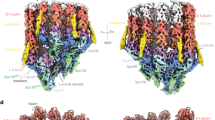
Abstract
Microtubule polarity and dynamic polymerization arise from the self-association properties of the αβ-tubulin heterodimer. For decades, it has remained unclear how the tubulin cofactors TBCD, TBCE, TBCC, and the Arl2 GTPase mediate the biogenesis of αβ-tubulin from individual α- and β-tubulins. Here, we use cryo-electron microscopy to determine structures of tubulin cofactors bound to αβ-tubulin. TBCD, TBCE, and Arl2 form a heterotrimeric cage-like assembly, we term TBC-DEG, around the αβ-tubulin heterodimer. The TBC-DEG-αβ-tubulin structures show that TBC-DEG wraps around β-tubulin while TBCE extends along α-tubulin. The TBC-DEG/TBCC-αβ-tubulin structures reveal that TBCC forms multi-domain interactions with Arl2 and TBCD to engage the αβ-tubulin intradimer-interface, promoting TBCE rotation while TBCD holds β-tubulin. TBCC engages the GTP-bound Arl2, multiple sites of TBCD, and the native αβ-tubulin intradimer interface near the α-tubulin N-site GTP. Together, these structures uncover transition states for αβ-tubulin biogenesis and degradation, suggesting a vise-like, GTP-hydrolysis-dependent mechanism in which TBCC binding to TBC-DEG modulates αβ-tubulin interfaces. Our studies provide structural evidence that tubulin cofactors act as enzymatic regulators that assemble the invariant αβ-tubulin architecture. By catalyzing α- and β-tubulin biogenesis and degradation, the TBC-DEG and TBCC assemblies regulate the polymerization competency of αβ-tubulin for microtubule formation.
Similar content being viewed by others
Data availability
Cryo-EM maps and models are available in the Electron Microscopy Database (EMDB) with the EMBD-IDs: EMD-47949, EMD-47954, EMD-47947, EMD-47948. The corresponding atomic coordinates for models are available at the Protein Data Bank (PDB) with the accession numbers, PDB-ID: 9EDT, 9EEB, 9EDR, 9EDS, respectively. The work also utilized the following coordinates for model building and comparisons: 1FFX, 4DRX, and 6GWD. Source data are provided with this paper.
References
Akhmanova, A. & Steinmetz, M. O. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16, 711–726 (2015).
Akhmanova, A. & Kapitein, L. C. Mechanisms of microtubule organization in differentiated animal cells. Nat. Rev. Mol. Cell Biol. 23, 541–558 (2022).
Gudimchuk, N. B. & McIntosh, J. R. Regulation of microtubule dynamics, mechanics and function through the growing tip. Nat. Rev. Mol. Cell Biol. 22, 777–795 (2021).
Cason, S. E. & Holzbaur, E. L. F. Selective motor activation in organelle transport along axons. Nat. Rev. Mol. Cell Biol. 23, 699–714 (2022).
Grossman-Haham, I. Towards an atomic model of a beating ciliary axoneme. Curr. Opin. Struct. Biol. 78, 102516 (2023).
Lowe, J., Li, H., Downing, K. H. & Nogales, E. Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 313, 1045–1057 (2001).
Downing, K. H. & Nogales, E. Tubulin and microtubule structure. Curr. Opin. Cell Biol. 10, 16–22 (1998).
Lundin, V. F., Leroux, M. R. & Stirling, P. C. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 35, 288–297 (2010).
Al-Bassam, J. Revisiting the tubulin cofactors and Arl2 in the regulation of soluble alphabeta-tubulin pools and their effect on microtubule dynamics. Mol. Biol. Cell 28, 359–363 (2017).
Radcliffe, P. A., Garcia, M. A. & Toda, T. The cofactor-dependent pathways for alpha- and beta-tubulins in microtubule biogenesis are functionally different in fission yeast. Genetics 156, 93–103 (2000).
Radcliffe, P. A., Hirata, D., Vardy, L. & Toda, T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol. Biol. Cell 10, 2987–3001 (1999).
Fleming, J. A., Vega, L. R. & Solomon, F. Function of tubulin binding proteins in vivo. Genetics 156, 69–80 (2000).
Szymanski, D. Tubulin folding cofactors: half a dozen for a dimer. Curr. Biol. 12, R767–R769 (2002).
Okumura, M., Sakuma, C., Miura, M. & Chihara, T. Linking cell surface receptors to microtubules: tubulin folding cofactor D mediates Dscam functions during neuronal morphogenesis. J. Neurosci. 35, 1979–1990 (2015).
Jin, S. et al. Drosophila tubulin-specific chaperone E functions at neuromuscular synapses and is required for microtubule network formation. Development 136, 1571–1581 (2009).
Pode-Shakked, B. et al. Microcephaly, intractable seizures and developmental delay caused by biallelic variants in TBCD: further delineation of a new chaperone-mediated tubulinopathy. Clin. Genet. 91, 725–738 (2017).
Flex, E. et al. Biallelic mutations in TBCD, encoding the tubulin folding cofactor D, perturb microtubule dynamics and cause early-onset encephalopathy. Am. J. Hum. Genet. 99, 962–973 (2016).
Tian, G., Huang, M. C., Parvari, R., Diaz, G. A. & Cowan, N. J. Cryptic out-of-frame translational initiation of TBCE rescues tubulin formation in compound heterozygous HRD. Proc. Natl. Acad. Sci. USA 103, 13491–13496 (2006).
