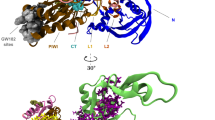
Abstract
The clustering of neurotransmitter receptors at appropriate postsynaptic sites is essential for controlling synaptic transmission. While most known mechanisms involve receptor binding with cytoplasmic scaffolds, recent evidence highlights the importance of extracellular interactions that directly target receptors. Using Caenorhabditis elegans, we identified a trans-synaptic complex that involves RIG-5 and ZIG-8, two adhesion molecules of the immunoglobulin (Ig) superfamily and orthologous to Drosophila DIPs and Dprs, and mammalian IgLONs. Our results show that RIG-5 and ZIG-8 are anchored in the pre- and postsynaptic membranes, respectively, and interact in vivo via their first Ig domains. Furthermore, ZIG-8 directly binds a α7-like acetylcholine receptor (AChR), known as ACR-16, via a cis-interaction between its Ig2 domain and the base of the extracellular AChR domain. This study provides direct evidence that trans-synaptic IgLON interactions can organize neurochemical synapses and suggests that the IgLONs may directly interact with ionotropic receptors in the mammalian nervous system.
Similar content being viewed by others
Data availability
Source data are provided with this paper. The plasmids and C. elegans strains generated in this study are available from the corresponding authors upon request. Source data are provided with this paper.
Code availability
The code used to perform analyses of synapse density is deposited on Zenodo, and is accessible via (https://doi.org/10.5281/zenodo.17571550)95.
References
Südhof, T. C. The cell biology of synapse formation. J. Cell Biol. 220, e202103052 (2021).
Sanes, J. R. & Zipursky, S. L. Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell 181, 536–556 (2020).
Wilkinson, B. & Coba, M. P. Molecular architecture of postsynaptic Interactomes. Cell. Signal. 76, 109782 (2020).
Tyagarajan, S. K. & Fritschy, J. M. Gephyrin: a master regulator of neuronal function?. Nat. Rev. Neurosci. 15, 141–56 (2014).
Yuzaki, M. Two classes of secreted synaptic organizers in the central nervous system. Annu Rev. Physiol. 80, 243–262 (2018).
Xu, D. et al. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron 39, 513–528 (2003).
O’Brien, R. J. et al. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product narp. Neuron 23, 309–323 (1999).
Elegheert, J. et al. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science 353, 295–9 (2016).
Matsuda, K. et al. Transsynaptic modulation of kainate receptor functions by C1q-like proteins. Neuron 90, 752–67 (2016).
Fossati, M. et al. Trans-synaptic signaling through the glutamate receptor Delta-1 mediates inhibitory synapse formation in cortical pyramidal neurons. Neuron 104, 1081–1094.e7 (2019).
Richmond, J. E. & Jorgensen, E. M. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat. Neurosci. 2, 791–7 (1999).
Gendrel, M., Rapti, G., Richmond, J. E. & Bessereau, J. L. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature 461, 992–6 (2009).
Rapti, G., Richmond, J. & Bessereau, J. L. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. Embo J. 30, 706–18 (2011).
Gally, C., Eimer, S., Richmond, J. E. & Bessereau, J. L. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature 431, 578–82 (2004).
Pinan-Lucarre, B. et al. C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature 511, 466–70 (2014).
Touroutine, D. et al. acr-16 encodes an essential subunit of the levamisole-resistant nicotinic receptor at the Caenorhabditis elegans neuromuscular junction. J. Biol. Chem. 280, 27013–21 (2005).
Zhou, X. et al. The HSPG syndecan is a core organizer of cholinergic synapses. J. Cell Biol. 220, e202011144 (2021).
Tu, H., Pinan-Lucarre, B., Ji, T., Jospin, M. & Bessereau, J. L. C. elegans Punctin Clusters GABA(A) receptors via neuroligin binding and UNC-40/DCC recruitment. Neuron 86, 1407–19 (2015).
Zhou, X. et al. The netrin receptor UNC-40/DCC assembles a postsynaptic scaffold and sets the synaptic content of GABAA receptors. Nat. Commun. 11, 2674 (2020).
Taylor, S. R. et al. Molecular topography of an entire nervous system. Cell https://doi.org/10.1016/j.cell.2021.06.023 (2021).
White, J. G., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode C. elegans. Philos. Trans. R. Soc. Lond. 314B, 1–340 (1986).
Zhang, L., Ward, J. D., Cheng, Z. & Dernburg, A. F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–84 (2015).
Yemini, E. et al. NeuroPAL: a multicolor atlas for whole-brain neuronal identification in C. elegans. Cell 184, 272–288.e11 (2021).
He, S., Cuentas-Condori, A. & Miller, D. M. NATF (native and tissue-specific fluorescence): a strategy for bright, tissue-specific GFP labeling of native proteins in Caenorhabditis elegans. Genetics 212, 387–395 (2019).
