Abstract
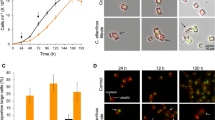
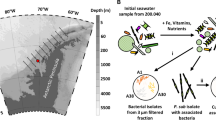
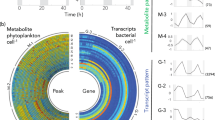
Diatoms are key contributors to global primary production, and have developed intricate partnerships with bacteria through long-term co-evolution. Here, we uncover a syntrophic relationship between the model obligate photoautotroph diatom Phaeodactylum tricornutum and the rod-shaped bacterium Loktanella vestfoldensis, which enables the diatom to indirectly utilize glucose. To be specific, growth of the diatom depends on the support of L. vestfoldensis for the supply of necessary carbon source when glucose serves as the sole carbon source, while L. vestfoldensis shows dependence on P. tricornutum when CO2 is the sole carbon source. Reanalysis of Tara Oceans metagenomic data shows frequent co-occurrence of Loktanella with diatoms including Chaetoceros and Thalassiosira, indicating the ecological relevance of this partnership. Co-culture with L. vestfoldensis supports robust growth of Chaetoceros muelleri and Thalassiosira pseudonana in the presence of glucose as the sole carbon source. Transcriptomic and metabolomic analyses reveal that P. tricornutum maintains a photoautotrophic metabolism in co-culture, as indicated by the up-regulation of genes involved in inorganic carbon concentration and photosynthesis, while the co-cultured bacterium likely supplies CO2 and growth-stimulating metabolites such as indole-3-acetic acid. Our findings demonstrate that bacterial-algal interactions may shape diatom adaptation to carbon changes and contribute to marine carbon cycling.
Similar content being viewed by others
Data availability
The raw data of RNA-Seq and 16S rRNA amplicon sequencing have been deposited in the Sequence Read Archive database under accession numbers PRJNA1276728 and PRJNA1330494. The Sanger sequencing data of 16S rRNA have been deposited in the Genbank database under accession numbers PX410800 (Janibacter anophelis) [https://www.ncbi.nlm.nih.gov/nuccore/PX410800] and PX410801 (Loktanella vestfoldensis) [https://www.ncbi.nlm.nih.gov/nuccore/PX410801]. The metabolomic data have been deposited on Metabolomics Workbench under Study ID ST004351 [https://doi.org/10.21228/M8WR9Z]. Source data are provided with this paper.
Code availability
The code used to produce the results is available at Figshare (https://doi.org/10.6084/m9.figshare.30178000).
References
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998).
Falkowski, P. G. et al. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360 (2004).
Bowler, C., Vardi, A. & Allen, A. E. Oceanographic and biogeochemical insights from diatom genomes. Ann. Rev. Mar. Sci. 2, 333–365 (2010).
Tréguer, P. et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37 (2018).
Reinfelder, J. R. Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Ann. Rev. Mar. Sci. 3, 291–315 (2011).
Nakajima, K., Tanaka, A. & Matsuda, Y. SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl. Acad. Sci. USA 110, 1767–1772 (2013).
Kikutani, S. et al. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 113, 9828–9833 (2016).
Cohen, N. R. Mixotrophic plankton foraging behaviour linked to carbon export. Nat. Commun. 13, 1302 (2022).
Ward, B. A. & Follows, M. J. Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux. Proc. Natl. Acad. Sci. USA 113, 2958–2963 (2016).
Kumar, M. et al. Mixotrophic growth of a ubiquitous marine diatom. Sci. Adv. 10, eado2623 (2024).
Amin, S. A., Parker, M. S. & Armbrust, E. V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684 (2012).
Ryther, J. H. & Guillard, R. R. L. Studies of marine planktonic diatoms: II. Use of Cyclotella nana Hustedt for assays of vitamin B12 in sea water. Can. J. Microbiol. 8, 437–445 (1962).
Foster, R. A. et al. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 5, 1484–1493 (2011).
