Abstract
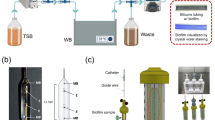
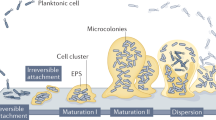
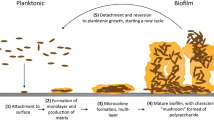
Microbial colonization and biofilm formation drive infection persistence and the spread of antimicrobial resistance, particularly under flow conditions typical of medical and natural environments. Here, we combine spontaneously buckled wrinkled topographies with microfluidic platforms to investigate the adhesion of Pseudomonas aeruginosa and Staphylococcus aureus across shear rates of 0.4-200 s−1. Wrinkled surfaces with tunable wavelengths (0.5-20 μm) are fabricated and characterized using optical, atomic force, and scanning electron microscopy. Sinusoidal wrinkles with a 2 μm wavelength reduce bacterial colonization by over 70% when oriented perpendicular to flow, while folded wrinkles of 5 μm achieve more than 90% reduction across broader shear regimes and suppress biofilm formation by over 85% relative to flat controls. These topographies retain antifouling performance under pulsatile flow. This work demonstrates a scalable, chemical-free strategy for passive biofilm control through geometric surface design, enabling durable antimicrobial materials for biomedical and industrial applications.
Similar content being viewed by others
Data availability
Additional data supporting this study are available in the Supplementary Information. A Source data file containing all data displayed in the figures and reported in the tables is provided with this paper and is accessible at https://doi.org/10.5281/zenodo.17303681. Any remaining data underlying this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Krajewski, S. et al. Bacterial interactions with proteins and cells relevant to the development of life-threatening endocarditis studied by use of a quartz-crystal microbalance. Anal. Bioanal. Chem. 406, 3395–3406 (2014).
Cheng, Y., Feng, G. & Moraru, C. I. Micro-and nanotopography sensitive bacterial attachment mechanisms: a review. Front. Microbiol. 10, 191 (2019).
Caldara, M., Belgiovine, C., Secchi, E. & Rusconi, R. Environmental, microbiological, and immunological features of bacterial biofilms associated with implanted medical devices. Clin. Microbiol. Rev. 35, e00221–20 (2022).
Hsu, L. C., Fang, J., Borca-Tasciuc, D. A., Worobo, R. W. & Moraru, C. I. Effect of micro-and nanoscale topography on the adhesion of bacterial cells to solid surfaces. Appl. Environ. Microbiol. 79, 2703–2712 (2013).
Tripathy, A., Sen, P., Su, B. & Briscoe, W. H. Natural and bioinspired nanostructured bactericidal surfaces. Adv. Colloid Interface Sci. 248, 85–104 (2017).
Skoog, S. A., Kumar, G., Narayan, R. J. & Goering, P. L. Biological responses to immobilized microscale and nanoscale surface topographies. Pharmacol. Ther. 182, 33–55 (2018).
Luan, Y. et al. Bacterial interactions with nanostructured surfaces. Curr. Opinion Colloid Interface Sci. 38, 170–189 (2018).
Lee, S. W., Phillips, K. S., Gu, H., Kazemzadeh-Narbat, M. & Ren, D. How microbes read the map: effects of implant topography on bacterial adhesion and biofilm formation. Biomaterials 268, 120595 (2020).
Ivanova, E. P. et al. Impact of nanoscale roughness of titanium thin film surfaces on bacterial retention. Langmuir 26, 1973–1982 (2010).
Mainwaring, D. E. et al. The nature of inherent bactericidal activity: insights from the nanotopology of three species of dragonfly. Nanoscale 8, 6527–6534 (2016).
Elbourne, A., Crawford, R. J. & Ivanova, E. P. Nano-structured antimicrobial surfaces: from nature to synthetic analogues. J. Colloid Interface Sci. 508, 603–616 (2017).
Tuson, H. H. & Weibel, D. B. Bacteria–surface interactions. Soft Matter 9, 4368–4380 (2013).
Vasudevan, R., Kennedy, A. J., Merritt, M., Crocker, F. H. & Baney, R. H. Microscale patterned surfaces reduce bacterial fouling-microscopic and theoretical analysis. Colloids Surf. B Biointerfaces 117, 225–232 (2014).
Hong, S.-H. et al. Surface waves control bacterial attachment and formation of biofilms in thin layers. Sci. Adv. 6, eaaz9386 (2020).
Pellegrino, L., Kriem, L. S., Robles, E. S. & Cabral, J. T. Microbial response to micrometer-scale multiaxial wrinkled surfaces. ACS Appl. Mater. Interfaces 14, 31463–31473 (2022).
Hizal, F. et al. Impact of 3D hierarchical nanostructures on the antibacterial efficacy of a bacteria-triggered self-defensive antibiotic coating. ACS Appl. Mater. Interfaces 7, 20304–20313 (2015).