Schmitt, C. et al. Specific expression of channelrhodopsin-2 in single neurons of Caenorhabditis elegans. Plos One 7, e43164 (2012).
Liu, Y. et al. Nicotine induces abnormal motor coupling through sensitization of a mechanosensory circuit in Caenorhabditis elegans. PLOS Biol. 23, e3003423 (2025).
Pereira, L. et al. A cellular and regulatory map of the cholinergic nervous system of C. elegans. Elife 4, e12432 (2015).
Xuan, Z. et al. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife 6, e29276 (2017).
Özkan, E. et al. An extracellular interactome of immunoglobulin and lrr proteins reveals receptor-ligand networks. Cell 154, 228–239 (2013).
Ranaivoson, F. M. et al. A proteomic screen of neuronal cell-surface molecules reveals IgLONs as structurally conserved interaction modules at the synapse. Structure 27, 893–906.e9 (2019).
Cheng, S. et al. Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence. Proc. Natl. Acad. Sci. 116, 201818631 (2019).
Lobb-Rabe, M. et al. Neuronal wiring receptors Dprs and DIPs Are GPI anchored and this modification contributes to their cell surface organization. eneuro 11, ENEURO.0184-23.2023 (2024).
McNamee, C. J., Youssef, S. & Moss, D. IgLONs form heterodimeric complexes on forebrain neurons. Cell Biochem Funct. 29, 114–119 (2011).
Venkannagari, H. et al. Highly conserved molecular features in iglons contrast their distinct structural and biological outcomes. J. Mol. Biol. 432, 5287–5303 (2020).
Birtley, J. R. et al. Inactivating mutations and X-ray crystal structure of the tumor suppressor OPCML reveal cancer-associated functions. Nat. Commun. 10, 3134 (2019).
Schwarz, V., Pan, J., Voltmer-Irsch, S. & Hutter, H. IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Dev. 4, 13 (2009).
Feresten, A. H. et al. wrk-1 and rig-5 control pioneer and follower axon navigation in the ventral nerve cord of Caenorhabditis elegans in a nid-1 mutant background. Genetics 223, iyac187 (2022).
Bénard, C. Y., Blanchette, C., Recio, J. & Hobert, O. The Secreted immunoglobulin domain proteins ZIG-5 and ZIG-8 cooperate with L1CAM/SAX-7 to maintain nervous system integrity. Plos Genet 8, e1002819 (2012).
Noviello, C. M. et al. Structure and gating mechanism of the α7 nicotinic acetylcholine receptor. Cell 184, 2121–2134 (2021).
Morud, J. et al. Deorphanization of novel biogenic amine-gated ion channels identifies a new serotonin receptor for learning. Curr. Biol. 31, 4282–4292.e6 (2021).
Albuquerque, E. X., Pereira, E. F., Alkondon, M. & Rogers, S. W. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol. Rev. 89, 73–120 (2009).
Letsinger, A. C., Gu, Z. & Yakel, J. L. α7 nicotinic acetylcholine receptors in the hippocampal circuit: taming complexity. Trends Neurosci. 45, 145–157 (2022).
Stone, T. W. Relationships and Interactions between Ionotropic Glutamate Receptors and Nicotinic Receptors in the CNS. Neuroscience 468, 321–365 (2021).
Garção, P., Oliveira, C. R., Cunha, R. A. & Agostinho, P. Subsynaptic localization of nicotinic acetylcholine receptor subunits: a comparative study in the mouse and rat striatum. Neurosci. Lett. 566, 106–110 (2014).
Conroy, W. G., Liu, Z., Nai, Q., Coggan, J. S. & Berg, D. K. PDZ-containing proteins provide a functional postsynaptic scaffold for nicotinic receptors in neurons. Neuron 38, 759–71 (2003).
Parker, M. J., Zhao, S., Bredt, D. S., Sanes, J. R. & Feng, G. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J. Neurosci. 24, 378–388 (2004).
Gómez-Varela, D., Schmidt, M., Schoellerman, J., Peters, E. C. & Berg, D. K. PMCA2 via PSD-95 controls calcium signaling by α7-containing nicotinic acetylcholine receptors on aspiny interneurons. J. Neurosci. 32, 6894–6905 (2012).
Baer, K. et al. PICK1 interacts with alpha7 neuronal nicotinic acetylcholine receptors and controls their clustering. Mol. Cell Neurosci. 35, 339–55 (2007).
Bondarenko, V., Chen, Q., Tillman, T. S., Xu, Y. & Tang, P. Unconventional PDZ Recognition Revealed in α7 nAChR-PICK1 complexes. ACS Chem. Neurosci. 15, 2070–2079 (2024).