Parvari, R. et al. Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat. Genet. 32, 448–452 (2002).
Martin, N. et al. A missense mutation in Tbce causes progressive motor neuronopathy in mice. Nat. Genet. 32, 443–447 (2002).
Lewis, S. A., Tian, G. & Cowan, N. J. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 7, 479–484 (1997).
Tian, G. et al. Pathway leading to correctly folded beta-tubulin. Cell 86, 287–296 (1996).
Hoff, K. J., Neumann, A. J. & Moore, J. K. The molecular biology of tubulinopathies: Understanding the impact of variants on tubulin structure and microtubule regulation. Front. Cell Neurosci. 16, 1023267 (2022).
Cai, X. B. et al. Whole-exome sequencing identified ARL2 as a novel candidate gene for MRCS (microcornea, rod-cone dystrophy, cataract, and posterior staphyloma) syndrome. Clin. Genet 96, 61–71 (2019).
Chen, C. L. et al. Novel Compound Heterozygous Variants in TBCD gene associated with infantile neurodegenerative encephalopathy. Children (Basel) https://doi.org/10.3390/children8121140 (2021).
Miyake, N. et al. Biallelic TBCD mutations cause early-onset neurodegenerative encephalopathy. Am. J. Hum. Genet. 99, 950–961 (2016).
Tian, G., Bhamidipati, A., Cowan, N. J. & Lewis, S. A. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the alpha/beta-tubulin heterodimer. J. Biol. Chem. 274, 24054–24058 (1999).
Nithianantham, S. et al. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alphabeta-tubulin pool for microtubule dynamics. Elife 4, e08811 (2015).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Punjani, A. & Fleet, D. J. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 213, 107702 (2021).
Campanacci, V. et al. Selection and characterization of artificial proteins targeting the tubulin alpha subunit. Structure https://doi.org/10.1016/j.str.2018.12.001 (2018).
Seong, Y., Kim, H., Byun, K., Park, Y. W. & Roh, S. H. Structural dissection of alphabeta-tubulin heterodimer assembly and disassembly by human tubulin-specific chaperones. Science 390, eady2708 (2025).
Wethekam, L. C. & Moore, J. K. Tubulin isotype regulation maintains asymmetric requirement for alpha-tubulin over beta-tubulin. J. Cell Biol. https://doi.org/10.1083/jcb.202202102 (2023).
Tian, G. & Cowan, N. J. Tubulin-specific chaperones: components of a molecular machine that assembles the alpha/beta heterodimer. Methods Cell Biol. 115, 155–171 (2013).
Bhamidipati, A., Lewis, S. A. & Cowan, N. J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096 (2000).
Andresen, M., Schmitz-Salue, R. & Jakobs, S. Short tetracysteine tags to beta-tubulin demonstrate the significance of small labels for live cell imaging. Mol. Biol. Cell 15, 5616–5622 (2004).
Castoldi, M. & Popov, A. V. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32, 83–88 (2003).
Pecqueur, L. et al. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. USA 109, 12011–12016 (2012).
Nakane, T., Kimanius, D., Lindahl, E. & Scheres, S. H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife 7, e36861 (2018).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 874 (2021).
Casanal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1069–1078 (2020).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D. Struct. Biol. 74, 814–840 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Acknowledgements
We thank the Bay Area Cryo-EM consortium led by Prof. Eva Nogales (Molecular Cell Biology, UC-Berkeley) and the UC-Davis BioEM facility for Cryo-EM data collection support. Large Scale Cryo-EM data were collected at UC-Davis and the Cryo-EM facility at UC-San Francisco with support from Dr Alexander Mysanikov (Biochemistry and Biophysics, UC-San Francisco). We thank Dr. Camille Scott and the UC-Davis High-Performance Computing Facility (HPCCF) for computational HPC infrastructure building and support. We thank Dr Stanley Nithianantham (Molecular Cellular Biology, UC-Davis) for the preliminary biochemical studies. We thank Prof Jeffrey K Moore (Cell Developmental Biology, University of Colorado, Anschutz Medical Campus) for advice and suggestions and for the critical reading of this manuscript. We thank Prof Richard McKenney, Prof Jonathan Scholey (Molecular Cellular Biology, UC-Davis), and Prof Ahmet Yildiz (Molecular Cell Biology, UC-Berkeley) for comments on the manuscript. J.A.B. acknowledges funding support from the National Institutes of Health (GM110283 and GM158334).
Author information
Authors and Affiliations
Contributions
A.T. purified and assembled TBC-DEG/TBCC-αβ-tubulin complexes, collected cryo-EM data, determined and refined all single particle cryo-EM structures, built, refined, and validated all models for assemblies, prepared figures, wrote and edited the manuscript. Z.W. purified and assembled TBC-DEG-αβ-tubulin complexes, collected cryo-EM data, and determined initial single particle cryo-EM structures. B.S. built and refined initial models. F.G. supported cryo-EM grid preparation, cryo-EM screening, and large-scale cryo-EM data collection. J.A.B. planned and managed the project, trained scientists, obtained funding for the project, prepared assemblies, prepared figures, and wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taheri, A., Wang, Z., Singal, B. et al. Cryo-EM structures of the tubulin cofactors reveal the molecular basis of alpha/beta-tubulin biogenesis. Nat Commun (2025). https://doi.org/10.1038/s41467-025-68142-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-68142-0