Chang, Y.-R., Weeks, E. R. & Ducker, W. A. Surface topography hinders bacterial surface motility. ACS Appl. Mater. Interfaces 10, 9225–9234 (2018).
Mok, R., Dunkel, J. & Kantsler, V. Geometric control of bacterial surface accumulation. Phys. Rev. E 99, 052607 (2019).
Zheng, S. et al. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 9, 643722 (2021).
Surapaneni, V. A. et al. Groovy and gnarly: surface wrinkles as a multifunctional motif for terrestrial and marine environments. Integr. Comp. Biol. 62, 749–761 (2022).
Demenego, G. et al. Neurodevelopmental origins of structural and psychomotor defects in CXCR4-linked primary immunodeficiency. Neuron 113, 2636–2655 (2025).
Rusconi, R., Guasto, J. S. & Stocker, R. Bacterial transport suppressed by fluid shear. Nat. Phys. 10, 212–217 (2014).
Secchi, E. et al. The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat. Commun. 11, 2851 (2020).
Lee, Y. K., Won, Y.-J., Yoo, J. H., Ahn, K. H. & Lee, C.-H. Flow analysis and fouling on the patterned membrane surface. J. Membr. Sci. 427, 320–325 (2013).
Wang, Q. & Zhao, X. A three-dimensional phase diagram of growth-induced surface instabilities. Sci. Rep. 5, 8887 (2015).
Allen, J. The classification of cross-stratified units. with notes on their origin. Sedimentology 2, 93–114 (1963).
Efimenko, K. et al. Nested self-similar wrinkling patterns in skins. Nat. Mater. 4, 293–297 (2005).
Genzer, J. & Groenewold, J. Soft matter with hard skin: from skin wrinkles to templating and material characterization. Soft Matter 2, 310–323 (2006).
Bayley, F. A., Liao, J. L., Stavrinou, P. N., Chiche, A. & Cabral, J. T. Wavefront kinetics of plasma oxidation of polydimethylsiloxane: limits for sub-μm wrinkling. Soft Matter 10, 1155–1166 (2014).
Nania, M., Foglia, F., Matar, O. K. & Cabral, J. T. Sub-100 nm wrinkling of polydimethylsiloxane by double frontal oxidation. Nanoscale 9, 2030–2037 (2017).
Pellegrino, L., Khodaparast, S. & Cabral, J. T. Orthogonal wave superposition of wrinkled, plasma-oxidised, polydimethylsiloxane surfaces. Soft Matter 16, 595–603 (2020).
Pellegrino, L., Tan, A. & Cabral, J. T. Ripple patterns spontaneously emerge through sequential wrinkling interference in polymer bilayers. Phys. Rev. Lett. 128, 058001 (2022).
Nakazawa, K. et al. A human septin octamer complex sensitive to membrane curvature drives membrane deformation with a specific mesh-like organization. J. Cell Sci. 136, jcs260813 (2023).
Li, B., Cao, Y.-P., Feng, X.-Q. & Gao, H. Mechanics of morphological instabilities and surface wrinkling in soft materials: a review. Soft Matter 8, 5728–5745 (2012).
Stroock, A. D. et al. Chaotic mixer for microchannels. Science 295, 647–651 (2002).
Pyke, K. E., Dwyer, E. M. & Tschakovsky, M. E. Impact of controlling shear rate on flow-mediated dilation responses in the brachial artery of humans. J. Appl. Physiol. 97, 499–508 (2004).
Zöttl, A. et al. Dynamics of individual Brownian rods in a microchannel flow. Soft Matter 15, 5810–5814 (2019).
Shen, Y., Siryaporn, A., Lecuyer, S., Gitai, Z. & Stone, H. A. Flow directs surface-attached bacteria to twitch upstream. Biophys. J. 103, 146–151 (2012).
Leighton, T. L., Buensuceso, R. N., Howell, P. L. & Burrows, L. L. Biogenesis of Pseudomonas aeruginosa type IV pili and regulation of their function. Environ. Microbiol. 17, 4148–4163 (2015).
Duvernoy, M.-C. et al. Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. Nat. Commun. 9, 1120 (2018).
Roberge, N. A. & Burrows, L. L. Building permits-control of type IV pilus assembly by pilB and its cofactors. J. Bacteriol. 206, e00359–24 (2024).
Lecuyer, S. et al. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys. J. 100, 341–350 (2011).
Parente, R. et al. A multilayered imaging and microfluidics approach for evaluating the effect of fibrinolysis in Staphylococcus aureus biofilm formation. Pathogens 12, 1141 (2023).
Baba, T., Bae, T., Schneewind, O., Takeuchi, F. & Hiramatsu, K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190, 300–310 (2008).
Forson, A. M., van der Mei, H. C. & Sjollema, J. Impact of solid surface hydrophobicity and micrococcal nuclease production on Staphylococcus aureus newman biofilms. Sci. Rep. 10, 12093 (2020).