Li, Z.-L. et al. A novel effect of PDLIM5 in α7 nicotinic acetylcholine receptor upregulation and surface expression. Cell. Mol. Life Sci. 79, 64 (2022).
Palumbo, T. B. & Miwa, J. ulieM. Lynx1 and the family of endogenous mammalian neurotoxin-like proteins and their roles in modulating nAChR function. Pharmacol. Res. 194, 106845 (2023).
Paramonov, A. S. et al. Structural diversity and dynamics of human three-finger proteins acting on nicotinic acetylcholine receptors. Int. J. Mol. Sci. 21, 7280 (2020).
Carrillo, R. A. et al. Control of synaptic connectivity by a network of Drosophila IgSF cell surface proteins. Cell 163, 1770–1782 (2015).
Cheng, S. et al. Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. Elife 8, e41028 (2019).
Cosmanescu, F. et al. Neuron-subtype-specific expression, interaction affinities, and specificity determinants of DIP/Dpr cell recognition proteins. Neuron 100, 1385–1400.e6 (2018).
Wang, Y. et al. Systematic expression profiling of dprs and DIPs reveals cell surface codes in Drosophila larval motor and sensory neurons. Development 149, dev200355 (2022).
Ashley, J. et al. Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity. Elife 8, e42690 (2019).
Tan, L. et al. Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163, 1756–1769 (2015).
Venkatasubramanian, L. et al. Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-α and Dpr10. Elife 8, e42692 (2019).
Barish, S. et al. Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. Plos Genet 14, e1007560 (2018).
Xu, S. et al. Interactions between the Ig-superfamily proteins DIP-α and Dpr6/10 regulate assembly of neural circuits. Neuron 100, 1369–1384.e6 (2018).
Xu, C. et al. Control of synaptic specificity by establishing a relative preference for synaptic partners. Neuron 103, 865–877.e7 (2019).
Dombrovski, M. et al. Molecular gradients shape synaptic specificity of a visuomotor transformation. Nature 644, 453–462 (2025).
Sharma, K. et al. Cell type– and brain region–resolved mouse brain proteome. Nat. Neurosci. 18, 1819–1831 (2015).
Fearnley, S., Raja, R. & Cloutier, J.-F. Spatiotemporal expression of IgLON family members in the developing mouse nervous system. Sci. Rep. 11, 19536 (2021).
Jagomäe, T. et al. Alternative promoter use governs the expression of IgLON Cell adhesion molecules in histogenetic fields of the embryonic mouse brain. Int. J. Mol. Sci. 22, 6955 (2021).
Salluzzo, M. et al. The role of IgLON cell adhesion molecules in neurodegenerative diseases. Genes 14, 1886 (2023).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature https://doi.org/10.1038/s41586-022-04434-5 (2022).
Sabater, L. et al. A novel non-rapid-eye movement and rapid-eye-movement parasomnia with sleep breathing disorder associated with antibodies to IgLON5: a case series, characterisation of the antigen, and post-mortem study. Lancet Neurol. 13, 575–586 (2014).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Loh, K. H. et al. Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166, 1295–1307.e21 (2016).
Oostrum et al. The proteomic landscape of synaptic diversity across brain regions and cell types. Cell 186, 5411–5427.e23 (2023).
Peng, J. et al. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry*. J. Biol. Chem. 279, 21003–21011 (2004).
Miyata, S. et al. Biochemical and ultrastructural analyses of iglon cell adhesion molecules, kilon and obcam in the rat brain. Neuroscience 117, 645–658 (2003).
Zacco, A. et al. Isolation, biochemical characterization and ultrastructural analysis of the limbic system-associated membrane protein (LAMP), a protein expressed by neurons comprising functional neural circuits. J. Neurosci. 10, 73–90 (1990).
Zhang, Z. et al. The schizophrenia susceptibility gene OPCML regulates spine maturation and cognitive behaviors through eph-cofilin signaling. Cell Rep. 29, 49–61.e7 (2019).
Hashimoto, T., Maekawa, S. & Miyata, S. IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct. 27, 496–498 (2009).
Hashimoto, T., Yamada, M., Maekawa, S., Nakashima, T. & Miyata, S. IgLON cell adhesion molecule Kilon is a crucial modulator for synapse number in hippocampal neurons. Brain Res 1224, 1–11 (2008).
Wit, J. de & Ghosh, A. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 4–4 (2016).
Merrion, H. G. et al. Dynamic extracellular interactions with AMPA receptors. Proc. Natl. Acad. Sci. 122, e2517436122 (2025).
Gao, S. et al. Excitatory motor neurons are local oscillators for backward locomotion. eLife 7, e29915 (2018).
Nawrocka, W. I. et al. Nematode extracellular protein interactome expands connections between signaling pathways. Preprint at bioRxiv https://doi.org/10.1101/2024.07.08.602367 (2024).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Zuryn, S. & Jarriault, S. Deep sequencing strategies for mapping and identifying mutations from genetic screens. Worm 2, e25081 (2013).
