. Cell (in press) https://doi.org/10.1016/S0092-8674(25)01478-3 (2025).
Jo, A., Li, S., Shin, J. W., Zhao, X. & Cho, Y. Structure basis for shaping the Nse4 protein by the Nse1 and Nse3 dimer within the Smc5/6 complex. J. Mol. Biol. 433, 166910 (2021).
Li, Q. et al. Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms. Nat. Struct. Mol. Biol. 31, 1532–1542 (2024).
Zheng, F., Georgescu, R., Yao, N. Y., Li, H. & O’Donnell, M. E. Cryo-EM structures reveal that RFC recognizes both the 3′- and 5′-DNA ends to load PCNA onto gaps for DNA repair. eLife 11, e77469 (2022).
Yao, N. et al. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 278, 50744–53 (2003).
Kochaniak, A. B. et al. Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 284, 17700–10 (2009).
Peng, X. P. et al. Acute Smc5/6 depletion reveals its primary role in rDNA replication by restraining recombination at fork pausing sites. PLoS Genet. 14, e1007129 (2018).
Xue, X. et al. Restriction of replication fork regression activities by a conserved SMC complex. Mol. Cell 56, 436–45 (2014).
Agashe, S. et al. Smc5/6 functions with Sgs1-Top3-Rmi1 to complete chromosome replication at natural pause sites. Nat. Commun. 12, 2111 (2021).
Mangione, R. M. et al. DNA lesions can frequently precede DNA:RNA hybrid accumulation. Nat. Commun. 16, 2401 (2025).
Whalen, J. M., Dhingra, N., Wei, L., Zhao, X. & Freudenreich, C. H. Relocation of collapsed forks to the nuclear pore complex depends on sumoylation of DNA repair proteins and permits Rad51 association. Cell Rep. 31, 107635 (2020).
Horigome, C. et al. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 30, 931–45 (2016).
Ryu, T. et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17, 1401–11 (2015).
Liu, H. W. et al. The SMC hinge is a selective gate for obstacle bypass. Nat. Commun. 16, 10457 (2025).
Oravcova, M. et al. The Nse5/6-like SIMC1-SLF2 complex localizes SMC5/6 to viral replication centers. Elife 11, e79676 (2022).
Pebernard, S., Wohlschlegel, J., McDonald, W. H., Yates, J. R. 3rd & Boddy, M. N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell Biol. 26, 1617–30 (2006).
Raschle, M. et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 348, 1253671 (2015).
Chen, H., Lisby, M. & Symington, L. S. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 50, 589–600 (2013).
Nguyen, B. et al. Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 426, 3246–3261 (2014).
de Laat, W. L. et al. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 12, 2598–609 (1998).
Kolpashchikov, D. M. et al. Polarity of human replication protein A binding to DNA. Nucleic Acids Res. 29, 373–9 (2001).
Iftode, C. & Borowiec, J. A. 5’ −> 3’ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry 39, 11970–81 (2000).
Canal, B. et al. The DNA replication checkpoint limits Okazaki fragment accumulation to protect and restart stalled forks. Mol. Cell 85, 2462–2473.e6 (2025).
Chhetri, G. et al. PAF15-PCNA assembly exhaustion governs lagging strand replication and replisome integrity. bioRxiv https://doi.org/10.1101/2025.03.15.641049 (2025).
Li, S. et al. Molecular basis for Nse5-6 mediated regulation of Smc5/6 functions. Proc. Natl. Acad. Sci. USA 120, e2310924120 (2023).
Kelman, Z. & O’Donnell, M. Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res. 23, 3613–20 (1995).
Finkelstein, J., Antony, E., Hingorani, M. M. & O’Donnell, M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 319, 78–87 (2003).
Henricksen, L. A., Umbricht, C. B. & Wold, M. S. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–32 (1994).
Chua, G. N. L. et al. Differential dynamics specify MeCP2 function at nucleosomes and methylated DNA. Nat. Struct. Mol. Biol. 31, 1789–1797 (2024).
Chang, J. T. H. Kymographs of Smc5/6 binding. Zenodo https://zenodo.org/records/17792658 (2025).
Watters, J. Scripts for single-molecule C-Trap data analysis. Zenodo https://zenodo.org/records/7618698 (2023).
Acknowledgements
We thank John Watters in the Shixin Liu Laboratory for technical support, Sophia Park in the Xiaolan Zhao Laboratory for helpful comments. J.T.C. was supported by an NCI F30 fellowship (F30CA275379). J.T.C. and B.J.K. were supported by a Medical Scientist Training Program grant to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program (T32GM152349) from the National Institutes of Health. V.M.B. and J.Z. acknowledge support from a training grant from NIGMS awarded to the Molecular Biophysics Program at Weill Cornell Graduate School (T32GM132081). G.N.L.C. was supported by an NIH F31 fellowship (F31MH132306). M.E.O. was supported by NIH (R01GM115809) and Howard Hughes Medical Institute. X.Z. was supported by NIH (R35GM145260) and Memorial Sloan-Kettering Cancer Center Core Grant P30 CA 008748?. S. Liu was supported by the Alfred P. Sloan Foundation, the Marlene Hess Center for Research in Women’s Health and Biomedicine at Rockefeller University, and NIH (R01GM149862).
Author information
Authors and Affiliations
Contributions
X.Z. and S.Liu oversaw the project. J.T.C., V.M.B., and G.N.C. performed single-molecule experiments and analyzed the data. S.Li, V.M.B., and J.Z. prepared protein constructs and performed biochemical experiments. B.J.K. assisted with single-molecule experiments. M.E.O. and E.C.B. provided key reagents. J.T.C., V.M.B., X.Z., and S.Liu wrote the paper with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Maxim Molodtsov, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chang, J.TH., Miller-Browne, V., Chua, G.N.L. et al. Molecular determinants of Smc5/6 association with DNA junctions. Nat Commun (2026). https://doi.org/10.1038/s41467-025-67999-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67999-5