Desbois, A. P., Mearns-Spragg, A. & Smith, V. J. A fatty acid from the diatom Phaeodactylum tricornutum is antibacterial against diverse bacteria, including multi-resistant Staphylococcus aureus (MRSA). Mar. Biotechnol. 11, 45–52 (2009).
Paul, C. & Pohnert, G. Interactions of the algicidal bacterium Kordia algicida with diatoms: regulated protease excretion for specific algal lysis. PLoS ONE 6, e21032 (2011).
Yu, G. et al. Mitochondrial phosphoenolpyruvate carboxylase contributes to carbon fixation in the diatom Phaeodactylum tricornutum at low inorganic carbon concentrations. N. Phytol. 235, 1379–1393 (2022).
Cerón Garcı́a, M. C. et al. Mixotrophic growth of the microalga Phaeodactylum tricornutum: influence of different nitrogen and organic carbon sources on productivity and biomass composition. Process Biochem. 40, 297–305 (2005).
Vuong, T. T., Kwon, B. R., Eom, J. I., Shin, B. K. & Kim, S. M. Interaction between marine bacterium Stappia sp. K01 and diatom Phaeodactylum tricornutum through extracellular fatty acids. J. Appl. Phycol. 32, 71–82 (2020).
Zhang, W. et al. Heterotrophic modification of Phaeodactylum tricornutum Bohlin. Algal Res. 72, 103137 (2023).
Zheng, Y., Quinn, A. H. & Sriram, G. Experimental evidence and isotopomer analysis of mixotrophic glucose metabolism in the marine diatom Phaeodactylum tricornutum. Microb. Cell Factor. 12, 1–17 (2013).
Schreiber, V. et al. The central vacuole of the diatom Phaeodactylum tricornutum: identification of new vacuolar membrane proteins and of a functional di-leucine-based targeting motif. Protist 168, 271–282 (2017).
Ødum, M. T. et al. DeepLoc 2.1: multi-label membrane protein type prediction using protein language models. Nucleic Acids Res. 52, W215–W220 (2024).
Di Costanzo, F., Di Dato, V. & Romano, G. Diatom-bacteria interactions in the marine environment: complexity, heterogeneity, and potential for biotechnological applications. Microorganisms 11, 2967 (2023).
Sittmann, J. et al. Bacterial diketopiperazines stimulate diatom growth and lipid accumulation. Plant Physiol. 186, 1159–1170 (2021).
Andrew, S., Wilson, T., Smith, S., Marchetti, A. & Septer, A. N. A tripartite model system for Southern Ocean diatom-bacterial interactions reveals the coexistence of competing symbiotic strategies. ISME Commun. 2, 97 (2022).
Abd-El-Aziz, A. et al. Bacteria-microalgae interactions from an evolutionary perspective and their biotechnological significance. Biotechnol. Adv. 82, 108591 (2025).
Le Chevanton, M. et al. Screening and selection of growth-promoting bacteria for Dunaliella cultures. Algal Res. 2, 212–222 (2013).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Durham, B. P. et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc. Natl. Acad. Sci. USA 112, 453–457 (2015).
Schäfer, H., Abbas, B., Witte, H. & Muyzer, G. Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42, 25–35 (2002).
Amin, S. A. et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101 (2015).
Deng, Y. et al. Dynamic diatom-bacteria consortia in synthetic plankton communities. Appl. Environ. Microbiol. 88, e01619–22 (2022).
Töpel, M. et al. Complete genome sequence of Loktanella vestfoldensis strain SMR4r, a novel strain isolated from a culture of the chain-forming diatom Skeletonema marinoi. Genome Announc. 6, e01558–17 (2018).
Johansson, O. N. et al. Friends with benefits: exploring the phycosphere of the marine diatom Skeletonema marinoi. Front. Microbiol. 10, 1828 (2019).