Minevich, G., Park, D. S., Blankenberg, D., Poole, R. J. & Hobert, O. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–69 (2012).
Ghanta, K. S. & Mello, C. C. Melting dsDNA donor molecules greatly improves precision genome editing in caenorhabditis elegans. Genetics 216, 643–650 (2020).
Katz, M., Corson, F., Iwanir, S., Biron, D. & Shaham, S. Glia Modulate a Neuronal Circuit for Locomotion Suppression during Sleep in C. elegans. Cell Rep. 22, 2575–2583 (2018).
Gao, S. et al. The NCA sodium leak channel is required for persistent motor circuit activity that sustains locomotion. Nat. Commun. 6, 6323 (2015).
Meng, J. et al. A tonically active master neuron modulates mutually exclusive motor states at two timescales. Sci. Adv. 10, eadk0002 (2024).
Frokjaer-Jensen, C. et al. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11, 529–34 (2014).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform 18, 529 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Cizeron, M., Granger, L., BÜlow, H. E. & Bessereau, J.-L. Specific heparan sulfate modifications stabilize the synaptic organizer MADD-4/Punctin at C. elegans neuromuscular junctions. Genetics 218, iyab073 (2021).
Jourdren, L., Delaveau, T., Marquenet, E., Jacq, C. & Garcia, M. CORSEN, a new software dedicated to microscope-based 3D distance measurements: mRNA–mitochondria distance, from single-cell to population analyses. RNA 16, 1301–1307 (2010).
Mialon, M. & Weinreb, A. NeuraCelLab_synapse-density-analysis. https://doi.org/10.5281/zenodo.17571550 (2025).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021 10, 463034 (2022). 04.
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Schrödinger & L. L. C. The PyMOL Molecular Graphics System, Version 3.0.4. (2015).
Acknowledgements
We thank members of the Bessereau lab for providing feedback, Mélissa Cizeron for advice on image analysis, Driss Laabid and Lina Boumasmoud for technical assistance, and Océane Romatif, Delphine Le Guern and Camille Vachon for strains. We are grateful to Elise Forgues, Cecile Chatras, Mathieu Ben Abu and Manon Courtieux for their help during their respective internships. We thank Pierre-Jean Corringer for critical reading of the manuscript, Mei Zhen and Jun Meng for providing information on neuronal promoters, Alexander Gottschalk for sharing plasmids and Eviatar Yemini for advice on NeuroPAL. We thank the Caenorhabditis Genetics Center (CGC), funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing strains. We thank the SFR Biosciences (University Lyon 1 CNRS UAR 3444 INSERM US8, ENS de Lyon), Matthieu Caron and Francesca Palladino for access to equipment. We thank Le Centre d’Imagerie Quantitative Lyon-Est (LyMIC-CIQLE, Lyon, France) imaging facility for support and access to equipment, and Camilla Luccardini for technical assistance. Some strains were generated by SEGiCel (SFR Santé Lyon Est CNRS UAR 3453, Lyon, France) with the support of CNRS and IBiSA. Some graphical elements in Figs. 1d and 6b were provided by Servier Medical Art (https://smart.servier.com), licensed under CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). This work was supported as follows: M.M.: fellowship from the French Ministry of Research; BPL: LABEX Cortex (ANR-11-LABX-0042) of University Lyon 1, within the program “Investissements d’Avenir” (ANR-11-IDEX-0007) and Fondation pour la Recherche Médicale grant (FRM-MND-202411019867); EÖ: National Institutes of Health, National Institute of Neurological Disorders and Stroke grant R01 NS139060 grant; JLB: Équipe FRM 2023 EQU202303016267, ANR Synapunct ANR-22CE16-0024-01, ERC_Adg C.NAPSE #695295, ANR-11-LABX-0042/ANR-11-IDEX-0007.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.M., J.-L.B. and B.P.-L.; Methodology: M.M., A.W., E.Ö., J.-L.B. and B.P.-L.; Investigation: M.M., L.P., L. G. and B.P.-L.; Formal analysis: M.M., L.P., E.Ö. and B.P.-L.; Visualization: M.M., E.Ö., J.-L.B. and B.P.-L.; Writing – Original Draft: M.M., J.-L.B. and B.P.-L.; Writing – Review & Editing: M.M., A.W., E.Ö., J.-L.B. and B.P.-L.; Funding Acquisition: E.Ö., J.-L.B. and B.P.-L.; Supervision: J.-L.B. and B.P.-L.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mialon, M., Patrash, L., Granger, L. et al. A trans-synaptic IgLON adhesion molecular complex directly contacts and clusters a nicotinic receptor. Nat Commun (2026). https://doi.org/10.1038/s41467-025-68141-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68141-1