Parsons, J. B., Westgeest, A. C., Conlon, B. P. & Fowler Jr, V. G. Persistent methicillin-resistant Staphylococcus aureus bacteremia: host, pathogen, and treatment. Antibiotics 12, 455 (2023).
Liesenborghs, L., Verhamme, P. & Vanassche, T. Staphylococcus aureus, master manipulator of the human hemostatic system. J. Thromb. Haemost. 16, 441–454 (2018).
McAdow, M. et al. Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog. 7, e1002307 (2011).
Belgiovine, C. et al. Interaction of bacteria, immune cells, and surface topography in periprosthetic joint infections. Int. J. Mol. Sci. 24, 9028 (2023).
Gammie, A. et al. International continence society guidelines on urodynamic equipment performance. Neurourol. Urodyn. 33, 370–379 (2014).
Secchi, E. et al. The structural role of bacterial eDNA in the formation of biofilm streamers. Proc. Natl. Acad. Sci. USA 119, e2113723119 (2022).
Savorana, G., Słomka, J., Stocker, R., Rusconi, R. & Secchi, E. A microfluidic platform for characterizing the structure and rheology of biofilm streamers. Soft Matter 18, 3878 - 3890 (2022).
Lee, S. W., Phillips, K. S., Gu, H., Kazemzadeh-Narbat, M. & Ren, D. How microbes read the map: effects of implant topography on bacterial adhesion and biofilm formation. Biomaterials 268, 120595 (2021).
Nicolle, L. E. Catheter associated urinary tract infections. Antimicrob. Resist. Infect. Control 3, 1–8 (2014).
Werneburg, G. T. Catheter-associated urinary tract infections: current challenges and future prospects. Res. Rep. Urol. 14, 109–133 (2022).
Marschall, J., Carpenter, C. R., Fowler, S. & Trautner, B. W. Antibiotic prophylaxis for urinary tract infections after removal of urinary catheter: meta-analysis. Bmj 346, f3147 (2013).
Holmes, A. H. et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187 (2016).
Dean, B. & Bhushan, B. Shark-skin surfaces for fluid-drag reduction in turbulent flow: a review. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 368, 4775 - 4806 (2010).
Gomez, S. et al. Substrate stiffness impacts early biofilm formation by modulating Pseudomonas aeruginosa twitching motility. eLife 12, e81112 (2023).
Ariel, G. et al. Swarming bacteria migrate by Lévy Walk. Nat. Commun. 6, 8396 (2015).
Lauta, F. C., Pellegrino, L. & Rusconi, R. Macrophages on the wrinkle: exploring microscale interactions with substrate topography. Biophys. Rev. 5, 022001 (2024).
Vinci, V. et al. Breast implant surface topography triggers a chronic-like inflammatory response. Life Sci. Alliance 7, e202302132 (2024).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Acknowledgements
This work was supported by EU funding within the Italian Ministry of University and Research (MUR) PNRR Extended Partnership initiative on Emerging Infectious Diseases (project no. PE00000007, INF-ACT) to R.R., EU HORIZON-TMA-MSCA-PF-EF investigating microbial colonization and removal on dynamic patterned surfaces (grant no. 101110029, MOBILE) to L.P., and by a grant from the Italian Ministry of University and Research (MUR), Dipartimenti di Eccellenza 2023-2027 (I.232/2016, art. 1, commi 314-337) to V.C., R.C., and F.M.. E.D.I. and M.C. are grateful to Humanitas University Office of Information Technology for the computing resources maintenance. L.P., V.C., R.C., and F.M. gratefully acknowledge the support of the ISIS@MACH ITALIA Research Infrastructure, the hub of ISIS Neutron and Muon Source (UK), [MUR official registry U. 0008642.28-05-2020-16th April 2020]. IM@IT is listed in the Italian Ministry of University and Research’s Piano Nazionale delle Infrastrutture di Ricerca (PNIR 2021–2027) “in the broader notion of ISIS”, and ISIS Facility and IM@IT are jointly listed in high-priority RI’s (see Table 6, page 30, note 38, PNIR in 2021–2027).
Author information
Authors and Affiliations
Contributions
L.P. and R.R. conceived and designed the project. L.P. conducted the microfluidic experiments and performed data analysis. L.P., G.S., and E.S. carried out numerical simulations and interpreted the results. L.P., V.C., R.C., and F.M. performed and analyzed AFM measurements. M.C. and E.D.I. conducted and analyzed SEM measurements. S.L. and C.B. contributed to data analysis and interpretation. V.V., M.K., and R.R. acquired funding and supervised the research. L.P. and R.R. wrote the original draft, and all authors contributed to reviewing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks James Henderson and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pellegrino, L., Savorana, G., Cassina, V. et al. Reduction of bacterial colonization on buckling-induced wrinkled surfaces under fluid shear. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68078-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68078-5