Roth, M. S. et al. Regulation of oxygenic photosynthesis during trophic transitions in the green alga Chromochloris zofingiensis. Plant Cell 31, 579–601 (2019).
Mazur, H., Konop, A. & Synak, R. Indole-3-acetic acid in the culture medium of two axenic green microalgae. J. Appl. Phycol. 13, 35–42 (2001).
Barbosa-Filho, J. M. et al. Botanical study, phytochemistry and antimicrobial activity of Tabebuia aurea:(with 1 table & 1 figure). Phyton 73, 221–228 (2004).
Judge, V. et al. Isonicotinic acid hydrazide derivatives: synthesis, antimicrobial activity, and QSAR studies. Med. Chem. Res. 21, 1451–1470 (2012).
Murata, M. et al. Loliolide, a carotenoid metabolite, is a potential endogenous inducer of herbivore resistance. Plant Physiol. 179, 1822–1833 (2019).
Castañeda, I. S., Werne, J. P., Johnson, T. C. & Powers, L. A. Organic geochemical records from Lake Malawi (East Africa) of the last 700 years, part II: Biomarker evidence for recent changes in primary productivity. Palaeogeogr. Palaeoclimatol. Palaeoecol. 303, 140–154 (2011).
Van Tol, H. M., Amin, S. A. & Armbrust, E. V. Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 11, 31–42 (2017).
Skoog, A. & Benner, R. Aldoses in various size fractions of marine organic matter: Implications for carbon cycling. Limnol. Oceanogr. 42, 1803–1813 (1997).
Benner, R. Chemical composition and reactivity. Biogeochemistry of Marine Dissolved Organic Matter 59–90 (2002). https://doi.org/10.1016/B978-012323841-2/50005-1
De Martino, A., Meichenin, A., Shi, J., Pan, K. & Bowler, C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J. Phycol. 43, 992–1009 (2007).
Guillard, R. R. Culture of phytoplankton for feeding marine invertebrates. in Culture of Marine Invertebrate Animals (eds Smith, W.L., Chanley, M.H.) 29–60 (Springer, 1975).
DeLong, E. F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89, 5685–5689 (1992).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Callahan, B. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Wickham, H. Data Analysis. In: ggplot2. https://doi.org/10.1007/978-3-319-24277-4_9 (Use R! Springer, Cham, 2016).
Becker, R. A. & Wilks, A. R. Constructing a Geographical Database.AT&T Bell Laboratories Statistics Research Report (1997). https://www.researchgate.net/publication/2444672_Constructing_a_Geographical_Database
Tackmann, J., Matias Rodrigues, J. F. & von Mering, C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. Cell Syst. 9, 286–296 (2019).
Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2, e107 (2023).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Bowler, C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Tsugawa, H. et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 12, 523–526 (2015).
Xia, J., Psychogios, N., Young, N. & Wishart, D. S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660 (2009).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2016).
Van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K. & Van der Werf, M. J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 7, 1–15 (2006).
Rohart, F., Gautier, B., Singh, A. & Lê Cao, K. A. MixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752 (2017).
Acknowledgements
This work was funded by the National Key Research and Development Program of China with No. 2023YFA0914400 (2023YFA0914402 to H.H.) and National Natural Science Foundation of China with No. 42376131 (to H.H.). We acknowledge the Metabolomics Workbench (https://www.metabolomicsworkbench.org, supported by NIH grants U2C-DK119886 and OT2-OD030544) for hosting the data and metadata for this study (Study ID: ST004351).
Author information
Authors and Affiliations
Contributions
H.H. designed the research. C.L., W.Y., and Y.P. performed experiments and data analysis. C.L. and H.H. drafted the manuscript. H.H. revised the manuscript. H.H. acquired the financial support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Sean Anderson, Xin Lin, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, C., Yin, W., Pan, Y. et al. Interactions with bacteria shape diatom adaptation to carbon concentration changes. Nat Commun (2025). https://doi.org/10.1038/s41467-025-68050-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68050-3